Abstract
It is well established that amyloid proteins play a primary role in neurodegenerative diseases. Alzheimer’s, Parkinson’s, type II diabetes, and Creutzfeldt-Jakob’s diseases are part of a wider family encompassing more than 50 human pathologies related to aggregation of proteins. Although this field of research is thoroughly investigated, several aspects of fibrillization remain misunderstood, which in turn slows down, or even impedes, advances in treating and curing amyloidoses. To solve this problem, several research groups have chosen to focus on short fragments of amyloid proteins, sequences that have been found to be of great importance for the amyloid formation process. Studying short peptides allows bypassing the complexity of working with full-length proteins and may provide important information relative to critical segments of amyloid proteins. To this end, efficient biophysical tools are required. In this review, we focus on two essential types of spectroscopic techniques, i.e., vibrational spectroscopy and its derivatives (conventional Raman scattering, deep-UV resonance Raman (DUVRR), Raman optical activity (ROA), surface-enhanced Raman spectroscopy (SERS), tip-enhanced Raman spectroscopy (TERS), infrared (IR) absorption spectroscopy, vibrational circular dichroism (VCD)) and solid-state nuclear magnetic resonance (ssNMR). These techniques revealed powerful to provide a better atomic and molecular comprehension of the amyloidogenic process and fibril structure. This review aims at underlining the information that these techniques can provide and at highlighting their strengths and weaknesses when studying amyloid fragments. Meaningful examples from the literature are provided for each technique, and their complementarity is stressed for the kinetic and structural characterization of amyloid fibril formation.
Keywords: Amyloid fragments, Raman spectroscopy, IR spectroscopy, VCD spectroscopy, ssNMR, Polymorphism
Introduction
Although amyloid diseases have been studied for a long time, a lot remains to be understood. A tremendous amount of work is done to try and eradicate these diseases from modern societies, where longer life expectancies directly correlate with more diseased persons (Selkoe 2015). The first descriptions of Parkinson’s (PD) and Alzheimer’s (AD) diseases, the two most widespread amyloid pathologies, go back to 1817 and 1906, respectively (Alzheimer 1906; Parkinson 2002), and to date there is still no cure for these diseases. With more than 50 amyloid-related diseases affecting and killing millions of people around the world and with several unanswered questions (Eisenberg and Jucker 2012), finding ways of either preventing or curing them is amongst the most urgent challenges of the twenty-first century.
The common point between these diseases is an abnormal misfolding of a specific protein. A majority of the proteins involved in these diseases exist in a globular form in their non-pathological state, but today no consensus exists regarding how, nor why, these proteins start to misfold, aggregate, and eventually form amyloid fibrils. This issue is particularly complex as the causes of aggregation may be multiple. For sporadic forms, the impairment of protein functions and subsequent cellular death can be explained by number of chemical, biological, and physiological reactions, such as the presence of reactive oxidative species, post-translational modifications, protein over-expression, membrane modifications, protein clearance impairment, and intra- and extra-cellular environment modifications.
Whereas amyloid diseases are most often sporadic, familial forms related to genetic factors and protein mutations also exist, notably in AD (Sherrington et al. 1995) and PD (Valente et al. 2004), while Huntington’s disease (HD) is a hereditary-only disease (Walker 2007). Selkoe and Hardy recently reviewed the advancements over the last 25 years in the knowledge and comprehension of AD. Amongst the remaining questions are the following: which forms of Aβ are toxic and what is its spreading mechanism (Selkoe and Hardy 2016). These questions extend to other amyloid proteins.
Several avenues of therapeutic development of amyloidoses are being explored, as the pathways of toxicity leading to cellular death are most certainly proper to each disease. Some think that accelerating fibril formation past early-stage toxic oligomers and stabilizing them could be a strategy to prevent the formation of more toxic species (Chen et al. 2010) while others propose that destabilizing fibrils could solve the problem (Johnson et al. 2012). Another aspect of the research focuses on molecules able to maintain proteins in their globular or monomeric form, thus preventing the disease simply from appearing (Collier et al. 2017; Hoyer et al. 2008). More generally, Eisele et al. reviewed novel therapeutic approaches which are at different stages of development for several amyloid diseases, and discussed the potential of these disease-modifying strategies for mitigating these amyloidoses (Eisele et al. 2015).
Amyloid fibrils have been assumed to be the putative species responsible of amyloid diseases but oligomers have been identified as an element of the fibrillization pathway that is also cytotoxic, and possibly more deleterious than fibrils (Stefani 2012). Protein aggregation leads to the appearance of oligomers, protofibrils, and mature fibrils, a hallmark of amyloid diseases. A precise molecular characterization of each peptide and protein at various steps along the fibrillization process is needed to eventually consider a possible cure or to synthesize biomarkers for early-stage disease identification.
Study of amyloid peptides and proteins has revealed to be complicated by the fibrillary polymorphism that most of them exhibit. This issue, combined with the high molecular weight of most amyloid peptides and proteins, complicates spectroscopic investigations. A remedy consists in studying fragments of these amyloid peptides and proteins. Peptides with short amino acid sequences corresponding to amyloidogenic stretches of their parent protein can be used to bypass the complexity of working with the full-length proteins, and has proven to be useful for spectroscopic investigations and to better understand pathological fibrillization. Short peptides can be synthesized in large quantities and can selectively be isotopically labeled during synthesis. Moreover, certain amino acid regions of pathological proteins exhibit particular properties, propensities, and/or pathogenicities, so that the study of protein fragments represents a research avenue able to provide insightful information regarding amyloidoses. For example, the segment 71–82 of α-synuclein (AS) has revealed to be of great importance regarding its pathogenicity in PD (Giasson et al. 2001), which justified structural studies carried out on the peptide AS71–82 (Bédard et al. 2014). Another example is the identification of the fragment 106–126 of the prion protein (PrP) as a key stretch for fibrillization in Gerstmann-Sträussler-Scheinker’s disease (Tagliavini et al. 1993).
Amongst the multitude of techniques suited for the study of amyloid peptides and proteins, two families of techniques are particularly adapted for a morphological, structural, and dynamical description of amyloid fibrils: vibrational spectroscopy (Raman spectroscopy, infrared spectroscopy (IR), vibrational circular dichroism (VCD)) and solid-state NMR (ssNMR). Combined, these techniques allow a detailed structural characterization, from atomic level to quaternary structure, through intermediary hierarchical steps. This review highlights the advantages of these techniques, as well as their inherent limitations, for the study of amyloid peptides and proteins. Their complementarity is also discussed. Examples of the contemporary literature using these particular techniques for the study of amyloid peptides and protein fragments are given. It should be mentioned that X-ray diffraction (XRD) and microelectron diffraction are also very relevant techniques for the study of amyloid crystals, providing crucial structural characterizations of amyloid fragments (Guenther et al. 2018; Rodriguez et al. 2015; Soriaga et al. 2016). XRD has even been used early on to uncover structural details of fibers (Astbury et al. 1935). These techniques are however not in the scope of the present article and excellent reviews exist on this subject (Eisenberg and Jucker 2012; Eisenberg and Sawaya 2017).
Morphology and challenges to overcome
Morphology
Almost all peptides and proteins can form amyloid fibrils in appropriate conditions. Therefore, a great diversity of amino acid sequences share common amyloid structural features. The fibril diameter and length are around 10 nm and a few hundreds of nanometers, respectively. They assemble with a cross β-sheet core: β-strands run perpendicularly to the fibril long axis, with an interstrand distance of ~ 4.7 Å, a structure described for the first time in 1935 (Astbury et al. 1935). Amyloid fibrils are highly favorable energetically and extremely stable. β-strands are hydrogen-bonded and the protofilaments formed are held together mostly via sidechain-sidechain interactions, resulting in a highly dense and interdigitated packing. Depending on the amino acids, the nature of these interactions varies: they can be of hydrophobic, electrostatic, or even of aromatic (π-stacking) nature. Some residues containing an amide group, such as glutamine and asparagine, are able to stabilize the fibril via the formation of hydrogen bonds with the sidechain of the alike neighboring residue, a characteristic most often observed in in-register parallel configuration of the β-sheets (Eisenberg and Jucker 2012).
Polymorphism
Many amyloid peptides and proteins exhibit polymorphism, which complicates structural analyses and comprehension of amyloid formation. Nonetheless, attempts to formalize and classify fibrillar morphologies allow finding common features and redundant properties. For instance, the three-dimensional description of fibrils lead to classify cross β-sheet cores as steric zippers (Eisenberg and Sawaya 2017).
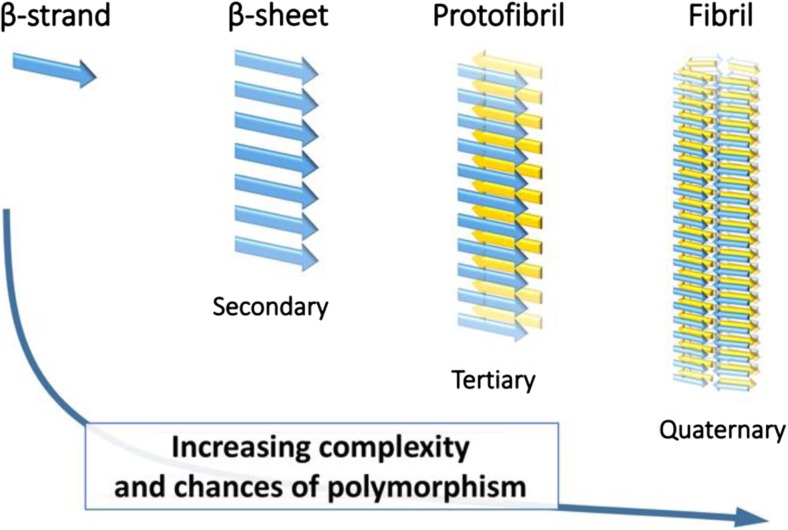
Several parameters account for amyloid fibril polymorphism, including concentration, temperature, mechanical agitation, and pH. Studies have even proven the existence of polymorphs grown simultaneously in the exact same conditions (Hoyer et al. 2002; Petkova et al. 2005). This characteristic adds a level of complexity, since even the smallest molecular variations may have an impact all along the amyloid formation cascade, as illustrated in Fig. 1. Polymorphism has been, and still is, a source of problems, from both structural characterization and biological relevance points of view (Meier et al. 2017; Tycko 2015). For example, amyloid-β1–40 (Aβ1–40) and AS polymorphs, involved in AD and PD, respectively, displayed different toxicity towards cell lineages (Bousset et al. 2013; Gath et al. 2014; Petkova et al. 2005). For structural characterization, a tight control of the incubation conditions is of primary importance in order to obtain a unique type of fibril polymorph, to facilitate analysis and result interpretation.
Fig. 1.
Typical hierarchical description of the amyloid formation steps, from the single β-strand to the mature fibril (from secondary to quaternary structure). Molecular variations occurring at any level affect all the subsequent formation steps and thus are sources of fibril polymorphism. Figure based on a similar figure reported by Fitzpatrick et al. 2013
Vibrational techniques
Vibrational techniques can be divided into two fundamental families: those based on Raman scattering and those based on the absorption of mid-IR light. The former techniques include conventional Raman spectroscopy, deep-UV resonance Raman (DUVRR), surface-enhanced Raman spectroscopy (SERS), tip-enhanced Raman spectroscopy (TERS), and Raman optical activity (ROA). The latter techniques encompass conventional IR (transmission, attenuated total reflection (ATR) and reflection-absorption spectroscopy), VCD, and two-dimensional IR (2D-IR). These tools can provide dynamical and structural description of amyloid fibrillogenesis and fibrils. Whereas Raman spectroscopy is based on inelastic scattering resulting from changes in bond polarizability, IR spectroscopy relies on the absorption of light due to changes in molecular dipole moments. This difference makes these two types of techniques complementary since some vibrations may be only active or may be reasonably intense in one of the two techniques, and not in the other. This traditional picture is however challenged due to the rise of more recent, derived techniques since each one may enhance specific vibrational modes and may have its own specific application, with its advantages and drawbacks.
Raman spectroscopy
Conventional Raman spectroscopy is not much used to investigate the secondary structure of amyloid proteins. Although the spectral contribution due to water is smaller than in IR spectroscopy, it still overlaps and obscures the amide I region so that solvation in D2O may be necessary. Even in this case, higher concentrations are required to obtain reasonable signal-to-noise ratios, especially for amide bands. Then, a few works have been dealing with the backbone conformation of amyloid proteins. An interesting example is however provided by the study of AS in several media that proved the strength of conventional Raman for secondary structure determination of unfolded proteins (Maiti et al. 2004). Nevertheless, conventional Raman cannot discriminate without ambiguity parallel from antiparallel β-sheets, a limitation that represents a pitfall for the investigation of amyloid peptides.
By contrast, vibrations due to amino acid sidechains are particularly apparent, so that Raman scattering can fruitfully be used to investigate specific and localized interactions. As an example, the effect of Cu(II) on the N-terminal part of the PrP was investigated using the octapeptide PHGGGWGQ. Upon pH modification from a neutral to a weakly acidic state, the Cu(II) binding site would be modified to form His-Cu-His bridges that in turn would be responsible for protein aggregation (Miura et al. 1999). Miura et al. also studied the Aβ1–16 fragment, corresponding to the N-terminal part of Aβ, to probe interactions with Fe(III), a known aggregation inducer. A preferential interaction of Fe(III) with Tyr10 was observed in both Aβ1–16 and Aβ1–40, and to a lesser extent with Asp/Glu residues (Miura et al. 2001). Such studies are part of the global issue of the role played by metal ions in amyloid diseases (Viles 2012). In this context, being able to probe the role of specific residues when coordinating to metal ions via Raman spectroscopy is obviously an asset.
Raman spectroscopy has been derived in a variety of techniques, such as DUVRR, ROA, SERS, and TERS. DUVRR is based on a different physical principle than conventional Raman: rather than being in the visible domain, the laser has a wavelength in the UV domain (typically ~ 200 nm), which causes resonant electronic transitions (Oladepo et al. 2012). DUVRR is then particularly efficient to probe amide chromophore vibration modes (Chi et al. 1998; Lednev et al. 2005), and provides a signal enhancement between 106 and 108 when compared to conventional Raman spectroscopy (Oladepo et al. 2012). An excellent review recently compiled studies on full-length amyloid peptides and proteins using Raman techniques (Kurouski et al. 2015b).
In contrast to conventional Raman, DUVRR can discriminate parallel from antiparallel β-sheets. As a matter of fact, the group of Asher developed a semi-empirical equation based on the amide III band frequency allowing the determination of the dihedral angle psi (ψ) of protein and peptide amide groups (Mikhonin et al. 2006). Popova et al. applied this method to Aβ1–40 and to the Aβ34–42 fragment. As their secondary structure was known to be parallel and antiparallel, respectively, this study supported the reliability of Asher’s group formula to determine secondary structure elements (Popova et al. 2010) and to distinguish parallel and antiparallel β-sheets by their torsion angle ψ. Moreover, the combination of DUVRR with hydrogen/deuterium (H/D) exchange made it possible to distinguish the structure of exchangeable (water-accessible or exposed) and non-exchangeable (solvent-inaccessible, then located in the amyloid core) amide groups.
Asher’s group focused on polyglutamine (polyQ) fibrils formed by the peptide D2Q10K2. This peptide can adopt different forms in solution: NDQ10, a β-hairpin-like structure and DQ10, a PPII-like (polyproline II) structure. By combining different deuteration schemes of both fibrils and fibril preparation, various Raman bands were distinguished and assigned. Notably, H/D exchange of fibrils prepared in H2O allowed identification of the rigid cross-β core of the fibrils, while D/H exchange of fibrils prepared in D2O removed the amide III band of any component associated with rigid structures, thus highlighting the flexible/unordered structure components. Using their method developed earlier (Mikhonin et al. 2006) in conjunction with molecular dynamics, they showed that NDQ10 and DQ10 peptides preferentially adopt an antiparallel β-sheet structure that seemed thermodynamically more stable than the usual parallel arrangement commonly found in amyloid fibrils. Moreover, the authors determined the χ3 dihedral angle of Gln sidechains using a similar method than for ψ. The results indicated that Gln sidechains would be planar and highly interdigitated, thus forming a stable steric zipper (Punihaole et al. 2016).
Raman optical activity (ROA) relies on the variation in the intensity of the scattered light between left- and right-circularly polarized incident light by optically active molecules (Barron et al. 2000). ROA has mainly been used to probe the prefibrillar states of amyloid proteins, more particularly disordered structures. This conformational state was often (inadequately) referred to random coil. As a matter of fact, ROA is sensitive to 31-helix structure, also called PPII helix. This structure, which is considered as a regular secondary structure, actually appeared to be an essential component of “random coil” proteins. This structural element also appeared as an essential structure in amyloidogenesis. Blanch et al. witnessed a high content of PPII helix in amyloid prefibrillar intermediates of a fragment of the N-terminal part of the ovine prion protein, and discussed its amyloidogenic potential (Blanch et al. 2004). The in-solution structure of the amyloid fragment Aβ1–28 has also been described as adopting a PPII conformation via a combination of IR, and isotropic and anisotropic Raman spectroscopy (Eker et al. 2004).
The aim of SERS is to enhance the Raman signal by placing molecules in the vicinity of metal nanoparticles or on a metal surface, which induce plasmonic resonance and strong signal enhancement when interacting with the incident light (Dornhaus et al. 1980). This technique was applied to study the influence of Cu(II) on the Aβ1–16 fragment using a combination of SERS, electronic circular dichroism and scanning tunneling microscopy (STM). The data lead to conclude that Cu(II) stabilizes Aβ1–16, induces a change of conformation from PPII to β-sheet, and a stabilization of β-sheets through binding to the imidazole rings of His13 and His14 of adjacent β-strands (Yugay et al. 2016). TERS is similar to SERS, but offers a typical ~ 10–15 nm spatial resolution thanks to the coupling with a scanning probe microscope (atomic force microscope (AFM) or STM). TERS has been used to study the fragments Aβ16–22 and CGNNQQNY of yeast prion protein Sup35 (Krasnoslobodtsev et al. 2016; Paulite et al. 2013). In the latter study, analysis of the amide III bands demonstrates that fibrils formed at pH 2.0 were composed of parallel β-sheets, while fibrils formed at pH 5.6 displayed an antiparallel β-sheet structure, along with some random coil and α-helix. Moreover, TERS allowed identification of variations in the exposed secondary structures along the axis of the fibrils, with a lateral resolution of 7 nm. To date, SERS and TERS have only been sparsely used to investigate amyloid fibrils.
IR and VCD: Amide I Band
The region of main interest of IR techniques is the amide I band (also named amide I′ band when the backbone amide groups are deuterated) which extends from 1700 to 1600 cm−1. This band enables the determination of peptide and protein global secondary structures, as more or less narrow frequency ranges correspond to specific secondary structures. The amide I band arises mostly from the stretching vibration of the peptide bond C = O, combined with the out-of-phase CN stretching vibration, the CCN deformation and the NH in-plane bend. Deuterium oxide is often used as solvent to avoid the overlap between the amide I band and the O–H bending vibration of water. Deuteration of the backbone NH results in a slight shift (~ 5–10 cm−1) of the amide I band. In the case of amyloid peptides and proteins, the secondary structure of interest is most often, but not only, the β-sheet. IR is particularly sensitive to this structure and is efficient to follow the kinetics of aggregation.
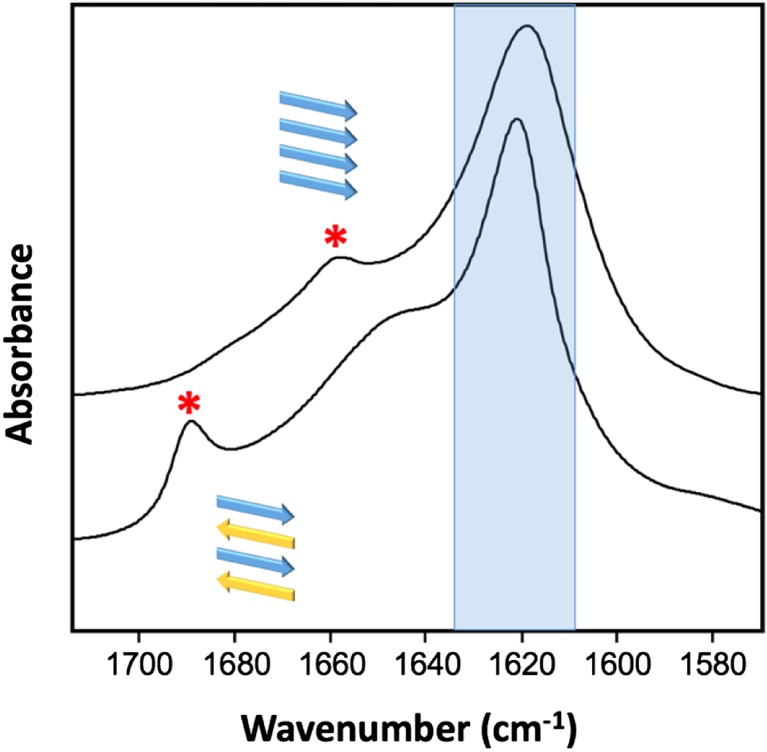
β-Sheets resulting from aggregation/amyloidogenesis are often associated with two narrow and well-separated spectral components, an intense one at low wavenumbers (1615–1635 cm−1) and a small one at higher wavenumbers (1655–1695 cm−1). This splitting of the amide I β-sheet band is due to transition dipole coupling (TDC): the resonance interaction of adjacent amide groups (oscillators) depends on the distance that separates them and on their relative orientation. These spectral components correspond to out-of-phase and in-phase vibrations of β-sheet-coupled oscillators (Krimm and Bandekar 1986). The arrangement of β-sheets in an antiparallel configuration leads to a large splitting of ~ 60 cm−1, and can reach up to 70 cm−1. For a parallel conformation of the strands, a smaller splitting of 30–40 cm−1 is observed (Schweitzer-Stenner and Measey 2010). The spectral difference in the two types of β-sheets is illustrated in Fig. 2.
Fig. 2.
Typical IR spectra of parallel (upper spectrum) and antiparallel (lower spectrum) intermolecular β-sheets. The major component is at low wavenumbers (blue-shaded area) and the less intense one at higher wavenumbers (red asterisks). Figure based on a similar figure reported by Sarroukh et al. 2013
For a given arrangement (parallel or antiparallel), the β-sheet components exhibit a certain position variability. The reason lies in the fact that the amide I vibration depends on many parameters: in-register or out-of-register ordering, length, number and twist of β-strands (Welch et al. 2013b). The number of possible configurations, coupled with the complexity of the three-dimensional arrangement of amyloid fibrils, adds to the overall structural diversity of amyloid fibrils.
Infrared spectroscopy—IR
Conventional IR spectroscopy was used concomitantly with the advances on different amyloid proteins and peptides. Works on amyloid proteins and fibrils using conventional IR then go back to the 70s (Glenner et al. 1974; Termine et al. 1972). They precede molecular analyses of amyloid deposits obtained ex vivo, showing that they are mostly composed of a single type of protein (Glenner and Wong 1984; Prusiner et al. 1984). Then, the sequencing of amyloid proteins allowed the study of specific fragments, notably of Aβ and IAPP (islet amyloid polypeptide, then-called amylin) in the group of Lansbury in the early 90s. They assessed the structure of two fragments derived from the Aβ peptide (Aβ26–33 and Aβ34–42) in aqueous and organic solvents and concluded that while both peptides form antiparallel β-sheets with a cross-β core in the solid-state, Aβ34–42 seemed to play a more important role in the amyloidogenic process (Halverson et al. 1990).
In the same group, a study paved the way to a very detailed characterization of amyloid structures using 13C-labeled amino acids on the fragments Aβ34–42 and IAPP20–29. This method makes it possible to precisely probe which amino acids are included in the β-sheets, thanks to characteristic bandshifts of 13C-labeled amide I groups. Usually, three spectra are acquired for each labeled position: (i) a spectrum of the unlabeled peptide, (ii) a spectrum with labeled-only peptide and (iii) a spectrum with isotopically diluted labeled peptides (dilution between 1:2 and 1:5 labeled/unlabeled). The difference in the position of the 13C component between spectra (ii) and (iii) can indicate a specific secondary structure. Also, the shift of the 13C component to lower frequencies is accompanied with a decoupling between amide bonds which results in a shift of the 12C amide I band to higher frequencies. This type of study allows the characterization of specific segment(s) involved in the β-sheets (Ashburn et al. 1992; Halverson et al. 1991). 13C-labeling was also used to study the dynamical rearrangement of β-sheets during the nucleation process of PrP109–122, a peptide from the PrP involved in Creutzfeldt-Jakob’s disease and “mad cow” disease (Petty and Decatur 2005). For a review highlighting the possibilities offered by this type of experiment and for theoretical considerations, see Decatur (2006) and Welch et al. (2013a).
Isotopic labeling can be exploited even further, with the use of doubly labeled carbonyls 13C = 18O, as it induces an even larger redshift and the disappearance of spectral overlap issue. This method has been applied to study the effect of deamidation on specific residues of human IAPP (hIAPP) by 2D-IR spectroscopy (Dunkelberger et al. 2012). Recent experimental and computational studies of Aβ1–40 and Aβ1–42 highlighted the strength of this technique for distinguishing fibrillar oligomers from fibrils, thanks to spectral features hidden in standard IR. More particularly, the presence of a low-wavenumber component at 1610 cm−1, persistent to disaggregation by sodium dodecyl sulfate micelles, unlike the 1625 cm−1 component, led the authors to the conclusion that the 1610 cm−1 band corresponds to fibrils while the one at 1625 cm−1 would correspond to fibrillar oligomers (Lomont et al. 2018). We can regret that 2D-IR is not as commonly used as standard IR yet, given its strength in revealing structural details, as demonstrated in existing studies.
Nonetheless, standard IR is also a powerful technique on its own, the main advantages of which are (i) the possibility of working with either protein solutions or suspensions of membranes and proteins (in a wide range of pH and ionic strength), (ii) rapidity of acquisition, allowing investigation of kinetic experiments, (iii) and the possibility to record spectra as a function of temperature. IR might be considered quite demanding regarding sample quantity, as protein and peptide concentrations may be above 2.5 mg mL−1. Therefore, working with synthetic peptides may be an advantage as they may be more easily available in higher amount than recombinant proteins. One of the major drawbacks of IR is the overlap of secondary structure band components in the amide I region, but this problem can be overcome by isotopic labeling and/or 2D-IR as explained above. However, β-sheets are amongst the easiest structures to observe, due to their band narrowness, position at lower wavenumbers than other structures and their high absorption coefficient, thus facilitating the study of amyloid peptides and proteins. IR is then a powerful technique to investigate aggregation kinetics. Other secondary structures may be more difficult to analyze as they may overlap, exhibit spectral variations depending on the aqueous environment and because their assignment is less ascertained (Jackson and Mantsch 1995).
Conventional IR can probe insoluble samples, which is quite common amongst amyloids, by using attenuated total reflection IR (ATR-IR). Solid samples could be probed as well. ATR-IR was used by Berthelot et al. to study the kinetics of the yeast amyloid HET-s218–289 and one of its mutant. ATR-IR allowed the identification of secondary structure changes when pH increased from 2 to 7.4: for HET-s218–289, a transition from random coil to parallel β-sheets was observed, while its mutant M8 instantly changes from a random coil conformation to antiparallel β-sheets (Berthelot et al. 2010). For a thorough review on ATR-IR applied to amyloid proteins, see Sarroukh et al. (2013).
Variations of pH are accounted to be amongst the possible causes inducing protein misfolding. Perálvarez-Marín et al. probed the effect of photoinduced pH jumps (from 8.5 to < 6.0) on the aggregation rate of Aβ1–28. An antiparallel arrangement of the β-sheets was observed, with a transition state corresponding to oligomers with the same antiparallel structure (Perálvarez-Marín et al. 2008). Guo et al. also explored the effect of pH on the structure of Aβ fragments (Aβ1–25, Aβ25–35, and Aβ33–42). While Aβ1–25 forms antiparallel β-sheets at pH 4.6 and 7.4, Aβ33–42 adopts a parallel arrangement, with also some antiparallel β-sheets at pH 7.4. The particularity of these two fragments is their ability to adopt, along with β-sheets, a rather high proportion of random coil conformation at pH 7.4 (38 and 28% for Aβ1–25 and Aβ33–42, respectively). By contrast, the structure of Aβ25–35 is somehow pH-independent, and conserves the same highly ordered parallel β-sheet structure regardless of pH. The authors cautiously conclude that Aβ25–35 can form fibrils on its own, while the other fragments would form oligomeric structures. Therefore, perspective regarding the full-length peptide would be that the presence of antiparallel β-sheets is synonym of oligomers while a parallel β-sheet structure would lead to amyloid fibrils (Guo and Wang 2012).
The parallel β-sheet structure of Aβ25–35 has been confirmed using IR, this configuration being also adopted in the presence of anionic model membranes (Labbé et al. 2013). IR has indeed the potential to allow structural studies of amyloid proteins and peptides in a more physiological relevant membrane context. Another example is the above-mentioned peptide AS71–82, that also adopts a parallel β-sheet structure upon interactions with negatively charged model membranes (Bédard et al. 2014). Its conformation is mainly disordered in the absence of membranes or in the presence of neutral (zwitterionic) membranes although some β-sheets are also detected (Bédard et al. 2014). The parallel β-sheet structure of AS71–82 looks similar to the one of fibrils formed by the parent protein AS (Conway et al. 2000; Uversky et al. 2001).
Even if the structural characterization of amyloid peptides/proteins upon interactions with model membranes lies beyond the scope of the present review, it is noteworthy that it is also of primary importance, as membranes are thought to influence and accelerate the oligomerization and fibrillization in several diseases (Galvagnion 2017; Murphy 2007). Membranes are also important for their effect on the local concentration of amyloid polypeptides (molecular crowding) (Andreasen et al. 2015).
Spectral overlap of secondary structures is often mentioned as a problem in IR studies. However, this drawback can be partly overcome by cautious analyses (Jackson and Mantsch 1995; Surewicz et al. 1993). Spectral deconvolution is most often used to quantify secondary structures, while the calculation of second derivatives allows identification of the different peak contributions, and thus the secondary structures present, in the overlapped spectra (Kong and Yu 2007). Juszczyk et al. highlighted the structural insights provided by second derivatives applied to the study of Aβ11–28 and its clinically relevant variants to identify helix-containing structures of would-be monomeric species in the aggregation process of all the mutants studied. Here, the problems caused by spectral overlap, and especially in the α-helix/310-helix region, were overcome with a better peak identification provided by second derivatives (Juszczyk et al. 2009).
IR has also the advantage of being sensitive to H/D exchanges, notably in the amide I/I′ and amide II/II′ regions. This kind of experiment allows kinetics analysis of fibrillogenesis, when undeuterated peptides or proteins are allowed to fibrillate in a deuterated solvent. Although, to the best of our knowledge, it has not been applied to amyloid fragments, a study demonstrating the capacity of such experiments has been carried on the protein insulin (Dzwolak et al. 2006).
IR perspectives
It should be noted that new techniques employing IR for the study of amyloid peptides and proteins were recently developed. Lately, nano-IR (also called infrared nanospectroscopy) emerged, and represents to IR what TERS is to Raman spectroscopy: it combines an IR spectrometer with an AFM and appears to be a valuable tool to characterize precise molecular variations with a 30-nm lateral resolution (unique amyloid fibrils) (Amenabar et al. 2013; Henry et al. 2018). Also, in-cell IR is now made possible by coupling an IR microscope with a microfluidic live-cell incubator, and could be applied to the study of amyloids (Gelfand et al. 2015). Moreover, in situ IR microspectroscopy of patients-retrieved cells seems a promising way of establishing early diagnostics of amyloid diseases (Ami et al. 2016).
Vibrational circular dichroism spectroscopy—VCD
VCD differs from conventional IR spectroscopy in that VCD is sensitive to the chiral property of biomolecules. It is homolog of ROA for IR spectroscopy. The signal corresponds to the difference in absorbance of a left- and a right-circularly polarized light (ΔA = AL-AR). The ΔA signal is however approximatively five orders of magnitude smaller than the corresponding “unpolarized” absorbance A, and thus requires quite longer acquisition times in order to obtain a good signal-to-noise ratio (Polyanichko 2007). For the same reason, VCD requires rather high minimal sample concentrations (~ 20 mg mL−1) (Polyanichko 2007). However, amyloid fibrils display an intense signal in the amide I region, compared to “regular” secondary structures of globular proteins. This peculiar feature was first underlined in the group of Nafie a decade ago (Ma et al. 2007), with experiments conducted on the fibrillization of insulin. This study highlighted the high sensitivity of VCD for this type of structure, which then sparked the interest of the “amyloid community” for VCD.
Explanations for this unusual high intensity reside in the intrinsic morphology of amyloid fibrils: their high structural order induces a stacking of amide groups along a same axis, thus making intrasheet vibrational couplings very strong and thereby inducing an intense VCD signal. The most favorable parameters enhancing the signal are an in-register parallel orientation of the β-strands, along with a twisted morphology of the fibril (Measey and Schweitzer-Stenner 2011).
VCD spectroscopy has almost the same advantages as standard IR (possibility of kinetics and temperature-controlled experiments) but the foremost advantage of VCD over IR is that the signal relies on the chirality of molecules. When applied to amyloid peptide and protein fibrils, two types of signals are most often observed: almost-symmetrical couplets of opposite direction, one with a negative-positive couplet (“normal” signal), and another with a positive-negative couplet (“reversed” signal) corresponding to left- and right-twisted fibril morphologies, respectively (Fig. 3). Left-twisted structures are usually more common than right-twisted ones, due to the natural chirality of amino acids inducing a slight left-twist along β-strands (Volpatti et al. 2013).
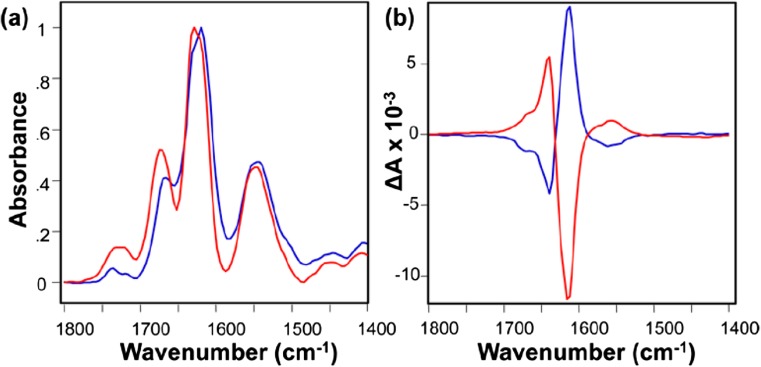
Fig. 3.
IR amide I bands (a) and corresponding VCD spectra (b) of HET-s218–289 amyloid fibrils at pH 2.0 (red) and pH 6.0 (blue). Fibrils initially formed at pH 2.0 exhibited a left-handed twist, and when the pH was increased to 6.0, the chirality of the fibrils reversed, as proved by the inversion of the VCD signal. This example highlights the complementarity of VCD to IR, where spectra at both pHs look very alike. Reprinted with permission from Shanmugasundaram et al. 2015. Copyright © 2015 American Chemical Society
VCD is particularly appropriate to examine amyloid fibrillization and has thus been used extensively for kinetics study. VCD can in turn apprehend the polymorphism of amyloid fibrils as exemplified by the changes of supramolecular chirality of insulin and the peptide HET-s218–289 upon pH modification (Kurouski et al. 2012; Shanmugasundaram et al. 2015). In these two cases, the signal completely reverses upon increase in pH, demonstrating an inversion of the fibril twist, probably due to protonation/deprotonation of sidechains, thus rearranging to a more energetically favorable configuration. Chirality inversion of HET-s218–289 upon pH elevation is illustrated in Fig. 3. Nonetheless, the core structure of the fibrils remains the same upon this pH modification, as revealed by the highly similar DUVRR spectra at pH 2.0 and 6.0 (Shanmugasundaram et al. 2015).
A more extensive study has been conducted with the aim of linking supramolecular chirality, VCD signal and polymorphism in amyloids (Kurouski et al. 2014). The comparison of the VCD signals of five proteins and peptides at different pHs: insulin, lysozyme, apo-α-lactalbumin, HET-s218–289 and transthyretin105–115 (TTR105–115) allowed assigning morphology to the different spectral features. In almost all cases, the normal VCD spectra correspond to left-twisted fibrils, while ‘reversed’ signals correspond to flat tape-like fibrils. The correlation between spectra and morphology was assessed by scanning electron microscopy (SEM) and AFM. The authors proposed pathways of fibrillary assembly for both signals: for left-twisted fibrils to form, left-twisted protofilaments would braid together, while the fibrils at the origin of the reversed signal would be constituted of laterally associated right-twisted protofilaments. The authors concluded that VCD probes chirality at the filament level, and not at the fibril level, and that this pH-controlled polymorphism could be a general feature of amyloid proteins and peptides.
Aggregates of polyQ stretches are known to be related to several neurodegenerative diseases, including HD (Bates 2003; Ross 2002). Kurouski et al. studied polyQ peptides of different lengths in order to identify the critical sequence length responsible for aberrant aggregation. While the IR spectra of Q18 (stretch of 18 glutamine residues), Q25, Q26 and Q41 are almost identical and characteristic of a β-sheet-rich secondary structure, the corresponding VCD spectra vary dramatically between Q18, Q25 and Q26 on one side, and Q41 on the other. The latter peptide, despite exhibiting the same peaks as the formers, has an intensity one order of magnitude higher. The authors, after experiments conducted with Q30, Q35 and Q40 peptides, concluded that a threshold exists between Q30 and Q35, a critical length responsible for a change in the supramolecular chirality, causing a large increase in the VCD signal. It remains however to be confirmed whether this change in supramolecular structure is somehow related to pathogenicity. The authors also used DUVRR to gain insights into the core structure of polyQ fibrils. The DUVRR spectra obtained in water for Q18, Q26 and Q41 fibrils are almost identical, consistently with the IR spectra. However, upon H/D exchange, the DUVRR spectrum of Q41 fibrils displays a more intense amide II′ band than for Q26 fibrils. This difference suggested that despite being more stable, the twist along Q41 fibrils makes them particularly prone to H/D exchange (Kurouski et al. 2013).
VCD has also been used to study two fragments of the IAPP peptide, namely IAPP21–27 and IAPP28–33, and to compare the results of solution-formed fibrils and microcrystals of the same sequences (Kurouski et al. 2015a). The main goal was to assess and validate the structures obtained previously via X-ray diffraction in the group of Eisenberg (Wiltzius et al. 2008). For each peptide, IR and VCD spectra of both the fibrils and the microcrystals were very similar (aside from sign and magnitude), validating the steric zipper models previously described, and confirming that the interaction most responsible for the enhanced VCD signal is the parallel orientation of β-sheets.
Aside from working in different aqueous conditions (ionic strength, concentration, pH), VCD also allows working in different media other than aqueous solutions. It is noteworthy that it is also possible to work with dry amyloid films or suspensions in organic solvents. For example, Shanmugam and Polavarapu worked on Aβ25–35 in various organic solvents and in different physical states (liquids, gels and films) to unravel the different structures Aβ25–35 can adopt in specific environments: methanol for membrane-like environment, acidic conditions to study the aggregated form, and dimethyl sulfoxide for the solution form of Aβ25–35 (Shanmugam and Polavarapu 2004). Their results highlight the different structures accessible for Aβ25–35, and maybe more importantly, provide insights into the polymorphism of a short 11-residue long peptide. One can thus imagine the problem caused by full-length amyloid protein polymorphism for their characterization.
As for IR, isotopic labeling for VCD is useful to decrease spectral overlap. The physical basis is the same as for IR, where two types of experiments are needed, a first one (i) with isotopic dilution, usually between a 1:2 and 1:4 ratio (isotopically labeled/unlabeled), and another one (ii) with uniquely isotopically labeled peptides. The comparison of the two corresponding 13C = O (or 13C = 18O) bands in (i) and (ii) allows one to determine the secondary structure of the labeled residue. If a large shift is observed, it indicates a strong coupling of the 13C-labeled carbonyls, as predicted for highly ordered structures such as β-sheets. Shanmugam and Polavarapu demonstrated the usefulness of isotopic labeling by applying it to the structural investigation of amyloid fragment Aβ16–22. Using a set of four peptides with 13C-labeled carbonyls positioned at residues 17 and 21 (Aβ16–22, Aβ16–22:13C-17, Aβ16–22:13C-21, Aβ16–22:13C-17/21), they managed to describe precisely the structure of Aβ16–22 as antiparallel β-sheets. In this case, VCD served as a more structure-sensitive tool than IR for validating their model (Shanmugam and Polavarapu 2011). For a more extensive description of 13C-labeling aiming at the structural characterization of amyloid fibrils, see Welch et al., where they compare experimental and DFT-based (density functional theory) results for four different peptides, including the above-mentioned example (Welch et al. 2013a). Since VCD is not yet a commonly used technique, DFT has revealed to be a valuable tool for result interpretation. As VCD is sensitive to several structural features (mostly the degree of twist, β-strand length and number of protofilament in the fibril), DFT was used to probe the effect of their interplay on VCD signal (Measey and Schweitzer-Stenner 2011; Welch et al. 2013a, b).
VCD has revealed to be a powerful tool to study amyloid peptides and proteins, especially in fibrillary form. It is noteworthy that VCD, more than being an efficient technique capable of probing fibrillogenesis and chiral features, highlights the chiral facet of polymorphism and is undoubtedly the most adapted technique to study this aspect. As demonstrated by Kurouski et al., VCD is able to detect supramolecular chirality features that lie below the direct limit of detection of imaging techniques such as SEM or AFM (Kurouski et al. 2014). The attractiveness of VCD is rather recent and consequently, it is not as widespread as conventional IR, but one can imagine that progressively more laboratories will acquire VCD spectrometers and expertise, given the strength demonstrated by this technique for amyloid studies.
Like ROA, VCD is also very sensitive to PPII structure. It is actually the only technique that can distinguish true random coil from PPII (Dukor and Keiderling 1991; Paterlini et al. 1986).
Solid-state nuclear magnetic resonance—ssNMR
Of the techniques presented herein, ssNMR is indubitably the most powerful, but it also comes with its drawbacks: ssNMR is a costly technique due to the necessary isotopic labeling of residues, and the high sample quantities required. Several pulse sequences of different complexity can be applied to the study of amyloid peptides and proteins, from one-dimensional (1D) spectra (e.g., 13C spectra) to multi-dimensional spectra with several nuclei involved (chemical shifts and/or dipolar couplings of 13C, 15N and 1H usually). The structural features revealed by ssNMR mainly consist of secondary structure information, intra- and intermolecular distance and dihedral angle measurements, which combined altogether allow the determination of tertiary and quaternary structures. The use of fragments is particularly suited for ssNMR where selectively labeled peptides greatly facilitate the interpretation of 1D and multi-dimensional spectra.
In ssNMR, the distribution of molecular orientations due to the lack of isotropic tumbling leads to spectra with broad and often overlapped peaks, due to a phenomenon called chemical shift anisotropy (CSA). While CSA can be a way to obtain structural and dynamical information on the sample, most ssNMR pulse sequences tend to cancel the CSA interaction via magic-angle spinning (MAS). Since MAS tends to cancel homo- and heteronuclear couplings, ssNMR pulse sequences are generally designed to reintroduce specific dipolar couplings, which contain the internuclear distance information.
Technological advances such as higher magnetic fields and higher MAS frequencies allowed the elaboration of more complex pulse sequences leading to more advanced structural characterizations. Recently, a study demonstrated the possibility of working with 0.7 mm probes and sample quantity < 500 μg at spinning rates over 100 kHz in high magnetic fields with the aim of assigning alpha protons in fully protonated and 13C, 15N-labeled proteins. This method, which aims at facilitating structural studies of biomolecular assemblies, has been applied to five proteins in different forms, and notably to HET-s218–289, to establish a proof of concept (Stanek et al. 2016). To illustrate the high-resolution obtained with these proton-detected pulse sequences, Fig. 4 displays representative 2D cross-sections of 15N- and 13C′-linked Hα-detected spectra of four proteins, including HET-s218–289.
Fig. 4.
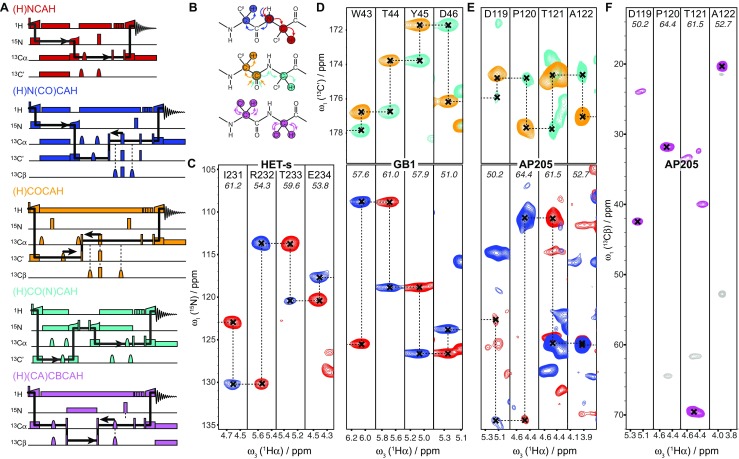
a Simplified correlation pulse sequence schemes, with indirect chemical shift evolution periods indicated by black arrows. b Intramolecular magnetization flows corresponding to the color-matching correlation pulse sequences presented in (a). 2D cross-sections of 15N- (blue and red) and 13C′-linked (orange and cyan) Hα-detected spectra of c HET-s218–289, d GB1, e AP205CP and the respective intra-residue Cβ-Cα-Hα correlations for AP205 (f). Reprinted with permission from Stanek et al. 2016. Copyright © 2016 by John Wiley & Sons, Inc.
Recently, Smith et al. also used similar proton-detected (2D and 3D) pulse sequences and fast MAS (60 kHz) to assign HN and Hα of HET-s218–289 (Smith et al. 2017). In order to increase spectral resolution, a set of differently-deuterated samples was used. These HN and Hα assignments were facilitated by the use of previously-obtained Cα, Cβ, C′, and N assignments used for the determination of the three-dimensional structure of HET-s218–289, in the same group. Their results indicate that HET-s218–289 adopts a left-handed β-solenoid conformation with β-strands parallel to each other (Van Melckebeke et al. 2010). However, corrections due to deuterium effects must have been applied for Cα, Cβ and N resonances. Such assignments of HN and Hα allowed a better and more precise structural and dynamical description of HET-s218–289.
As for IR, the first ssNMR studies of amyloid peptides and proteins began concomitantly with the progressive identification and sequencing of the polypeptides and proteins responsible for amyloid diseases. Amongst the early studies of amyloid fragments, ssNMR was used alongside IR for the study of IAPP20–29 (Ashburn et al. 1992). Besides the structural information provided by IR 13C amide bands (see the IR section), the carbonyl 13C chemical shifts obtained for each peptide are indicative of the structural rigidity and allowed the identification of the β-sheet-only stretch, reaching similar conclusions as those obtained by IR. The structural model proposed for IAPP20–29 was later refined with intercarbon distance measurements via rotational resonance (R2) experiments, thus allowing a two-dimensional description of the β-sheet pleated peptide IAPP20–29 (Griffiths et al. 1995). Rotational resonance is a MAS ssNMR technique that uses specific dipolar recoupling to obtain internuclear distances. Since the discovery of the R2 effect (Andrew et al. 1966), several pulse sequences have been implemented in order to extend the distance measurement possibilities: to name a few, the REDOR (rotational-echo double-resonance) pulse sequence and its close cousin TEDOR (transferred-echo double-resonance) were both designed to measure distances between unlike nuclei such as 13C and 15N (Gullion and Schaefer 1989; Hing et al. 1992), while R2 is generally a technique for alike nuclei (13C-13C typically). A greater diversity of complementary pulse sequences means a better structural characterization thanks to a wider range of measurable distances, such as intra- and interstrand as well as intra- and interprotofilaments distance measurements.
Similarly, the peptide Aβ34–42 was described with an unusual cis amide bond conformation via R2 measurements (Spencer et al. 1991), and later with a refined antiparallel β-sheet structure thanks to a combination of R2 measurements and 13C chemical shifts of carbonyl carbons and α-carbons (Lansbury Jr et al. 1995). Aβ16–22 was also studied by ssNMR, and forms antiparallel β-sheets all along the seven residues (Balbach et al. 2000). However, the fragment Aβ10–35 has been described with a parallel in-register structure with 13C-13C distances measured by the DRAWS (dipolar recoupling in a windowless sequence) pulse sequence, and has been found to be more representative of the parent Aβ peptides (Benzinger et al. 1998), the parallel configuration of the β-sheets having later been found in several polymorphs of the full Aβ peptides (Colvin et al. 2016; Petkova et al. 2005; Ravotti et al. 2016; Wälti et al. 2016; Xiao et al. 2015). Aβ peptides being indubitably the most studied amyloid peptides, it is to be noted that a recent review compiled the latest structural characterization studies carried on Aβ1–42. Notably, the authors underlie the encouraging similarities in the structures obtained in different laboratories, thereby emphasizing the strength of ssNMR for the study of amyloid fibrils, and more specifically the reliability of such studies. However, amongst the issues addressed in this review, the importance of the sample preparation control in order to work on monomorphs is particularly stressed (Meier et al. 2017).
ssNMR was also used to determine the structure of a fragment of the amyloid protein transthyretin, responsible for several neuropathies: TTR105–115. The authors used a combination of 2D and 3D spectra to obtain a total of 76 constraints on dihedral angles (41 constraints for 19 dihedral angles) and 13C-15N distance measurements (35 distances between 3 and 6 Å) (Jaroniec et al. 2004). To obtain such precise measurements, MAS ssNMR was used on uniformly 13C-15N-labeled peptides in consecutive stretches of four amino acids. Most of the 13C-15N distances were obtained with the pulse sequence 3D zf-TEDOR (3D z-filtered-TEDOR) (Jaroniec et al. 2002a). This pioneering work showed early on the strength of MAS ssNMR for precise amyloid fibril structural characterization. Prior to this refinement study, the same authors had used a large set of chemical shifts of 15N, 13CO, 13Cα, and 13Cβ to predict quantitatively the backbone tension angles, as well as backbone atom distances (13C-15N) with the REDOR pulse sequence (Jaroniec et al. 2002b).
The peptide IAPP20–29 was also characterized by ssNMR in two research groups that obtained different results. While Nielsen et al. described an antiparallel structure for IAPP20–29 with face-to-back contacts (Nielsen et al. 2009), Madine et al., almost at the same time, identified two forms of the same peptide, a parallel and an antiparallel configuration (Madine et al. 2008). Madine et al. used the DARR pulse sequence (dipolar-assisted rotational resonance (Takegoshi et al. 2001)) to identify 13C-13C cross-peaks that allowed, when combined with R2, the assignment of chemical shifts to certain residues in the antiparallel form. Similarly, Nielsen et al. used the DARR pulse sequence to determine long-range contacts between several amino acids, which allowed them to refine, along with the symmetry information obtained via R2, their model of IAPP20–29. These two studies underline the complexity brought to structural studies by amyloid fibril polymorphism, and particularly to ssNMR studies where result interpretation can be prone to ambiguities.
A fragment of the yeast protein Sup35 (GNNQQNY7–13) has also been studied via ssNMR. This fragment is of interest mainly because it can be perceived as a model peptide of the amyloidogenic process. van der Wel et al. used the DARR pulse sequence to identify and assign 13C chemical shifts in 2D spectra, and R2 width to measure intermolecular 13C = O/13C = O distances. 13C-15N distances were determined with 1D REDOR and 2D TEDOR pulse sequences. The authors identified three fibril conformers, each of which displayed in-register parallel β-sheets, but with variations along the strand backbone, revealed by differences in their 13C and 15N chemical shifts (van der Wel et al. 2010).
The group of Sharpe is particularly interested in the prion protein (PrP). They developed models for three fragments of this protein: PrP106–126, PrP244–249 and PrP245–250 (Walsh et al. 2009; Yau and Sharpe 2012). PrP106–126 is a peptide that shares many features with its parent protein, and is able to turn cellular PrP (PrPc) into its scrapie form (PrPSc), and perhaps the best argument to study this peptide is the ability of monoclonal antibodies to recognize both fibrils formed by PrP106–126 and PrPSc fibrils (Jones et al. 2009). PrP106–126 has been described as adopting a class 1 steric zipper configuration, with parallel β-strands stacking along the fibril long axis and protofilaments oriented in an antiparallel manner. 13C-13C distances were obtained via PITHIRDS (constant-time recoupling with π-pulses lasting one-third of the MAS rotation period (Tycko 2007)) for alike 13C atoms (interstrand distance) while long-range 13C-13C interactions (up to 6 Å) were probed with 13C 2D correlation spectra with long spin diffusion times, and complemented with R2 distance measurements to determine the tertiary and quaternary structures. Similarly, the structures of the two cytotoxic peptides PrP244–249 and PrP245–250 were determined with the use of basically the same pulse sequences (13C spin diffusion, R2, PITHIRDS and TEDOR) and found to adopt a class 1 steric zipper configuration for PrP244–249 and classes 2 and 3 for PrP245–250.
In the structural determinations presented herein, most of them used the chemical shift difference between the 13C chemical shifts of carbonyl, α- and β-carbons and the 13C chemical shift of an unordered peptide for secondary structure determination (Wishart and Sykes 1994). Moreover, most of the backbone angular constraints were predicted with the TALOS program algorithm based on the backbone 13C and 15N chemical shifts (Cornilescu et al. 1999).
Future developments in ssNMR
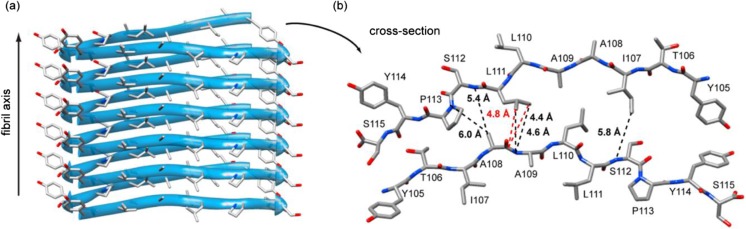
ssNMR studies are often time-consuming but higher magnetic fields increase spectral resolution and sensitivity, and so decrease the acquisition time. Higher MAS frequencies also increase the resolution, but then the sample quantity is limited due to the need to work with smaller rotors (i.e., 0.7 mm), which can be convenient to reduce the costs of labeled peptides/proteins required. A third possibility is the use of dynamic nuclear polarization (DNP). This technique uses the large polarization of biradical molecule electrons which is transferred by microwave irradiation to the nuclei of interest at low temperatures (90–100 K) (Maly et al. 2008). A probing study showing the efficiency of DNP-enhanced ssNMR was carried out on the amyloid fragment TTR105–115, in the laboratory of Griffin (Debelouchina et al. 2013). The authors reached the same conclusions as a decade earlier (Jaroniec et al. 2004) but obtained a total of 111 constraints against 76 previously. This study highlights more particularly the time saved by the use of DNP: when a buildup curve for a 13C-13C distance measurement (via DQF-DRAWS, double-quantum filtered DRAWS) takes 3.5 days without DNP at room temperature, the DNP-enhanced acquisition time is lowered to 1.5 h. Moreover, this article compiles the major distance measurements and the associated pulse sequences necessary for an amyloid fibril structural description: intrasheet contacts (parallel/antiparallel, in- or out-of-register), intersheet contacts (head-to-head or head-to-tail) and protofilament arrangement. An example of structural reconstruction at the protofilament level is displayed in Fig. 5.
Fig. 5.
Lateral view (a) and cross-section (b) of TTR105–115 protofilament. β-Sheets are parallel and in-register along the fibril axis (a) and stacked in an antiparallel manner at the protofilament interface (b). Distances indicated in (b) were obtained via TEDOR measurements (black) and R2TRW (red). Reprinted with permission from Debelouchina et al. 2013. Copyright © 2013 American Chemical Society
For a more extensive review of amyloid protein studies conducted by ssNMR, see Tycko (2006).
Concluding remarks
Fragments of amyloid peptides and proteins have revealed to be valuable tools to study the amyloidogenic process. While they do not always reflect the exact behavior of their parent protein, fragments have proven to be very useful when it comes to structural and dynamical description of amyloid fibrils. The stretches chosen often constitute the amyloidogenic core of the protein fibrils, and their study thus gives insight into the amyloidogenic process, the folding dynamics, and more specifically precise interactions between residues. Raman, IR, VCD, and ssNMR are versatile biophysical tools adapted for the structural characterization of amyloids. Each of them presents advantages and completes each other. Specific residue interactions and conformational elements may be probed using Raman scattering. IR and VCD are useful for fast secondary structure analysis, and supramolecular chirality in the case of VCD. More extensive and precise structural characterizations can be conducted by IR and VCD with the help of isotopic labeling of specific residues. ssNMR on the other hand is more complex but a lot more structural information can be obtained. ssNMR also stands out from the other techniques by its ability to detect polymorphs formed simultaneously. Modern multi-dimensional pulse sequences, coupled with technological advances, allow precise structural characterization, such as torsional angles, short- and long-range interactions, and distances in the 1–10 Å range.
Although not the main object of the present review, it is noteworthy to briefly mention the usefulness of imaging techniques and their added value in most studies. Even though high-resolution AFM can be used as a stand-alone technique, as for the description of the morphological inversion of insulin fibrils (Usov et al. 2013), it is most often used complementarily to other biophysical tools (Kurouski et al. 2014). Transmission electron microscopy (TEM) is probably the most widespread technique to routinely assess amyloid fibril morphologies alongside structural characterization, and was used in the majority of the works presented herein. Cryo-EM and STEM (cryo-electron microscopy and scanning transmission electron microscopy) are probably the most powerful imaging techniques, as they can be used for fibril reconstruction and mass-per-length measurements, respectively (Goldsbury et al. 2011; Jiménez et al. 2002). Used together, these techniques (AFM, STEM and cryo-EM), and coupled with ssNMR, have allowed the full structural characterization and reconstruction of TTR105–115 (Fitzpatrick et al. 2013).
Altogether, these biophysical tools, when applied to appropriate and relevant amyloid peptides, can provide insights into the molecular interactions taking place in amyloid fibrils and during amyloidogenesis. Such insights and a better “global” comprehension of these phenomena constitute an invaluable cornerstone to bring our knowledge of amyloid diseases to the next level, and thereby might offer new avenues of research for a currently-lacking efficient medication. Moreover, this type of research helps in better understanding the process of peptide assembly and the structures formed, which in turn may contribute to the development of new rationally-designed nanostructured materials for biomedical, optoelectronic, food science or other purposes.
Abbreviations
- 2D-IR
Two-dimensional infrared
- 3D zf-TEDOR
Three-dimensional z-filtered transferred-echo double-resonance
- A
Absorbance
- AD
Alzheimer’s disease
- AFM
Atomic force microscopy
- AS
α-Synuclein
- ATR-IR
Attenuated total reflection infrared
- Aβ
Amyloid-β
- Cryo-EM
Cryo-electron microscopy
- CSA
Chemical shift anisotropy
- DARR
Dipolar-assisted rotational resonance
- DFT
Density functional theory
- DNP
Dynamic nuclear polarization
- DQF-DRAWS
Double-quantum filtered dipolar recoupling in a windowless sequence
- DRAWS
Dipolar recoupling in a windowless sequence
- DUVRR
Deep-UV resonance Raman
- HD
Huntington’s disease
- IAPP
Islet amyloid polypeptide
- IR
Infrared
- MAS
Magic-angle spinning
- PD
Parkinson’s disease
- PITHIRDS
Constant-time recoupling with π-pulses lasting one-third of the MAS rotation period
- PolyQ
Polyglutamine
- PPII
Polyproline II helix
- PrP
Prion protein
- R2
Rotational resonance
- REDOR
Rotational-echo double-resonance
- ROA
Raman optical activity
- SEM
Scanning electron microscopy
- SERS
Surface-enhanced Raman spectroscopy
- ssNMR
Solid-state nuclear magnetic resonance
- STEM
Scanning transmission electron microscopy
- STM
Scanning tunneling microscopy
- TDC
Transition dipole coupling
- TEDOR
Transferred-echo double-resonance
- TEM
Transmission electron microscopy
- TERS
Tip-enhanced Raman spectroscopy
- TTR
Transthyretin
- VCD
Vibrational circular dichroism
- XRD
X-ray diffraction
Funding
The authors would like to acknowledge funding from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Fonds de recherche du Québec - Nature et Technologies (FRQNT), the Regroupement québécois de recherche sur la fonction, l’ingénierie et les applications des protéines (PROTEO), the Centre de recherche sur les matériaux avancés (CERMA) and the Centre québécois sur les matériaux fonctionnels (CQMF). B.M. would like to acknowledge graduate scholarships from Bionano (NSERC) and PROTEO.
Compliance with ethical standards
Benjamin Martial declares that he has no conflict of interest. Thierry Lefèvre declares that he has no conflict of interest. Michèle Auger declares that she has no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Alzheimer A. Über einen eigenartigen schweren Erkrankungsprozess der Hirnrinde. Neurologisches Centralblatt. 1906;23:1129–1136. [Google Scholar]
- Amenabar I, et al. Structural analysis and mapping of individual protein complexes by infrared nanospectroscopy. Nat Commun. 2013;4:2890. doi: 10.1038/ncomms3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ami D, et al. In situ characterization of protein aggregates in human tissues affected by light chain amyloidosis: a FTIR microspectroscopy study. Sci Rep. 2016;6:29096. doi: 10.1038/srep29096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen M, Lorenzen N, Otzen D. Interactions between misfolded protein oligomers and membranes: a central topic in neurodegenerative diseases? Biochim Biophys Acta Biomembr. 2015;1848:1897–1907. doi: 10.1016/j.bbamem.2015.01.018. [DOI] [PubMed] [Google Scholar]
- Andrew ER, Clough S, Farnell LF, Gledhill TD, Roberts I. Resonant rotational broadening of nuclear magnetic resonance spectra. Phys Lett. 1966;21:505–506. [Google Scholar]
- Ashburn TT, Auger M, Lansbury PT. The structural basis of pancreatic amyloid formation: isotope-edited spectroscopy in the solid state. J Am Chem Soc. 1992;114:790–791. [Google Scholar]
- Astbury WT, Dickinson S, Bailey K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J. 1935;29:2351–2360. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbach JJ, et al. Amyloid fibril formation by Aβ16-22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- Barron LD, Hecht L, Blanch EW, Bell AF. Solution structure and dynamics of biomolecules from Raman optical activity. Prog Biophys Mol Biol. 2000;73:1–49. doi: 10.1016/s0079-6107(99)00017-6. [DOI] [PubMed] [Google Scholar]
- Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- Bédard L, Lefèvre T, Morin-Michaud É, Auger M. Besides fibrillization: putative role of the peptide fragment 71–82 on the structural and assembly behavior of α-synuclein. Biochemistry. 2014;53:6463–6472. doi: 10.1021/bi5008707. [DOI] [PubMed] [Google Scholar]
- Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer’s β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K, Lecomte S, Géan J, Immel F, Cullin C. A yeast toxic mutant of HET-s(218-289) prion displays alternative intermediates of amyloidogenesis. Biophys J. 2010;99:1239–1246. doi: 10.1016/j.bpj.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanch EW, Gill AC, Rhie AGO, Hope J, Hecht L, Nielsen K, Barron LD. Raman optical activity demonstrates poly(L-proline) II helix in the N-terminal region of the ovine prion protein: implications for function and misfunction. J Mol Biol. 2004;343:467–476. doi: 10.1016/j.jmb.2004.08.058. [DOI] [PubMed] [Google Scholar]
- Bousset L, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Armstrong AH, Koehler AN, Hecht MH. Small molecule microarrays enable the discovery of compounds that bind the Alzheimer’s Aβ peptide and reduce its cytotoxicity. J Am Chem Soc. 2010;132:17015–17022. doi: 10.1021/ja107552s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z, Chen XG, Holtz JSW, Asher SA. UV resonance Raman-selective amide vibrational enhancement: quantitative methodology for determining protein secondary structure. Biochemistry. 1998;37:2854–2864. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- Collier TJ, et al. Nortriptyline inhibits aggregation and neurotoxicity of alpha-synuclein by enhancing reconfiguration of the monomeric form. Neurobiol Dis. 2017;106:191–204. doi: 10.1016/j.nbd.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin MT, et al. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KA, Harper JD, Lansbury PT. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Debelouchina GT, et al. Higher order amyloid fibril structure by MAS NMR and DNP spectroscopy. J Am Chem Soc. 2013;135:19237–19247. doi: 10.1021/ja409050a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur SM. Elucidation of residue-level structure and dynamics of polypeptides via isotope-edited infrared spectroscopy. Acc Chem Res. 2006;39:169–175. doi: 10.1021/ar050135f. [DOI] [PubMed] [Google Scholar]
- Dornhaus R, Benner RE, Chang RK, Chabay I. Surface plasmon contribution to SERS. Surf Sci. 1980;101:367–373. [Google Scholar]
- Dukor RK, Keiderling TA. Reassessment of the random coil conformation: vibrational CD study of proline oligopeptides and related polypeptides. Biopolymers. 1991;31:1747–1761. doi: 10.1002/bip.360311409. [DOI] [PubMed] [Google Scholar]
- Dunkelberger EB, Buchanan LE, Marek P, Cao P, Raleigh DP, Zanni MT. Deamidation accelerates amyloid formation and alters amylin fiber structure. J Am Chem Soc. 2012;134:12658–12667. doi: 10.1021/ja3039486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzwolak W, Loksztejn A, Smirnovas V. New insights into the self-assembly of insulin amyloid fibrils: an H−D exchange FT-IR study. Biochemistry. 2006;45:8143–8151. doi: 10.1021/bi060341a. [DOI] [PubMed] [Google Scholar]
- Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discov. 2015;14:759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DS, Sawaya MR. Structural studies of amyloid proteins at the molecular level. Annu Rev Biochem. 2017;86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- Eker F, Griebenow K, Schweitzer-Stenner R. Aβ1-28 fragment of the amyloid peptide predominantly adopts a polyproline II conformation in an acidic solution. Biochemistry. 2004;43:6893–6898. doi: 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AWP, et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc Natl Acad Sci U S A. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagnion C. The role of lipids interacting with alpha-synuclein in the pathogenesis of Parkinson’s disease. J Parkinsons Dis. 2017;7:433–450. doi: 10.3233/JPD-171103. [DOI] [PubMed] [Google Scholar]
- Gath J, Bousset L, Habenstein B, Melki R, Böckmann A, Meier BH. Unlike twins: an NMR comparison of two α-synuclein polymorphs featuring different toxicity. PLoS One. 2014;9:e90659. doi: 10.1371/journal.pone.0090659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand P, Smith RJ, Stavitski E, Borchelt DR, Miller LM. Characterization of protein structural changes in living cells using time-lapsed FTIR imaging. Anal Chem. 2015;87:6025–6031. doi: 10.1021/acs.analchem.5b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Murray IVJ, Trojanowski JQ, Lee VM-Y. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Eanes ED, Bladen HA, Linke RP, Termine JD. β-Pleated sheet fibrils. A comparison of native amyloid with synthetic protein fibrils. J Histochem Cytochem. 1974;22:1141–1158. doi: 10.1177/22.12.1141. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, et al. Amyloid structure and assembly: insights from scanning transmission electron microscopy. J Struct Biol. 2011;173:1–13. doi: 10.1016/j.jsb.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JM, Ashburn TT, Auger M, Costa PR, Griffin RG, Lansbury PT. Rotational resonance solid-state NMR elucidates a structural model of pancreatic amyloid. J Am Chem Soc. 1995;117:3539–3546. [Google Scholar]
- Guenther EL, et al. Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2. Nat Struct Mol Biol. 2018;25:311–319. doi: 10.1038/s41594-018-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson (1969) 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Guo Y, Wang J. Spectroscopic evidence for polymorphic aggregates formed by amyloid-β fragments. ChemPhysChem. 2012;13:3901–3908. doi: 10.1002/cphc.201200611. [DOI] [PubMed] [Google Scholar]
- Halverson K, Fraser PE, Kirschner DA, Lansbury PT. Molecular determinants of amyloid deposition in Alzheimer’s disease: conformational studies of synthetic β-protein fragments. Biochemistry. 1990;29:2639–2644. doi: 10.1021/bi00463a003. [DOI] [PubMed] [Google Scholar]
- Halverson KJ, Sucholeiki I, Ashburn TT, Lansbury PT. Location of β-sheet-forming sequences in amyloid proteins by FTIR. J Am Chem Soc. 1991;113:6701–6703. [Google Scholar]
- Henry S, Bercu NB, Bobo C, Cullin C, Molinari M, Lecomte S. Interaction of Aβ1-42 peptide or their variant with model membrane of different composition probed by infrared nanospectroscopy. Nanoscale. 2018;10:936–940. doi: 10.1039/c7nr07489a. [DOI] [PubMed] [Google Scholar]
- Hing AW, Vega S, Schaefer J. Transferred-echo double-resonance NMR. J Magn Reson (1969) 1992;96:205–209. [Google Scholar]
- Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. Dependence of α-synuclein aggregate morphology on solution conditions. J Mol Biol. 2002;322:383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- Hoyer W, Grönwall C, Jonsson A, Ståhl S, Härd T. Stabilization of a β-hairpin in monomeric Alzheimer’s amyloid-β peptide inhibits amyloid formation. Proc Natl Acad Sci U S A. 2008;105:5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Mantsch HH. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit Rev Biochem Mol Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon−nitrogen distances in uniformly 13C, 15N-labeled solids. J Am Chem Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc Natl Acad Sci U S A. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci U S A. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci U S A. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Connelly S, Fearns C, Powers ET, Kelly JW. The transthyretin amyloidoses: from delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J Mol Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, et al. An antibody to the aggregated synthetic prion protein peptide (PrP106–126) selectively recognizes disease-associated prion protein (PrPSc) from human brain specimens. Brain Pathol. 2009;19:293–302. doi: 10.1111/j.1750-3639.2008.00181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczyk P, Kołodziejczyk AS, Grzonka Z. FTIR spectroscopic studies on aggregation process of the β-amyloid 11–28 fragment and its variants. J Pept Sci. 2009;15:23–29. doi: 10.1002/psc.1085. [DOI] [PubMed] [Google Scholar]
- Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- Krasnoslobodtsev AV, Deckert-Gaudig T, Zhang Y, Deckert V, Lyubchenko YL. Polymorphism of amyloid fibrils formed by a peptide from the yeast prion protein Sup35: AFM and tip-enhanced Raman scattering studies. Ultramicroscopy. 2016;165:26–33. doi: 10.1016/j.ultramic.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm S, Bandekar J (1986) Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem 38:181–364. 10.1016/S0065-3233(08)60528-8 [DOI] [PubMed]
- Kurouski D, Dukor RK, Lu X, Nafie LA, Lednev IK. Spontaneous inter-conversion of insulin fibril chirality. Chem Commun. 2012;48:2837–2839. doi: 10.1039/c2cc16895b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurouski D, Kar K, Wetzel R, Dukor RK, Lednev IK, Nafie LA. Levels of supramolecular chirality of polyglutamine aggregates revealed by vibrational circular dichroism. FEBS Lett. 2013;587:1638–1643. doi: 10.1016/j.febslet.2013.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurouski D, et al. Is supramolecular filament chirality the underlying cause of major morphology differences in amyloid fibrils? J Am Chem Soc. 2014;136:2302–2312. doi: 10.1021/ja407583r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurouski D, Handen JD, Dukor RK, Nafie LA, Lednev IK. Supramolecular chirality in peptide microcrystals. Chem Commun. 2015;51:89–92. doi: 10.1039/c4cc05002a. [DOI] [PubMed] [Google Scholar]
- Kurouski D, Van Duyne RP, Lednev IK. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review. Analyst. 2015;140:4967–4980. doi: 10.1039/c5an00342c. [DOI] [PubMed] [Google Scholar]
- Labbé J-F, Lefèvre T, Guay-Bégin A-A, Auger M. Structure and membrane interactions of the β-amyloid fragment 25-35 as viewed using spectroscopic approaches. Phys Chem Chem Phys. 2013;15:7228–7239. doi: 10.1039/c3cp44623a. [DOI] [PubMed] [Google Scholar]
- Lansbury Jr PT, et al. Structural model for the β-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- Lednev IK, Ermolenkov VV, He W, Xu M. Deep-UV Raman spectrometer tunable between 193 and 205 nm for structural characterization of proteins. Anal Bioanal Chem. 2005;381:431–437. doi: 10.1007/s00216-004-2991-5. [DOI] [PubMed] [Google Scholar]
- Lomont JP, Rich KL, Maj M, Ho J-J, Ostrander JS, Zanni MT. Spectroscopic signature for stable β-amyloid fibrils versus β-sheet-rich oligomers. J Phys Chem B. 2018;122:144–153. doi: 10.1021/acs.jpcb.7b10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, et al. Vibrational circular dichroism shows unusual sensitivity to protein fibril formation and development in solution. J Am Chem Soc. 2007;129:12364–12365. doi: 10.1021/ja074188z. [DOI] [PubMed] [Google Scholar]
- Madine J, Jack E, Stockley PG, Radford SE, Serpell LC, Middleton DA. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20−29 of amylin revealed by solid-state NMR and X-ray fiber diffraction. J Am Chem Soc. 2008;130:14990–15001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- Maiti NC, Apetri MM, Zagorski MG, Carey PR, Anderson VE. Raman spectroscopic characterization of secondary structure in natively unfolded proteins: α-synuclein. J Am Chem Soc. 2004;126:2399–2408. doi: 10.1021/ja0356176. [DOI] [PubMed] [Google Scholar]
- Maly T, et al. Dynamic nuclear polarization at high magnetic fields. J Chem Phys. 2008;128:052211. doi: 10.1063/1.2833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measey TJ, Schweitzer-Stenner R. Vibrational circular dichroism as a probe of fibrillogenesis: the origin of the anomalous intensity enhancement of amyloid-like fibrils. J Am Chem Soc. 2011;133:1066–1076. doi: 10.1021/ja1089827. [DOI] [PubMed] [Google Scholar]
- Meier BH, Riek R, Böckmann A. Emerging structural understanding of amyloid fibrils by solid-state NMR. Trends Biochem Sci. 2017;42:777–787. doi: 10.1016/j.tibs.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. Peptide secondary structure folding reaction coordinate: correlation between UV Raman amide III frequency, Ψ Ramachandran angle, and hydrogen bonding. J Phys Chem B. 2006;110:1928–1943. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- Miura T, Hori-i A, Mototani H, Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- Miura T, Suzuki K, Takeuchi H. Binding of iron(III) to the single tyrosine residue of amyloid β-peptide probed by Raman spectroscopy. J Mol Struct. 2001;598:79–84. [Google Scholar]
- Murphy RM. Kinetics of amyloid formation and membrane interaction with amyloidogenic proteins. Biochim Biophys Acta Biomembr. 2007;1768:1923–1934. doi: 10.1016/j.bbamem.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, et al. Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy. Angew Chem Int Ed. 2009;48:2118–2121. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- Oladepo SA, Xiong K, Hong Z, Asher SA, Handen J, Lednev IK. UV resonance Raman investigations of peptide and protein structure and dynamics. Chem Rev. 2012;112:2604–2628. doi: 10.1021/cr200198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. An essay on the shaking palsy. J Neuropsychiatr Clin Neurosci. 2002;14:223–236. doi: 10.1176/jnp.14.2.223. [DOI] [PubMed] [Google Scholar]
- Paterlini MG, Freedman Teresa B, Nafie Laurence A. Vibrational circular dichroism spectra of three conformationally distinct states and an unordered state of poly(L-lysine) in deuterated aqueous solution. Biopolymers. 1986;25:1751–1765. doi: 10.1002/bip.360250915. [DOI] [PubMed] [Google Scholar]
- Paulite M, Blum C, Schmid T, Opilik L, Eyer K, Walker GC, Zenobi R. Full spectroscopic tip-enhanced Raman imaging of single nanotapes formed from β-amyloid(1–40) peptide fragments. ACS Nano. 2013;7:911–920. doi: 10.1021/nn305677k. [DOI] [PubMed] [Google Scholar]
- Perálvarez-Marín A, Barth A, Gräslund A. Time-resolved infrared spectroscopy of pH-induced aggregation of the Alzheimer Aβ1–28 peptide. J Mol Biol. 2008;379:589–596. doi: 10.1016/j.jmb.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau W-M, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Petty SA, Decatur SM. Intersheet rearrangement of polypeptides during nucleation of β-sheet aggregates. Proc Natl Acad Sci U S A. 2005;102:14272–14277. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyanichko AM. Vibrational circular dichroism and its applications to protein studies. In: Uversky VN, Permyakov EA, editors. Methods in protein structure and stability analysis: vibrational spectroscopy. New York: Nova Science Publishers, Inc.; 2007. pp. 267–302. [Google Scholar]
- Popova LA, Kodali R, Wetzel R, Lednev IK. Structural variations in the cross-β core of amyloid β fibrils revealed by deep UV resonance Raman spectroscopy. J Am Chem Soc. 2010;132:6324–6328. doi: 10.1021/ja909074j. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Groth DF, Bolton DC, Kent SB, Hood LE. Purification and structural studies of a major scrapie prion protein. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- Punihaole D, Workman RJ, Hong Z, Madura JD, Asher SA. Polyglutamine fibrils: new insights into antiparallel β-sheet conformational preference and side chain structure. J Phys Chem B. 2016;120:3012–3026. doi: 10.1021/acs.jpcb.5b11380. [DOI] [PubMed] [Google Scholar]
- Ravotti F, Wälti MA, Güntert P, Riek R, Böckmann A, Meier BH. Solid-state NMR sequential assignment of an amyloid-β(1–42) fibril polymorph. Biomol NMR Assign. 2016;10:269–276. doi: 10.1007/s12104-016-9682-y. [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Sarroukh R, Goormaghtigh E, Ruysschaert J-M, Raussens V. ATR-FTIR: a “rejuvenated” tool to investigate amyloid proteins. Biochim Biophys Acta Biomembr. 2013;1828:2328–2338. doi: 10.1016/j.bbamem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Schweitzer-Stenner R, Measey TJ. Simulation of IR, Raman and VCD amide I band profiles of self-assembled peptides. Spectroscopy. 2010;24:25–36. [Google Scholar]
- Selkoe DJ. Chapter 67—Alzheimer disease. In: Pascual JM, editor. Rosenberg’s molecular and genetic basis of neurological and psychiatric disease. 5. Boston: Academic Press; 2015. pp. 753–768. [Google Scholar]
- Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Molecular Medicine. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam G, Polavarapu PL. Structure of Aβ(25-35) peptide in different environments. Biophys J. 2004;87:622–630. doi: 10.1529/biophysj.104.040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam G, Polavarapu PL. Isotope-assisted vibrational circular dichroism investigations of amyloid β peptide fragment, Aβ(16–22) J Struct Biol. 2011;176:212–219. doi: 10.1016/j.jsb.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Shanmugasundaram M, Kurouski D, Wan W, Stubbs G, Dukor RK, Nafie LA, Lednev IK. Rapid filament supramolecular chirality reversal of HET-s (218–289) prion fibrils driven by pH elevation. J Phys Chem B. 2015;119:8521–8525. doi: 10.1021/acs.jpcb.5b04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Smith AA, Ravotti F, Testori E, Cadalbert R, Ernst M, Böckmann A, Meier BH. Partially-deuterated samples of HET-s(218–289) fibrils: assignment and deuterium isotope effect. J Biomol NMR. 2017;67:109–119. doi: 10.1007/s10858-016-0087-0. [DOI] [PubMed] [Google Scholar]
- Soriaga AB, Sangwan S, Macdonald R, Sawaya MR, Eisenberg D. Crystal structures of IAPP amyloidogenic segments reveal a novel packing motif of out-of-register beta sheets. J Phys Chem B. 2016;120:5810–5816. doi: 10.1021/acs.jpcb.5b09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer RGS, Halverson KJ, Auger M, McDermott AE, Griffin RG, Lansbury PT. An unusual peptide conformation may precipitate amyloid formation in Alzheimer’s disease: application of solid-state NMR to the determination of protein secondary structure. Biochemistry. 1991;30:10382–10387. doi: 10.1021/bi00107a004. [DOI] [PubMed] [Google Scholar]
- Stanek J, et al. NMR spectroscopic assignment of backbone and side-chain protons in fully protonated proteins: microcrystals, sedimented assemblies, and amyloid fibrils. Angew Chem Int Ed. 2016;55:15504–15509. doi: 10.1002/anie.201607084. [DOI] [PubMed] [Google Scholar]
- Stefani M. Structural features and cytotoxicity of amyloid oligomers: implications in Alzheimer’s disease and other diseases with amyloid deposits. Prog Neurobiol. 2012;99:226–245. doi: 10.1016/j.pneurobio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Surewicz WK, Mantsch HH, Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993;32:389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- Tagliavini F, et al. Synthetic peptides homologous to prion protein residues 106-147 form amyloid-like fibrils in vitro. Proc Natl Acad Sci U S A. 1993;90:9678–9682. doi: 10.1073/pnas.90.20.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- Termine JD, Eanes ED, Ein D, Glenner GG. Infrared spectroscopy of human amyloid fibrils and immunoglobulin proteins. Biopolymers. 1972;11:1103–1113. doi: 10.1002/bip.1972.360110512. [DOI] [PubMed] [Google Scholar]
- Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usov I, Adamcik J, Mezzenga R. Polymorphism complexity and handedness inversion in serum albumin amyloid fibrils. ACS Nano. 2013;7:10465–10474. doi: 10.1021/nn404886k. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in α-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- van der Wel PCA, Lewandowski JR, Griffin RG. Structural characterization of GNNQQNY amyloid fibrils by magic angle spinning NMR. Biochemistry. 2010;49:9457–9469. doi: 10.1021/bi100077x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, AB E, Loquet A, Böckmann A, Meier BH. Atomic-resolution three-dimensional structure of HET-s(218−289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- Viles JH. Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord Chem Rev. 2012;256:2271–2284. [Google Scholar]
- Volpatti LR, Vendruscolo M, Dobson CM, Knowles TPJ. A clear view of polymorphism, twist, and chirality in amyloid fibril formation. ACS Nano. 2013;7:10443–10448. doi: 10.1021/nn406121w. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Walsh P, Simonetti K, Sharpe S. Core structure of amyloid fibrils formed by residues 106–126 of the human prion protein. Structure. 2009;17:417–426. doi: 10.1016/j.str.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Wälti MA, et al. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc Natl Acad Sci U S A. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WRW, Keiderling TA, Kubelka J. Structural analyses of experimental 13C edited amide I’ IR and VCD for peptide β-sheet aggregates and fibrils using DFT-based spectral simulations. J Phys Chem B. 2013;117:10359–10369. doi: 10.1021/jp405613r. [DOI] [PubMed] [Google Scholar]
- Welch WRW, Kubelka J, Keiderling TA. Infrared, vibrational circular dichroism, and Raman spectral simulations for β-sheet structures with various isotopic labels, interstrand, and stacking arrangements using density functional theory. J Phys Chem B. 2013;117:10343–10358. doi: 10.1021/jp4056126. [DOI] [PubMed] [Google Scholar]
- Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic structure of the cross-β spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Xiao Y, et al. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau J, Sharpe S. Structures of amyloid fibrils formed by the prion protein derived peptides PrP(244–249) and PrP(245–250) J Struct Biol. 2012;180:290–302. doi: 10.1016/j.jsb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Yugay D, et al. Copper ion binding site in β-amyloid peptide. Nano Lett. 2016;16:6282–6289. doi: 10.1021/acs.nanolett.6b02590. [DOI] [PubMed] [Google Scholar]