Abstract
Several morphology- and polymerase chain reaction (PCR)-based methods for chromosome 1p 19q deletion status assessment are available. Important prerequisites for all molecular techniques concern tissue quality and selection of regions of interest. The most common methods for diagnostic 1p 19q assessment are fluorescence in situ hybridization and PCR-based microsatellite analysis. While the latter requires the use of autologous blood samples, more advanced techniques such as array comparative genomic hybridization, multiplex ligation-dependent probe amplification or real-time PCR are independent from autologous DNA samples. However, due to high technical demand and experience required their applicability as diagnostic tests remains to be shown. On the other hand, chromogenic in situ hybridization evolves as attractive alternative to FISH. Herein, the available test methods are reviewed and outlined, their advantages and drawbacks being discussed in detail.
KEYWORDS : 1p 19q deletion, analytical performance, clinical performance, oligodendroglial tumors
Practice points.
1p 19q testing has become diagnostic standard in diffuse gliomas.
For routine diagnostic assessments, FISH, PCR-based microsatellite analysis and MLPA are still widely used techniques with novel techniques upcoming.
Concordance of test results across different platforms suggest validity, but intra- and interlaboratory comparisons and consensus recommendations for test evaluation shall be further scrutinized.
Rationale for 1p 19q testing
Technical advances in genomic, proteomic and methylation profiling have resulted in the identification of a large number of molecular markers of putative prognostic and/or predictive value in gliomas (see Box 1). Increasingly, clinical trials are incorporating tissue-based analyses to study the value of these markers aiming at the development of personalized treatment approaches. Despite the huge number of proposed candidate markers, only few have translated into routine clinical use, so far. Whether and how fast molecular markers translate from bench to bedside largely depends on their clinical utility comprising analytical and clinical performances (see Box 2) [1].
Box 1. . Biomarker definition in the clinical context.
Biomarkers in the clinical context are defined as objectively measurable patient-related factors which provide clinically meaningful disease-related information with regard to diagnosis, prognosis, therapy decisions and patient follow-up [1].
Box 2. . Clinical utility of a molecular marker.
The clinical utility of a molecular marker is based on its analytical and clinical (prognostic/predictive) performances. Based on different levels-of-evidence the reliability of a marker is rated from unclear performance (suggested by small case series or single-center reports) to robust performance (confirmed by one or more adequately designed round robin tests or prospective clinical trials) [1].
With regard to gliomas, prominent examples include the IDH1 mutation in diffuse gliomas [2,3], BRAF gene fusion in pilocytic astrocytomas [3], MGMT promoter methylation status in glioblastomas [4] and codeletion of chromosomal arms 1p 19q in oligodendroglial tumors [5,6].
Among the above-mentioned molecular markers, the 1p 19q codeletion probably constitutes the best characterized and most extensively studied marker, so far [6]. The combined deletion is mediated by a balanced whole-arm translocation of chromosomes 1 and 19, leading to the formation of two derivative chromosomes. One of these derivative chromosomes being composed of 1p and 19q (der [1, 19][p10; q10]) is typically lost [7,8]. The 1p 19q deletion is the genetic hallmark of oligodendrogliomas with a prevalence of above 80% among pure oligodendrogliomas and roughly 40% among oligoastrocytomas [9,10]. Virtually all 1p 19q codeleted tumors harbor an accompanying IDH1/IDH2 mutation [11,12]. In contrast, they practically never show EGFR amplification and/or 10q loss, which is frequent in glioblastomas [13,14]. Interestingly, among 1p 19q codeleted tumors, tumors with polysomies have relatively worse outcome compared with those without polysomy [15].
Clinical performance
Two large prospective randomized clinical trials have meanwhile provided evidence for a strong positive prognostic value of the combined deletion [16,17]. Patients with 1p 19q codeleted tumors show increased overall survival and are more likely to respond to chemotherapy [16,17]. In addition to its value as prognostic and predictive marker, there is also a role as a diagnostic aid in cases of morphologically uncharacteristic oligodendroglioma mimicks [18].
In summary, the clinical interest in this marker has increased over the last decade and has led to the implementation of its testing in the majority of neuropathology laboratories [6].
Indications for 1p 19q testing
Information on the 1p 19q status is of special relevance within the setting of clinical trials, where it serves as an important stratification factor (e.g., European Organization for Research and Treatment of Cancer (EORTC) trial 22033–26033, EORTC 26053–22054 CATNON versus EORTC 26081-NCCTG N0577 CODEL) [6];
Information on the 1p 19q status is helpful in the routine clinical setting for prognostic assessment and aids in making therapeutic decisions [19–22];
1p 19q testing is a useful diagnostic aid in morphologically challenging cases to substantiate the diagnosis of an oligodendroglioma [18]. However, according to the current WHO 2007 consensus criteria, the neuropathological diagnosis of an oligodendroglioma remains morphology-based irrespective of the deletion status [23];
1p 19q testing is useful in case of pure oligodendroglioma and mixed oligoastrocytoma, whereas it cannot be generally recommended in pure astrocytomas as the prevalence of the deletion is rare [24];
Repeated 1p 19q testing in case of tumor recurrence seems not useful as the deletion typically constitutes an early genetic event [25];
1p 19q testing is of secondary importance in pediatric and adolescent oligodendroglial tumors as the prevalence of the deletion is rare in those cases [25].
Overview of available test methods
The 1p 19q deletion status can be analyzed with various molecular-genetic methods including FISH, comparative genomic hybridization (CGH), chromogenic in situ hybridization (CISH), PCR-based microsatellite analysis, real-time comparative quantitative PCR and multiplex ligation-dependent probe amplification (MLPA). The two most common methods for routine diagnostic use are FISH and PCR-based microsatellite analysis.
General remarks on tissue selection
Whereas fresh-frozen (FF) tissue is ideal for a broad spectrum of molecular analyses, its storage and handling are complex and cost intense [26]. In contrast, formalin-fixed paraffin-embedded (FFPE) specimens are easy to handle and economic to store, but their applicability for molecular methods is restricted [26]. Formalin fixation inevitably leads to excessive molecular cross-linking compromising DNA, RNA and protein integrities [26]. Even though FF tissue warrants optimal DNA, RNA and protein qualities, samples are mostly used for research purposes. On a practical, routine diagnostic level the majority of tumor samples are only available as FFPE tissues from pathology archives [27]. To address this issue, molecular-genetic techniques including array CGH or MLPA have been optimized and further adapted for the use on FFPE tissues with promising results [28]. Novel tissue fixatives such as RCL2 or HOPE, which warrant better preservation of DNA, RNA and protein structures, are thus evolving as promising alternatives and deserve further attention [26,29].
Irrespective of the molecular-genetic technique chosen, a high content of viable tumor tissue with limited areas of necrosis and low contamination from non-neoplastic cells is optimal. Therefore, histological review of selected tissue areas is of high relevance. Nevertheless, some techniques including PCR-based approaches are more susceptible to contamination with normal tissues compared with more robust techniques such as FISH/CISH.
Fluorescence in situ hybridization
• Methodological background
FISH is a general cytogenetic technique, which is used to detect the presence or absence of specific DNA sequences on chromosomes. FISH uses fluorescently labeled locus-specific probes, which bind to complementary DNA sequences. The hybridization result is then visualized by fluorescence microscopy and morphologically evaluated.
• Material & methods
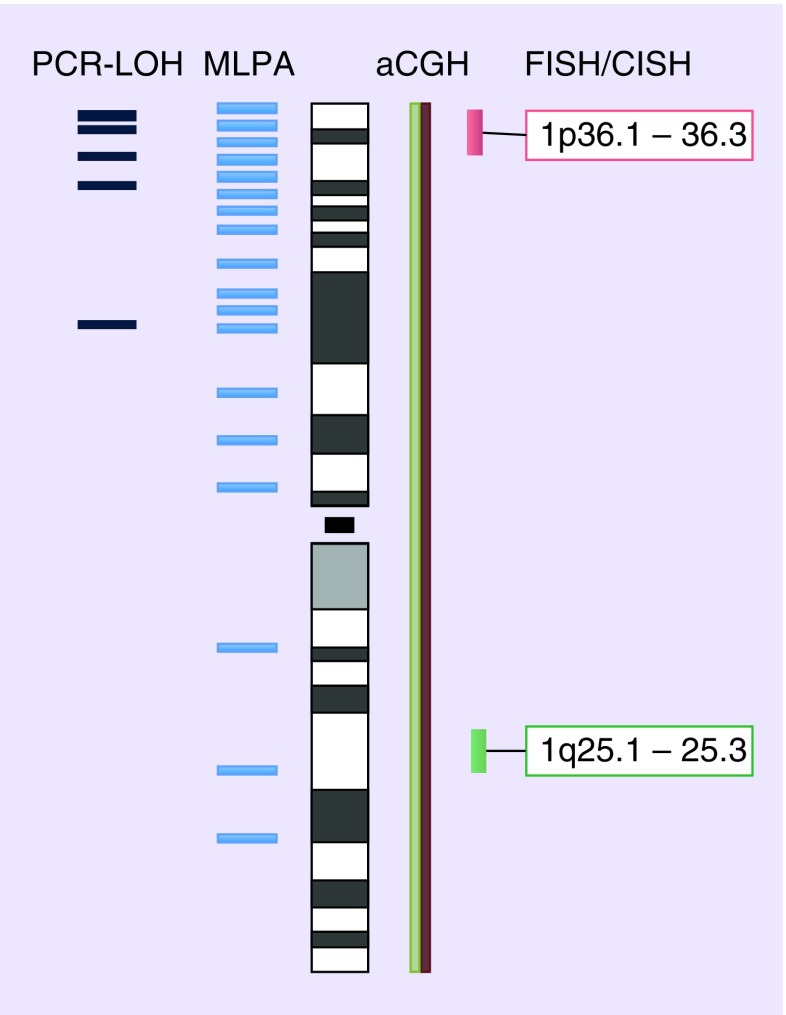
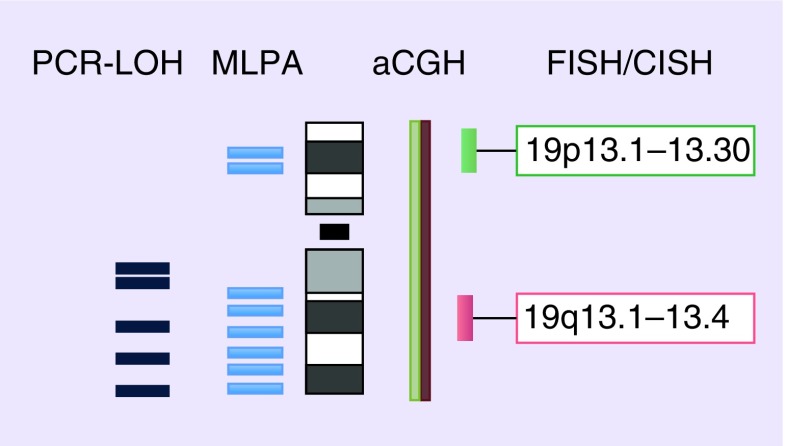
FISH is an approved method for molecular genetic testing on routinely available FFPE tissues [30]. It can be performed on single isolated cells or tissue sections. It does not require reference tissues such as autologous blood samples. Working steps include tissue pretreatment, probe hybridization, washing procedures and morphological evaluation [6]. Many companies provide ready-to-use FISH kits containing all necessary reagents for pretreatment and washing procedures as well as locus-specific DNA probes, hybridization protocols and technical support. Recently, a consensus protocol of the Medical Universities of Vienna and Innsbruck (MUV/MUI protocol) has been made publicly available [6]. Briefly, chromosomes 1 and 19 are tested separately using dual-color FISH probes combining target-specific probes hybridizing to subtelomeric regions of 1p36 and 19q13 with reference/control probes on 1q and 19p, respectively (see Figures 1 & 2).
Figure 1. . Evaluated regions on chromosome 1.
aCGH: Array comparative genomic hybridization; CISH: Chromogenic in situ hybridization; LOH: Loss of heterozygosity; MLPA: Multiplex ligation-dependent probe amplification.
Figure 2. . Evaluated regions on chromosome 19.
aCGH: Array comparative genomic hybridization; CISH: Chromogenic in situ hybridization; LOH: Loss of heterozygosity; MLPA: Multiplex ligation-dependent probe amplification.
• Evaluation
The hybridization result is evaluated with the use of a fluorescence microscope equipped with appropriate filters for DAPI as well as the two fluorophores (dual color probes) utilized. As the fluorescence signals fade over time, digital images are necessary for documentation and archiving purposes. Signal ratios of 100–200 nonoverlapping nuclei are assessed separately for chromosomes 1 and 19 [31]. Whereas normal nuclei show a diploid signal ratio of 2/2, a nucleus is considered to display a deletion if the target signal is 0 or 1 in relation to normal or excess control signals (e.g., 2/0, 2/1, 3/0, 3/1) (see Figure 3 ). With regard to chromosomal polysomies, FISH cannot resolve whether a relative loss of the target (4/2, 5/3) corresponds to a hemizygous deletion in presence of reduplication or not. The fraction of nuclei displaying a deletion or relative deletion in presence of polysomy are summed and expressed as percentages. If the fraction of ‘deleted’ nuclei exceeds a certain cut-off, the tumor is considered ‘1p 19q deleted’. So far, there is no consensus on the exact cut-off levels ranging from as low as 20% to as high as 70% [32,33]. Empiric analysis points toward maximal sensitivity and specificity with cut-offs lower than 0.75 per probe pair or at least 40% of tumor nuclei showing relative deletion of both 1p36 and 19q13. In this respect, the presence of large necrotic areas and/or high contamination from non-neoplastic cells, for example, endothelial cells or microglia is a major issue and requires histological review of selected tissue areas [6,24,34].
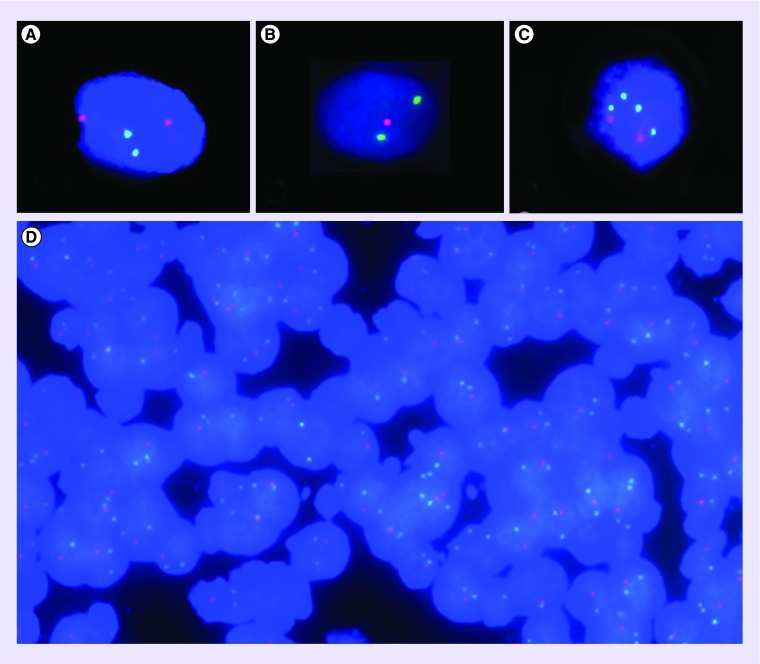
Figure 3. . Representative 1p 19q FISH images.
Target signal: red; control signal: green; DAPI counterstained. (A) Normal diploid signal ratio (2 controls/2 targets), magnification 63×. (B) Deletion status (signal ratio 2 controls/1 target), magnification 63×. (C) Imbalance with relative loss (4 control/2 target signals), magnification 63×. (D) FISH on a 4-micron-thick tissue section (magnification 40×) displaying a case with deletion status (signal ratio 2 controls/1 target) of the majority of nuclei.
• Advantages
FISH allows straightforward morphological evaluation of numerical genomic abnormalities at the cytogenetic level [32];
As FISH preserves the architecture of fixed tissue sections and cytological features, the evaluation is morphology based, which is intuitive to pathologists in general;
Due to numerous FISH applications in general pathology, it is a widely available technique, which has been already implemented in many histology laboratories.
• Shortcomings
FISH probes are typically at least 20 Kb in size and small intragenic events remain undetected by this technique [35] One probe typically targets telomeric regions at 1p36 and thus, in contrast to array CGH, are unable to discriminate between full-arm and partial deletions pertaining to this region, which is of relevance as both are associated with distinct clinical outcomes [36];
FISH is unable to proof loss of heterozygosity (LOH) in presence of polysomy [6];
FISH requires a high labor input and the evaluation is time consuming;
FISH necessitates expensive equipment (fluorescence microscope, digital camera for documentation and archival storage, reagents and solutions);
If FISH is performed on fixed sections nuclear truncation artifacts might occur, which can be avoided when using touch preparations [37] or isolated cell nuclei 31;
In a small fraction of cases hybridization failures with absent or only weak signals may occur [38];
There is no consensus on exact cut-off levels which define a deletion 32.
• Analytical performance
FISH is very sensitive for detecting whole-arm codeletions. A number of studies indicate excellent concordance of test results with PCR-based microsatellite analysis and CGH [32,37,39–42]. However, the sensitivity is lower compared with PCR-based microsatellite analysis because some high-grade gliomas have random interstitial deletions on multiple chromosomes including 1p36 and 19q13 [34,36].
Comparative genomic hybridization
• Methodological background
Comparative genomic hybridization allows for analysis of copy number changes, in other words, losses, deletions, gains and amplifications. It is based on the in situ hybridization of differentially labeled tumor and reference DNA to normal human metaphase chromosomes. It has a resolution of 5–10 megabases but requires the use of reference DNA. The differentially labeled fluorescence signals are compared along entire chromosomes for identification of differences between the two DNA sources. A higher intensity of the reference DNA in a specific chromosomal region indicates loss of genetic material in the tumor DNA. The more specific form of array CGH uses DNA microarrays instead of metaphase spreads. Array CGH allows for high-resolution locus-specific DNA measures [43].
• Materials & methods
CGH requires the isolation of DNA from tumor tissue (FF tissue preferred, but FFPE tissues also potentially possible) and reference tissue (mostly leukocytes of healthy male and female donors). DNA is subsequently differentially labeled with fluorophores and competitively cohybridized to metaphase spreads from phytohemagglutinin-stimulated peripheral blood lymphocytes obtained from healthy normal male donors or commercial providers. Instead of metaphase spreads, commercially available oligonucleotide platforms are used for array CGH. Whereas the majority of CGH arrays are intended for research purposes, first array CGH analysis are offered for the routine diagnostic setting [44].
• Evaluation
The evaluation of hybridization results requires an adequately equipped fluorescence microscope (see FISH). High spatial resolution images are then processed by dedicated and commercially available CGH softwares. A ‘relative copy number karyotype’ which presents chromosomal areas of deletions or amplifications is generated by averaging the ratios of several metaphases and plotting them along an ideogram, a diagram identifying chromosomes based on banding patterns. Interpretation of ratio profiles depends on fixed or statistical thresholds. Copy number deviations between tumor and reference DNA are defined as loss with ratios <0.8 and gain >1.2.
• Advantages
• Shortcomings
CGH is labor intense and requires an expert person skilled in chromosome identification [39];
Care must be taken to avoid DNA contamination at any step;
Inability to detect structural chromosomal aberrations without copy number changes with CGH;
Inconsistencies in visualization and imaging softwares as well as interpretation parameters may compromise reproducibility and interlaboratory comparisons.
• Analytical performance
Several research reports have used array CGH technology for the detection of 1p 19q loss in oligodendrogliomas [39,45–47]. Additionally, high concordance rates of CGH results with PCR-based microsatellite analysis have been found [48]. Still, the feasibility of array CGH for diagnostic use requires further evaluation [49].
Single nucleotide polymorphism array
• Methodological background
Single nucleotide polymorphisms (SNPs) are the most common types of genetic variation and are highly conserved within a population. SNP arrays have originally been designed to genotype human DNA at thousands of SNPs across the genome simultaneously [50]. However, meanwhile important applications include not only genetic linkage studies assessing individual disease susceptibility but also the detection and characterization of copy number variations including LOH [50].
• Materials & methods
SNP arrays can be performed on FF or FFPE tissues [51]. Commercially available probe-based SNP array platforms include those of Affymetrix® (CA, USA) and Illumina® (CA, USA). Both use different chemistries, although they share several aspects [50]. Fluorescence signal intensity after hybridization of target DNA to nucleotide probe sequences depends upon the amount of target DNA and affinity between target and probe [50,52]. Thereby, SNP arrays allow for simultaneous screening of several millions of genetic markers with a genotyping accuracy of over 99.5% according to the manufacturer's specifications.
• Evaluation
Analysis of SNP array raw data necessitates the use of advanced software algorithms which enable the detection of copy-number variation. Loss of heterozygosity (LOH) is typically detected by comparing tumor DNA to matched normal DNA of the same individual.
• Advantages
Like CGH, SNP arrays constitute a genome-wide screening method allowing for simultaneous detection of DNA copy number variations at multiple sites with high resolution;
Major advantages of SNP arrays compared with CGH are its ability to distinguish deletions from copy-neutral LOH (uniparental disomy) as well as the identification of small segmental deletions [53].
• Shortcomings
Several shortcomings that apply for other molecular techniques also apply for SNP arrays, in other words, contamination of tumor tissue from normal preexisting cells is a concern, there is a need for autologous blood samples, and it constitutes a cost- and labor-intense technique that requires expertise in software analysis;
However, a major limiting factor is that commercially available SNP arrays are intended for research use only and not yet approved for diagnostic procedures.
• Analytical performance
Several studies have reported concordant or even superior findings for SNP arrays compared with microsatellites [53–56]. With regard to 1p 19q, few studies using SNP arrays are available including case reports [57,58] or larger case series [53,59,60].
Chromogenic in situ hybridization
• Methodological background
CISH enables genetic analysis in the context of tissue morphology. It uses differentially labeled DNA probes to localize a specific DNA or RNA sequence in tissue specimens.
• Materials & methods
CISH utilizes conventional peroxidase or alkaline phosphatase reactions visualized under standard bright-field microscopy, and is applicable to FFPE sections, metaphase chromosome spreads and fixed cells. 1p 19q CISH probes equivalent to 1p 19q FISH probes have recently become commercially available.
• Evaluation
The evaluation of signal ratios is morphology-based and analogous to conventional FISH.
• Advantages
CISH warrants permanent staining results and, thus, does not require image documentation;
The evaluation of the hybridization result is morphology-based using light microscopy and allows simultaneous multi-investigator evaluation [61];
Identical locus-specific hybridization probes exclude the possibility of discrepant test results due to different hybridization sites [42];
CISH uses only standard methods, which are already present in histology labs;
Minimum amount of training required and moderate overall costs [42].
• Shortcomings
• Analytical performance
So far, experiences with CISH are limited to a single center report, which found high concordance between FISH and CISH results (93%, 39/42 cases). Validation of discrepant findings in the remaining few cases by repeated FISH and independent PCR-based microsatellite analysis further substantiated the CISH results, thereby suggesting superiority of CISH-based testing [42].
PCR–based microsatellite analysis
• Methodological background
Polymerase chain reaction (PCR)-based LOH analysis is based on the amplification of multiple microsatellite markers composed of short tandem repeats on the chromosomes of interest. Based on the comparison with paired nontumor DNA (usually autologous blood leukocytes, but microdissection of bordering nontumor tissue is also possible [49]) polymorphisms are identified and amplicons are scored as heterozygous (informative), homozygous or indeterminate. A deletion is defined as reduction from heterozygosity to homozygosity [27].
• Materials & methods
Working steps include DNA extraction from tumor and paired nontumor tissue, DNA amplification, as well as a gel or CE system, and visual inspection using autoradiographs, silver stainings or fluorescent labels for automated sequencing. For PCR-based microsatellite analysis FF tissue is preferred although it can also be performed on FFPE tissues. PCR-based microsatellite analysis can also be applied in cases, where no autologous control DNA is available but the technique is then less robust as it is more prone to contamination from normal tissue [39]. Microsatellite markers are usually selected based on amplicon size and high heterozygosity score (usually up to four or five microsatellites extending from 1p22 to 1p36.22 and from 19q13.31 to 19q13.32, respectively). So far, no consensus exists with regard to the exact number and location of microsatellite markers. However, the primer set, which has been used by the EORTC study ‘primary chemotherapy with temozolomide versus radiotherapy in patients with low-grade gliomas after stratification for genetic 1p 19q loss: a Phase II study’ and already applied to a large series of routine diagnostic patients has been published along with the detailed protocol [49].
• Evaluation
The evaluation is optimally done together with paired normal DNA as reference in order to determine whether the germline is homozygous (noninformative) or heterozygous (informative) at a specific locus and hence, whether homozygous amplicons from the tumor represent LOH. Peak reductions of 50% for one allele can be easily detected [49]. Data from several highly polymorphic loci are combined to conclude that there is a high likelihood of loss of heterozygosity if all microsatellite loci show only one allele (single allele pattern) in tumor compared with control DNA [41]. So far, there is no consensus on the exact number of microsatellite markers to use and how to interpret partial deletions.
• Advantages
• Shortcomings
PCR-based microsatellite analysis requires autologous nontumor DNA as control; obtaining and storing blood samples from each patient imposes a logistical challenge in practice [49], but more advanced techniques can overcome this (see the ‘More advanced PCR-based methods’ section);
There is no consensus on exact numbers of microsatellite markers and interpretation of partial deletions;
As for every other technique the infiltration zone may not yield reliable results due to high non-neoplastic cell load [49];
There may be a potential difference in heterozygosity scores at tested loci across different ethnic populations [41];
There may be difficulties of extracting sufficient tumor DNA from small amounts of FFPE tissues.
• Analytical performance
Excellent concordance of PCR-based microsatellite analysis with FISH and CGH results has been shown by multiple works on large case series [32,37,39–41].
• More advanced PCR-based methods
In order to overcome the need for autologous blood samples as a major limitation factor for PCR-based microsatellite marker analysis, more advanced PCR-based assays have been developed such as quantitative microsatellite analysis based on real-time quantitative PCR [27], multiplex PCR [41] and multiplex ligation-dependent probe amplification MLPA [62]. All of these methods are independent from autologous blood samples.
Real-time PCR analysis is based on selected marker and reference genes. It uses a multigene-containing recombinant DNA standard that allows quantitative analysis of absolute ratios of marker to reference gene copy numbers in sample DNA (normal ratio 1/1, deletion <0.8 amplification >1.2) [63]. While real-time PCR (rtPCR) depends on copy number (reference genes compared with marker genes) rather than polymorphisms, theoretically all loci should be informative [27]. MLPA is a multiplex PCR-based method which is able to detect changes in copy number, DNA methylation and point mutations simultaneously. It recognizes target sequences of only 50–100 nucleotides in length (high-resolution gene dosage assay) thereby allowing analysis of highly fragmented DNA. MLPA facilitates the amplification process and detects multiple targets with a single primer pair [64]. MLPA is, thus, a reliable, cost-effective and robust method, which can be performed using a standard thermocycler and CE equipment [65]. Primer sets containing approximately 40 MLPA probes and nine control fragments are commercially available (e.g., Salsa MLPA®). However, the kits are distributed for research purposes only and, so far, not CE/US FDA certified for routine diagnostic use. In any case, intrasample and intersample normalization need be performed.
• Advantages
No need to use paired nontumoral DNA (blood leukocytes) from the patient;
Percentage of tumor cells up to at least 50% is sufficient;
Large intrageneic deletions can be identified.
• Shortcomings
Like with microsatellite analysis there is no consensus on the definition of a deletion status; a recent work using MLPA suggests at least four probes for 1p and two probes for 19q with normalized values under 0.75 [64];
Amplification products can be noisy, as multiple lengths of PCR products can be produced from cells even with homozygous loci [63];
Inherited with PCR-based molecular assays is the inability to detect copy neutral events such as chromosomal translocations [63];
Both, rtPCR and MLPA rely on the stability of reference genes – therefore the choice of reference genes residing on relatively stable areas of cancer-specific genomes is important [63].
• Analytical performance
For both methods, rtPCR and MLPA, high concordance of results was found for CGH [66], FISH [62,63] and PCR-LOH [32,64].
Conclusion
A number of molecular genetic techniques are available for 1p 19q deletion status assessment. The most widely used techniques for routine diagnostic use are FISH and PCR-based microsatellite analysis. Both techniques are suitable for archival FFPE tissues and have their unique advantages and disadvantages. Probably, the most important advantage of FISH is its morphology-based evaluation, which is intuitive to pathologists. PCR-based LOH analysis in contrast, is a rapid and efficient technique but is dependent on autologous DNA samples extracted from non-neoplastic tissue. Although both techniques show high concordance of results and are generally considered valid tests, still, important issues remain to be addressed: intra- and inter-laboratory comparisons have to be further scrutinized and consensus on the test evaluation need to be defined for both methods. The choice of which test to use also depends on the available laboratory infrastructure and the experience of the technical and academic staff. More advanced molecular-genetic methods such as array CGH, SNP arrays, MLPA and rtPCR have been mostly used for research purposes, so far. Relevant issues include cost efficiency and a high level of training and experience for the staff. Their applicability as diagnostic tests will have to be evaluated in the future. Among the various test methods, CISH evolves as a potentially interesting alternative to FISH, as it is easy to implement and independent from fluorescence microscopy (Table 1).
Table 1. . Overview of common molecular techniques used to detect 1p 19q loss.
| Technique | FISH | CISH | aCGH | PCR-based LOH | MLPA |
|---|---|---|---|---|---|
| Tissue | FF, FFPE | FF, FFPE | FF (FFPE) | FF, FFPE | FF (FFPE) |
| Material/tissue | Isolated nuclei or tissue sections | Isolated nuclei or tissue sections | 1 µg DNA | 50 ng tumor and control DNA | 50 ng DNA |
| Tissue prerequisites | Solid tumor (infiltration zone)† | Solid tumor (infiltration zone)† | Solid tumor | Solid tumor | Solid tumor (infiltration zone) |
| DNA resolution | 100–500 bp | 100–500 bp | 100–200 bp | 10–100 bp | 1–40 bp |
| Number of investigated loci | 1–3 loci | 1–3 loci | Up to 500,000 loci | 1–5 loci | Up to 45 loci |
| Amount of training required | High | Medium | High | Low | Low |
| Commercial availability | Yes | Yes | Yes | Yes | Yes |
| Costs | High | Low | High | Medium | Medium |
| Evaluation of results | Morphology based | Not morphology based | |||
| Routine diagnostic use | + | + | ± | + | ± |
| Analytical performance | High | Promising | High | High | High |
†If morphologically identifiable.
aCGH: Array comparative genomic hybridization; CISH: Chromogenic in situ hybridization; FF: Fresh frozen; FFPE: Formalin-fixed paraffin embedded; LOH: Loss of heterozygosity; MLPA: Multiplex ligation-dependent probe amplification.
Adapted with permission from LWW [67].
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Berghoff AS, Stefanits H, Woehrer A, Heinzl H, Preusser M, Hainfellner JA. Clinical neuropathology practice guide 3–2013: levels of evidence and clinical utility of prognostic and predictive candidate brain tumor biomarkers. Clin. Neuropathol. 2013;32(3):148–158. doi: 10.5414/NP300646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl Cancer Inst. 1998;90(19):1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 6.Woehrer A, Sander P, Haberler C, et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice – a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS) Clin. Neuropathol. 2011;30(2):47–55. doi: 10.5414/npp30047. [DOI] [PubMed] [Google Scholar]
- 7.Griffin CA, Burger P, Morsberger L, et al. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J. Neuropathol. Exp. Neurol. 2006;65(10):988–994. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 9.Fallon KB, Palmer CA, Roth KA, et al. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J. Neuropathol. Exp. Neurol. 2004;63(4):314–322. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 10.Fuller CE, Perry A. Molecular diagnostics in central nervous system tumors. Adv. Anat. Pathol. 2005;12(4):180–194. doi: 10.1097/01.pap.0000175117.47918.f7. [DOI] [PubMed] [Google Scholar]
- 11.Labussiere M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 12.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012;226(1):7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idbaih A, Criniere E, Marie Y, et al. Gene amplification is a poor prognostic factor in anaplastic oligodendrogliomas. Neuro Oncol. 2008;10(4):540–547. doi: 10.1215/15228517-2008-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idbaih A, Marie Y, Lucchesi C, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int. J. Cancer. 2008;122(8):1778–1786. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 15.Snuderl M, Eichler AF, Ligon KL, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin. Cancer. Res. 2009;15(20):6430–6437. doi: 10.1158/1078-0432.CCR-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J. Clin. Oncol. 2012;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J. Clin. Oncol. 2013;31(3):344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 18.Fuller CE, Schmidt RE, Roth KA, et al. Clinical utility of fluorescence in situ hybridization (FISH) in morphologically ambiguous gliomas with hybrid oligodendroglial/astrocytic features. J. Neuropathol. Exp. Neurol. 2003;62(11):1118–1128. doi: 10.1093/jnen/62.11.1118. [DOI] [PubMed] [Google Scholar]
- 19.Hainfellner JA, Heinzl H. Neuropathological biomarker candidates in brain tumors: key issues for translational efficiency. Clin. Neuropathol. 2010;29(1):41–54. doi: 10.5414/npp29041. [DOI] [PubMed] [Google Scholar]
- 20.Tabatabai G, Stupp R, van den Bent MJ, et al. Molecular diagnostics of gliomas: the clinical perspective. Acta Neuropathol. 2010;120(5):585–592. doi: 10.1007/s00401-010-0750-6. [DOI] [PubMed] [Google Scholar]
- 21.Gorlia T, Delattre JY, Brandes AA, et al. New clinical, pathological and molecular prognostic models and calculators in patients with locally diagnosed anaplastic oligodendroglioma or oligoastrocytoma. A prognostic factor analysis of European Organisation for Research and Treatment of Cancer Brain Tumour Group Study 26951. Eur. J. Cancer. 2013;49(16):3477–3485. doi: 10.1016/j.ejca.2013.06.039. [DOI] [PubMed] [Google Scholar]
- 22.Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J. Clin. Oncol. 2013;31(3):328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis DN, Ohgaki H, Wiestler D, Cavanee WK, editors. WHO Classification of Tumours of the Central Nervous System. IARC Press; Lyon, France: 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark KH, Villano JL, Nikiforova MN, Hamilton RL, Horbinski C. 1p/19q testing has no significance in the workup of glioblastomas. Neuropathol. Appl. Neurobiol. 2013;39(6):706–17. doi: 10.1111/nan.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horbinski C, Miller Cr, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2011;21(1):57–73. doi: 10.1111/j.1750-3639.2010.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun M, Menon R, Nikolov P, et al. The HOPE fixation technique – a promising alternative to common prostate cancer biobanking approaches. BMC Cancer. 2011;11:511. doi: 10.1186/1471-2407-11-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nigro JM, Takahashi MA, Ginzinger DG, et al. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am. J. Pathol. 2001;158(4):1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braggio E, McPhail ER, Macon W, et al. Primary central nervous system lymphomas: a validation study of array-based comparative genomic hybridization in formalin-fixed paraffin-embedded tumor specimens. Clin. Cancer Res. 2011;17(13):4245–4253. doi: 10.1158/1078-0432.CCR-11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preusser M, Plumer S, Dirnberger E, Hainfellner JA, Mannhalter C. Fixation of brain tumor biopsy specimens with RCL2 results in well-preserved histomorphology, immunohistochemistry and nucleic acids. Brain Pathol. 2010;20(6):1010–1020. doi: 10.1111/j.1750-3639.2010.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock C, Ambros IM, Mann G, Gadner H, Amann G, Ambros PF. Detection of Ip36 deletions in paraffin sections of neuroblastoma tissues. Genes Chromosomes Cancer. 1993;6(1):1–9. doi: 10.1002/gcc.2870060103. [DOI] [PubMed] [Google Scholar]
- 31.Gelpi E, Ambros IM, Birner P, et al. Fluorescent in situ hybridization on isolated tumor cell nuclei: a sensitive method for 1p and 19q deletion analysis in paraffin-embedded oligodendroglial tumor specimens. Mod. Pathol. 2003;16(7):708–715. doi: 10.1097/01.MP.0000076981.90281.BF. [DOI] [PubMed] [Google Scholar]
- 32.Jha P, Sarkar C, Pathak P, et al. Detection of allelic status of 1p and 19q by microsatellite-based PCR versus FISH: limitations and advantages in application to patient management. Diagn. Mol. Pathol. 2011;20(1):40–47. doi: 10.1097/PDM.0b013e3181e961e9. [DOI] [PubMed] [Google Scholar]
- 33.Senetta R, Verdun Di Cantogno L, Chiusa L, et al. A “weighted” fluorescence in situ hybridization strengthens the favorable prognostic value of 1p/19q codeletion in pure and mixed oligodendroglial tumors. J. Neuropathol. Exp. Neurol. 2013;72(5):432–441. doi: 10.1097/NEN.0b013e3182901f41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horbinski C, Nikiforova MN, Hobbs J, et al. The importance of 10q status in an outcomes-based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction-based microsatellite loss of heterozygosity analysis of oligodendrogliomas. J. Neuropathol. Exp. Neurol. 2012;71(1):73–82. doi: 10.1097/NEN.0b013e318240fa65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol. 2002;12(1):67–86. doi: 10.1111/j.1750-3639.2002.tb00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Idbaih A, Kouwenhoven M, Jeuken J, et al. Chromosome 1p loss evaluation in anaplastic oligodendrogliomas. Neuropathology. 2008;28(4):440–443. doi: 10.1111/j.1440-1789.2008.00863.x. [DOI] [PubMed] [Google Scholar]
- 37.Scheie D, Andresen PA, Cvancarova M, et al. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am. J. Surg. Pathol. 2006;30(7):828–837. doi: 10.1097/01.pas.0000213250.44822.2e. [DOI] [PubMed] [Google Scholar]
- 38.Kouwenhoven MC, Gorlia T, Kros JM, et al. Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: a report from EORTC study 26951. Neuro Oncol. 2009;11(6):737–746. doi: 10.1215/15228517-2009-011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burger PC, Minn AY, Smith JS, et al. Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin-embedded sections. Mod. Pathol. 2001;14(9):842–853. doi: 10.1038/modpathol.3880400. [DOI] [PubMed] [Google Scholar]
- 40.Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18(28):4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 41.Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD. Molecular diagnosis of oligodendroglioma in paraffin sections. Lab. Invest. 2003;83(3):419–428. doi: 10.1097/01.lab.0000059948.67795.ef. [DOI] [PubMed] [Google Scholar]
- 42.Lass U, Hartmann C, Capper D, et al. Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas. Brain Pathol. 2013;23(3):311–318. doi: 10.1111/bpa.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallioniemi Op, Kallioniemi A, Piper J, et al. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10(4):231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 44.www.ascendgenomics.com Ascend Genomics.
- 45.Cowell JK, Barnett GH, Nowak NJ. Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa) J. Neuropathol. Exp. Neurol. 2004;63(2):151–158. doi: 10.1093/jnen/63.2.151. [DOI] [PubMed] [Google Scholar]
- 46.Koschny R, Holland H, Koschny T, Vitzthum HE. Comparative genomic hybridization pattern of non-anaplastic and anaplastic oligodendrogliomas – a meta-analysis. Pathol. Res. Pract. 2006;202(1):23–30. doi: 10.1016/j.prp.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Kitange G, Misra A, Law M, et al. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer. 2005;42(1):68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- 48.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am. J. Pathol. 1999;155(2):375–386. doi: 10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartmann C, Mueller W, Lass U, Kamel-Reid S, von Deimling A. Molecular genetic analysis of oligodendroglial tumors. J. Neuropathol. Exp. Neurol. 2005;64(1):10–14. doi: 10.1093/jnen/64.1.10. [DOI] [PubMed] [Google Scholar]
- 50.Laframboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res. 2009;37(13):4181–4193. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lips EH, Dierssen JW, Van Eijk R, et al. Reliable high-throughput genotyping and loss-of-heterozygosity detection in formalin-fixed, paraffin-embedded tumors using single nucleotide polymorphism arrays. Cancer Res. 2005;65(22):10188–10191. doi: 10.1158/0008-5472.CAN-05-2486. [DOI] [PubMed] [Google Scholar]
- 52.Zheng HT, Peng ZH, Li S, He L. Loss of heterozygosity analyzed by single nucleotide polymorphism array in cancer. World J. Gastroenterol. 2005;11(43):6740–6744. doi: 10.3748/wjg.v11.i43.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harada S, Henderson LB, Eshleman JR, et al. Genomic changes in gliomas detected using single nucleotide polymorphism array in formalin-fixed, paraffin-embedded tissue: superior results compared with microsatellite analysis. J. Mol. Diagn. 2011;13(5):541–548. doi: 10.1016/j.jmoldx.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindblad-Toh K, Tanenbaum DM, Daly MJ, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol. 2000;18(9):1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 55.Dumur Ci, Dechsukhum C, Ware JL, et al. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81(3):260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 56.Wang ZC, Lin M, Wei LJ, et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64(1):64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 57.Hiniker A, Hagenkord JM, Powers MP, Aghi MK, Prados MD, Perry A. Gliosarcoma arising from an oligodendroglioma (oligosarcoma) Clin. Neuropathol. 2013;32(3):165–170. doi: 10.5414/NP300577. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez FJ, Perry A, Rosenblum MK, et al. Disseminated oligodendroglial-like leptomeningeal tumor of childhood: a distinctive clinicopathologic entity. Acta Neuropathol. 2012;124(5):627–641. doi: 10.1007/s00401-012-1037-x. [DOI] [PubMed] [Google Scholar]
- 59.Idbaih A, Ducray F, Dehais C, et al. SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PLoS ONE. 2012;7(10):e45950. doi: 10.1371/journal.pone.0045950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reyes-Botero G, Dehais C, Idbaih A, et al. Contrast enhancement in 1p/19q-codeleted anaplastic oligodendrogliomas is associated with 9p loss, genomic instability, and angiogenic gene expression. Neuro Oncol. 2013;16(5):662–670. doi: 10.1093/neuonc/not235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elliott K, Hamilton PW, Maxwell P. Fluorescence (FISH) and chromogenic (CISH) in situ hybridisation in prostate carcinoma cell lines: comparison and use of virtual microscopy. Br. J. Biomed. Sci. 2008;65(4):167–171. doi: 10.1080/09674845.2008.11732823. [DOI] [PubMed] [Google Scholar]
- 62.Natte R, van Eijk R, Eilers P, et al. Multiplex ligation-dependent probe amplification for the detection of 1p and 19q chromosomal loss in oligodendroglial tumors. Brain Pathol. 2005;15(3):192–197. doi: 10.1111/j.1750-3639.2005.tb00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaturbedi A, Yu L, Linskey ME, Zhou YH. Detection of 1p19q deletion by real-time comparative quantitative PCR. Biomark. Insights. 2012;7:9–17. doi: 10.4137/BMI.S9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franco-Hernandez C, Martinez-Glez V, de Campos JM, et al. Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation-dependent probe amplification versus loss of heterozygosity. Cancer Genet. Cytogenet. 2009;190(2):93–96. doi: 10.1016/j.cancergencyto.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 65.Homig-Holzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn. Mol. Pathol. 2012;21(4):189–206. doi: 10.1097/PDM.0b013e3182595516. [DOI] [PubMed] [Google Scholar]
- 66.Jeuken J, Cornelissen S, Boots-Sprenger S, Gijsen S, Wesseling P. Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J. Mol. Diagn. 2006;8(4):433–443. doi: 10.2353/jmoldx.2006.060012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Dijk MC, Rombout PD, Boots-Sprenger SH, et al. Multiplex ligation-dependent probe amplification for the detection of chromosomal gains and losses in formalin-fixed tissue. Diagn. Mol. Pathol. 2005;14(1):9–16. doi: 10.1097/01.pas.0000146701.98954.47. [DOI] [PubMed] [Google Scholar]