Abstract
DNA has traditionally been used for programmable design of nanostructures exploiting its sequence-defined supramolecular recognition. However, control on larger lengths scales or even hierarchical materials translating to the macroscale remain difficult to construct. Here, we show that the polymer character of ssDNA can be activated via a nucleobase-specific lower critical solution temperature (LCST), providing unique access to mesoscale structuring mechanisms on larger length scales. We integrate both effects into ssDNA multiblock copolymers coding sequences for phase-separation, hybridization, and functionalization. Kinetic pathway guidance using temperature ramps balances the counteracting mesoscale phase-separation during heating with nanoscale duplex recognition during cooling, yielding a diversity of complex all-DNA colloids with control over internal dynamics and of their superstructures. Our approach provides a facile and versatile platform to add mesostructural layers into hierarchical all-DNA materials. The high density of addressable ssDNA blocks opens routes for applications like gene delivery, artificial evolution, or spatially encoded (bio)materials.
Structural length scales in self-assembly are specified by building block sizes, and the length and time scales of the underlying force fields.1 Under appropriate dynamics, these translate towards higher hierarchical levels with emergent properties. Increased structural and functional complexity occur in systems with competing and synergistic interactions interfacing on multiple levels, but, the resulting energy landscapes become non-trivial and hard to navigate for rational material design. Ultimately, though, simple design rules based on kinetic pathway guidance need to be devised to establish well-defined structure formation pathways and reliably program new hierarchical materials with useful functionality.2
For molecular systems, sequence-specific recognition of DNA duplexes provides a powerful platform to program intricate 2D/3D nanostructures3–7 with emergent functionalities.8–10 Combination with synthetic polymers has allowed the design of self-assemblies and hydrogels, and homogeneous hydrogels formed solely through duplexes have been reported.11–16 However, self-assembly in all-DNA systems (i.e., without using large polymer or colloid building blocks9,17–19) remains difficult to transduce from duplex formation through additional hierarchical levels to the macroscale because hybridization-driven structuration slows down on larger length scales.7,20,21 This limitation requires to identify mesoscale interactions, orthogonal to hybridization, and operating quickly on larger length scales, to drive additional mesostructuration and form all-DNA materials structured hierarchically from the sequence to the macroscopic level.7
Unlike DNA, self-assembly in the world of synthetic polymers relies on an entirely different concept, and uses phase-separation to induce mesostructuration.22–24 Thermo-responsive polymers exhibiting Lower Critical Solution Temperatures (LCSTs) are important for switchable self-assemblies or hydrogels, enabling applications from tissue engineering to photonics and actuators.24–29 Whereas increasing success is realized for hierarchical design on larger length scales,24,28 the recent progress towards sequence-controlled synthetic polymers has not translated yet into similar programmability, addressability and structural complexity offered by DNA assembly.30
Here, we integrate the self-assembly paradigms from these two worlds into all-DNA self-assembling systems. We demonstrate that combining reversible phase-separation with hybridization enables fast and efficient, kinetically controlled formation of addressable and hierarchical all-DNA micro- and macrostructures. At the core of our approach is the critical discovery that purine-containing single-stranded DNA (ssDNA) sequences undergo phase-separation during heating, similar to LCST-type polymers. Orchestration of these antagonistic interactions, phase-separation at high temperature and hybridization at low temperature, using controlled heating pathways adds mesoscale structural layers to all-DNA self-assembly. We combine both effects into multiblock-type ssDNA, featuring sequences coding for phase-separation and hybridization, and reveal how balancing the competing interactions enables structuration on hierarchical length scales. Using single-step heating ramps, we showcase our concept for the formation of addressable homogeneous and core/shell microgels, as well as capsules, that consist only of DNA. We further demonstrate control over their physical state and present examples of protocell-like DNA capsules entrapping a liquid DNA core in a hydrogel DNA shell. We also illustrate the potential for all-DNA materials design with the assembly of addressable multi-compartment and cellular DNA hydrogels. Finally, we reach to new functionality with the design of thermoplasmonic DNA/Au-NP protocell-like delivery particles, which eject their genetic content upon laser actuation.
The Phase-Separation ssDNA Multiblock Copolymers
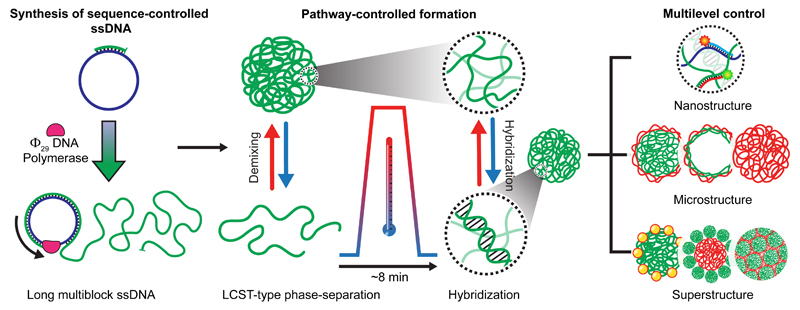
Figure 1 summarizes the process from ssDNA synthesis to hierarchical superstructure assembly. The integration of phase-separation and duplex hybridization requires ssDNAs with blocky architectures, defined length and compositional control. We synthesized those utilizing Rolling Circle Amplification (RCA)31 and, critically, avoided in-situ crystallization of MgP2O7 (using pyrophosphatase) to prevent complexation with ssDNA, suppressing so-called “nanoflowers” (Supplementary Note 1, Supplementary Fig. 1). Overall, RCA provides tailor-made starting materials for scale-up and simplification of self-assembled all-DNA materials.16,32,33
Figure 1. Kinetic pathway guidance towards multilevel control of hierarchical all-DNA structures exploiting competing interactions.
(From left to right) Synthesis of sequence-controlled, multiblock ssDNAs via RCA. Thermally induced mesoscale phase-separation of specific ssDNAs during heating and antagonistic duplex formation during cooling allow the formation of complex metastable microstructures and their co-assembled superstructures with hierarchical design and control.
The foundation of this work rests upon the discovery and comprehensive elucidation of a new composition-dependent heat-induced phase-separation of ssDNA. For double-stranded DNA (dsDNA), it is known that isothermal dsDNA compaction requires counterions of trivalent or higher charge.34 Few reports discuss temperature-promoted phase-separation of dsDNA, and explain it via strand displacement, inter-duplex bridges or triplex formation.35–37 However, we focus on ssDNA, for which such mechanisms cannot apply and we show that LCST considerations familiar from synthetic polymers26 can explain the behaviour. LCST systems can be distinguished into two main categories: Many neutral polymers, such as poly(N-isopropyl acrylamide), majorly expel the water upon heating, resulting in solid particles that flocculate, whereas polyelectrolytes often form hydrated coacervates.38–40 Our experiments suggest that ssDNA phase-separation belongs to the coacervate group (Supplementary Note 2; Supplementary Fig. 2).
First, we focus on the effect of composition on the LCST behaviour using four multiblock ssDNAs, each containing 20-base runs of one of the four DNA nucleobases (Table 1). The two main sequences relevant for this work are p(A20-i) and p(T20-j), but in the interest of a broader perspective, we also synthesized p(G20-l) and p(C20-l). All ssDNAs contain additional barcode sequences (i,j,k,l) to bind functional units or dyes.
Table 1. Repeat sequences of the ssDNA materials.
| Namea | Repeat block sequencesc |
|---|---|
| p(A20-i) | [AAAAAAAAAAAAAAAAAAAA-TTAGGATAGATATACGGGTTC]12-50 |
| p(T20-j) | [TTTTTTTTTTTTTTTTTTT-GATTTTAGAGGATCGTGTGGTTTTAC]10-60 |
| p(A20-i-XL) | [AAAAAAAAAAAAAAAAAAAA-TTAGGATAGATATACGGGTTC-CGGATGCGCATCCG]7-14 |
| p(T20-k-XL) | [TTTTTTTTTTTTTTTTTTTT-CTAAGTACAGTCAGGTAAGCTA-CGGATGCGCATCCG]6-16 |
| p(C20-l)b | [CCCCCCCCCCCCCCCCCCCC-CGAGATGTCACT]2-10 |
| p(G20-l)b | [GGGGGGGGGGGGGGGGGGGG-CGAGATGTCACT]4-20 |
| p(A20-l)b | [AAAAAAAAAAAAAAAAAAAA -CGAGATGTCACT]2-10 |
i, j, k, l are barcode sequences that allow functionalization and are used to localize dyes with complementary strands. XL is a palindromic sequence for self-hybridizing duplex formation.
Synthesised via ligation rather than RCA, because RCA cannot be performed due to the strong CG base pairing (and their unusually high content).
The lower case number denotes the range of degree of polymerization obtained from gel electrophoresis (Supplementary Note 1, Supplementary Fig. 1).
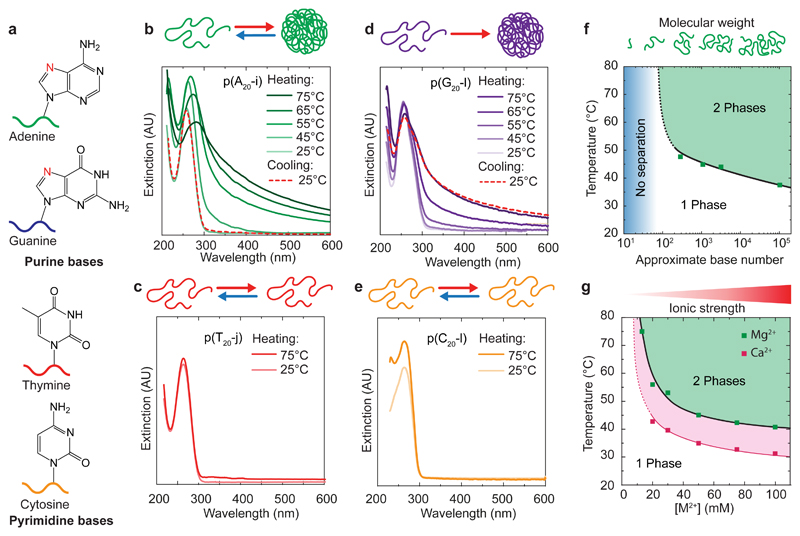
Both purine-rich p(A20-i) and p(G20-l) phase-separate at high temperature in presence of 50 mM Mg2+ (MgAc2) as indicated by the appearance of scattering in UV-Vis spectra (Fig. 2b,d). The phase-separation is reversible for p(A20-i), but not for p(G20-l), which is likely due to G-quadruplex formation preventing the re-dissolution of the DNA-rich phase upon cooling. In contrast, pyrimidine-rich p(T20-j) or p(C20-l) do not phase-separate at high temperatures (Fig. 2c,e). These results unambiguously confirm that phase-separation of ssDNA at high temperature is bound to large purine contents. Due to its easy synthesis and absence of kinetically trapped aggregates during cooling, below, we focus on adenine-containing structures (with their thymine partners) to explore the parameter space for the phase-separation behaviour and possibilities for structuration.
Figure 2. LCST behaviour of ssDNA.
(a) Simplified molecular structures of the four DNA nucleobases. (b,d) Temperature-dependent UV-Vis spectra of 0.1 g/L solutions of (b) p(A20-i) and (d) p(G20-l) in 50 mM MgAc2 with schemes of their aggregation. (c,e) UV-Vis spectra at 25 and 75°C of 0.1 g/L solutions of p(T20-j) and p(C20-l) show absence of temperature-induced aggregation in 100 mM MgAc2. (f,g) Evolution of the Tcp of 0.06 g/L solutions of p(A20-i) with (f) molecular weight of the DNA strands (50 mM MgAc2; Supplementary Fig. 1) and (g) as a function of counterion concentration (MgAc2, CaCl2). Solid and dotted lines are guides to the eye. The upturns at low molecular weight and low MgAc2 and CaCl2 concentrations represent the absence of cloud points below 100 bases and at low salinity (see also Supplementary Notes 3&4).
Phase-separation in polymers depends on the degree of polymerization.26 This complies with our results on ssDNA. Neither pure A20, nor single A20-l phase-separate, but ligated p(A20-l) oligomers do (Supplementary Fig. 3). Correspondingly, RCA products p(A20-i) with decreased length, obtained by thermal cleavage, present systematically increased cloud points (Tcp) and remain soluble below ca. 100 nucleobases (Fig. 2f, Supplementary Notes 1&3). Moreover, specifically synthesized, narrowly dispersed polyadenine homopolymers of widely different length fully confirm this trend, and allow to place the critical length for phase-segregation to ca. 75-100 nucleobases at 50 mM Mg2+ (Supplementary Note 4, Supplementary Fig. 4). In the following, we routinely use ssDNA with approximately 1000 nucleobases to facilitate handling and speed-up dynamics during heating ramps.
Finally, we discuss the influence of counterions. Not surprisingly, p(A20-i) does not show a Tcp in Mg2+-free TE buffer (pH=8; 10 mM TRIS, 1 mM EDTA) due to the electrostatic stabilization from the phosphate backbone. The LCST-type behaviour starts to occur with a Tcp ≈ 75°C at 17.5 mM Mg2+, which lowers down to a Tcp ≈ 40°C at 100 mM Mg2+ (Fig. 2g). Exchange to CaCl2 leads to similar reversible phase-transitions, but with Tcps typically 10°C lower than for MgAc2, reaching down to human body temperature. Larger earth metals (BaCl2 or SrCl2) do not lead to temperature-induced phase-separation (≤100 mM), while transition metals (ZnCl2, MnCl2) induce aggregation at room temperature (RT) (≥ 20 mM). Only 10 mM of MgAc2 is necessary to trigger p(G20-l) phase-separation at 65°C, while both p(T20-j) and p(C20-l) remain soluble up to 500 mM MgAc2 (Supplementary Fig. 3).
To summarize, selective ssDNA phase-separation is due to a hydrophilic/hydrophobic transition controlled by composition, length and counterions, as common for LCST-type polymers. While non-specific charge screening plays a role, also specific interactions between the counterions and the purine residues, such as between the Mg2+/Ca2+ hydration shell with the N7 residue (in red Fig. 2a), and dehydration at high temperature, need to be considered.41,42 Mg2+ is known to bind preferably to purine bases (G>A>C>U/T),41 assisting in higher counterion binding and thus lowering electrostatic stabilization. Additionally, purines feature increased intermolecular interactions to stabilize a collapsed form, such as increased π-stacking (10 versus 6 aromatic electrons in pyrimidines) and non-canonical purine/purine hydrogen bonding (e.g., G-quadruplexes and parallel duplexes).43,44
Pathway-Controlled Formation of Metastable all-DNA Microgels
Focusing on multiblock ssDNAs containing large proportions of oppositely behaving adenine and thymine repeats, we developed rational design strategies to kinetically trap complex microparticle architectures. The process generally involves heating 0.05-0.9 g/L ssDNA solutions until 95°C for 5 min, before cooling to RT (at 3°C/s). Such heating cycles are quick (ca. 8 min) and structures formed via phase-separation during heating can be trapped by appropriate hybridization during cooling. This results in metastable microparticles, whose architectures are determined by the sequence of self-assembly transitions, and the synchronization of the dynamics of both events. The tunable Tcp of the pA-rich ssDNA, as well as the adjustable melting temperature, Tm, of involved complementary sequences allow tunability of both events. Post-functionalization with fluorescent oligomers (Atto488-i*, Atto565-j*) visualizes the compartments containing the barcode domains (i,j) in confocal laser scanning microscopy (CLSM).
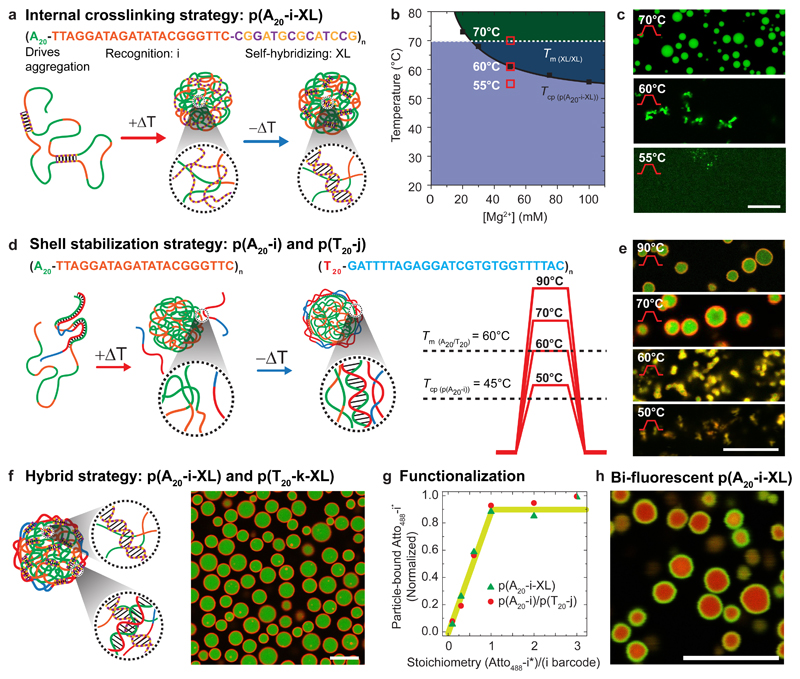
The first strategy exploits the addition of a palindromic (self-complementary) XL sequence into p(A20-i), resulting in p(A20-i-XL). The XL runs serve to freeze the DNA-rich phase formed at high temperature during cooling (Fig. 3a). Hence, they are designed with a Tm(XL) ≈ 69°C above the Tcp ≈ 60°C (at 50 mM Mg2+) of p(A20-i-XL), which itself is slightly above pure p(A20-i) (45°C) due to an overall lower pA content (Fig. 3b). Important differences can be observed after heating p(A20-i-XL) to different temperatures during the ramps (Fig. 3c). Heating to 55°C, thus below the Tcp, does not result in defined structures. The ssDNA remains dissolved, and the palindromic sequences form intramolecular loops. Structures appear upon reaching the Tcp (60°C), however, they remain ill-defined until the temperature reaches the Tm(XL) (69°C) of the palindromic domains, where duplex exchange dynamics and melting allow for a reorganization of the aggregates. This leads to the formation of spheres due to minimization of interfacial energy. Upon cooling, the re-hybridization of the XL domains (fast) occurs at temperatures above the dissolution (slow) of the phase-segregated particles, thereby leading to persistent intermolecular duplex crosslinks, trapping the DNA microparticles before the system returns in the miscibility domain. This strategy forms homogeneous DNA microgels, and mostly serves to shed light on mechanistic details and kinetic balances. Critically, those and all following colloids remain trapped in their metastable configuration for months, but can be dissolved to the individual ssDNA strands by removing MgAc2 and heating (Supplementary Note 5, Supplementary Fig. 5).
Figure 3. Pathway-controlled formation of tunable and addressable metastable all-DNA microgel architectures.
(a-c) Pathway for microgel stabilization via internal crosslinking. (b) Phase diagram overlaying phase-separation of p(A20-i-XL) (solid line; dark green) and hybridization domains (dashed line; light blue) of the palindromic sequence (XL) as function of MgAc2. (c) CLSM at RT of a 0.3 g/L p(A20-i-XL) dispersion in 50 mM MgAc2 after heating for 5 min at 55, 60 and 70°C and post-functionalized with Atto488–i*. (d-e) Core/shell microgels with liquid cores via shell crosslinking. CLSM at RT of a dispersion of 0.2 g/L p(A20-i) and 0.05 g/L p(T20-i) in 50 mM MgAc2 after heating 5 min to 50, 60, 70 and 90°C and post-functionalized with Atto488–i* and Atto568–j*. The p(T20-j) shell strands can be introduced in smaller amounts than the p(A20-i) core strands, as they only need to stabilize the particle interface. 4:1 is an empirical optimization leading to minimum free p(T20-j). (f) Double-crosslinked core/shell microgels merging internal and interfacial hybridization in a hybrid strategy based on 0.5 g/L p(A20-i-XL) and 0.1 g/L p(T20-k-XL), post-functionalized with Atto488–i* and Atto647–k*. (g) All barcode domains (i,j,k,l) can be quantitatively addressed by their corresponding partners (i*,j*,k*,l*) as measured by fluorescence spectroscopy after centrifugation and dissolution of functionalized particles (see Methods). (h) Kinetic control over barcode functionalization of homogeneous 0.1 g/L p(A20-i-XL) microgels via the successive addition of 0.2 eq Atto488–i* (green channel), followed by 0.8 eq of Atto647–i* (green channel) allows preparing core/shell compartmentalized microgels from homogeneous ones. Scale bars: 10 μm. Solid lines in (b) and (g) serve as guide to the eye.
The second strategy opens the design space towards higher structural complexity by using binary systems based on p(A20-i) and p(T20-j). Those lack palindromic sequences, but are hetero-complementary in the A20/T20 runs (Fig. 3d). Fig. 3e reveals that the structure formation follows qualitatively similar considerations as for p(A20-i-XL). Ill-defined aggregates form close to the Tcp of 45°C of p(A20-i), and grow until the A20/T20 duplexes melt at Tm(A20/T20) ≈ 60°C, whereupon defined particles evolve that expel the soluble and unbound p(T20-j). Upon cooling, re-hybridization between A20/T20 occurs at the boundary of the p(A20-i) cores. This yields crosslinked shells that freeze in their dynamics during cooling, thereby preventing dissolution of the p(A20-i) cores. The difference of Tm(A20/T20) to Tcp(pA20-i) of ca. 15 °C is important for arresting the dynamics of A20/T20 duplex exchange before p(A20-i) starts to resolubilize. Both trapping strategies can also be merged into a binary self-hybridizing system, p(A20-i-XL)/p(T20-j-XL), which results in core/shell microgels with a double gel structure, crosslinked within the core and the shell via palindromic sequences and via A20/T20 duplexes at the interface (Fig. 3f).
A key advantage of our systems is the selective addressability and functionalization of the individual compartments, which we routinely exploit for CLSM by binding fluorescent oligomers to the barcode domains (i,j,k,l). The degree of functionalization can be controlled stoichiometrically, as the bound fluorescent dye-barcodes* equal the used ratio of dye-barcode*/barcode and level off once all barcodes are saturated (Fig. 3g). Under appropriate kinetics, it is even possible to post-compartmentalize homogeneous colloids with spatial functionalities (Fig. 3h). For instance, p(A20-i-XL) microgels allow comparably slow diffusion of dye-barcodes*, and can thus be post-compartmentalized into bi-fluorescent core/shell microgels via sequential hybridization of 0.2 eq. Atto488–i* (green) and 0.8 eq. of Atto647–i* (red) addressing the same barcode sequences.
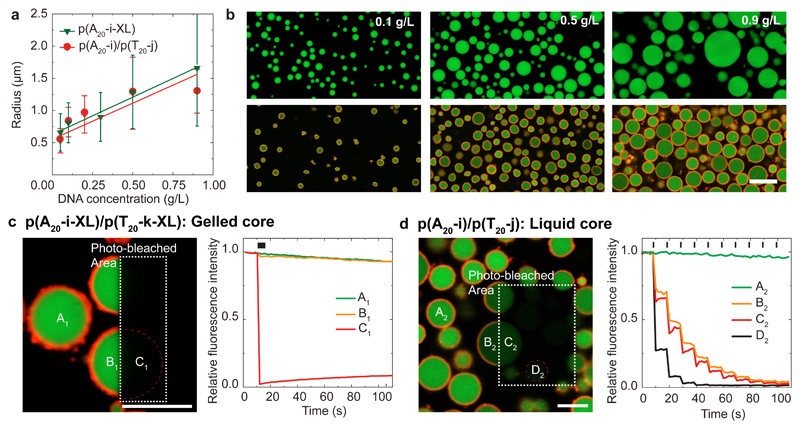
Overall, all of these examples set the scene to design and control metastable, hierarchical and all-DNA microparticles with complex architectures, controlled dynamics (liquid/gel) and to functionalize them using different strategies. The average particle size is controlled by the DNA concentration (Fig. 4a,b). Larger particles form at higher concentrations, which is reminiscent of binodal decomposition characterized by a nucleation and growth process. Yet, as inherent to phase-separation processes, it is not easily possible to control the dispersity.
Figure 4. Control over microgel size and physical state in protocell-like particles.
(a) Average particle sizes (z-section and standard deviations of 1000 particles) as function of the ssDNA concentration and the corresponding linear fits. (b) CLSM of particles formed at increasing DNA concentration (from left to right): top p(A20-i-XL) and bottom p(A20-i)/p(T20-j). (c) CLSM of self-crosslinked p(A20-i-XL)/p(T20-i-XL) core/shell microgels after photo-bleaching the rectangle. Corresponding fluorescence during photo-bleaching at position A1, B1, and C1 shows absence of core (and shell) dynamics. (d) CLSM of p(A20-i)/p(T20-j) core/shell microgels after the third photo-bleaching step in the rectangle. The corresponding fluorescence intensities at positions A2, B2, C2 and D2 (during 10 sequential photo-bleaching steps) demonstrate the cores to be liquid (B2, C2 partly bleached) and the shells to be crosslinked (only bleached in the irradiated area). Black dashes on top indicate the photo-bleaching steps (Supplementary Videos 1-2). Scale bars: 5 μm.
Of particular importance is the ability to control the physical state of the final core/shell microgels as elucidated by fluorescence recovery after photo-bleaching (FRAP) experiments for p(A20-i-XL)/p(T20-j-XL) and p(A20-i)/p(T20-j) (Fig. 4c,d). The crosslinked p(A20-i-XL)/p(T20-j-XL) cores and shells show no fluorescence recovery within the exposed area after a single photo-bleaching step (Fig. 4c; Supplementary Video 1). In striking contrast, the p(A20-i)/p(T20-j) core/shell microgels display near-simultaneous bleaching of the irradiated and the non-irradiated core halves of single particles and repeated photo-bleaching leads to a step-wise bleaching of the full particle core (Fig. 4d; Supplementary Video 2). This confirms a liquid-like dynamic within the cores. The shells only undergo photo-bleaching in the irradiated area, confirming their duplex-based hydrogel state. Such liquid/gel particles feature an intriguing morphology with liquid DNA encapsulated under high osmotic pressure inside a solid DNA hydrogel shell – being a much sought-after architecture in protocell research and difficult to prepare with other means to date.45,46
Capsule Formation
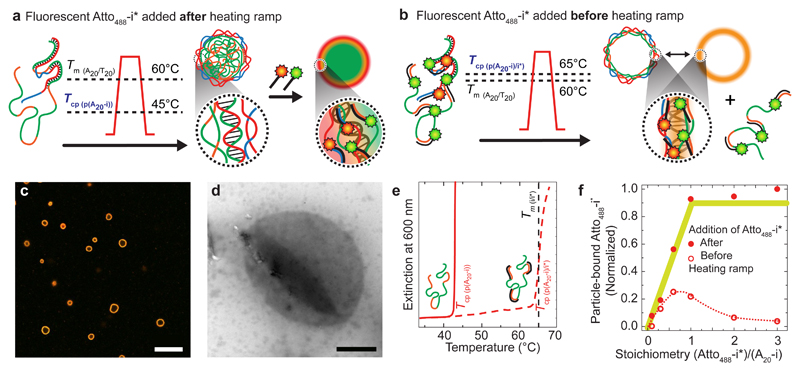
Core/shell microgels form when p(T20-j) coats the phase-separated p(A20-i) cores well above their dissolution temperature, so that DNA-rich p(A20-i) phases remain kinetically trapped during cooling due to frozen A20/T20 duplex shells (Tm > Tcp). We hypothesized that larger morphological diversity might be accessible by matching Tcp and Tm. To address this challenge, we exploit the fact that the hybridization of p(A20-i) with ssDNA oligomers (i* or Atto488-i*) increases the Tcp((pA20-i)/i*) closely to the Tm(i/i*) located at 64°C (Fig. 5b,e). This is due to the fact that i/i* duplexes stabilize p(A20-i) against phase-separation until melting of i/i*. Since the altered Tcp((pA20-i)/i*) ≈ 65°C is slightly above the Tm(A20/ T20) ≈ 60°C of the duplexes stabilizing the shell, the re-dissolution of the p(A20-i) cores (structure loss) and hybridization on the surface (structure formation) during cooling superimpose closely. Strikingly, the high dynamics of both processes lead to the selective formation of hollow capsules, appearing orange in CLSM due to co-localization of red- and green-emitting Atto488–i* and Atto565-j* for p(A20-i) and p(T20-j), and corroborated by a collapsed character in transmission electron microscopy (TEM, Fig. 5c,d).
Figure 5. Single-step capsule formation via matching the dynamics of core/shell hybridization and core dissolution during cooling.
(a) The labelling with complementary fluorescent oligomers after particle formation (standard protocol) results in core/shell microgels (see CLSM in Fig. 3e and 4b). (b) In contrast, the presence of complementary dye-barcode* oligomers (Atto488-i* and Atto565-j*) added before particle formation leads to an increase of the Tcp of p(A20-i), which results in the formation of capsules (with ejection of p(A20-i)/Atto488-i*). (c-d) CLSM and TEM visualize all-DNA capsules (scale bars are 10 and 1 μm). Further grey scale analysis, a TEM of a solid particle and z-stack CLSM image can be found in Supplementary Fig. 6. (e) Addition of i* to p(A20-i) shifts the Tcp of the p(A20-i) from ca. 45 to 65°C (0.06 g/L), hence close to the Tm(i/i*) ≈ 64°C as seen by temperature-dependent UV-Vis spectroscopy. (f) DNA capsules entrap less fluorescent Atto488-i* compared to core/shell microgels, confirming the release of p(A20-i)/i* into the solution (removed by cleaning with centrifugation after the heating ramp, see Supplementary Note 6, Supplementary Fig. 6). Lines in (f) serve as guide to the eye.
To understand whether the core material (p(A20-i)) ends up in a thicker shell or is released into the solution, we measured the amount of fluorescent Atto488–i* trapped inside the capsules and compared it to standard core/shell microgels above (Supplementary Note 6; Fig. 5e). Indeed, only sub-stoichiometric amounts of Atto488–i* are found for the capsules, confirming that some p(A20-i) escapes from the phase-segregated particles and is removed with its Atto488–i* partners during purification (details and further gel electrophoresis of the supernatant in Supplementary Fig. 6). This demonstrates that subtle effects in balancing Tm and Tcp can lead to unforeseen, but defined morphologies, further broadening the possibilities for all-DNA microcompartment design.
Supra-Particulate Assemblies and Cellular Hydrogels
Beyond using the addressable barcodes for fluorescent labelling, we will next capitalize on their presence for constructing supra-particulate assemblies and materials (Supplementary Note 7). We first realize supra-particulate structures by functionalization (and fluorescent labelling) of two fractions of p(A20-i-XL) microgels with complementary oligomers (i*-R1 and i*-R1*; Fig. 6a). Subsequent mixing at RT leads to millimetre-scale aggregates consisting of thousands of particles held together by R/R* hybridization (Fig. 6b). Core/satellite structures or aggregates with tuned size ratios are accessible by adjusting the ratio and sizes of the particles mixed (Fig. 6c). This demonstrates that the diversity of DNA-driven colloid assembly known in literature6,47,48 will be applicable to advance the present supra-particular assembly strategies in future.
Figure 6. Supra-particulate assemblies, cellular hydrogels, and light-triggered hybrid Au-NP/DNA colloids.
(a-b) p(A20-i-XL) particle batches functionalized with complementary oligomers p(A20-i-XL)/i*-R1 and p(A20-i-XL)/i*-R1* form millimetre-scale co-assembled supra-particulate hydrogels via R/R* recognition. (c) Asymmetric co-assemblies and core/satellite structures obtained by mixing particles of different size and with different ratios. Appropriate controls are presented in Supplementary Fig. 7. (d-e) Stepwise compaction and sintering of p(A20-i-XL)/p(T20-j-XL) particles yields cellular, compartmentalized hydrogels. (f) TEM (top) and CLSM (bottom) of capsules formed by p(A20-i)/Au-T20-NPs. (g,h) Core/shell particles formed by p(A20-i)/p(T20-j)/Au-T20-NPs (liquid core) or p(A20-i-XL)/Au-T20-NPs (solid core). (i,j) Photothermal heating via irradiation at 561 nm within the dashed rectangles triggers (i) ejection of entrapped ssDNA from the all-DNA protocells (Supplementary Videos 3), and (j) spatially selective erosion of p(A20-i-XL)/Au-T20-NPs particles. (k) Photothermal heating via irradiation at 561 nm within the dashed rectangles induces ejection of ssDNA content of p(A20-i)/p(T20-j)/Au-T20-NPs protocells within seconds as followed by spatially resolved tracking of the fluorescence intensity. Conditions: TE buffer with 50 mM MgAc2. Scale bars: (b) left to right: 200, 20, 2 μm, (c) 10 μm, (e) left to right: 500, 5, 5 μm, (f) top 1 μm, bottom 5 μm, (g-k) 1 μm.
Another unique feature of these metastable microgels is the heat-induced dynamization of the DNA crosslinks stabilizing the microgels. This can be used to create new types of unusual cellular hydrogels, as demonstrated for p(A20-i-XL)/p(T20-k-XL) particles. After compaction of the core/shell microgels using centrifugation, simple temperature treatments (60°C) sinter them together, finally leading to microgels adapting a space-filling pattern (Fig. 6d,e; Supplementary Fig. 7; Supplementary Note 7). Setting the temperature slightly below Tm(XL) ≈ 69°C prevents full melting, but realizes sufficient dynamics for XL reorientation. This strategy enables the design of cellular hydrogels that can easily be decorated with functional groups within the compartments.
Light-Responsive Ternary Au-NP/DNA Hybrid Colloids
Developing pathways for remote and spatiotemporal manipulation would promote the level of functionality and enable e.g. on-demand release of ssDNA. We focused on the integration of light triggers by co-assembling spherical Au-NPs (R = 10 nm; Supplementary Note 8) that serve as photo-thermal hotspots to locally melt DNA duplexes.49 To embed the Au-NPs into the shell, we hypothesized that a surface coating with HS-T20 (Au-T20-NPs) would allow replacing p(T20-j) ssDNA partly or fully during the heating ramps. Indeed, Au-T20-NPs remain non-aggregated during the heating ramps (Supplementary Fig. 8). In combination with complementary p(A20-i-XL), p(A20-i)/p(T20-j) and p(A20-i) ssDNA, Au-T20-NPs systematically end up around, or co-assemble with, the A20-rich phase during heating ramps. Co-assembly with p(A20-i-XL) results in solid core/shell microgels where the Au-NPs homogeneously localize on the surface during cooling (Fig. 6h), while addition to the non-self-crosslinking p(A20-i)/p(T20-j) binary system leads to core/shell hybrid protocells with liquid cores and hydrogel shells with entrapped Au-T20-NPs (Fig. 6g). Finally, complete exchange of the surface stabilizer p(T20-j) by Au-T20-NPs yields capsules stabilized by p(A20-i)/Au-T20-NPs hybrid hydrogel shells (Fig. 6f).
For proof-of-concept application, we elucidate the different responses to photothermal heating depending on the particle architecture. Upon laser excitation near the plasmon absorption peak of the Au-T20-NPs (λabs = 521 nm) in the CLSM, the solid p(A20-i-XL)/Au-T20-NPs hybrid colloids undergo rapid thermoplasmonic heating. This melts the local XL-crosslinked microgel cores as well as the A20/T20 duplexes anchoring the Au-NPs, eventually resulting in the ejection of the Au-T20-NPs, which stops the heating and self-limits the erosion to DNA material nearby (Fig. 6j). More interestingly, toward spatiotemporally controlled release of ssDNA (or genes in long term),16 the core/shell microgels entrapping a liquid p(A20-i) core and decorated with p(T20-j)/Au-T20-NPs shells display a localized capsule burst and release of the entrapped ssDNA (Fig. 6i). The high osmotic pressure and high mobility of the encapsulated, liquid DNA result in a release within seconds. Obviously, when equipped with NIR-absorbing Au-nanorods to approach the transparent tissue window, such hybrids could be viable for the development of new therapeutic carriers allowing light-triggered release of active molecules or genes deep within tissues.50,51
Conclusion
We discovered a new nucleobase-dependent LCST behaviour specific to ssDNA. By activating this polymer feature of DNA for DNA-based self-assembling systems, this work steps away from using classical hybridization as main structuring tool, and allows facile access towards larger length scales. We thereby provide a microstructuration platform, enabling the controlled formation of hierarchical all-DNA micro- and macrostructures without the need for moulds, templating colloids, or complexing polyelectrolytes, but simply by orchestrating kinetics of antagonistic mesoscale phase-separation and nanoscale hybridization events during heating cycles. The processes are controlled and versatile, yielding a rich palette of complex metastable colloids, their superstructures up to a compartmentalized hydrogel level, and even incorporating means for external spatiotemporal modulation via thermoplasmonic effects in DNA/Au-NP protocells.
Simplicity is a key: The rational design principles for facile and quick control over architecture and physicochemical properties, together with the high density of addressable sites for functionalization makes it a platform of choice for applications requiring mesostructural layers in DNA technology: prototypes of life-inspired materials, gene delivery, cell-free protein expression, in-vitro evolution and spatially encoded hydrogels for biomaterials. Furthermore, the inherent compatibility of our system with aptamers, DNAzymes, and strand displacement reactions will stir the design of multi-responsive, functional and out-of-equilibrium materials and systems, bringing them ultimately closer to mimic living entities.52,53 By merging the self-assembly paradigms of two fields (DNA and polymer sciences), we believe that our approach will cross-fertilize conceptual exchange, as well as self-assembly and materials design strategies from researchers with backgrounds in DNA nanoscience, polymer science and soft matter in general. The insights for the phase-separation of purine-based ssDNA provided herein motivate for a closer inspection in future combining experiments, simulations and theory to arrive at a quantitative understanding. Beyond that, LCST phenomena similar to the one introduced here could open new perspectives to understand the emergence of compartmentalization in the prebiotic RNA world and as a selection criteria in later evolutionary stages in the origin of life.45,54
Methods
Synthesis of circular ssDNA templates and Rolling circle amplification (RCA)
Linear ssDNA template and ligation strand (Integrated DNA Technology) are mixed at 1 μM in 100 μL of TE buffer (Sigma Aldrich 10 mM Tris(hydroxymethyl)aminomethane pH=8 and 1 mM of EDTA) containing 100 mM NaCl (Sigma Aldrich). The solution is heated to 85°C for 5 min and cooled down to 25°C at 0.01 °C/s. After hybridization 20 μL of 10 X commercial ligase buffer (Lucigen: 500 mM TRIS-HCI, 100 mM MgCl2, 50 mM dithiothreitol and 10 mM ATP), 70 μL of water and 10 μL of T4 Ligase (Lucigen: 2 U/μL) are introduced in the tube gently mixed and left to react for 3 hours at room temperature. The enzyme is then deactivated by heating the mixture for 20 min at 70°C.
Then 10 μL of Exonuclease I (Lucigen: 40 U/μL) and 10 μL Exonuclease III (Lucigen: 200 U/μL) are introduced and the mixture is left overnight at 37°C to remove ligation strands and non-circularized templates in solution. The enzymes are then deactivated by heating at 80°C for 40 min. The templates are purified by filtration over Amicon Ultracentrifugal filters with a 10 kDa cut-off (Merck Millipore) and rinced 3 times using 400 µL TE buffer in the same filter. The ssDNA concentrations are measured using a NanoDrop 2000 C (Fisher Scientific) and the solutions are diluted to 1 μM using TE buffer.
For RCA, 5 μL circularized template (1 μM) are mixed with 76 μL of ultrapure water, 1 μL of exonuclease resistant primer (10 μM in TE buffer), 10 μL of commercial 10 X polymerase buffer (Lucigen: 500 mM TRIS-HCl, 100 mM (NH4)2SO4, 40 mM Dithiothreitol, 100 mM MgCl2), 2 μL of Φ29 Polymerase (Lucigen: 10 U/μL), 1 μL of inorganic pyrophosphatase (Lucigen: 2 U/μL) and 5 μL of an adjusted dNTP mix (Jena bioscience: 100 mM, the mix contain pure dATP, dTTP, dCTP, dGTP solutions mixed in proportions corresponding to the expected product composition). The samples is kept for 60 hours at 30°C before heat-induced cleavage for 15 min at 95°C. The product is then purified by filtration over Amicon Ultracentrifugal filters with a 30 kDa cut-off (Merck Millipore) and rinced 3 times using 400 μL of TE buffer. Amount and purity of the ssDNA concentration are then measured using a NanoDrop 2000 C (Fisher Scientific).
For an overview of the process and gel electrophoresis (GEP) characterization of the starting materials see Supplementary Fig. 1.
Determination of the cloud point temperature (Tcp)
Temperature-dependent UV-Vis spectra were recorded on a Jasco V650 spectrophotometer coupled with a Huber Petite Fleur thermocontroller or on a ScanDrop (Jena Analytics) with Peltier heating. The DNA was diluted to ca. 0.06 g/L in TE buffer containing 50 mM of MgAc2 (or the specified amount of corresponding divalent cation salt). Heating ramps were performed on 50 μL of solution introduced in a 45 μL Quartz cuvett (Hellma Analytics) and heated at 1 °C/min, the extinction at 600 nm was recorded every 30 seconds. The cloud point temperature, Tcp, was determined as the onset of extinction increase at 600 nm.
General procedure for particle preparation
The particles are prepared by mixing the starting strands at the target concentration (typically 0.2 g/L) in TE buffer. For p(A20-i)/p(T20-j) or p(A20-i-XL)/p(T20-k-XL) particles, it is the concentration of A20 containing strand that is taken into account, while the T20 containing strand is added in a 1/4 ratio compared to the A20 containing strand. The solution is then heated to 95°C for 15 min for homogenization and thermal reduction of molecular weight (no phase-separation). The solution is then supplemented with 50 mM of MgAc2 (for phase-separation) and heated to 95°C for 5 min with heating and cooling ramps of 3°C/min. After particle formation, stoichiometric amounts of corresponding fluorescent dye-barcode* oligomers (Atto488-i*, Atto565-j* and/or Atto647-k*) are added to the particle solution and left 1 h at room temperature to hybridize with the particles before CLSM.
Quantification of the functionalization with Atto488-i*
To quantify the particle-bound Atto488-i* (Fig. 3g), we prepared two master DNA solutions of (i) core/shell type of p(A20-i) at 0.13 g/L (i.e. 10 μM of repeat sequence) mixed with p(T20-j) at 0.03 g/L (i.e. 2.5 μM of repeat sequence), and (ii) homogeneous microgels of p(A20-i-XL) at 0.17 g/L (i.e. 10 μM of repeat sequence) in TE buffer, which we reduced in molecular weight and homogenized at 95°C for 15 min. After splitting into 6 individual 10 μL fractions, we added 50 mM MgAc2 and formed the particles by heating the solutions for 5 min at 95°C. After cooling, 10, 30, 60, 100, 200, and 300 pmol of Atto488-i* were added (corresponding to 0.1, 0.3, 0.6, 1, 2, and 3 equivalents of the i-barcode sites) and allowed to hybridize for one hour. After cleaning via 2 cycles of centrifugation/re-dispersion in TE buffer containing 50 mM MgAc2 to remove the excess of unbound Atto488-i*, the buffer was exchanged to 20 μL of pure TE buffer (without Mg2+). Re-heating of the mixtures for 1 min to 95°C dissolves the particles and allows to perform scattering- and quenching-free fluorescence spectroscopy to quantify the amount of hybridized fluorophore-ssDNA (Atto488-i*).
The samples were then diluted 50 times in TE buffer to prevent self-quenching and saturation before fluorescence measurements on a Horiba Fluoromax-4 fluorometer using 45 μL Quartz cuvettes (Hellma Analytics) with excitation at 480 nm (3 nm slit) and fluorescence recording between 500 and 600 nm (2 nm slit). We report the fluorescence maximum at 522 nm normalized by the one obtained when 3 equivalents of Atto488-i* are added (i.e. full coverage of all barcode domains). The values reported therefore correspond to the proportion of fluorescent dye-barcode* marker, Atto488-i*, that have bound to barcode sites i inside the particles.
Bi-fluorescent core/shell microgels prepared by sequential addition of dye-barcode* ssDNA conjugates
The particles in Fig. 3h originate from a solution of 0.1 g/L p(A20-i-XL). After standard particle preparation protocol, 10 pmol of Atto488-i* are introduced to ca. 10 μL dispersion of p(A20-i-XL) microgels (ca. 60 pmol of i-site). The dispersion is immediately and vigorously mixed by pipetting back and forth and left 1 hour at room temperature. Later 50 pmol of Atto647-i* are added and the solution is mixed and left at room temperature for another hour before CLSM.
Photo-bleaching experiments
The particles are first imaged using low intensity. Photo-bleaching was achieved using 100 % intensity on both 488 nm and 561 lines (≤ 10 mW in the focal plane). Photo-bleaching (6 s) was used for the solid p(A20-i-XL)/p(T20-k-XL) microgels to fully deplete fluorescence in the region of interest. The liquid/solid core/shell p(A20-i)/p(T20-j) microgels were photo-bleached with repetitive single passes (1 s at each step) (see also Supplementary Videos 1 and 2). The data is presented in Fig. 4c,d.
Formation of hollow capsules
The capsules were prepared by mixing 0.4 g/L p(A20-i) with 0.1 g/L p(T20-j) in TE buffer (total volume 10 μL), and heating the mixture to 95°C for 15 min for homogenization and thermal reduction of molecular weight. The solutions were then supplemented with 1 μL of MgAc2 1 M (final concentration of 50 mM), as well as with stoichiometric amount of Atto488-i* (3 μL at 100 μM; final concentration 15 μM), Atto568-j* (0.6 μL at 100 μM; final concentration 3 μM) to saturate the barcode domains, and 5.6 μL of TE buffer (final volume 20 μL), the final concentration of p(A20-i) and p(T20-j) was 0.2 g/L (15 μM) and 0.05 g/L (3 μM). The standard heating ramp with 5 min at 95°C was then applied and the capsules were directly imaged with CLSM. The data is presented in Fig. 5.
Quantification of the amount of trapped p(A20-i) in p(A20-i)/p(T20-j) particles
Quantification of the trapped p(A20-i) from particle-bound Atto488-i* (Fig. 5f) relies on a protocol similar to the quantification of functionalization described above. In order to quantify the amount of p(A20-i) trapped inside p(A20-i)/p(T20-j) depending on whether the Atto488-i* was introduced before (capsules) or after (core/shell) the heating ramp, we prepared a master solution of p(A20-i) at 0.13 g/L (i.e. 10 μM of repeat sequences) mixed with p(T20-j) at 0.03 g/L (i.e. 2.5 μM of repeat sequence) in TE buffer, heated it at 95°C for 15 min (molecular weight reduction) and split this master solution in 12 individual 10 μL fractions. Subsequently, for 6 of them we only added 0.5 μL of 1 M MgAc2 solution, while for the other 6 we also added 10, 30, 60, 100, 200, and 300 pmol of Atto488-i* (between 1 and 3 μL of Atto488-i* at either 10 or 100 μM master solution in TE buffer). We then ramped all 12 fractions via 95°C for 5 min and cooled them down to room temperature. For the 6 samples without Atto488-i*, 10, 30, 60, 100, 200, and 300 pmol of Atto488-i* were added and the dispersion were left at room temperature for 1 hour. After cleaning the particles with 3 cycles of centrifugation/re-dispersion (12.5 krcf for 5 min; TE buffer containing 50 mM MgAc2) to remove free p(A20-i) and excess Atto488-i*, the particles were disassembled by adding 20 μL of pure TE buffer and heating the mixtures for 1 min to 95°C.
The samples were then diluted 50 times in TE buffer to prevent self-quenching and saturation before fluorescence measurements on a Horiba Fluoromax-4 fluorometer using 45 μL Quartz cuvettes (Hellma Analytics) with excitation at 480 nm (3 nm slit) and fluorescence recording between 500 and 600 nm (2 nm slit). We report the fluorescence maximum at 522 nm normalized by the one obtained when 3 equivalents of Atto488-i* are added (i.e. full functionalization in core/shell particles). The values plotted therefore correspond to the proportion of Atto488-i* bound to p(A20-i) strand which are trapped inside the p(A20-i)/p(T20-j) particles, since p(A20-i) in solution is washed away during the centrifugation step. The data confirms escape of p(A20-i) to the solution by finding lower amounts of fluorophores in the capsules compared to the core/shell microgels.
Hybridization-driven self-assembly for supra-colloidal assemblies
Three batches of p(A20-i-XL) microgels containing each 1 g/L (10 μL), 0.2 g/L (50 μL) and 0.1 g/L (100 μL) were prepared as described above. All volumes were set to 100 μL with a TE buffer containing 50 mM of MgAc, and Tween 80 was introduced to a final concentration of 0.1 wt% to avoid the particles to stick to the tube wall. Each batch was split in two 45 μL fractions which were each functionalized with 5 μL of one of the two following functionalization mixtures. The functionalization mixtures contained either 10 μM of Atto488-i* and 90 μM of i*-R1 or 10 μM of Atto647-i* and 90 μM of i*-R1*. Multiparticle hydrogels were obtained by mixing the two complementary particle batches in a 1:1 ratio and core/satellite structures by mixing them in a 1:20 ratio and shaken overnight at room temperature. The data is presented in Fig. 6a-c.
Cellular hydrogels
Cellular hydrogels are formed by two consecutive heating steps (Fig. 6d,e). A batch of p(A20-i-XL)/p(T20-k-XL) particles at 0.2 g/L was prepared as described above in 50 mM MgAc2 TE buffer. The particles were functionalized with stoichiometric amounts of Atto488-i* and Atto647-k* left at room temperature for 1 hour. After centrifugation down to the bottom of the tube (10 krcf, 3 min) the tube is heated to 60°C for 5 minutes and cooled back down to room temperature to induce stickiness between the particles. To further form the continuous gel the supernatant is removed from the sedimented particles the tube is tightly closed and a second heating ramp to 60°C for 5 min is applied. The hydrogel is gathered by adding 50 μL of TE buffer containing 50 mM of MgAc, and 0.1 wt% of Tween 80.
Synthesis of ssDNA coated gold nanoparticles (Au-NP)
Citrate-stabilized Au-NPs (Nanocomposix; R = 10 nm, 6 nM) were cleaned by centrifugation at 14.1 krcf for 5 min and redispersed to their initial volume in ultrapure water. Subsequently, thiol-functionalized ssDNA (10 μM, sequence T20-SH, see Table S1) were activated using 5 mM of Tris(2-carboxyethyl)phosphine in a citrate buffer (2.5 mM) at pH=5 for 30 min. The Au-NPs were then added (molar ratio ssDNA/Au-NPs was 1000:1) and the solution was incubated for 30 min at room temperature. An equivalent amount of TE buffer supplemented with 0.2 wt% of Tween 80 was then introduced in the previous mixture (increasing the pH to 7) and the solution was incubated for 2 more hours at room temperature. The ionic strength of the solution was then progressively increased over 3 hours using 1 M NaCl: first to 100 mM, to 200 mM after 1 hour, and to 300 mM after 2 hours. This last step increases the functionalization density of the ssDNA on the Au-NPs. Finally the particles were centrifuged at 14.1 krcf for 5 min and re-dispersed in water three times. The final particle concentration was typically around 10 nM.
Hybrid Au-NP/DNA particles
For hybrid structure formation we used 0.2 g/L of ssDNA strands, either p(A20-i-XL) or p(A20-i) (with or without 0.05 g/L of (T20-j)) and 5 nM of Au-NPs and we applied exactly the same protocol as for the standard particle formation. That is homogenization for 15 min at 95°C in TE, followed by addition of 50 mM MgAc and heating to 95°C for 5 min. The fluorescent dyes Atto488-i* and Atto565-j* were added afterwards. Corresponding particles are displayed in Fig. 6f-h.
Thermoplasmonic actuation of Au-NP/DNA hybrid particles
Thermoplasmonic actuation of Au-NP/DNA hybrid particles was performed using 561 nm laser (100 % of the maximum intensity ≤10 mW in the focal plane). We used the FRAP setup to image the particles (1 % of the maximum intensity) immediately before and after a single high power photo-thermal actuation step (1 s) in the depicted region of interest. To follow the release of the particle content we simultaneously imaged the particle at 488 nm (1 % maximum intensity) and actuated them at 561 nm (100 % of the maximum intensity) between t = 5 s and t = 10 s (Supplementary Video 3). The data is presented in Fig. 6i-k.
Instrumentation
Thermal ramps were performed on a Personal Thermocycler (Jena Analytics), DNA concentrations were measured using a NanoDrop 2000 C (Fisher Scientific) or a ScanDrop (Jena Analytic) spectrophotometer with a standard value of 33 μg/OD260. Confocal laser scanning microscopy (CLSM) was performed on Leica TCS SP8 microscope. Transmission electron microscopy (TEM) images were taken on a Zeiss LIBRA 120 using an acceleration voltage of 120 kV. Scanning electron micrograph (SEM) were recorded on Hitachi S4800 after coating the samples with about 8 nm Au/Pd (80:20) using a Leica EM ACE600 sputter coater.
Supplementary Material
Supplementary information is available in the online version of the paper.
Acknowledgments
This work was funded via the ERC Starting Grant “TimeProSAMat” (677960). S.L. is funded through a scholarship of the FCI. We thank A. Kuehne for critically reading the manuscript.
Footnotes
Author Contributions
R.M. and A.W. conceived the project, designed the experiments and analysed the data. R.M., S.L. and A.S. carried out the experiments. A.W. supervised the project. R.M. and A.W. wrote the manuscript.
Competing Financial Interests statement
The authors declare no competing financial interests.
Additional Information
Reprints and permission information is available online at www.nature.com/reprints.
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
Contributor Information
Rémi Merindol, Institute for Macromolecular Chemistry, Stefan-Meier-Str. 31, University of Freiburg, 79104 Freiburg, Germany.
Sebastian Loescher, Freiburg Materials Research Center, Stefan-Meier-Str. 21, University of Freiburg, 79104 Freiburg, Germany.
Avik Samanta, Freiburg Center for Interactive Materials and Bioinspired Technologies, Georges-Köhler-Allee 105, University of Freiburg, 79110 Freiburg, Germany.
Andreas Walther, Freiburg Institute for Advanced Studies (FRIAS), Albertstraße 19, University of Freiburg, 79104 Freiburg, Germany.
References
- 1.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Tschierske C. Development of structural complexity by liquid-crystal self-assembly. Angew Chem Int Ed. 2013;52:8828–8878. doi: 10.1002/anie.201300872. [DOI] [PubMed] [Google Scholar]
- 3.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 4.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones MR, Seeman NC, Mirkin CA. Programmable materials and the nature of the DNA bond. Science. 2015;347 doi: 10.1126/science.1260901. 1260901. [DOI] [PubMed] [Google Scholar]
- 7.Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- 8.Wilner OI, et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 9.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, et al. Reversible trapping and reaction acceleration within dynamically self-assembling nanoflasks. Nat Nanotechnol. 2016;11:82–88. doi: 10.1038/nnano.2015.256. [DOI] [PubMed] [Google Scholar]
- 11.Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF. Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly. Nat Chem. 2013;5:868–875. doi: 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- 12.Schnitzler T, Herrmann A. DNA Block Copolymers: Functional Materials for Nanoscience and Biomedicine. Acc Chem Res. 2012;45:1419–1430. doi: 10.1021/ar200211a. [DOI] [PubMed] [Google Scholar]
- 13.Alemdaroglu FE, Herrmann A. DNA meets synthetic polymers-highly versatile hybrid materials. Org Biomol Chem. 2007;5:1311–1320. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]
- 14.Um SH, et al. Enzyme-catalysed assembly of DNA hydrogel. Nat Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 15.Liao W-C, Willner I. Synthesis and applications of stimuli-responsive DNA-based nano- and micro-sized capsules. Adv Funct Mater. 2017 1702732. [Google Scholar]
- 16.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducrot E, He M, Yi GR, Pine DJ. Colloidal alloys with preassembled clusters and spheres. Nat Mater. 2017;16:652–657. doi: 10.1038/nmat4869. [DOI] [PubMed] [Google Scholar]
- 18.Qi H, et al. DNA-directed self-assembly of shape-controlled hydrogels. Nat Commun. 2013;4 doi: 10.1038/ncomms3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelikin AN, et al. A general approach for DNA encapsulation in degradable polymer microcapsules. ACS Nano. 2007;1:63–69. doi: 10.1021/nn700063w. [DOI] [PubMed] [Google Scholar]
- 20.Ong LL, et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature. 2017;552:72–77. doi: 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- 22.Groeschel AH, et al. Precise hierarchical self-assembly of multicompartment micelles. Nat Commun. 2012;3:710. doi: 10.1038/ncomms1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bates FS. Polymer-polymer phase-behavior. Science. 1991;251:898–905. doi: 10.1126/science.251.4996.898. [DOI] [PubMed] [Google Scholar]
- 24.Qiu HB, Hudson ZM, Winnik MA, Manners I. Multidimensional hierarchical self-assembly of amphiphilic cylindrical block comicelles. Science. 2015;347:1329–1332. doi: 10.1126/science.1261816. [DOI] [PubMed] [Google Scholar]
- 25.Stuart MAC, et al. Emerging applications of stimuli-responsive polymer materials. Nat Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 26.Aseyev V, Tenhu H, Winnik FM. Self Organized Nanostructures of Amphiphilic Block Copolymers II. Springer; Berlin Heidelberg: 2011. [Google Scholar]
- 27.Heuser T, Merindol R, Loescher S, Klaus A, Walther A. Photonic devices out of equilibrium: transient memory, signal propagation, and sensing. Adv Mater. 2017;29:9807–9814. doi: 10.1002/adma.201606842. [DOI] [PubMed] [Google Scholar]
- 28.Groeschel AH, et al. Guided hierarchical co-assembly of soft patchy nanoparticles. Nature. 2013;503:247–251. doi: 10.1038/nature12610. [DOI] [PubMed] [Google Scholar]
- 29.Lyon LA, Meng ZY, Singh N, Sorrell CD, John AS. Thermoresponsive microgel-based materials. Chem Soc Rev. 2009;38:865–874. doi: 10.1039/b715522k. [DOI] [PubMed] [Google Scholar]
- 30.Lutz JF, Ouchi M, Liu DR, Sawamoto M. Sequence-controlled polymers. Science. 2013;341:1238149. doi: 10.1126/science.1238149. [DOI] [PubMed] [Google Scholar]
- 31.Blanco L, et al. Highly efficient DNA-synthesis by the phage Φ29 DNA polymerase. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 32.Ali MM, et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]
- 33.Nelissen FHT, Goossens EPM, Tessari M, Heus HA. Enzymatic preparation of multimilligram amounts of pure single-stranded DNA samples for material and analytical sciences. Anal Biochem. 2015;475:68–73. doi: 10.1016/j.ab.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Estevez-Torres A, Baigl D. DNA compaction: fundamentals and applications. Soft Matter. 2011;7:6746–6756. [Google Scholar]
- 35.Bomboi F, et al. Re-entrant DNA gels. Nat Commun. 2016;7:1319. doi: 10.1038/ncomms13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duguid JG, Bloomfield VA. Aggregation of melted DNA by divalent metal ion-mediated cross-linking. Biophys J. 1995;69:2642–2648. doi: 10.1016/S0006-3495(95)80134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zozulya VN, Nesterov AB, Ryazanova OA, Blagoi YP. Conformational transitions and aggregation in poly(dA)-poly(dT) system induced by Na+ and Mg2+ ions. Int J Biol Macromolec. 2003;33:183–191. doi: 10.1016/j.ijbiomac.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Kohno Y, Saita S, Men YJ, Yuan JY, Ohno H. Thermoresponsive polyelectrolytes derived from ionic liquids. Polym Chem. 2015;6:2163–2178. [Google Scholar]
- 39.Plamper FA, Schmalz A, Ballauff M, Muller AHE. Tuning the thermoresponsiveness of weak polyelectrolytes by pH and light: Lower and upper critical-solution temperature of Poly(N,N-dimethylaminoethyl methacrylate) J Am Chem Soc. 2007;129:14538–14539. doi: 10.1021/ja074720i. [DOI] [PubMed] [Google Scholar]
- 40.Elbert DL. Liquid–liquid two-phase systems for the production of porous hydrogels and hydrogel microspheres for biomedical applications: A tutorial review. Acta Biomater. 2011;7:31–56. doi: 10.1016/j.actbio.2010.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Shabalin IG, Handing KB, Bujnicki JM, Minor W. Magnesium-binding architectures in RNA crystal structures: validation, binding preferences, classification and motif detection. Nucleic Acids Res. 2015;43:3789–3801. doi: 10.1093/nar/gkv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zavitsas AA. Aqueous solutions of calcium ions: hydration numbers and the effect of temperature. J Phys Chem B. 2005;109:20636–20640. doi: 10.1021/jp053909i. [DOI] [PubMed] [Google Scholar]
- 43.Yatsunyk LA, Mendoza O, Mergny J-L. “Nano-oddities”: unusual nucleic acid assemblies for DNA-based nanostructures and nanodevices. Acc Chem Res. 2014;47:1836–1844. doi: 10.1021/ar500063x. [DOI] [PubMed] [Google Scholar]
- 44.Rippe K, Fritsch V, Westhof E, Jovin TM. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. Embo J. 1992;11:3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Arco L, Li M, Mann S. Phagocytosis-inspired behaviour in synthetic protocell communities of compartmentalized colloidal objects. Nat Mater. 2017;16:857–863. doi: 10.1038/nmat4916. [DOI] [PubMed] [Google Scholar]
- 47.Park SY, et al. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Crystallization of DNA-coated colloids. Nat Commun. 2015;6:7253. doi: 10.1038/ncomms8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baffou G, Quidant R. Thermo-plasmonics: using metallic nanostructures as nano-sources of heat. Laser Photon Rev. 2013;7:171–187. [Google Scholar]
- 50.Mura S, Nicolas J, Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 51.Shanmugam V, Selvakumar S, Yeh CS. Near-infrared light-responsive nanomaterials in cancer therapeutics. Chem Soc Rev. 2014;43:6254–6287. doi: 10.1039/c4cs00011k. [DOI] [PubMed] [Google Scholar]
- 52.Willner I, Shlyahovsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem Soc Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 53.Merindol R, Walther A. Materials learning from life: concepts for active, adaptive and autonomous molecular systems. Chem Soc Rev. 2017;46:5588–5619. doi: 10.1039/c6cs00738d. [DOI] [PubMed] [Google Scholar]
- 54.Hyman AA, Weber CA, Juelicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.