Abstract
Protein aggregation is widely considered to be a nonspecific coalescence of misfolded proteins, driven by interactions between solvent-exposed hydrophobic surfaces that are normally buried within a protein's interior. Accordingly, abnormal interactions between misfolded proteins with normal cellular constituents has been proposed to underlie the toxicity associated with protein aggregates in many neurodegenerative disorders. Here we have used fluorescence resonance energy transfer and deconvolution microscopy to investigate the degree to which unrelated misfolded proteins expressed in the same cells coaggregate with one another. Our data reveal that in cells, protein aggregation exhibits exquisite specificity even among extremely hydrophobic substrates expressed at very high levels.
Aggregation of proteins into insoluble intracellular complexes and inclusion bodies is a common problem in bioengineering and is also intimately linked to the pathogenesis of most neurodegenerative diseases in man (1). Protein aggregation is widely viewed as nonspecific coagulation of incompletely folded or partially denatured polypeptides, driven by interaction among inappropriately exposed hydrophobic surfaces. According to this view, production of misfolded or denatured proteins has been suggested to be deleterious to cells by virtue of their ability to coaggregate with and thereby trap unrelated cellular proteins that may transiently display complementary surfaces (2–4). However, refolding studies of chemically denatured polypeptides suggest that protein aggregation in vitro is due to specific intermolecular interactions among defined domains within structured folding intermediates (5). Evidence of this specificity is found in the seeding behavior of amyloidogenic proteins (6) and in the selectivity of aggregate formation by model proteins (7). Although these studies suggest specificity in aggregation of purified denatured proteins in dilute solution, few studies have addressed the mechanism and specificity of protein aggregation in vivo. In cells, the vectorial nature of protein synthesis, the high protein concentration, the action of molecular chaperones and proteases, the potential for posttranslational modification, and the “molecular crowding” effect (8) are all factors likely to influence protein aggregation.
In cells, aggregated proteins are usually sequestered in discrete structures called inclusion bodies. Bacterial inclusion bodies are often highly enriched in a single aggregated protein species, suggesting that misfolded proteins do not coaggregate with cellular proteins. In contrast, inclusion bodies in mammalian cells are complex structures that contain many proteins, including molecular chaperones, components of the ubiquitin-proteasome system, centrosomal material, and cytoskeletal proteins (9). Inclusion bodies are also enriched in proteins involved in cell signaling, cell division, and apoptosis, suggesting that coaggregation of misfolded, damaged, or mutant proteins with normal cellular proteins could explain both the presence of multiple proteins in inclusion bodies and the toxicity associated with protein aggregation in many neurodegenerative diseases (2, 10, 11). The complexity of inclusion bodies in the mammalian cytoplasm could either be due to protein coaggregation or it could reflect a degree of complexity intrinsic to the mechanism of inclusion body formation.
We have previously reported that formation of cytoplasmic inclusion bodies requires an intact microtubule cytoskeleton, and that aggregated proteins accumulate near the microtubule organizing center in discrete pericentriolar inclusion bodies called aggresomes (12). Several recent reports support our proposal that aggresome formation is a general response of mammalian cells to the presence of protein aggregates and suggest a role for dynein/dynactin-mediated retrograde transport of aggregated proteins on cytoplasmic microtubules in inclusion body formation (13–15). One model to explain the presence of multiple, unrelated proteins in or near aggresomes is that misfolded proteins can coaggregate with cellular proteins, recruiting them into cytoplasmic inclusion bodies. Alternatively, the presence of multiple proteins in inclusion bodies could simply reflect the centrosome's function as the common terminus of cytoplasmic microtubules.
To discriminate between these models, we have developed a method that exploits fluorescence resonance energy transfer (FRET) to study the process of protein aggregation in vivo. This technique involves the transfer of energy from a fluorescent donor in its excited state to another excitable moiety, the acceptor, via nonradiative dipole–dipole interactions. This energy transfer process is highly sensitive to the distance and the orientation between the two fluorophores in question, typically occurring over a donor–acceptor separation of ≈10–100 Å, making FRET an ideal technique to study protein–protein interaction (16, 17). Most FRET studies have been conducted in vitro because of the inherent difficulty in labeling two proteins in vivo, but mutant forms of green fluorescent protein (GFP) from Aequoria Victoria, namely cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP), with spectral overlap suitable for FRET, have recently been developed (18). Using aggregation-prone proteins biosynthetically tagged with these fluorophores, our data indicate that in cells, protein aggregation is highly specific.
Experimental Procedures
Cells, Plasmids, and Transfections.
HEK293 cells were cultured in DMEM with 10% FBS and antibiotics. CFP and YFP fusions of P23H-rhodopsin, Q25, and Q103 were made by ligating the coding regions into CFP-N1 and YFP-N1 vectors (CLONTECH), respectively. CFP-ΔF508 and YFP-ΔF508 were generated by transferring a DNA fragment containing ΔF508 from GFP-ΔF508 (19) to the CFP-C1 and YFP-C1 vectors (CLONTECH), respectively. The HA-tagged T-cell receptor alpha chain (TCRα) construct has been described (20). Antibody to the HA tag was obtained from CRP Inc. All transfections were performed by using the calcium phosphate protocol (21). For FRET competition studies, cells were cotransfected with 1.36 μg of fluorescent P23H (P23H-CFP + P23H-YFP in equal proportion) and 6.8 μg of competition mix (competitor + empty vector in varying proportion). To correct for background YFP fluorescence, an identical set of cells was transfected with 0.68 μg of P23H-YFP and 0.68 μg of unlabeled P23H, along with the same amount of competitor/empty vector. For experiments in which proteasome activity was inhibited, ALLN (N-acetyl-Leu-Leu-norleucinal) was added 36 h posttransfection to a final concentration of 10 μg/ml. For microtubule disrupting experiments, nocodazole (10 μg/ml) was added 1 h before ALLN addition.
Fluorescence Microscopy.
Transfected HEK293 cells grown on glass coverslips were fixed in 4% paraformaldehyde. Conventional epifluorescence micrographs were obtained on a Zeiss Axiovert microscope with a 63× oil lens (NA1.4; Zeiss). Digital (12-bit) images were acquired with a cooled CCD (Princeton Instruments, Trenton, NJ) and processed by using METAMORPH software (Universal Imaging, Media, PA). The excitation filters used for conventional microscopy were 425DF20 (CFP), 490DF20 (YFP), and 570DF20 (Texas red). Emission filters were 475DF20 (CFP), 535DF20 (YFP), and 630DF25 (Texas red). The dichroics were: 440DCLP (CFP), 505 DCLP (YFP), and 595 DCLP (Texas red). Images in Figs. 1 G–I and 3C were acquired by using an Olympus microscope with 436 DF10 (CFP) and 500DF20 (YFP) filters for excitation and 470 DF30 (CFP) and 535 DF30 (YFP) filters for emission. Digital images (12 bit) were digitally deconvolved by using DELTAVISION hardware and software (Applied Precision, Issaquah, WA).
Figure 1.
Colocalization of aggregated proteins in cytoplasmic inclusion bodies. (A–F) Visualization of inclusion bodies in cells cotransfected with P23H-YFP and CFP-ΔF508 (A), TCRα (C), or Q103-CFP (E) by conventional epifluorescence microscopy. TCRα was detected by indirect immunofluorescence against an HA epitope bar. Inclusion bodies are denoted by arrows. (G–I) Visualization of inclusion bodies by digital deconvolution microscopy. (G) P23H-YFP (red) and ΔF508-CFP (green; single cell). (H) P23H-YFP (red) and Q103 (green; single cell). (I) Q103 -CFP (red) and ΔF508-YFP (green; two cells, each with one inclusion body). All cells were treated with ALLN to maximize protein aggregation. (Scale bars: A–F, 15 μm; G, 5 μm; H and I, 7.5 μm.)
FRET Measurements.
Fluorescence spectra were recorded on suspended cells (≈106 cells per ml) in a Spex fluorolog fluorometer with a Spex 1620 dual grating emission monochromator (Spex Industries, Metuchen, NJ). FRET measurements were made by exciting the donor (CFP) at 425 nm and monitoring emission between 450 and 600 nm. Slit widths were 2 mm for all experiments. All FRET spectra were corrected for background YFP fluorescence by subtracting the spectrum obtained from cells transfected with identical amounts of the corresponding YFP construct. For competition experiments, the ratio of fluorescence at 525 nm (YFP) to the fluorescence at 476 nm (CFP) was measured. This ratio was 0.42 ± 0.013 (SEM; n =5) for P23H-CFP alone and 0.649 ± 0.031 (SEM; n =5) for P23H-CFP cotransfected with P23H-YFP. These ratios were assigned FRET values of 0% and 100%, respectively. Energy transfer efficiency (E) can be calculated as:
 |
where FA (obs) is the observed acceptor emission due to FRET and FA (100%) is the acceptor emission that would be observed if there were 100% energy transfer (16, 17). Because FA (100%) is a constant for a given donor–acceptor pair (CFP and YFP, respectively, in this case), relative FRET efficiencies in this study were calculated as ratios of observed acceptor emissions.
Results
Protein Colocalization in Inclusion Bodies Does Not Necessarily Indicate Coaggregation.
To assess specificity in protein aggregation, we transiently coexpressed a panel of unrelated aggregation-prone proteins in HEK293 cells. The ΔF508 mutant of the cystic fibrosis transmembrane conductance regulator (CFTR) has been shown to quantitatively misfold (22) and to accumulate in aggresomes on overexpression (12). Like ΔF508, the P23H mutant of rhodopsin, which is linked to autosomal dominant retinitis pigmentosa (23), is a highly hydrophobic polytopic membrane protein that is unable to fold in the endoplasmic reticulum (24). We have recently shown that this protein is degraded by the ubiquitin proteasome system, that undegraded forms accumulate in aggresomes, and that presence of the GFP tag does not influence its intracellular trafficking or its biochemical behavior (M.E.I., R.S.R., and R.R.K., unpublished data). When coexpressed with CFP-ΔF508 and evaluated by conventional epifluorescence microscopy, P23H-YFP was colocalized in aggresomes in a pattern indistinguishable from that of ΔF508 (Fig. 1 A and B).¶ We also evaluated the intracellular distribution of aggregated forms of the alpha subunit of the T-cell receptor (TCRα). This type I integral membrane protein, when expressed in the absence of its oligomeric partners, is unable to fold (25) and, hence, is targeted to the cytosol for rapid ubiquitin and proteasome-dependent degradation (26, 27). Undegraded TCRα subunits form detergent-insoluble aggregates (27); these were sequestered in aggresomes indistinguishable from those formed by P23H (Fig. 1 C and D). Finally, we compared the localization of P23H in aggresomes with the distribution of a fragment of huntingtin containing Q103, an aggregation-promoting polyglutamine (polyQ) tract fused to CFP (Fig. 1 E and F). Previous studies have shown that huntingtin fragments containing expanded polyQ form large, dense cytoplasmic inclusion bodies when expressed in tissue culture cells (28). Our data (Fig. 1 E and F) confirm that Q103-CFP is indeed sequestered in singular cytoplasmic inclusion bodies that also contain P23H. However, unlike ΔF508 and TCRα, Q103-CFP inclusion bodies did not appear to align precisely with P23H (or ΔF508; data not shown). In some cases, there seemed to be cavities in the fluorescence from P23H-YFP that were filled by Q103-CFP. In other cases, there appeared to be an overlap in the regions occupied by the two proteins. To more clearly image these inclusion bodies, we used digital deconvolution microscopy to eliminate out-of-focus light from other focal planes (Fig. 1 G–I). Strikingly, we observed that the inclusion bodies composed of ΔF508 and P23H—which appeared homogeneous under conventional epifluorescence microscopy—were composed of smaller structures (Fig. 1G). Some of these particles [pseudocolored to reveal P23H (red) or ΔF508 (green)] overlapped to produce yellow particles, whereas other particles were distinctly red or green. Deconvolution revealed that Q103 localization was always distinct from that of P23H (Fig. 1H) or ΔF508 (Fig. 1I). In some cases, the inclusion bodies appeared to be composed of side-by-side regions enriched in the two proteins (Fig. 1H), whereas in other cases the membrane protein aggregates appeared to surround the Q103 aggregates (Fig. 1I). These observations suggest that Q103 does not coaggregate with P23H and ΔF508. However, the partial colocalization of P23H aggregates with aggregates of the membrane proteins ΔF508 and TCRα does not rule for or against coaggregation of P23H with the other membrane proteins.
P23H Does Not Coaggregate with Other Aggregation-Prone Proteins.
To investigate protein aggregation at higher resolution, we used FRET to evaluate the process of protein aggregation in living cells. To validate this approach, we recorded the fluorescence emission spectrum (excited for CFP at 425 nm) of a suspension of cells cotransfected with P23H-CFP and P23H-YFP and treated with the proteasome inhibitor ALLN to promote maximal aggregation. Under these conditions, the majority of cellular P23H, which is unable to fold, aggregated and was localized to cytoplasmic aggresomes (Fig. 1). The two peaks at 476 nm and 505 nm (Fig. 2A, solid line) correspond to CFP emission, whereas the shoulder at 525 nm reflects the sensitized emission from YFP due to energy transfer from CFP. This conclusion was confirmed in a control experiment where we performed the same measurement on a suspension of identically treated cells expressing only P23H-CFP mixed with cells expressing only P23H-YFP. Because the two fluorophores are present in different cells, no FRET can occur. Under these conditions (Fig. 2A, dashed line), the 525-nm shoulder is reduced and there is a corresponding increase in CFP emission. This reciprocal relationship between emission at the donor and acceptor wavelengths is a hallmark of FRET, reflecting both quenching of donor fluorescence and sensitized emission by the acceptor. Thus, we used the ratio of YFP fluorescence (at 525 nm) to CFP fluorescence (at 476 nm) to quantify the degree of FRET.
Figure 2.
Specificity of protein aggregation measured by FRET. (A) FRET in cells containing P23H inclusion bodies. Fluorescence emission spectra (excitation 425 nm) of a homogeneous suspension of cells cotransfected with equal amounts of P23H-CFP and P23H-YFP plasmid (solid line) compared with the spectrum of a suspension of a mixture of cells transfected with only P23H-CFP and cells transfected with only P23H-YFP (dashed line). Arrows indicate the emission peaks for CFP (476) and YFP (525) used to calculate the FRET value as described in the text. (B) P23H (open squares), but not ΔF508 (filled squares), can compete out FRET between P23H-CFP and P23H-YFP. The ratio of unlabeled competitor plasmid to the total labeled plasmid is plotted on the x axis. (C) Other aggregation-prone proteins (at a 4:1 plasmid ratio) do not decrease FRET between P23H-CFP and P23H-YFP.
We designed a FRET competition experiment to assess the degree to which P23H can coaggregate with other proteins. Coaggregation of an unlabeled aggregation-prone protein with P23H-C/YFP will reduce the average distance and, hence, FRET between P23H-CFP and P23H-YFP molecules. Coexpression of unlabeled P23H reduced the FRET value between P23H-CFP and P23H-YFP in a dose-dependent manner (Fig. 2B, open symbols). In contrast, coexpression of ΔF508 at similar levels did not significantly reduce P23H FRET, suggesting that the ΔF508 molecules (Fig. 2B, closed symbols), which are themselves largely aggregated under these conditions (12), do not interact with P23H. Using this approach we found no evidence for coaggregation between P23H and other aggregation-prone proteins including ΔF508, TCRα, and Q103—all of which colocalize to the same inclusion body as P23H (Fig. 2C). A trivial explanation for this observation is that the nonhomologous competitors have very different transfection efficiencies compared with P23H, so that on an average most cells that express P23H-CFP and P23H-YFP do not express the competitor. However, fluorescence microscopy revealed that 75–82% (n = 400) of cells expressing P23H also express the heterologous competitor. These data, therefore, establish that proteins localized to the same inclusion body do not necessarily coaggregate with each other.
Aggregation Is Independent of Inclusion Body Formation.
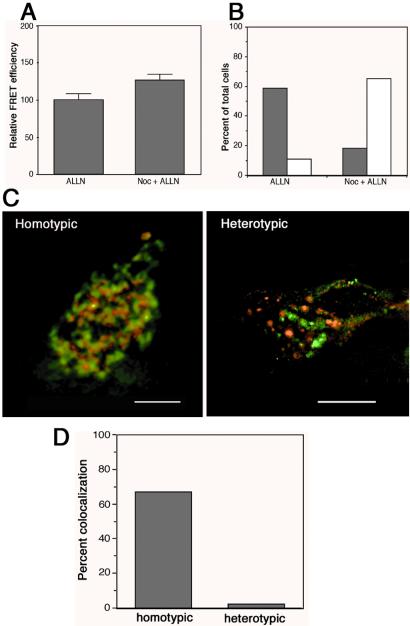
We and others have previously shown that disruption of microtubule-based retrograde transport prevents the formation of a single juxtanuclear inclusion body and results in the appearance of dispersed foci of misfolded protein throughout the cell (12–15). If inclusion bodies are the intracellular sites at which misfolded proteins aggregate, then disruption of microtubules with nocodazole would be predicted to decrease the extent of protein aggregation. To test this prediction, we assessed the effect of nocodazole treatment on FRET between P23H-CFP and P23H-YFP in proteasome-inhibited cells (Fig. 3A). However, we observed a nearly 25% increase in FRET in nocodazole-treated cells, suggesting that protein aggregation is enhanced rather than diminished in the absence of microtubules. A control experiment (Fig. 3B) confirmed that the presence of nocodazole caused a 3-fold decrease in the number of inclusion bodies. Thus, these data demonstrate that aggregation of P23H occurs independently of transport to pericentriolar inclusion bodies.
Figure 3.
Protein aggregation is independent of inclusion body formation. (A) Cells coexpressing P23H-CFP and P23H-YFP were treated with the proteasome inhibitor ALLN to enhance protein aggregation in the presence or absence of nocodazole to disrupt microtubules. Data represent mean ± SEM from three independent trials. (B) The fraction of cells from the experiment in A with a single inclusion body (shaded bars) or dispersed foci (open bars). A total of 400 cells were counted for each treatment group. (C) Intracellular localization (deconvolved microscopic images) of fusion proteins in dispersed foci from nocodazole + ALLN treated cells expressing the heterotypic aggregation pair YFP-ΔF508 (red) + P23H CFP (green) or the homotypic pair P23H-CFP (red) + P23H-YFP (green). (D) Quantification of colocalization in the experiment from C.
If protein aggregation is independent of inclusion body formation, then the distribution of coexpressed proteins in nocodazole-dispersed foci should reveal the extent of protein coaggregation. To test this prediction, CFP-ΔF508 and P23H-YFP were coexpressed in HEK293 cells and treated with nocodazole in the presence of ALLN. Dispersed foci of P23H (green) were nearly always distinct from foci of ΔF508 (red; Fig. 3C, Right), exhibiting less than 2% colocalization (Fig. 3D). In contrast, there was extensive (67%) overlap between dispersed foci of the homotypic pair, P23H-CFP and P23H-YFP (Fig. 3C, Left, and D). These data further confirm that the hydrophobic proteins P23H and ΔF508 do not coaggregate with one another, even under conditions in which inclusion body formation is suppressed.
Coaggregation of PolyQ Proteins.
While the preceding data suggest that unrelated proteins do not coaggregate, even when they share extensive hydrophobic character, they do not address whether or not coaggregation can occur between proteins that share other potential complementary “aggregation motifs.” One such motif that has been proposed to potentially mediate coaggregation of otherwise unrelated proteins is polyQ (3). Short polyQ stretches (<25 Q) are present in many normal proteins, including nuclear transcription factors and the wild-type versions of genes linked to dominantly inherited neurodegenerative diseases like Huntington's disease. However, proteins containing long (>35 Q) polyQ runs are highly prone to aggregate and have been suggested to coaggregate with and “recruit” otherwise unrelated proteins containing short polyQ stretches (3, 29). This type of interaction has been proposed to lead to a potential dominant interference of mutant huntingtin with the wild-type gene product (30).
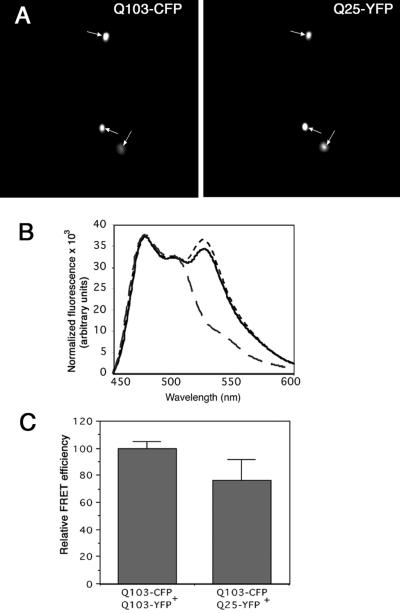
To test this hypothesis we used C/YFP fusions to evaluate the interaction between variants of a huntingtin fragment (exon 1) containing long (Q103) and short (Q25) glutamine homopolymers. A high fraction of cells expressing Q103 fusions alone spontaneously accumulate the huntingtin fusion into cytoplasmic inclusion bodies; this fraction is dramatically increased by inhibition of proteasome activity (31). By contrast, Q25 fusions remain diffusely cytoplasmic even on overexpression and exposure to proteasome inhibitors (31). When we coexpressed Q103-CFP and Q25-YFP in HEK293 cells, we found that a significant fraction of normally soluble Q25-YFP was sequestered into Q103-positive inclusion bodies (Fig. 4A). FRET measurements of Q103-CFP coexpressed with Q103-YFP revealed the presence of a strong signal from the sensitized emission of Q103-YFP, corresponding to Q103 aggregation (Fig. 4B). On the other hand, no FRET between Q25-CFP and Q25-YFP was detectable, consistent with Q25 being a soluble, monomeric protein. Strikingly, we observed a strong FRET signal between coexpressed Q103-CFP and Q25-YFP (Fig. 4B). The efficiency of energy transfer between Q103-CFP and Q25-YFP was ≈76% of that between Q103-CFP and Q103-YFP (Fig. 4C).
Figure 4.
Huntingtin Q103 coaggregates with a huntingtin fragment containing a nonpathogenic polyQ tract. (A) Inclusion bodies of Q103 colocalize with Q25. (B) fluorescence emission spectra, as in Fig. 2A, from cells cotransfected with equal amounts of Q25-CFP + Q25-YFP (large dashes), Q103-CFP + Q103-YFP (small dashes), or Q103-CFP + Q25-YFP (solid line). Spectra were normalized for CFP fluorescence. (C) Quantification of FRET efficiency between huntingtin fragments containing different glutamine repeat lengths.
Discussion
In this study, we have used two independent approaches to demonstrate specificity of protein aggregation in vivo. In one approach, deconvolution microscopy was used to assess the intracellular localization of different aggregated proteins expressed in the same cell. These data show that proteins aggregate into discrete foci that are homogeneous with respect to a particular protein and that aggregation is independent of inclusion body formation. In a second approach, FRET was used to assess the degree to which hydrophobic (ΔF508, TCRα) or hydrophilic (Q103) proteins can coaggregate with the very hydrophobic protein P23H rhodopsin. Our data strongly suggest that nonspecific aggregation between hydrophobic proteins does not occur and supports the view that protein aggregation is highly specific.
Several potential mechanisms could account for the specificity in protein aggregation observed in these studies. A trivial possibility is that proteins might not coaggregate because they are present in different cellular compartments or cytoplasmic regions. However, three of the proteins studied are integral membrane proteins that are unable to fold and are dislocated from the ER membrane to the cytoplasm by a common mechanism (refs. 12 and 27; M.E.I., R.S.R., and R.R.K., unpublished data); there is no evidence to suggest spatial restriction of these proteins. A second possibility is that proteins could segregate into separate aggregates if the kinetics of their aggregation were to differ substantially. Indeed, such an explanation could potentially account for the differences in the appearance of inclusion bodies formed from two hydrophobic proteins compared with inclusion bodies formed from hydrophobic proteins and Q103. However, there is no a priori reason to argue that two topologically similar proteins, P23H and ΔF508, should exhibit significant differences in aggregation kinetics. Instead, we propose that the observed specificity in aggregation among hydrophobic proteins reflects specific interactions between partially folded (misfolded) intermediates. Because P23H and ΔF508 are integral membrane proteins whose hydrophobic domains are not normally exposed to cytosol, it is likely that the intermediates leading to aggregation are different from those that are common to the native folding pathway. It is all of more surprising, therefore, that such exquisite specificity is maintained.
While our data demonstrate specificity in aggregation among hydrophobic proteins or between hydrophobic and hydrophilic proteins, they also show that specific coaggregation can occur between proteins, such as Q103 and Q25, that share a common aggregation-promoting motif, previously suggested by studies of colocalization and detergent solubility (32). However, because our constructs differ only in the number of glutamine residues, we cannot exclude the possibility that other parts of the proteins might contribute to or modulate this interaction. It is also not possible to extrapolate from these data to predict whether different pairs of otherwise unrelated proteins sharing limited polyQ domains would coaggregate with one another. Such interactions have been proposed to explain the colocalization of polyQ- containing cellular regulators like the CREB-binding protein (11, 32) and TATA binding protein (33), which bear short (≤19) polyQ tracts, to inclusion bodies in moribund neurons and in cellular models of neurodegenerative disease. Although our data do not rule for or against the existence of such interactions, they do explicitly show that colocalization to the same inclusion body cannot substitute for biochemical or biophysical evidence for interaction.
Finally, the finding that GFP and its variants fluoresce, even while participating in aggregates, illustrates that aggregated proteins have not necessarily adopted a completely non-native structure. Although our constructs are all artificial fusions, it is reasonable to extrapolate from these findings to propose that similar “partial” aggregation may be possible for naturally occurring multidomain proteins. This raises the possibility that, in addition to nonspecific coaggregation (which does not appear to occur) and “specific” coaggregation (as illustrated by polyQ-mediated interaction) a third mode of coaggregation—“native coaggregation”—may be possible. In this mode, binding partners of multidomain proteins could be recruited into an aggregate by virtue of their interaction with the native domains of aggregated proteins. Such interactions, if they exist, could provide another mechanism by which proteins aggregates disrupt cellular metabolism.
Acknowledgments
We thank J. Nathans for P23H rhodopsin, B. Stanton for GFP-CFTR, A. Tobin for huntingtin-GFP constructs and S. Palmieri for help with deconvolution microscopy. We acknowledge L.J.-P. Wong, J. Frydman, and members of the Kopito lab for helpful discussions, and M. Gelman and M. Bucci for critical comments on the manuscript. This work was funded by a grant from the National Institutes of Health (to R.R.K.).
Abbreviations
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- ALLN
N-acetyl-Leu-Leu-norleucinal
- TCRα
T-cell receptor alpha chain
Footnotes
Identical results were obtained with non-C/YFP variants (data not shown).
References
- 1.Fink A L. Folding Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 2.Trojanowski J Q, Lee V M. Ann NY Acad Sci. 2000;924:62–67. doi: 10.1111/j.1749-6632.2000.tb05561.x. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijn L I, Houseweart M K, Kato S, Anderson K L, Anderson S D, Ohama E, Reaume A G, Scott R W, Cleveland D W. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel R. Cell. 1996;86:699–702. doi: 10.1016/s0092-8674(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 6.Jarrett J T, Lansbury P T., Jr Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- 7.Speed M A, Wang D I, King J. Nat Biotechnol. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- 8.Minton A P. J Biol Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 9.Kopito R R. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 10.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. . (First Published May 23, 2000; 10.1073/pnas.100110097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 12.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notterpek L, Ryan M C, Tobler A R, Shooter E M. Neurobiol Dis. 1999;6:450–460. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Mata R, Bebok Z, Sorscher E J, Sztul E S. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabore A F, Wang W J, Russo S J, Beers M F. J Cell Sci. 2001;114:293–302. doi: 10.1242/jcs.114.2.293. [DOI] [PubMed] [Google Scholar]
- 16.Lakowicz J R. Principles of Fluorescence Spectroscopy. New York: Kluwer Academic/Plenum; 1999. [Google Scholar]
- 17.Clegg R M. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 18.Miyawaki A, Llopis J, Heim R, McCaffery J M, Adams J A, Ikura M, Tsien R Y. Nature (London) 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 19.Moyer B D, Loffing J, Schwiebert E M, Loffing-Cueni D, Halpin P A, Karlson K H, Ismailov I I, Guggino W B, Langford G M, Stanton B A. J Biol Chem. 1998;273:21759–21768. doi: 10.1074/jbc.273.34.21759. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Kopito R R. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 21.Graham F L, van der Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Ward C L, Kopito R R. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 23.Sung C H, Davenport C M, Hennessey J C, Maumenee I H, Jacobson S G, Heckenlively J R, Nowakowski R, Fishman G, Gouras P, Nathans J. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung C H, Schneider B G, Agarwal N, Papermaster D S, Nathans J. Proc Natl Acad Sci USA. 1991;88:8840–8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonifacino J S, Suzuki C K, Lippincott-Schwartz J, Weissman A M, Klausner R D. J Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Kaung G, Kobayashi S, Kopito R R. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 28.Cooper J K, Schilling G, Peters M F, Herring W J, Sharp A H, Kaminsky Z, Masone J, Khan F A, Delanoy M, Borchelt D R, et al. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 29.Preisinger E, Jordan B M, Kazantsev A, Housman D. Philos Trans R Soc London B. 1999;354:1029–1034. doi: 10.1098/rstb.1999.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 31.Wyttenbach A, Carmichael J, Swartz J, Furlong R A, Narain Y, Rankin J, Rubinsztein D C. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C C, Faber P W, Persichetti F, Mittal V, Vonsattel J P, MacDonald M E, Gusella J F. Somatic Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]