Summary
Drug combinations are synergistic when their combined efficacy exceeds the sum of the individual actions, but they rarely include ineffective drugs that become effective only in combination. We identified several “enabling pairs” of neutralizing and non-neutralizing anti-ebolavirus monoclonal antibodies, whose combination exhibited new functional profiles, including transforming a non-neutralizing antibody to a neutralizer. Sub-neutralizing concentrations of antibodies 2G4 or m8C4 enabled non-neutralizing antibody FVM09 (IC50 >1 μM) to exhibit potent neutralization (IC50 1–10 nM). While FVM09 or m8C4 alone failed to protect Ebola-virus-infected mice, a combination of the two antibodies provided 100% protection. Furthermore, non-neutralizers FVM09 and FVM02 exponentially enhanced the potency of two neutralizing antibodies against both Ebola and Sudan viruses. We identified a hotspot for the binding of these enabling antibody pairs near the interface of the glycan cap and GP2. Enabling cooperativity may be an underappreciated phenomenon for viruses, with implications for the design and development of immunotherapeutics and vaccines.
Keywords: Ebola virus, vaccine, filovirus, monoclonal antibody, antibody therapeutics, antibody synergy, cooperative neutralization, synergistic neutralization
Graphical Abstract
Highlights
-
•
We describe cooperative neutralization and in vivo protection
-
•
Cooperativity turns non-neutralizing ebolavirus antibodies into potent neutralizers
-
•
A hotspot for antibody cooperativity identified on Ebola virus glycoprotein
Howell et al. describe the phenomenon of “enabling, cooperative neutralization” where weakly neutralizing and/or non-neutralizing ebolavirus monoclonal antibodies exhibit potent neutralization when combined. The previously identified antibodies m8C4 and FVM09, which are non-protective alone, offer 100% protection against Ebola and Sudan virus challenge in mice when combined.
Introduction
The 2014 Ebola virus (EBOV) disease (EVD) epidemic claimed over 11,000 lives, underscoring the need for effective therapeutics for this deadly virus and other members of the family Filoviridae that have caused human outbreaks in the past—Sudan virus (SUDV), Bundibugyo virus (BDBV), and Marburg virus (MARV) (Rougeron et al., 2015). Monoclonal antibodies (mAbs) targeting the filovirus glycoprotein (GP) are among the most promising countermeasures for filoviruses (Zeitlin et al., 2016). Several studies have shown the superiority of specific combinations of mAbs targeting different EBOV GP epitopes compared to monotherapy (Olinger et al., 2012, Pettitt et al., 2013, Qiu et al., 2012a, Qiu et al., 2012b, Qiu et al., 2014). The combination of antibody therapy with adenovirus-expressed interferon (IFN)-α in nonhuman primates (NHPs) also resulted in enhanced efficacy (Qiu et al., 2013). While these studies focused on EBOV-specific therapies, we recently targeted other ebolaviruses and demonstrated improved neutralization efficacy by combining several pan-ebolavirus therapeutic mAbs (Holtsberg et al., 2015, Howell et al., 2016, Keck et al., 2015). These findings indicate that the engagement of multiple antibodies through different protective mechanisms can be highly effective in controlling filovirus infection.
The filovirus surface glycoproteins (GPs) consist of disulfide-linked subunits GP1 and GP2. The crystal structure of EBOV GP in complex with the neutralizing mAb KZ52 showed that three GP1 subunits assemble in a chalice-like structure surrounded by three GP2 subunits, which, with an N-terminal portion of GP1, form the base of the chalice (Lee et al., 2008) and anchor GP to the virus membrane. The endosomal receptor binding site (RBS) is positioned at the apex of the GP1 core and is largely concealed by a glycan cap at the rim of the chalice and by the highly glycosylated and disordered mucin-like domain (MLD). The best-characterized neutralizing epitope within EBOV GP is the so-called “base epitope” consisting of residues within GP1 and GP2 and recognized by mAbs KZ52 (Davidson et al., 2015, Lee et al., 2008) and by the ZMapp therapeutic mAb cocktail components 2G4 and 4G7 (Davidson et al., 2015, Murin et al., 2014, Pallesen et al., 2016, Tran et al., 2016). Recently, we and others have identified a number of neutralizing epitopes within the RBS (Flyak et al., 2015, Howell et al., 2016, Misasi et al., 2016), the glycan cap (Bornholdt et al., 2016, Flyak et al., 2016, Holtsberg et al., 2015), and the internal fusion loop (Bornholdt et al., 2016, Misasi et al., 2016).
A number of studies have identified synergistic pairs of neutralizing mAbs, targeting HIV (Li et al., 1998, Mascola et al., 1997, Miglietta et al., 2014, Pollara et al., 2014, Vijh-Warrier et al., 1996), Severe acute respiratory syndrome (SARS) coronavirus (ter Meulen et al., 2006), and hepatitis C virus (Carlsen et al., 2014). In all examples, synergy was observed between two or more antibodies that could each neutralize the virus independently. Here, we describe a novel phenomenon of cooperative neutralization by mAb pairs consisting of a neutralizing mAb and a non-neutralizing mAb. We identified several such pairs by screening a panel of mAbs for binding to ebolavirus glycoproteins and for their ability to neutralize one or more ebolaviruses. We observed neutralization by weakly or moderately neutralizing mAbs that was enhanced when they were paired with specific non-neutralizing mAbs targeting epitopes within the glycan cap of ebolavirus GP. We also found that an otherwise non-neutralizing pan-ebolavirus mAb acquired neutralizing activity in the presence of sub-neutralizing concentrations of a mAb that bound to an adjacent epitope. Finally, we demonstrated that cooperative activity extended to protection in vivo, as a pair of mAbs that were individually non-protective, provided full protection against EBOV infection when combined in a cocktail.
Results
Binding Enhancement between Antibodies against Ebolavirus GP
We first evaluated the potential competition between various mAbs for binding to EBOV GP using ELISA-based assays. ELISA plates coated with EBOV GP were pre-incubated with saturating concentrations of individual EBOV-specific mAbs, pan-ebolavirus mAbs (Holtsberg et al., 2015, Keck et al., 2015), or a non-binding, negative control mAb. The test mAbs (biotinylated or having a mouse Fc region) were then added at their EC50 concentrations, and relative binding was determined. We noted competition between mAbs with closely located epitopes (Table S1), for example, mAbs 13C6 and FVM04, as we have described (Keck et al., 2015), and among the GP base-binding mAbs 2G4, 4G7, and KZ52, consistent with previous reports (Murin et al., 2014). Interestingly, this study revealed that pre-incubation with the macaque-derived mAb FVM09 enhanced the binding of m8C4 and m2G4, by 99% and 41%, respectively, but decreased binding of KZ52. In addition, FVM20, whose epitope overlaps with that of FVM09 (Keck et al., 2015), also enhanced m8C4 and m2G4 binding, by 71% and 29%. When 2G4 or 4G7 was bound first to GP, the binding of FVM20 was enhanced nearly 400%.
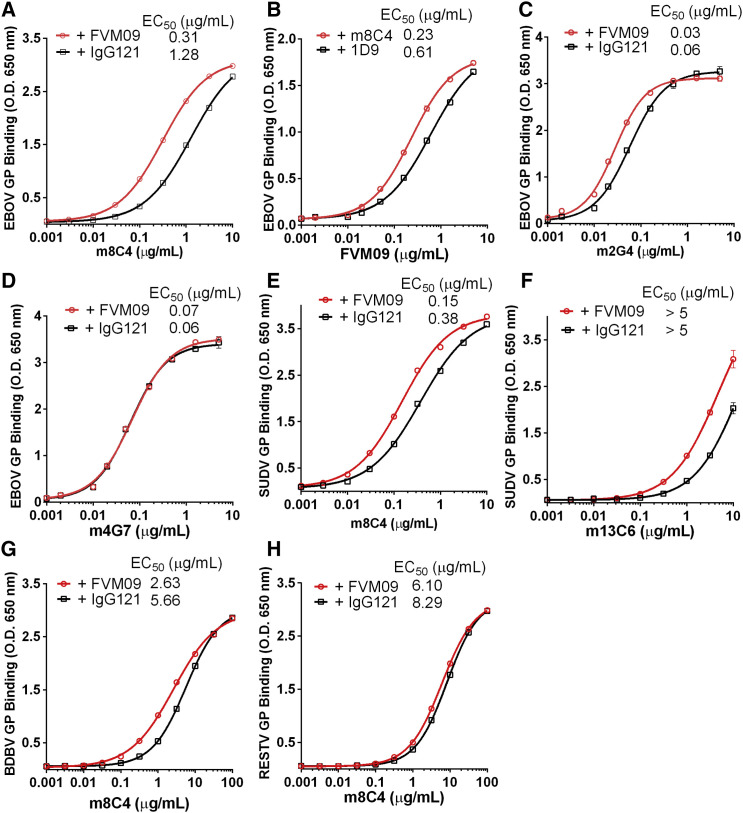
Since some of the mAbs tested exhibit pan-ebolavirus reactivity, including FVM09, FVM20, and m8C4 (Holtsberg et al., 2015, Keck et al., 2015), we also assayed for enhancement of mAb binding to SUDV GP and observed a modest increase of m8C4 binding to SUDV GP in the presence of FVM09 (31% higher) and FVM20 (12% higher) (Table S2). mAb 13C6 is a glycan cap binder and is primarily EBOV reactive but also binds weakly to SUDV GP. Binding of 13C6 to SUDV was enhanced by FVM09 and FVM20 (by 55% and 29%, respectively; Table S2). We then tested the binding enhancement over a full range of mAb concentrations. The binding of m8C4 to EBOV GP was strongly increased in the presence of a saturating concentration of FVM09, as was the binding of FVM09 in the presence of a saturating concentration of m8C4 (4- and 2.5-fold reduction of EC50, respectively) (Figures 1A and 1B). A similar, but lesser, increase in binding was observed for m2G4 with EBOV GP in the presence of FVM09 (2-fold reduction of EC50; Figure 1C), but not for binding of 4G7 (Figure 1D). The binding of m8C4 to SUDV GP was enhanced >2-fold in presence of FVM09 (Figure 1E), and an enhancement of the weak binding of m13C6 to SUDV GP was observed (Figure 1F). FVM09 also slightly enhanced the binding of m8C4 to BDBV GP (Figure 1G) but not RESTV GP (Figure 1H). Our results thus demonstrate multiple combinations of mAbs that enhance binding to EBOV, SUDV, and/or BDBV.
Figure 1.
Enhancement of Antibody Binding to GPΔTM
Shown are ELISA binding curves for mAbs m8C4 (A, E, G, and H), FVM09 (B), m2G4 (C), m4G7 (D), and m13C6 (F), to GPdTM of EBOV (A–D), SUDV (E and F), BDBV (G), or RESTV (H), in the presence of a fixed concentration (20 μg/mL) of FVM09 (A and C–H) or m8C4 (B) in comparison to 20 μg/mL of an irrelevant mAb (IgG121 or 1D9, in black). Each data point represents the average of three measurements ±SD. See also Tables S1 and S2.
Cooperative Neutralization of Ebolaviruses
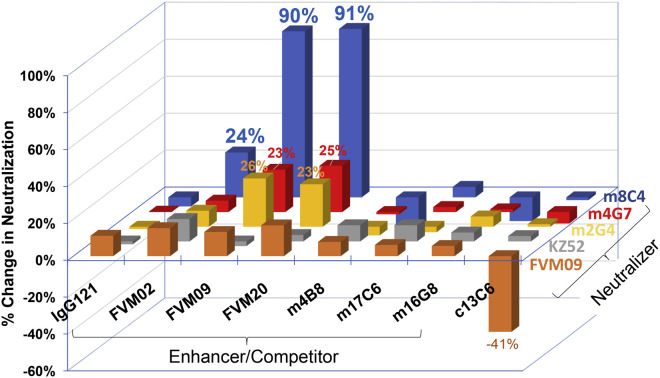
We next determined whether the enhancement of binding also extended to the ability of mAbs to neutralize EBOV. A similar matrix array enhancement experiment was performed for EBOV neutralization, using pseudotyped vesicular stomatitis virus (VSV) (VSV-EBOV GP-Luc) infection of Vero cells. MAbs m8C4, FVM04, KZ52, m2G4, or m4G7 were applied at their respective EC50 concentrations, and their ability to neutralize was determined in the absence or presence of individual non-neutralizing antibodies at saturating concentrations. Consistent with the mAb binding studies, m8C4-mediated neutralization was potentiated by more than 90% in the presence of FVM09 or FVM20 and was moderately increased by FVM02 but was not increased by m4B8, m17C6, m16G8, or c13C6 (Figure 2 ). For GP base-binding mAbs m2G4 and m4G7, a more moderate enhancement of 23%–26% was also observed in the presence of FVM09 or FVM20, while the other mAbs tested did not have any impact on neutralization. In contrast, for another base-binding mAb, KZ52, neutralization in the presence of any tested mAb remained within ±10% of the value seen with KZ52 alone. FVM04-mediated neutralization was slightly potentiated in the presence of FVM09, FVM02, and FVM20 (13%, 15%, and 17%, respectively) but was significantly reduced in the presence of c13C6, consistent with the overlapping binding footprints of these two mAbs on EBOV GP (Howell et al., 2016, Keck et al., 2015).
Figure 2.
Antibody Pairs Show Enhanced Neutralization of EBOV
Shown are the relative values for neutralization of VSV-EBOV GP-Luc by neutralizing mAbs FVM04, KZ52, m2G4, m4G7, and m8C4 used at their respective EC50 concentrations in the presence of an excess concentration (20 μg/mL) of the individual mAbs listed on the x axis. IgG121 is an irrelevant control antibody. Data are shown as a percentage increase or decrease (negative values) of neutralization in the presence of each antibody compared to no excess antibody. For example, for the m8C4/FVM09 pair the percentage change is calculated as follows: [(% neutralization in the presence of FVM09 and m8C4) – (% neutralization in the presence of m8C4 only)] divided by percentage neutralization in presence of m8C4 only. Since neutralizers were used at their respective EC50, the maximum possible change is 100%.
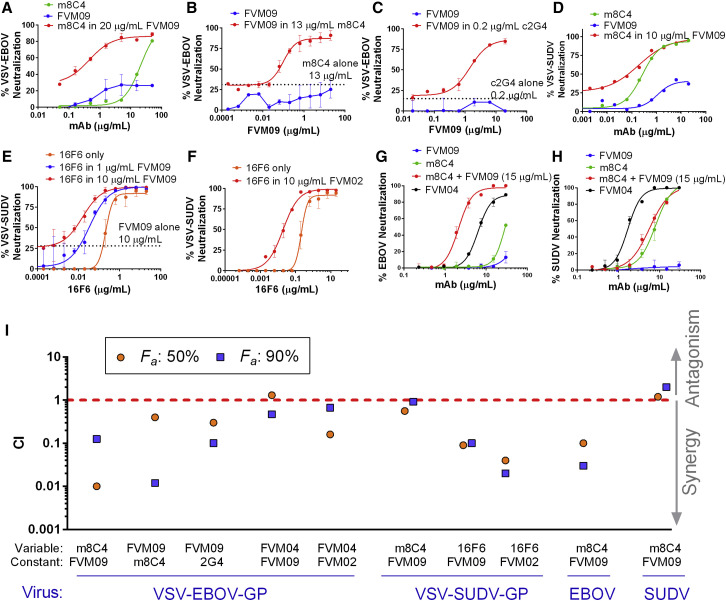
We then examined the potential for cooperative neutralization for one mAb over a wide concentration range, in the presence of a second non-neutralizing mAb at a constant, saturating concentration (20 μg/mL) or a neutralizing mAb at a sub-neutralizing concentration. While m8C4 by itself showed poor neutralization of VSV-EBOV GP-Luc (IC50 of ∼20 μg/mL), its neutralizing potency was dramatically augmented (∼70-fold reduction in IC50) in the presence of FVM09 (Figure 3 A). In the reverse experiment, FVM09, which is non-neutralizing by itself, exhibited strong dose-dependent neutralization in the presence of sub-neutralizing concentrations of m8C4 (Figure 3B) or 2G4 (Figure 3C).
Figure 3.
Neutralization of Pseudotyped VSV-EBOV-GP-Luc and VSV-SUDV-GP-Luc
(A) VSV-EBOV neutralization by m8C4 alone (green), FVM09 alone (blue), or m8C4 titrated into a fixed amount of FVM09 (20 μg/mL) (red).
(B) FVM09 is titrated into a fixed sub-neutralizing concentration of m8C4 (13 μg/mL) (red), which shows enhanced potency compared to FVM09 alone (blue) or m8C4 at 13 μg/mL (dotted line).
(C) FVM09 titrated into 0.2 μg/mL of c2G4 shows a clear neutralizing dose response (red) in contrast to FVM09 alone (blue). The dotted line shows percentage neutralization by 0.2 μg/mL c2G4 alone.
(D) The neutralization of VSV-SUDV-GP-Luc by m8C4 and FVM09. m8C4 alone (green) and FVM09 (blue) both show dose-dependent neutralization of virus, while the combination (red) appears to be merely additive.
(E) Neutralization by 16F6 alone (orange) or in the presence of 1 μg/mL FVM09 (blue) or 10 μg/mL FVM09 (red) is potentiated compared to 16F6 alone or FVM09 at 10 μg/mL (dotted line).
(F) Potentiation is also seen when 16F6 is titrated into 10 μg/mL FVM02 (red) compared to 16F6 neutralization alone (orange).
(G and H) PRNT assays were performed using live EBOV (G) and SUDV (H) treated with various concentrations of FVM09 (blue), m8C4 (green), or m8C4 in the presence of 15 μg/mL of FVM09 (red). FVM04 was used as a control pan-ebolavirus antibody.
(I) CI values are shown for the indicated combinations for 50% and 90% neutralization (Fa, fraction affected). Antibodies used at constant and variable concentrations and the virus used for the respective neutralization assay are shown on the x axis.
In (A)–(H), each data point represents the average of three measurements ± SD. See also Figures S1–S3.
FVM09 and m8C4 were also tested to determine whether the cooperativity shown in neutralizing EBOV also applied to neutralization of SUDV. m8C4 itself strongly neutralized VSV-SUDV GP-Luc, and FVM09 neutralized only weakly, but a combination of the two mAbs gave a neutralization pattern suggesting a merely additive improvement (Figure 3D), indicating that the FVM09-mediated potentiation of m8C4 was specific to EBOV. The anti-SUDV antibody 16F6 binds to a region within SUDV GP (Dias et al., 2011) homologous to the base epitope of EBOV GP targeted by 2G4 and KZ52. Since we observed enhanced neutralization of EBOV by FVM09 and 2G4, we examined whether FVM09 would also enhance the neutralization of SUDV by 16F6. In the presence of FVM09 (at 1 or 10 μg/mL), a strong left shift was observed in the 16F6 neutralization dose-response curve (Figure 3E). The neutralizing IC50 of 16F6 was reduced from 0.21 μg/mL to 0.03 and 0.02 μg/mL in the presence of 1 and 10 μg/mL of FVM09, respectively. Interestingly, the neutralizing activity of 16F6 was also potentiated in the presence of the fusion loop-binding mAb FVM02 (Figure 3F).
The combination of m8C4 and FVM09 was also tested using plaque reduction neutralization (PRNT) assays with live EBOV and SUDV under biosafety level 4 containment. FVM09 failed to neutralize EBOV at concentrations up to 30 μg/mL, and m8C4 was only weakly neutralizing (60% neutralization at 30 μg/mL; Figure 3G). However, in the presence of FVM09 at fixed concentration (15 μg/mL), m8C4 showed potent EBOV neutralizing activity with an IC50 of 2.1 μg/mL (Figure 3G). SUDV was effectively neutralized by m8C4 alone, and the addition of FVM09 did not affect the neutralizing potency of m8C4, confirming that the cooperativity of m8C4 and FVM09 is specific to EBOV (Figure 3H).
Quantitation of mAb Functional Synergy
The extent of the mAb synergy described above was quantified with CompuSyn software, which uses a mathematical approach developed by Chou and Talalay to quantify synergy (Chou, 2010, Chou and Talalay, 1984). This methodology defines two parameters—the combination index (CI) and the dose reduction index (DRI)—to describe the effect of drug combination. The CI provides a quantitative measure for additive effects (CI = 1), synergism (CI < 1), and antagonism (CI > 1). The DRI provides a measure of the potential fold dose reduction that can be achieved by drug combinations. CI and DRI values were calculated for EBOV neutralization over the full range of mAb concentrations (Figure S1) along with the values for 50% and 90% neutralization (fraction affected [Fa] 50 and Fa 90) (Figure 3I). This analysis revealed an extremely high degree of synergy between mAb pairs m8C4/FVM09, FVM09/m8C4, and FVM09/2G4, where the first mAb in each pair was tested at various concentrations, while the second was kept at a constant concentration (Figures S1A–S1C). The concentration-dependent CI values for m8C4/FVM09, FVM09/m8C4, and FVM09/2G4 ranged from 0.007 to 0.12, 0.012 to 1.7, and 0.13 to 1.1, and the DRI values ranged from 8 to 145, 0.5 to 84, and 0.6 to 10, respectively (Figures S1A–S1C). For SUDV neutralization, the degree of synergy between m8C4 and FVM09 was marginal (CI close to 1; Figure S1D), while clear synergy was evident when 16F6 and FVM09 were combined (CI as low as 0.1; Figures S1E and S1F). For the m8C4/FVM09 pair, analysis confirmed a synergistic neutralization of live EBOV (CI values as low as 0.0001 and DRI values as high as 56; Figure S1G) but only marginal neutralization enhancement of live SUDV (CI values mostly around 1 and DRI values between 1 and 3; Figure S1H). We also tested for synergy between FVM04 with FVM09 or FVM02 and found that the combination of FVM04/FVM09 exhibited marginal synergy (CI, 0.47–0.6; DRI, 1.5–2) and only at higher concentrations (Figures S2A–S2C). However, FVM02/FVM04 did result in synergy, which was more evident at lower concentrations (CI, 0.05–0.5 and DRI, 35–2 for FVM04 concentrations of 0.05–5 μg/mL; Figures S2D–S2F).
The Role of Bivalent Binding in Antibody Synergy
To determine whether the observed mAb synergy was dependent on either bivalent binding or the Fc portion of the mAbs, we generated purified Fab fragments of FVM09 and m8C4 following digestion with papain or ficin. FVM09 Fab bound readily to EBOV GP but at a lower level compared to the full immunoglobulin G (IgG) (Figure S3A). FVM09 Fab, as for full-length FVM09 (Figure 3B), displayed no neutralization of VSV-EBOV GP-Luc by itself but exhibited dose-dependent neutralization in the presence of m8C4 at a constant concentration (Figure S3B). This suggested that FVM09-mediated cooperative neutralization is independent of bivalent binding or the mAb Fc region.
Binding of m8C4 Fab to EBOV GP was drastically reduced as compared to full-length m8C4 (Figure S3C) and was only slightly enhanced in the presence of FVM09 Fab or full IgG (Figure S3C). Consistent with this low binding, the m8C4 Fab induced marginal neutralization, at only 20 μg/mL, and this neutralization was moderately enhanced by FVM09 (Figure S3D). For SUDV, the m8C4 Fab by itself showed moderate neutralization of VSV-SUDV GP-Luc, but this was strongly enhanced in the presence of FVM09. Taken together, these data indicate that bivalent binding and the Fc portion do not play a critical role in the observed cooperative neutralization. Nevertheless, bivalent configuration of full-length mAb is likely important for binding to the EBOV epitope with sufficient avidity to support maximal cooperative neutralization.
Relationship between m8C4 and FVM09 Epitopes
Since mAbs m8C4 and FVM09 demonstrated the strongest neutralization cooperativity, we identified the epitopes targeted by these two glycan cap-binding mAbs. We had previously shown, using overlapping peptides in competition experiments, that FVM09 binds to a linear epitope within the partially disordered β17-β18 loop of EBOV GP glycan cap (Keck et al., 2015), while m8C4 binds to a conformational epitope involving the glycan cap (Holtsberg et al., 2015). To further define the m8C4 and FVM09 epitopes, we employed “shotgun mutagenesis” to construct an Ala-scan library of EBOV GP in which 641 of 644 target GP residues were individually mutated (Davidson et al., 2015). HEK293T cells were transfected with the entire library in a 384-well array format, and binding of the mAbs to each clone was evaluated by high-throughput flow cytometry.
Epitope mapping of m8C4 identified EBOV GP residues R136 within the GP1 core and Q251 and F252 within the glycan cap as critical for m8C4 binding (Figure 4 A). Alanine substitutions at these residues reduced m8C4 binding to 2.5%, 12.9%, and 25.3% of wild-type, respectively, suggesting that these residues constitute key contact sites for m8C4 (Figure 4B). In contrast, FVM04 and FVM09 showed no reduction in binding to these mutants relative to wild-type GP; in fact, FVM09 exhibited higher binding to R136A than to wild-type GP. Similar analysis with FVM09 showed that the W288A, F290A, or W291A mutations within the β17-β18 loop essentially eliminated FVM09 binding to GP (to 0%, 6.2%, and 0% of wild-type reactivity, respectively; Figures 4C and 4D). Interestingly, the F290A and W291A loop mutations, while eliminating FVM09 binding, increased m8C4 binding by 3- to 4-fold (Figure 4F), suggesting an interaction between the two epitopes. In addition, we observed that an alanine substitution of E292, which is adjacent to the FVM09 epitope residues, increased binding of both FVM09 and m8C4 (Figure 4F). The key residues of the m8C4 epitope, Q251 and F252 within the glycan cap and R136 within the GP1 core, lie in close proximity on the outer side of GP1 and adjacent to the partially disordered β17-β18 loop (residues 279–302) (Lee et al., 2008, Zhao et al., 2016), which contains the FVM09 epitope (described below; Figure 4E). On the available EBOV GP structures, the β17-β18 loop partially masks the m8C4 epitope. The key contact residues for FVM09 (W288, F290, W291) and E292 are positioned at the bottom of the β17-β18 loop and interact with residues 510–512, which lie between the fusion loop and the base epitope (Figure 4E), as well as with GP1 residues near the GP1/GP2 interface. Mutation of E292, which is juxtaposed to K510, may remove constraints on the β17-β18 loop and allow better interaction with FVM09. Consistent with this notion, a K510A substitution also led to increased binding of both FVM09 and m8C4 by nearly 2-fold (Figure 4G). Mutation of several other residues in this region of GP2, including the disulfide bonded cysteines C511 and C556, also significantly increased binding by both FVM09 and m8C4 (Figure 4G).
Figure 4.
Epitope Mapping by Alanine Scanning Mutagenesis
A shotgun mutagenesis library was constructed for EBOV GP where 641 amino acids were individually mutated to alanine.
(A) The reactivity of m8C4 binding to each mutant was evaluated and compared to that of control mAb FVM04.
(B) When residues R136, Q251, and F252 are mutated, m8C4 binding is reduced 2.5%, 12.9%, and 25.3% compared to wild-type. Mutations do not decrease FVM04 (light gray) or FVM09 (black) binding. Error bars represent the average of three measurements ± SD.
(C) The reactivity of FVM09 binding to each mutant was evaluated and compared to control mAb FVM04.
(D) The mutation of residues W288, F290, and W291 reduced FVM09 binding to 0%, 6.2%, and 0% compared to wild-type, but did not significantly reduce FVM02 (dark gray) or FVM04 (light gray) binding.
(E) Cartoon representation of the FVM09 epitope (circled), which lies adjacent to the m8C4 binding site (red) on EBOV GP structure (PDB: 5JQ3). Magnified section shows the residues involved in FVM09 binding and interaction with GP2 (orange).
(F) This graph highlights the inverse relationship between m8C4 (red bar) and FVM09 (black bar) binding for several mutants within the respective epitopes. In particular, R136, Q251, and F252 dramatically impair m8C4 binding but increase FVM09 binding 200% compared to wild-type. In contrast, the W288, F290, and W291 mutants reduce FVM09 binding but enhance the binding of m8C4. Both FVM09 and m8C4 show enhanced binding to the E292A mutant.
(G) When GP2 residues adjacent to residues W288, F290, W291, and E292 are mutated, the binding of FVM09 and m8C4 to GP increases.
These data suggest an interplay between the m8C4 and FVM09 epitopes, possibly constrained by interactions with adjacent GP2 residues. Based on the binding data, we hypothesized that mutational impairment of FVM09 binding would mimic the enhancing effect of FVM09 binding on m8C4 neutralizing activity. To test this hypothesis, we generated a mutant GP with three mutations: W288A/F290A/W291A (EBOV-GP-AAA). While EBOV-GP-AAA failed to bind to FVM09, it exhibited moderately enhanced binding to m8C4 (2- to 3-fold higher than wild-type [WT] GP; Figure 5 A). Despite this moderate effect of the triple mutation on m8C4 binding, a pseudotyped VSV virus incorporating the triple mutations (VSV-EBOV GP-AAA-Luc) was neutralized by m8C4 over 600-fold more effectively than wild-type VSV-EBOV GP-Luc (Figure 5B). In contrast, the triple mutation caused only a modest reduction of KZ52-mediated neutralization compared to wild-type VSV-EBOV GP-Luc (Figure 5C), indicating that the enhanced neutralization is specific to m8C4.
Figure 5.
Mutations in the EBOV GP β17-β18 Loop Enhance Binding and Neutralization by mAb m8C4
(A) The ELISA reactivities of FVM09 and m8C4 with wild-type EBOV GPΔTM and mutant EBOV GPΔTM-AAA indicate that FVM09 fails to bind GP containing mutations W288A/F290A/W291A (GP-AAA; blue), while m8C4 shows enhanced binding to GP-AAA (green) compared to wild-type GP (orange).
(B) m8C4 shows dramatically increased neutralization of pseudotyped VSV-EBOV GP-AAA (green) compared to wild-type VSV-EBOV GP (orange).
(C) Control antibody KZ52 maintained similar neutralization potency with both VSV-EBOV GP-AAA (green) and wild-type VSV-EBOV GP (orange).
These data strongly suggest that the wild-type β17-β18 loop inhibits the neutralizing activity of m8C4 by masking its epitope and that the epitope can be unmasked through binding of this loop by mAbs such as FVM09 and FVM20, or by displacement of the loop by the W288A, F290A, or W291A mutations. The reduction (albeit modest) of KZ52 neutralization activity with VSV-EBOV GP-AAA may also be related to displacement of this loop, as it has been proposed that KZ52 makes contact with residues in the β17-β18 loop (Lee et al., 2008).
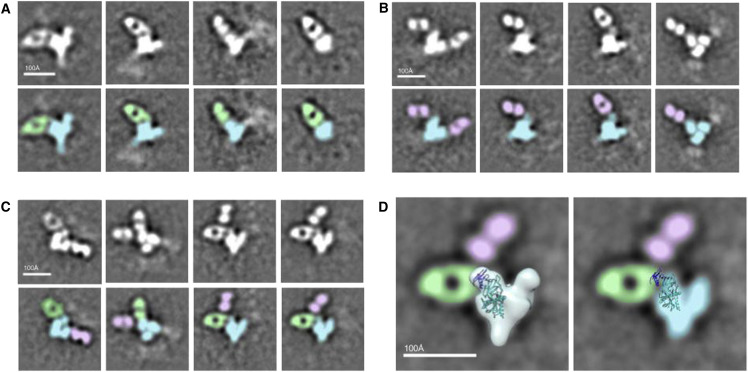
Electron Microscopy Analysis of GP Binding by mAbs m8C4 and FVM09
To further understand the relationship between synergistic epitopes, we conducted negative stain EM of EBOV GP in complex with m8C4 and FVM09 Fabs alone and in combination. Reference free 2D class averages show that m8C4 and FVM09 bind to a similar region of GP but at distinct angles of approach (Figures 6A and 6B). The data are consistent with FVM09 Fab binding to a flexible loop in the glycan cap, while m8C4 binds to an adjacent epitope, and it is possible for both antibodies to bind concurrently to the same protomer (Figures 6C and 6D). These results agree with alanine scan mapping that identified the FVM09 epitope on the β17-β18 loop and the m8C4 epitope residues within the glycan cap partially occluded by the loop. Despite the clear Fab densities in the class averages, we were not able to obtain a 3D reconstruction, which is likely due to high flexibility of the antibodies bound to these epitopes.
Figure 6.
Negative Stain Images of Ebola GP-Antibody Complexes
(A) EBOV GPΔMuc is shown in complex with m8C4 Fab. Reference free 2D class averages (top row) and corresponding false colored 2D class averages (bottom row) with m8C4 Fab (green) and GP (blue) are shown.
(B) EBOV GPΔMuc is shown in complex with FVM09 Fab. Reference free 2D class averages (top row) and corresponding false colored 2D class averages (bottom row) with FVM09 Fab (pink) are shown.
(C) m8C4 and FVM09 Fabs are shown in complex together with EBOV GPΔMuc. Reference free 2D class averages (top row) and corresponding false colored 2D class averages (bottom row) are shown.
(D) The left-hand panel shows Ebola GP trimer volume map and crystal structure (3CSY) of a protomer of EBOV GPΔMuc superimposed onto representative 2D class averages from (C). In the right-hand panel, the volume map was removed to highlight the Fab-GP interaction. The antibodies m8C4 and FVM09 appear to bind flexible epitopes within or proximal to the glycan cap (dark blue).
Cooperative Protection In Vivo
To determine whether the observed synergy of mAb neutralization in vitro would translate to in vivo protection, we tested the efficacy of FVM09, m8C4, and their combination in a stringent mouse model of EBOV infection using a single intraperitoneal injection of antibody at 2 days post-infection (dpi). In the first study, mice were treated with either 25 mg/kg of individual mAbs or 10 mg/kg of each in combination. Under this regimen, mice treated with m8C4 exhibited 30% survival, while all FVM09-treated mice, and nine out of ten control mice, died within 4–11 dpi (Figure 7 A). The level of protection afforded by m8C4 was not significant in comparison to the no treatment group (p = 0.0534) as determined by log-rank (Mantel-Cox) test. In contrast, all mice treated with a combination “cocktail” of the two mAbs survived the challenge (Figure 7A). The protection provided by the cocktail was highly significant when compared to individual mAbs m8C4 or FVM09 or control (p < 0.0001). Mice receiving the cocktail also did not develop severe disease as evident by their health scores and lack of weight loss (Figure 7A).
Figure 7.
Post-exposure Efficacy of the FVM09/m8C4 Cocktail in a Mouse Model of EBOV
Mice were infected (i.p.) with 100 PFUs of EBOV (A and B) or SUDV (C).
(A) MA-EBOV challenged mice were treated at 2 dpi with 25 mg/kg m8C4 alone (green), FVM09 alone (blue), or a combination of m8C4 and FVM09 (10 mg/kg each antibody) (red).
(B) MA-EBOV challenged mice were treated at 2 dpi with a lower dose (10 mg/kg) of m8C4 + control IgG (green), FVM09 + control IgG (blue), or m8C4 + FVM09 at the same dose level (red).
(C) SUDV-challenged mice were treated with m8C4 + FVM09 in combination at 5 mg/kg each on days 1 and 3 post-infection.
Since a lower dose of individual mAbs was used in the cocktail, to exclude the unlikely possibility that these mAbs alone would be more protective at lower doses, in a follow-up study each mAb was used at 10 mg/kg both in cocktail and monotherapy while keeping the total antibody dose at 20 mg by the addition of control IgG to the monotherapy arms. This experiment confirmed that the mAbs were not individually protective at low doses. Mice treated with 10 mg/kg of either m8C4 or FVM09 alone showed 10% and 0% survival, respectively, while 100% of mice treated with the cocktail survived the challenge (p < 0.0001) (Figure 7B).
We had previously shown that m8C4 alone (10 mg/kg) gave partial protection against SUDV infection in A129 (IFNR−/−) mice when administered three times starting 24 hr before infection and repeated at 1 and 3 dpi (Holtsberg et al., 2015). Here, we used the same model to test the post-exposure efficacy of the cocktail. Mice receiving m8C4 and FVM09 at 5 mg/kg each at 1 and 3 dpi (n = 7) were fully protected from SUDV infection, while five out of six control animals succumbed to infection (Figure 7C). However, in contrast to the EBOV study, SUDV-infected mice treated with the cocktail exhibited severe but transient signs of disease and weight loss before recovery (Figure 7C).
Discussion
In this study, we describe a novel phenomenon of functional cooperativity between mAbs targeting ebolavirus glycoproteins. We specifically identified two adjacent epitopes within the EBOV GP glycan cap that, when bound, facilitate cooperative neutralization. Furthermore, we observed a similar phenomenon for another set of mAbs for neutralizing SUDV. These data demonstrate that cooperative neutralization exists for several ebolavirus-specific antibody pairs and identify a previously unknown mechanism for antibody targeting of viruses, since the potentiating effect of non-neutralizing antibodies are rarely, if ever, evaluated in concert with neutralizing antibodies.
Synergistic neutralization has been described for several mAbs targeting HIV (Laal et al., 1994, Li et al., 1998, Mascola et al., 1997, Vijh-Warrier et al., 1996, Xu et al., 2001). This includes triple and quadruple combinations of mAbs against gp120 and gp41 (Li et al., 1998). These mAbs were each neutralizing when used singly, and their combination led to CI values as low as 0.2 and DRI values as high as 24 for triple mAb cocktails (Li et al., 1998). Other synergistic combinations of HIV antibodies reached a CI of 0.2 and DRI of 490 in the most potent combinations (Vijh-Warrier et al., 1996). While the data presented here are reminiscent of these reported synergies among HIV antibodies, a unique aspect of our finding is the identification of non-neutralizing antibodies that convert poorly neutralizing antibodies into potent neutralizers. While FVM09 by itself has almost no EBOV neutralizing activity, its combination with the poorly neutralizing mAb m8C4 showed a 70-fold reduction in neutralization IC50, and the FVM09/m8C4 cocktail exhibited a CI as low as 0.01 and a DRI as high as 145. In the presence of sub-optimal concentrations of m8C4 or the GP base binder 2G4, FVM09 behaved as a neutralizing mAb with a saturable dose-response curve. Similarly, we showed that FVM09, which poorly neutralizes SUDV, and the non-neutralizing GP2 mAb FVM02, strongly potentiate the neutralizing activity of the SUDV-specific mAb 16F6. Because of the dramatic effect of mAb combinations (extremely high DRI and low CI) and the fact that the combination results are qualitatively different from those obtained with individual mAbs (transforming non-neutralizers to neutralizers), we refer to this phenomenon as “enabling, cooperative neutralization” (i.e., effective neutralization is dependent on both mAbs while one mAb can be a non-neutralizer).
The observed mAb cooperativities may be related to triggering of an “induced epitope,” i.e., an epitope that is either formed or exposed upon binding of GP by another antibody. FVM09 and FVM20 may cause a conformational change that leads to better access of m8C4 and 2G4 to their respective epitopes. Extensive interactions of the GP1 β17-β18 loop with the GP1 core and a region of GP2 close to the base epitope suggests that FVM09-like loop-binding antibodies may “peel” the β17-β18 loop off from the main GP body, allowing more effective neutralization by GP base mAbs such as 2G4. Consistent with this hypothesis, our EM data suggest that this loop, when bound by FVM09, is flexible and appears to the side and above the GP trimer. This notion is also consistent with the observed competition between FVM09 and KZ52, since KZ52 appears to make contacts with the β17-β18 loop, and displacement of this loop would be expected to reduce KZ52 binding. In contrast, 2G4, which showed cooperativity with FVM09, binds to the GP base epitope at a sharper angle than KZ52 (Murin et al., 2014), which suggests that 2G4 may not contact the β17-β18 loop.
Despite the allure of this model, cooperative neutralization, at least for m8C4/FVM09 and FVM09/2G4, cannot be explained entirely by increased epitope exposure and subsequent mutual binding enhancement. Combining FVM09 and m8C4 in a cocktail increased their reciprocal binding by only 3- to 4-fold over the individual mAb values yet resulted in a 70-fold enhanced neutralization and CI values <0.001. Similarly, combining 2G4 and FVM09 resulted in only a modest binding enhancement, but the otherwise non-neutralizing FVM09 acquired a qualitatively novel property, a strong neutralizing potency similar to some of the most potent EBOV neutralizers. The qualitatively distinct ability of the mAb combination to neutralize EBOV may result from a combination of binding enhancement and cooperativity at a functional level, the mechanism of which remains to be determined.
A limitation of the binding studies presented here is that they are performed in the absence of virus target cells, where neutralization occurs. It is possible that binding enhancement is actually greater when the virus binds to its cellular attachment factors due to conformational changes that may be triggered cooperatively by interactions between GP with these factors and with FVM09-like antibodies. It is striking that the highest degree of cooperativity was observed between mAbs with adjacent epitopes. The enhanced neutralization may result from two mAbs giving more efficient occupancy and masking of critical regions of the glycoprotein compared to that provided by a single mAb. These different mechanisms are not mutually exclusive and may all contribute to the observed functional cooperativity.
The data presented here have significant implications for both immunotherapy and vaccine development. The enabling, cooperative neutralization of viruses by specific mAbs may be feasible for a variety of other viral pathogens and can be readily discovered using a matrix neutralization assay as described here. The use of two synergistic mAbs as a therapeutic, rather than a combination of antibodies with additive effects, may significantly reduce the dose required for effective treatment of viral infections, overcoming a major impediment in the development of anti-infective antibody therapy. However, at least for treatment of chronic infection, a caveat to this approach is the vulnerability to viral escape, as escape from one antibody may render the cocktail ineffective. These findings could also have implications for vaccine design, which could be exploited to promote cooperativity through epitope-specific antigen resurfacing.
Experimental Procedures
Protein Production
Filovirus proteins were produced and purified as previously described (Keck et al., 2015, Howell et al., 2016). Briefly, the coding regions of EBOV GPΔTM or SUDV GPΔTM were inserted into baculovirus transfer vectors (pFastBac, Invitrogen) and used to transfect Sf9 insect cells. The recombinant baculoviruses containing the GPΔTM were recovered from the supernatant and amplified in Sf9 cells. The final virus was used to infect Sf9 cells, and the protein was purified from the supernatant 3 dpi. The supernatant was concentrated and mixed with Ni Sepharose 6 Fast Flow beads (GE Life Sciences) overnight at 4°C. The next day, the beads were separated by centrifugation and packed into a Bio-Rad Econo column. The column was washed with PBS, 20% glycerol, 0.2% Tween 20, and 10 mM imidazole. Protein was eluted with the same buffer containing 500 mM imidazole and was dialyzed into PBS containing 10% glycerol, arginine, and glutamic acid. All proteins were analyzed by SDS-PAGE and western blot for purity, and protein concentrations were determined using the bicinchoninic acid (BCA) assay. Proteins were aliquoted and stored at −80°C for long-term storage.
Antibody Production and Purification
Antibodies were produced in HEK293 cells expressing the appropriate IgH and IgL following transfection with expression plasmids using polyethylene amine (Polysciences). Antibodies were purified from culture supernatants using Protein A chromatography columns as previously described (Keck et al., 2015, Howell et al., 2016). mAbs 2G4, 4G7, and 13C6 were generated in N. benthamiana plants that are genetically modified to produce homogeneous mammalian N-glycans of the GnGn glycoform. Plants were grown for 4 weeks in an enclosed growth room at 20°C–23°C before expression vectors for IgH and IgL were introduced by vacuum infiltration as described (Hiatt et al. 2014). After 7 days, the mAbs were extracted from leaf tissue and purified by Protein A chromatography. Endotoxins were removed using an acrodisc unit with Mustang Q membrane (Pall Life Sciences). Mouse mAbs were produced and purified as previously described (Holtsberg et al., 2015).
Generation of Fab Fragments
Fab versions of mAbs FVM09 and m8C4 were generated using the Pierce Fab Kit and the Pierce Mouse IgG1 Fab kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, FVM09 was digested with agarose-bound papain for 4 hr at 37°C before purification by Protein A column chromatography to remove undigested IgG and Fc from the reaction. The unbound fraction containing FVM09 Fab was dialyzed overnight in PBS at 4°C before analysis by non-reducing SDS-PAGE. Fab m8C4 was generated in the same manner except that mAb digestion was performed using ficin-bound agarose in 25 mM cysteine.
Enhanced Binding ELISAs
Antigens were diluted to 1 μg/mL in 1 × Dulbecco’s phosphate-buffered saline (DPBS) and coated onto Nunc 96-well maxisorb plates (Thermo Fisher Scientific) and incubated overnight at 4°C. The next day, plates were washed with 1 × DPBS + 0.02% Tween 20 before adding blocking buffer for 1 hr at room temperature. After blocking, m8C4, FVM09, or irrelevant control IgG or 1D9 was added to each well at 20 μg/mL for 30 min before adding a concentration range of the detection antibody for a further 30 min. After both antibodies were bound, the plates were washed, and secondary antibody, either anti-mouse or anti-human, was added for 1 hr at room temperature. After 1 hr, the plates were washed, and 100 μL of 3,3',5,5'-Tetramethylbenzidine (TMB) substrate was added for 15 min before reading at 650 nm. Softmax was used to collect data, and the data were analyzed in Prism and fit to a four-parameter logistic (4PL) curve.
Generation of Recombinant VSV-GP Pseudotyped Virus
VSV-EBOV GP-Luc, VSV-SUDV GP Luc, and VSV-EBOV GP-AAA Luc pseudotyped viruses were generated as previously described (Keck et al., 2015, Howell et al., 2016). Briefly, the glycoprotein plasmid was transfected into 293T cells in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S) overnight before infecting the cells with VSV-ΔG at an MOI of 3 in serum-free DMEM for 1 hr. After 1 hr, the infected cells were washed three times with 1 × DPBS, before exchanging the media to DMEM supplemented with 1% FBS and P/S and allowing the virus to propagate overnight. The supernatants containing the virus were collected, centrifuged at 1,200 rpm for 10 min to clarify, and then aliquoted and stored at −80°C. A plaque assay was performed to obtain viral titers.
Recombinant VSV-GP Pseudotyped Neutralization Assays
Vero cells (ATCC) were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% FBS and P/S at 37°C and 5% CO2. Cells were seeded at 60,000 cells/well in a 96-well black, flat-bottom tissue culture plates. The following day, antibodies were diluted in serum-free EMEM supplemented with P/S and mixed with vesicular stomatitis virus lacking G protein and expressing the appropriate filovirus GP (VSV-GP) (for EBOV, SUDV, or EBOV GP-AAA) for 1hr at room temperature. After 1 hr, 100 μL of the virus and antibody was added to the plated Vero cells at an MOI of 0.04. The plates were incubated for 1 hr at 37°C before the addition of 100 μL of EMEM supplemented with 2% FBS and P/S and overnight incubation. The next day, media and virus were removed from the plates before adding 30 μL of 1 × passive cell lysis buffer (Promega). Cells were allowed to lyse for 30 min at room temperate before the addition of 30 μL of luciferase activating reagent (Promega). Luminesce was read immediately on a BioMek plate reader. Data were fit to a 4PL curve using Prism GraphPad software. Percent neutralization was calculated based on wells containing virus without antibodies.
PRNT Assay
MAbs FVM09, m8C4, or FVM09 in combination with m8C4 or control mAb FVM04 were added at the indicated concentrations to 100 plaque-forming units (PFUs) of EBOV (Kikwit-95) or SUDV (Boniface) at 37°C, 5% CO2 for 1 hr. The virus and antibody mixture was then added to Vero cells and overlaid with 1% agarose (Seakem) and one part 2 × Eagle basal medium (EBME) containing 30 mM HEPES and 5% FBS. A second overlay of 5% neutral red was added 6–9 days later and incubated for another 24 hr before counting plaques.
Alanine Scanning Mutagenesis
Shotgun mutagenesis was performed as previously described (Howell et al., 2016). Briefly, a library containing EBOV GP Mayinga with residues 33–676 mutated to alanine was generated and transfected into HEK293T cells in 384-well plates. Cells expressing the GP mutants were incubated with each primary antibody and then with Alexa-Fluor-488-conjugated secondary antibody. Binding was detected using an Intellicyt high-throughput flow cytometer (Intellicyt). Background was subtracted from control wells, and relative binding to each mutant was compared to wild-type controls. Mutants were identified as critical only if they lost binding to the test antibody but not to the internal controls (Davidson and Doranz, 2014).
ELISA and VSV Pseudotyped Neutralization of EBOV GPdTM-AAA Mutant
The F290A/W291A/E292A mutations were introduced into the EBOV GPΔTM pCAGGS vector. The protein was expressed and purified as described for the wild-type protein. To compare the binding profiles of FVM09 and m8C4 to wild-type EBOV GP and EBOV GP-AAA mutant, 100 ng/well of wild-type or mutant protein was coated on 96-well Nunc MaxiSorb plates overnight at 4°C. The next day, plates were washed with 1 × DPBS + 0.02% Tween 20 and then blocked with 200 μL of blocking buffer for 1 hr at room temperature. The blocking buffer was washed off before the addition of FVM09 or m8C4. Antibodies were allowed to bind for 1 hr at room temperature before secondary anti-human (FVM09) or anti-mouse (m8C4) was added to detect mAb binding. After 1 hr at room temperature, plates were washed and 100 μL of TMB was added for 15 min before reading on a VersaMax plate reader at 650 nm. Data were fit to 4PL curve using GraphPad Prism 6.
Cryoelectron Microscopy
IgG of m8C4 and FVM09 was digested into Fab by incubation with papain for 4 hr at 37°C and was subsequently purified using a protein A column followed by an S200i column (GE Healthcare). A molar excess of each Fab was added to EBOV GPΔMuc and incubated overnight at 4°C. The complexes were then purified over SEC column S200i and pipetted onto carbon covered 400 copper mesh grids and negatively stained with 1% uranyl formate. The grids were imaged on a Technai T12 Spirit at 120 keV with Tietz TemCam-F416 complementary metal oxide semiconductor (CMOS) camera. Data collection was set at 52,000× magnification at a nominal defocus of 1.5 μM operating through Leginon (Suloway et al., 2005). Using Appion (Lander et al., 2009), micrographs were processed with Dogpicker (Voss et al., 2009) and sorted into 2D class averages in XMIPP (Sorzano et al., 2004). Isolated Fabs and unrecognizable classes were removed, and final 2D classes were generated showing different views of the complexes. Interpretation of data and figures was done using UCSF Chimera (Goddard et al., 2007).
Mouse Challenge Studies
Mouse Challenge Studies with EBOV
Female BALB/c mice at 6–8 weeks of age were purchased from Charles River Laboratories. Mice were challenged intraperitoneally (i.p.) with 100 PFUs of mouse-adapted EBOV (Mayinga strain) (maEBOV). At 2 dpi, mice were treated with the indicated mAbs or PBS by the i.p. route. Mice were observed a minimum of once per day, and pan weights and clinical signs of disease were recorded through day 14. Observations were increased to a minimum of twice daily upon onset of clinical signs of disease. Moribund and surviving mice were humanely euthanized on the basis of Institutional Animal Care and Use Committee (IACUC)-approved criteria. Mice were observed for a minimum of 21 days after exposure.
Mouse Challenge Study with SUDV
IFNAR−/− mice aged 4 weeks (B6.129S2-Ifnar1tm1Agt/Mmjax) on the C57BL/6 background were purchased from Jackson Laboratory. Mice were challenged i.p. with 1,000 PFUs of SUDV and treated at 1 and 3 dpi with the indicated antibodies by i.p. injection. Mice were observed daily for lethality or clinical signs of disease for 21 days. Moribund and surviving mice were humanely euthanized on the basis of IACUC-approved criteria.
Ethics Statement
Animal research using mice was conducted under a protocol approved by the US Army Medical Research Institute of Infectious Diseases (USAMRIID) Institutional Animal Care and Use Committee (IACUC) in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals. The USAMRIID facility is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International and adheres to the principles stated in the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011).
Author Contributions
K.A.H., J.M.B., C.B., E.D., A.M., X.Q., E.D., H.L.T., H.V., S.H., A.K., and A.S.H. performed characterization, epitope mapping, and animal experiments. F.W.H. and S.S. performed protein and antibody production. H.L.T. performed purification of complexes, and F.W.H., A.B.W., B.J.D., J.M.D., and M.J.A. were involved in the design of the experiments and data analysis and interpretation. All authors reviewed and analyzed data. M.J.A. and K.A.H. wrote the paper. All authors reviewed the paper.
Acknowledgments
This work was supported by a contract (HDTRA1-13-C-0015) from US Defense Threat Reduction Agency (DTRA) and a grant (R43AI124765) from National Institute of Allergy and Infectious Diseases (NIAID) to M.J.A., and NIH contract HHSN272201400058C to B.J.D. This work was also partially supported by Public Health Agency of Canada (PHAC). J.M.B., A.I.K., A.S.H., and J.M.D. acknowledge support from DTRA, CB4088. A.B.W. and X.Q. acknowledge support from NIAID grant U19AI109762. The authors thank Dr. Gary Kobinger (PHA Canada) for providing 2G4 and 4G7 antibodies. M.J.A., F.W.H., S.S., H.V., and K.A.H. are employees of Integrated Biotherapeutics; B.J.D., E.D., C.B., and A.M. are employees of Integral Molecular; M.J.A. is a founder and shareholder of Integrated Biotherapeutics; F.W.H., S.S., and H.V. own stock options in Integrated Biotherapeutics; and B.J.D. is a founder and shareholder of Integral Molecular. Opinions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the US Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense.
Published: April 11, 2017
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.03.049.
Supplemental Information
References
- Bornholdt Z.A., Turner H.L., Murin C.D., Li W., Sok D., Souders C.A., Piper A.E., Goff A., Shamblin J.D., Wollen S.E. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 2016;351:1078–1083. doi: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen T.H., Pedersen J., Prentoe J.C., Giang E., Keck Z.Y., Mikkelsen L.S., Law M., Foung S.K., Bukh J. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology. 2014;60:1551–1562. doi: 10.1002/hep.27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Davidson E., Doranz B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E., Bryan C., Fong R.H., Barnes T., Pfaff J.M., Mabila M., Rucker J.B., Doranz B.J. Mechanism of binding to ebola virus glycoprotein by the ZMapp, ZMAb, and MB-003 cocktail antibodies. J. Virol. 2015;89:10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J.M., Kuehne A.I., Abelson D.M., Bale S., Wong A.C., Halfmann P., Muhammad M.A., Fusco M.L., Zak S.E., Kang E. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak A.I., Ilinykh P.A., Murin C.D., Garron T., Shen X., Fusco M.L., Hashiguchi T., Bornholdt Z.A., Slaughter J.C., Sapparapu G. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak A.I., Shen X., Murin C.D., Turner H.L., David J.A., Fusco M.L., Lampley R., Kose N., Ilinykh P.A., Kuzmina N. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell. 2016;164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hiatt A., Whaley K.J., Zeitlin L. Plant-derived monoclonal antibodies for prevention and treatment of infectious disease. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.AID-0004-2012. AID-0004-2012. [DOI] [PubMed] [Google Scholar]
- Holtsberg F.W., Shulenin S., Vu H., Howell K.A., Patel S.J., Gunn B., Karim M., Lai J.R., Frei J.C., Nyakatura E.K. Pan-ebolavirus and Pan-filovirus mouse monoclonal antibodies: Protection against Ebola and Sudan viruses. J. Virol. 2015;90:266–278. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K.A., Qiu X., Brannan J.M., Bryan C., Davidson E., Holtsberg F.W., Wec A.Z., Shulenin S., Biggins J.E., Douglas R. Antibody treatment of Ebola and Sudan virus infection via a uniquely exposed epitope within the glycoprotein receptor-binding site. Cell Rep. 2016;15:1514–1526. doi: 10.1016/j.celrep.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck Z.Y., Enterlein S.G., Howell K.A., Vu H., Shulenin S., Warfield K.L., Froude J.W., Araghi N., Douglas R., Biggins J. Macaque monoclonal antibodies targeting novel conserved epitopes within filovirus glycoprotein. J. Virol. 2015;90:279–291. doi: 10.1128/JVI.02172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laal S., Burda S., Gorny M.K., Karwowska S., Buchbinder A., Zolla-Pazner S. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 1994;68:4001–4008. doi: 10.1128/jvi.68.6.4001-4008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.W. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Katinger H., Posner M.R., Cavacini L., Zolla-Pazner S., Gorny M.K., Sodroski J., Chou T.C., Baba T.W., Ruprecht R.M. Synergistic neutralization of simian-human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 1998;72:3235–3240. doi: 10.1128/jvi.72.4.3235-3240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Louder M.K., VanCott T.C., Sapan C.V., Lambert J.S., Muenz L.R., Bunow B., Birx D.L., Robb M.L. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J. Virol. 1997;71:7198–7206. doi: 10.1128/jvi.71.10.7198-7206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglietta R., Pastori C., Venuti A., Ochsenbauer C., Lopalco L. Synergy in monoclonal antibody neutralization of HIV-1 pseudoviruses and infectious molecular clones. J. Transl. Med. 2014;12:346. doi: 10.1186/s12967-014-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J., Gilman M.S., Kanekiyo M., Gui M., Cagigi A., Mulangu S., Corti D., Ledgerwood J.E., Lanzavecchia A., Cunningham J. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin C.D., Fusco M.L., Bornholdt Z.A., Qiu X., Olinger G.G., Zeitlin L., Kobinger G.P., Ward A.B., Saphire E.O. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc. Natl. Acad. Sci. USA. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council . National Academies Press; 2011. Guide for the Care and Use of Laboratory Animals, Eighth Edition. [Google Scholar]
- Olinger G.G., Jr., Pettitt J., Kim D., Working C., Bohorov O., Bratcher B., Hiatt E., Hume S.D., Johnson A.K., Morton J. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Murin C.D., de Val N., Cottrell C.A., Hastie K.M., Turner H.L., Fusco M.L., Flyak A.I., Zeitlin L., Crowe J.E., Jr. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat. Microbiol. 2016;1:16128. doi: 10.1038/nmicrobiol.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J., Zeitlin L., Kim D.H., Working C., Johnson J.C., Bohorov O., Bratcher B., Hiatt E., Hume S.D., Johnson A.K. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci. Transl. Med. 2013;5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- Pollara J., Bonsignori M., Moody M.A., Liu P., Alam S.M., Hwang K.K., Gurley T.C., Kozink D.M., Armand L.C., Marshall D.J. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J. Virol. 2014;88:7715–7726. doi: 10.1128/JVI.00156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Audet J., Wong G., Pillet S., Bello A., Cabral T., Strong J.E., Plummer F., Corbett C.R., Alimonti J.B., Kobinger G.P. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 2012;4:138ra81. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- Qiu X., Fernando L., Melito P.L., Audet J., Feldmann H., Kobinger G., Alimonti J.B., Jones S.M. Ebola GP-specific monoclonal antibodies protect mice and guinea pigs from lethal Ebola virus infection. PLoS Negl. Trop. Dis. 2012;6:e1575. doi: 10.1371/journal.pntd.0001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Wong G., Fernando L., Audet J., Bello A., Strong J., Alimonti J.B., Kobinger G.P. mAbs and Ad-vectored IFN-α therapy rescue Ebola-infected nonhuman primates when administered after the detection of viremia and symptoms. Sci. Transl. Med. 2013;5:207ra143. doi: 10.1126/scitranslmed.3006605. [DOI] [PubMed] [Google Scholar]
- Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J.B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V., Feldmann H., Grard G., Becker S., Leroy E.M. Ebola and Marburg haemorrhagic fever. J. Clin. Virol. 2015;64:111–119. doi: 10.1016/j.jcv.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano C.O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J.R., Scheres S.H., Carazo J.M., Pascual-Montano A. XMIPP: A new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.E., Nelson E.A., Bonagiri P., Simmons J.A., Shoemaker C.J., Schmaljohn C.S., Kobinger G.P., Zeitlin L., Subramaniam S., White J.M. Mapping of ebolavirus neutralization by monoclonal antibodies in the ZMapp cocktail using cryo-electron tomography and studies of cellular entry. J. Virol. 2016;90:7618–7627. doi: 10.1128/JVI.00406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijh-Warrier S., Pinter A., Honnen W.J., Tilley S.A. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J. Virol. 1996;70:4466–4473. doi: 10.1128/jvi.70.7.4466-4473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss N.R., Yoshioka C.K., Radermacher M., Potter C.S., Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Smith-Franklin B.A., Li P.L., Wood C., He J., Du Q., Bhat G.J., Kankasa C., Katinger H., Cavacini L.A. Potent neutralization of primary human immunodeficiency virus clade C isolates with a synergistic combination of human monoclonal antibodies raised against clade B. J. Hum. Virol. 2001;4:55–61. [PubMed] [Google Scholar]
- Zeitlin L., Whaley K.J., Olinger G.G., Jacobs M., Gopal R., Qiu X., Kobinger G.P. Antibody therapeutics for Ebola virus disease. Curr. Opin. Virol. 2016;17:45–49. doi: 10.1016/j.coviro.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ren J., Harlos K., Jones D.M., Zeltina A., Bowden T.A., Padilla-Parra S., Fry E.E., Stuart D.I. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.