Abstract
CDC20/CDH1 activates the anaphase-promoting complex (APC) and targets various substrates for degradation, thereby allowing the ordered progression through mitosis and G1. We have found multiple functional CDH1 homologues in the chick. The transcripts of these novel genes are differentially localized to proliferating, differentiated, and postmitotic tissues. All four proteins bind and form a complex with APC in vitro and in cultural cells and have quantitatively different activities in mediating ubiquitination of various APC substrates. Our results suggest that multiple CDH1s may temporally and spatially regulate APC activity both within and outside of the cell cycle.
Keywords: anaphase-promoting complex‖ubiquitin‖degradation
Regulated proteolysis is essential for cell-cycle progression, signal transduction, and development (1, 2). Ubiquitin protein ligases regulate the timing and substrate preferences in protein degradation. Several types of ubiquitin protein ligases have been identified, including the anaphase-promoting complex (APC)/cyclosome (3, 4), the SCF complex (5, 6), HECT domain proteins (7), Ring finger E3s (8), and the VBC complex (9). The functions and enzymology of APC and SCF complexes are relatively well understood. SCF E3s are composed of four major components, including Skp1, Cullin, Rbx1, and F-box proteins, whereas the APC contains 11 subunits (10). The APC may be an evolutionary distant relative of the SCF. There is similarity between two of the core components of these E3s: (i) APC2 and Cdc53 and (ii) APC11 and Rbx1(1). Four kinds of substrate-specific factors have been found for SCF complexes in different organisms that all bear the F-box motif. They are Cdc4, Grr1, Slim/β-TRCP, and Skp2.
CDC20 and CDH1, like F-box proteins for SCF ligases, are substrate-specific activators of the APC. They associate with the APC in a cell cycle-dependent manner and target distinct sets of substrates for degradation by the 26S proteosome (11, 12). In metaphase, CDC20 activates APC, leading to the degradation of the anaphase inhibitor Securin/Pds1 (13–15), resulting in sister chromatid separation. In late mitosis, CDH1 triggers APC to degrade the mitotic cyclins and the spindle-associated protein Ase1, which leads to mitotic exit (12, 16). In addition, APC targets the mitotic protein kinase Plk, mitosin (CENP-F), CDC20, an inhibitor of DNA replication Geminin, and the chromokinesin Xkid for degradation by the 26S proteasome. Moreover, the following observations have suggested roles for the APC outside of mitosis. Both CDH1 and the APC complex are expressed in postmitotic cells and are able to target cyclin B for ubiquitination in these cells (17, 18). The APC is also active in muscle precursor cells as judged by the instability of a cyclin B-chloramphenicol acetyltransferase fusion protein (19). Furthermore, biochemical studies have shown that CDH1 is tightly associated with APC and maintains APC in an active state during G1 (11). Nevertheless, it is not known how APC is involved in these processes and specifically what proteins might be ubiquitinated and how substrate specificity is achieved. As it has been shown in the SCF pathway, multiple substrate-specific activators might provide part of the mechanism for temporal and spatial regulation of the APC ligase during the cell cycle and throughout embryonic development. Thus, the existence of additional substrate recognition factors in the APC pathway might be predicted (1).
In this article, we show that APC is regulated by at least four activators in chick and that these activators function in circumstances outside of mitosis. One of these proteins is expressed in the nervous system, whereas the other is found in limb buds and somites during chicken embryogenesis. They all associate with APC in vitro and in cultured cells and have quantitatively different substrate profiles against known APC substrates. These results suggest that APC associates with diverse substrate-specific activators and has a role outside of the cell cycle.
Materials and Methods
Cloning of Chicken CDH1 Homologues.
The chicken CDH1 homologue cDNA were isolated by screening an embryonic stage cDNA library (stage 15–20) with a human CDH1 probe (20) and hybridization at low stringency. Four clones that contain complete ORFs of 1.36 kb, 1.2 kb, 1.5 kb, and 1.4 kb were isolated from positive clones and sequenced completely. Sequence comparisons were performed by using FASTA and PILE-UP from the GCG software package. The four clones, respectively, encode proteins with 83%, 76%, 95%, and 62% identity to human CDH1. We named these four chicken CDH1 homologues ChkCDH1-A (83%), ChkCDH1-B (76%), ChkCDH1-C (95%), and ChkCDH1-D (62%). These cDNAs were then subcloned in a modified version of pCS2 vector for in vitro translation and transfection in chicken fibroblast cells.
In Situ Hybridization in Chicken Embryo.
Whole mount in situ hybridization of chicken embryos from stage 15–20 was carried out as described (21) by using digoxigenin-11 UTP-labeled probes (Boehringer). A mixture of 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate 4-toluidine salt was used as substrate for the color reaction. Antisense probes for ChkCDH1-A, ChkCDH1-B, ChkCDH1-C, and ChkCDH1-D were synthesized with T3 RNA polymerase by using EcoRI-linearized plasmid as template. Images were obtained by using transillumination on a Zeiss stereomicroscope and were captured by video using a 3-color video rate CCD camera controlled by northern exposure software (Phase 3 Image System).
Northern Blot Analysis.
Total RNA was extracted from early chicken embryos by using the RNeasy kit (Qiagen, Chatsworth, CA). Thirty micrograms of total RNA was separated by denaturing formaldehyde electrophoresis and transferred by the TurboBlotter device (Schleicher & Schuell). Blots were prehybridized at 42°C with Ultra-hyb (Ambion, Austin, TX) and hybridized with ≈1 Mcpm/ml [32P]dCTP-labeled single-strand DNA probe generated by PCR, washed to high stringency, and exposed to a phosphorimage plate. The abundance of 28S ribosomal RNA was used as a loading control for each tissue type.
Interaction Assays for Chicken CDH1 Homologues and APC in Vitro and in Vivo.
For in vitro interaction assay, 35S-labeled myc-tagged chicken CDH1 homologous proteins were synthesized in the TNT expression system (Promega). Approximately 20 μl of normalized in vitro translated chicken CDH1 homologous proteins were added to 20-μl chicken fibroblast cell (CEF) extracts following the addition of 5 μl of anti-CDC27 antibody covalently coupled to protein A beads, control antibody beads (polyclonal anti-myc antibody protein A beads), or protein A beads only. One-fourth of the interaction assay mixture was saved from each reaction and detected by Western blot with anti-myc antibody (9E10, Santa Cruz Biotechnology) as the control of the abundance for the added 35S-labeled myc-tagged chicken CDH1 homologous proteins. The interaction reaction mixture was incubated at 4°C for 4 h with agitated shaking. The precipitated complexes of the antibody beads were extensively washed with XB buffer (22). IP complexes were dissociated by 2× sample buffer, and the samples were resolved by SDS/PAGE. The interaction signal was detected by phosphorimage (Bio-Rad).
For in vivo interaction analysis, chicken CDH1 constructs were transiently transfected into CEF cells by Effectin methods (Qiagen). Transfected cells were extracted with lysis buffer (11). Five-microliter polyclonal anti-myc antibody beads were added into 1-ml extracts and incubated at 4°C for 4 h. IP complexes were extensively washed with wash buffer (23) and eluted with 2× sample buffer. Samples were resolved by SDS/PAGE and blotted with anti-CDC27 antibody.
Preparation of Synchronized Chicken Fibroblast Cell Extracts.
Chicken fibroblast cells (CEF) were cultured for 24 h in DMEM supplemented with 10% chicken serum in 5% CO2. For synchronization, CEF cells were grown in the presence of 2 mM thymidine (Sigma) for 18 h, washed with PBS, and grown in fresh medium without thymidine for 8 h. Thymidine (2 mM) was then added to block cells at G1/S. After 18 h, cells were washed with PBS and harvested by scraping.
Harvested cells were resuspended in a hypotonic buffer (20 mM Hepes, pH 7.5/5 mM KCl/1.5 mM MgCl2/1 mM DTT/1× protease mixture/energy regeneration mixture) for 30 min to allow cells to swell. Cells were frozen and thawed followed by homogenization with a Dounce homogenizer. Cell lysate was spun at 100,000 × g at 4°C for 1 h. The clear supernatant was used for purification of APC.
In Vitro APC Activation and Ubiquitination Assays.
Interphase APC was immunoprecipitated from synchronized interphase CEF cell extracts with anti-CDC27 antibodies coupled to protein A beads and washed five times with XB containing 500 mM NaCl and 0.5% Nonidet P-40 detergent and two times with XB. Interphase APC beads were incubated, respectively, for 1 h with the four in vitro translated ChkCDH1 proteins to achieve the activity for APC. Approximately 100 ng of in vitro translated CDH1 protein was used for each reaction. Abundance of CDH1 proteins among the reactions was detected by anti-myc antibodies. Activated APC beads were subsequently washed three times with XB followed by addition of ubiquitination mixture (50 μg/ml Ubcx/1.25 mg/ml ubiquitin/200 μg/ml recombinant E1/0.1 mg/ml cycloheximide/2 μM ubiquitin aldehyde) with one-tenth the volume of 35S-labeled in vitro translated substrates. Ubiquitination was reflected in the formation of high molecular polyubiquitin conjugates. We compared the amount of polyubiquitin conjugates formed by each activation reaction with that for human CDH1, which was taken as 100% activity.
Results
Identification of Multiple CDH1 Homologues in Chicken.
We initially attempted to study the G1 function for APCCDH1 by generating a CDH1 null mutant cell line in chicken B cells (23). This work led to the identification of multiple CDH1 homologues in chicken. cDNAs for the four chicken CDH1 homologues were isolated by screening an embryonic cDNA library (stage 15–20) with a human CDH1 probe (11). Four clones that contain complete ORFs of 1.36 kb, 1.2 kb, 1.5 kb, and 1.4 kb were isolated from positive clones and sequenced completely (Fig. 1). The four chicken CDH1 homologues were named ChkCDH1-A, ChkCDH1-B, ChkCDH1-C, and ChkCDH1-D and, respectively, encode proteins with 83%, 76%, 95%, and 62% identity to human CDH1 (Fig. 1B). We subcloned these four clones into an SP6 expression vector and expressed them by coupled transcription translation system. In vitro translation of these cDNAs yielded proteins of 53 kDa, 48 kDa, 55 kDa, and 53 kDa, respectively. Sequence comparison showed that ChkCDH1-B, ChkCDH1-C, and ChkCDH1-D are relatively well conserved at the amino terminus, whereas ChkCDH1-A was not (Fig. 1A). ChkCDH1-A encodes a protein 42 aa residues shorter on the amino terminus, whereas CDH1-B is shorter by about 97 residues on the carboxyl terminus as compared with human CDH1. On the carboxyl terminus, there are 7 WD40 repeats. WD40 (1), (2), and (3) blocks are relatively well conserved among ChkCDH1-A, ChkCDH1-B, and ChkCDH1-D, whereas WD40 (4), (5), (6), and (7) blocks are quite diverse, especially in the case of ChkCDH1-D. Phylogenetic alignment of WD40 repeat proteins of CDC20 and CDH1 from Drosophila to human revealed that the CDH1 family is less conserved than the CDC20 family (Fig. 1C).
Figure 1.
Sequence alignment of the four novel chicken CDH1 homologues with human CDH1 and human CDC20. (A) Hs, Homo sapiens; Chk, chicken. The alignment was performed by using megaalign (DNAstar, Madison, WI) by the clustal method. Residues that are conserved in the amino terminus among those components are shaded and boxed in black. The identity of the seven WD40 repeats among those components is indicated. (B) Schematic drawing of the gene structural comparison of the four novel chicken CDH1 homologues, human CDH1, and human CDC20. Identical regions among those components are shaded in black. (C) Phylogenetic tree of CDH1 and CDC20 WD40 repeats protein family. Hs, H. sapiens; Chk, chicken; Mm, Mus musculus; Xl, Xenopus laevis; Dm, D. melanogaster; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae.
We have searched the expressed sequence tags and the completed genome databases for other CDH1 homologues. Five CDH1 homologous candidates were identified from the Drosophila melanogaster genome database. The GenBank accession numbers for the candidate clones of multiple Drosophila CDH1 homologues are AL121853, AL121813, AC017306, AC023715, and AC020592. Furthermore, one new CDH1 candidate was cloned by PCR from a human fetal thymus full-length cDNA library. Sequence comparison showed high similarity with ChkCDH1-D (data not shown). In sum, four chicken CDH1 homologues have been identified through a conventional hybridization screening, whereas our computer searches and PCR cloning results have demonstrated the existence of multiple CDH1s in flies and human.
Expression of the Multiple Chicken CDH1 in Chicken Embryos and Tissues.
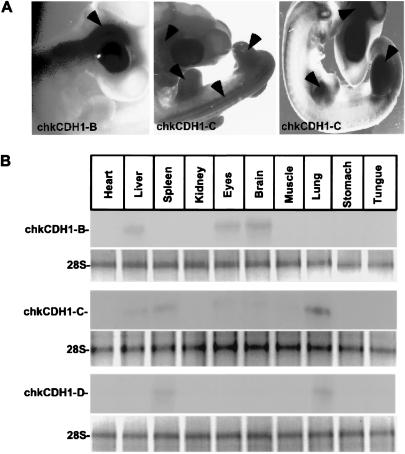
Fizzy-related (Fzr), a homologue of CDH1 in Drosophila, has been localized to the salivary gland of Drosophila (24). In Xenopus, CDH1 transcripts appear after the mid-blastula transition during the early embryogenesis (25). CDH1 protein is expressed in muscle, stomach, brain, thymus, spleen, ovary, and lung of the mouse (18) and in the lymphocyte system in humans (26). To determine the distribution of the multiple chicken CDH1 genes, we have carried out whole mount in situ hybridization and Northern blot analysis during chicken embryogenesis by using antisense RNA probes and single-strand DNA probes, respectively (21). As shown in Fig. 2A, ChkCDH1-B, which shares 76% identity with hCDH1, is expressed throughout the nervous system, whereas ChkCDH1-C, which is 95% identical to human CDH1, is expressed principally in the limb buds, somites, eyes, and neural tube. We saw no specific localization of ChkCDH1-A or ChkCDH1-D. There are also multiple ChkCDH1s in different tissues, as judged by Northern blots from18-day chicken embryos. As shown in Fig. 2B, we found that ChkCDH1-B is expressed in brain, eyes, and liver; ChkCDH1-C is mainly expressed in lung, spleen, and liver and, to a lesser extent, in brain, eye, and muscle. The transcripts of ChkCDH1-D are only found in spleen and lung. ChkCDH1-A is not detected in the above tissues probably because of its lower level of expression in these tissues (Fig. 2B).
Figure 2.
Expression pattern of chicken CDH1 homologues in embryos and tissues. (A) Specific expression pattern for two of the four chicken CDH1 homologues was observed from stage 15–20 in chicken embryogenesis by in situ hybridization using antisense ChkCDH1 homologous probes. As shown in this figure, ChkCDH1-B is expressed exclusively in the nervous system, whereas ChkCDH1-C is expressed in the limb bud, somite, and brain. (B Upper) Northern blot of RNA isolated from chicken embryos. RNA blots were hybridized by using single-strand ChkCDH1 homologous DNA probes and washing with high stringency. As shown, ChkCDH1-B is expressed in brain, eyes, and liver. ChkCDH1-C is broadly expressed in lung, spleen, liver, eyes, and brain. ChkCDH1-D is expressed in spleen and lung. (Lower) Loading abundance for the RNA samples was indicated by 28S rRNA.
The Multiple CDH1s Associate with APC in Vitro and in Vivo.
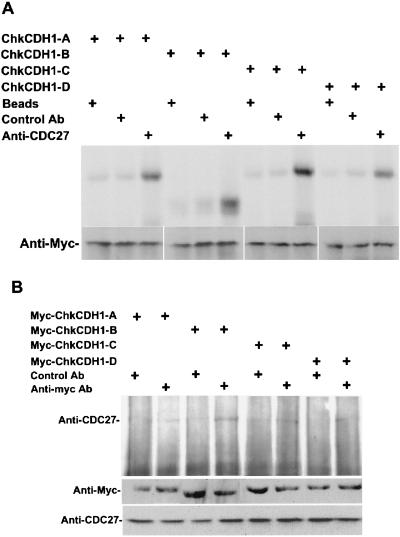
CDC20 and CDH1 associate with APC in a cell cycle-dependent manner. CDC20 is tightly associated with APC in mitosis and is degraded after mitosis. CDH1 binds to APC from the end of mitosis through G1 as determined in both yeast and vertebrates. To test the association of the multiple ChkCDH1s with APC, we have assayed binding in vitro as well as by coimmunoprecipitation in chicken fibroblast cells. In the in vitro interaction assays, 35S-labeled myc-tagged chicken CDH1 translated in vitro was incubated with chicken fibroblast cell extract followed by immunoprecipitation of interphase APC with anti-CDC27 antibodies (CDC27 is a core APC subunit). ChkCDH1 proteins were then detected by autoradiography. As shown in Fig. 3A, all four chkCDH1 homologues were specifically coimmunoprecipitated with the APC complex. The intensity of the signal suggests that the binding affinity of ChkCDH1-B and ChkCDH1-C for APC may be stronger than ChkCDH1-A and ChkCDH1-D.
Figure 3.
ChkCDH1 homologues interact with anaphase-promoting complex in vitro and in vivo. (A Upper) Incubation of 35S-labeled in vitro translated myc-ChkCDH1 homologues and anti-CDC27 antibody coupled to protein A beads in CEF cell extracts. As shown, ChkCDH1-A, -B, -C, and -D interact with APC in CEF extracts. (Lower) Western blot of myc-ChkCDH1 homologues, assayed for interaction with APC. (B Upper) Coimmunoprecipitation of CDC27 with transfected myc-ChkCDH1 homologues in chicken fibroblast cells. (Lower) Western blot analysis of ChkCDH1 homologous proteins in transfected chicken fibroblast cells; CDC27 protein is used as loading control.
To confirm the in vitro interaction results, we assayed complex formation in chicken fibroblast cells. Myc-tagged ChkCDH1 constructs were transiently transfected into CEF cells, and the various ChkCDH1s were immunoprecipitated with anti-myc antibody. Interaction with APC by Western blotting was detected with anti-CDC27 antibody after normalization for level of expression. In all cases, CDC27 coimmunoprecipitated with ChkCDH1, as judged by a Western blot with anti-CDC27 antibodies (Fig. 3B). Consistent with the in vitro analysis, ChkCDH1-B and ChkCDH1-C seem to bind more tightly to the APC, whereas ChkCDH-A and ChkCDH1-D bind less tightly in CEF cells.
Different CDH1s Show Different Substrate Specificities in Ubiquitination Assays.
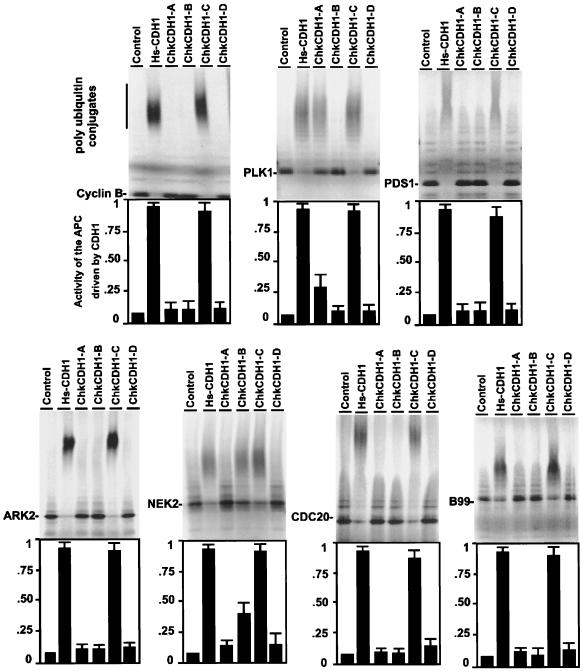
To test whether the novel CDH1 proteins can activate APC, we developed an in vitro ubiquitination assay using APC purified from somatic chicken cells. In this system, inactive or weakly active APC was purified from somatic cell extracts of double thymidine-arrested CEF cells. These S phase cells should not possess active APC (11). In vitro synthesized multiple chicken CDH1 proteins were then incubated individually with interphase APC. Activated APC was subsequently added to various 35S-labeled APC substrates in the presence of purified E1, Ubcx, ubiquitin, and an energy-regenerating mix. Activity was detected by the substrate conjugation to ubiquitin in the form of high molecular weight polyubiquitin conjugates. The level of polyubiquitin conjugation was used as the readout of the activity of an individual novel CDH1 protein for specific substrates.
Previous reports have provided a list of putative APC substrates (10, 22). We have tested the role of the multiple CDH1 proteins on the ubiquitination of cyclin B, Securin/Pds1, PlK (polo-like kinase), Nek2 (a NimA-related kinase), Ark2 (aurora-related kinase 2), CDC20, and B99 (a p53-induced protein). In the ubiquitination assays, the same amount of chicken CDH1 was used to activate the APC (the normalized abundance of the multiple CDH1 proteins was adjusted by using anti-myc antibodies; data not shown). The activity of human CDH1 for the APC was used as positive control (100%) to evaluate the ability of the multiple chicken CDH1 proteins to activate the APC. As shown in Fig. 4, ChkCDH1-C has a broad spectrum for targeting the APC substrates cyclin B, securin/Pds1, Plk, Nek2, Ark2, CDC20, and B99. It has the same target range and the same potency in activating the APC as human CDH1 does. ChKCDH1-A and ChkCDH1-B have very different substrate specificities. ChKCDH1-A converts Plk to form polyubiquitin conjugates (25% activity compared with human CDH1) but no appreciable activity against any other substrate. ChkCDH1-B targets Nek2 for ubiquitination (35% activity compared with human CDH1) but no activity against any other substrate. ChkCDH1-D shows no activity in the assay.
Figure 4.
In vitro activity and specificity of chicken homologues for ubiquitination of putative APC substrates. Interphase APC beads were incubated with in vitro translated ChkCDH1 proteins to activate for APC. Shown is the resource of 35S-labeled in vitro translated substrates. Cyclin B, amino-terminal human cyclin B1; PLK1, human polo-like kinase; PDS1, human securin; NEK2, human NimA-related kinase; CDC20, human CDC20; ARK2, human aurora-related kinase 2; and B99, human p53-induced protein.
Discussion
Ubiquitin-dependent proteolysis plays an important role in cell-cycle regulation, gene expression, receptor-mediated signal transduction, antigen presentation, viral pathogenesis, and stress response (1, 2, 10). So far, six types of E3s have been identified including the APC/cyclosome (3, 4), the SCF complex (5, 6), HECT domain proteins (7), Ring finger E3s (8), the VBC complex (9), and smurfs (27). In the case of the SCF complex, its broad substrate specificities are conferred by multiple F-box proteins and kinases that operate in developmental pathways throughout the cell cycle or in quiescent cells. These pathways are thought to be constitutively on with substrate availability regulated by phosphorylation. However, only a few substrate systems are understood, and the complete story of F-box regulation could be much more complicated. The association of the APC with distinct WD40 repeat proteins also creates diverse substrate specificities used in the regulation of progression through mitosis and G1. A potential difference between recognition by the SCF and APC involves the requirement for phosphorylation of their target substrates. In the case of Pds1 or cyclin B, the substrates are directly recognized by the substrate activator CDC20 without any apparent phosphorylation requirement.
Our initial studies of the G1 function of CDH1 led us to the identification of multiple CDH1 genes in chick. The existence of multiple functional homologues in metazoans is further suggested by the identification of additional CDH1-related sequences in Drosophila, as well as in human. Four novel proteins show differential expression pattern during the chicken embryogenesis and substrate specificities in in vitro ubiquitination assays. One putative homologue, ChkCDH1-D, recognizes none of the known substrates. Our results suggest a possible mechanism for temporal and spatial regulation of APC in and outside of the cell cycle.
The existence of multiple substrate-specific activators with different patterns of expression suggest temporal and spatial regulation of APC activity outside of the cell cycle similar to the SCF complex. In particular, the presence of a neuron-specific CDH1 with narrow substrate specificity suggests a specific developmental role for the APC in this tissue. Recently, a possible role of the APC in postmitotic neurons was further suggested by the expression of both CDH1 and the APC in brain (17, 18). Finally, several new APC substrates have been identified by small pool expression cloning (N. Ayad and M.W.K., unpublished results) and by searching for APC substrate consensus sequences in databases. Such screening should now be repeated for the different CDH1 homologues described here. In particular, there are no known substrates for ChkCDH1-D. Together, these observations suggest new roles of the APC in development and differentiation.
Acknowledgments
We thank C. J. Tabin for providing chicken embryonic cDNA library and A. Grapin-Botton and R. Davis for their assistance in in situ hybridization and microscopy. We are grateful to N. Ayad, C. M. Pfleger, H. Zou, and S. Olaf for critical discussions. Y.W. is the recipient of a Helen Hay Whitney Postdoctoral Fellowship. This work was supported by National Institutes of Health Grants GM26875-17 and GM39023-08 (to M.W.K.).
Abbreviation
- APC
anaphase-promoting complex
Footnotes
References
- 1.Peters J M. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 2.Koepp D M, Harper J W, Elledge S J. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 3.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 6.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 8.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin W G, Maher E R. Trends Genet. 1998;14:423–426. doi: 10.1016/s0168-9525(98)01558-3. [DOI] [PubMed] [Google Scholar]
- 10.Jackson P K, Eldridge A G, Freed E, Furstenthal L, Hsu J Y, Kaiser B K, Reimann J D. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 11.Fang G, Yu H, Kirschner M W. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 12.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 13.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou H, McGarry T J, Bernal T, Kirschner M W. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhao N, Lai F, Fernald A A, Eisenbart J D, Espinosa R, Wang P W, Le Beau M M. Genomics. 1998;53:184–190. doi: 10.1006/geno.1998.5473. [DOI] [PubMed] [Google Scholar]
- 18.Gieffers C, Peters B H, Kramer E R, Dotti C G, Peters J M. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandeis M, Hunt T. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 20.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 21.Grapin-Botton A, Cambronero F, Weiner H L, Bonnin M A, Puelles L, Le Douarin N M. Mech Dev. 1999;84:41–53. doi: 10.1016/s0925-4773(99)00069-6. [DOI] [PubMed] [Google Scholar]
- 22.Pfleger C M, Kirschner M W. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 23.Wan Y, Kurosaki T, Huang X Y. Nature (London) 1996;380:541–544. doi: 10.1038/380541a0. [DOI] [PubMed] [Google Scholar]
- 24.Sigrist S J, Lehner C F. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 25.Lorca T, Castro A, Martinez A M, Vigneron S, Morin N, Sigrist S, Lehner C, Doree M, Labbe J C. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C X, Fisk B C, Wadehra M, Su H, Braun J. Blood. 2000;96:259–263. [PubMed] [Google Scholar]
- 27.Zhu H, Kavsak P, Abdollah S, Wrana J L, Thomsen G H. Nature (London) 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]