Abstract
Zebu cattle (Bos taurus indicus) are highly adapted to tropical regions. However, females reach puberty after taurine heifers, which affects the economic efficiency of beef cattle breeding in the tropical regions. The aims of this study were to establish associations between the haplotype alleles of the bovine genome and age at first calving (AFC) in the Nelore cattle, and to identify the genes and quantitative trait loci (QTL) related to this phenotype. A total of 2,273 Nelore cattle (995 males and 1,278 females) genotyped using the Illumina BovineHD BeadChip were used in the current study. The association analysis included females with valid first calving records as well as open heifers. Linkage disequilibrium (LD) analysis among the markers was performed using blocks of 5, 10, and 15 markers, which were determined by sliding windows shifting one marker at a time. Then, the haplotype block size to be used in the association study was chosen based on the highest r2 average among the SNPs in the block. The five HapAlleles most strongly associated with the trait (top five) were considered as significant associations. The results of the analysis revealed four genomic regions related to AFC, which overlapped with 20 QTL of the reproductive traits reported previously. Furthermore, there were 19 genes related to reproduction in those regions. In conclusion, the use of haplotypes allowed the detection of chromosomal regions associated with AFC in Nelore cattle, and provided the basis for elucidating the mechanisms underlying this trait.
2. Introduction
Indicine cattle (Bos taurus indicus) are well-adapted to tropical environments, because of attributes like heat tolerance and partial tick resistance. However, as a general rule, indicine heifers present inferior reproductive performances, regarding the onset of puberty, in comparison with taurine cattle (Bos taurus taurus) [1]. Such late onset of puberty negatively affects the economic efficiency of beef cattle breeding and restricts the genetic improvement of cattle [2], and is, thus, becoming a key concern in the tropical countries. In Brazil, where beef production is primarily based on indicine breeds, Nelore heifers selected for growth reach the onset of puberty at about 23 months [3]. Anticipating this stage would increase the producer’s profitability and can benefit the world beef supply, because Nelore cattle have huge importance in the global beef market [4].
Age at first calving (AFC) is usually utilized in breeding programs as an indicator trait of female sexual precocity, because this information can be easily obtained; the use of this information is directly related to shortening generation intervals and increasing genetic gains [5, 6]. However, selection cannot easily affect AFC because it is a sex-limited trait, is measured after maturity, and its heritability range from low to moderate (0.09 to 0.28) [7–11]. For these type of traits, genome-wide information might allow improving genetic gains through genomic selection [12]. In addition, genome-wide information could be used in association studies (GWAS) to identify candidate regions associated with AFC, and improve our understanding of the genetic basis of sexual precocity in indicine cattle. This knowledge could then be used to improve the accuracy of genomic predictions by using more informative markers and discarding those that generate noise during the predictions.
The use of individual single nucleotide polymorphisms (SNPs) to perform GWAS have some limitations because of the small effect of single markers, which usually do not reach stringent significance thresholds, and also because of the incomplete linkage disequilibrium (LD) between the SNPs and causal variants [13, 14]. On the contrary, the use of a block of SNPs (haplotypes) may provide more robust association analysis, because it improves the resolution of associations and facilitates the approximation of associated markers and possible causal mutations by increasing the LD [15–17].
The aim of this study was to scan for genomic regions associated with AFC in a Nelore cattle population using haplotype allele information, in order to highlight the QTL/genes and genetic mechanisms underlying this trait.
3. Material and methods
3.1. Ethical statement
The present study was exempt of the local ethical committee evaluation as genomic DNA was extracted from stored hair and semen samples of animals from commercial herds.
3.2. Samples
The genotypes used in the current study were provided by the Zebu Genome Consortium (ZGC). The full data consisted of 2,273 Nelore samples (995 males and 1,278 females) of different Brazilian herds, born between 1968 and 2008. All samples were genotyped with Illumina BovineHD (~777,000 SNPs).
The association analyses were carried out by considering deregressed estimated breeding values (dEBV) for AFC as response variables, according to Garrick et al. [18].
Prior to deregression, the estimated breeding values (EBV) was obtained for the dataset by considering both the calving and the noncalving (open heifers) females. Open heifers had predicted records that were obtained by adding a penalty of two months to the maximum value of AFC in their respective contemporary groups. Consideration of open heifers was aimed to avoid bias in the estimation of genetic parameters and EBVs [19, 20]. This procedure is based on the assumption that open heifers could have calved if they were bred for longer breeding seasons [20, 21]. The contemporary groups were defined by the concatenation of herd code, year and season of birth, management group identifications (from birth to weaning and from weaning to yearling), and farm (birth, weaning and yearling).
Variance components and EBV were obtained using the DMU software [22]. In the analysis of AFC, a single-trait animal model was fitted, including the fixed effect of contemporary groups and the age of dam as covariates (linear and quadratic effects), as well as a random direct genetic effect and a random residual term. For each animal, the reliabilities of the EBVs were computed based on the corresponding estimates of prediction error variance.
Only genotyped animals that had dEBV with associated reliabilities greater than 25% were kept for further analysis (S1 Dataset), such, that a set of 1,189 animals was considered for association analyses (S2 Dataset). The dEBV average for this samples was equal to -2 (ranging from -46.7 to 60.6) days and reliabilities equal to 0.59 (ranging from 0.25 to 0.94).
3.3. Quality control
Genomic data were subjected to quality control (QC) measures, including the maintenance of autosomal markers with minor allele frequencies (MAF) >2%, Hardy-Weinberg equilibrium (HWE) significance at P > 10−5 based on Fisher’s exact test, and an SNP call rate > 95%. The following filters were used for sample exclusion: identity by state (IBS) analysis > 95% and the samples in which less than 90% of the genotypes were determined were discarded. The QC was conducted using the GenABEL package, version 1.8–0 [23] of the R software (version 3.2.1) [24].
3.4. Genotype phasing
Haplotype assembly requires the identification of the chromosome (maternal or paternal) in which a certain allele is located. Therefore, the haplotype phase was determined using the SHAPEIT software version 2.r837 [25], considering all genotyped animals (N = 2,273).
3.5. Linkage disequilibrium
LD between markers was analyzed to determine the number of SNPs per haplotype block, because high-LD markers minimize the occurrence of recombination within the blocks [26]. Therefore, LD was estimated as the squared correlation of allelic frequencies (r2), following Hill and Robertson [27]:
| (1) |
where, freq.A, freq.a, freq.B and freq.b denote the frequencies of A, a, B, and b alleles, respectively; and freq.AB, freq.ab, freq.Ab and freq.aB denote the frequencies of the haplotypes AB, ab, Ab and aB in the population, respectively.
LD between markers was performed using blocks of 5, 10, and 15 markers with a sliding window for each SNP. The window size that resulted in stronger LD was chosen to perform the association study. LD was analyzed using the Plink software, version 1.9 [28, 29].
3.6. Haplotypes
With the phased haplotypes of autosomal chromosomes, the haplotypes were constructed with blocks size chosen based on high r² between the SNPs in the LD analysis and sliding window of one marker. The R package GHap v1.2.1 [30] was used to build the haplotype blocks (HapBlock) and identify alleles (HapAllele). Therefore, genotypes were scored as 2, 1, or 0, corresponding with the presence of two copies, one copy, or the absence of the HapAllele, respectively. The genomic position in the association analysis was based on the average distance between the first and last SNPs in a HapBlock. HapAlleles with allele frequency lower than 3% were excluded.
3.7. Association analysis
Since dEBV reliability varies across individuals, weighted analyses are required to account for heterogeneous variances. The weights were obtained as proposed by Garrick et al. [18]:
| (2) |
where h2 is the heritability of the trait (0.092) estimated using the complete database of AFC recording, r2 is the dEBV reliability, and c is a constant that can assume values from 0 to 1. We assumed that c was equal to 0.5.
Statistical analyses were performed using the ghap.lmm and ghap.assoc functions of the GHap package. First, the variance was estimated using the maximum-likelihood method and the following mixed linear model:
| (3) |
where y is the vector of dEBV, X is the incidence matrix relating elements in y to fixed effects b (intercept), Z is the incidence matrix for random effects u (animal), and u is a vector of random effects ~ N (0, Kσu2), where K is HapAllele relationship matrix, and e is a random residual vector (with variance-covariance Wσe2), assuming independence of the residuals.
The estimated residuals from the model (3) were considered as adjusted observations for covariance and polygenic effects, these values were adjusted via Genomic Control [31] and were used in the least squares regression analysis to test the association between each haplotype allele and the phenotype.
3.8. Analysis of genomic regions
A genome-wide threshold level at 5 × 10−5 was selected for this study, as used by Lander and Kruglyak [32]. For Hapalleles with distances less than 1 Mb or those that overlapped, only the one with the lower P value was chosen. The 1 Mb sequence windows that flanked the significant regions of the top five haplotypes were explored using the bovine genome assembly of UMD version 3.1.
The BioMart tool (Ensembl release 89) [33] and the Cattle QTLdb (release 32) [34] were used to search for candidate gene and cataloged QTL, respectively, surrounding the regions of significant haplotype alleles. The results associated with the examined genes were revised using available scientific literature; the Mouse Genome Informatics (MGI) [35], String v.10.5 [36], and GeneCards: The Human Gene Database [37] provided information regarding the contribution and involvement of orthologous genes associated with AFC.
Due to the large number of genes in the region of these HapAlleles and a involvement of some of them in various metabolic pathways, we have discussed only those that have been previously described in association with reproductive traits, as well as performed by Xu et al. [38].
4. Results
After filtering through quality control analyses, 511,375 SNPs and 1,189 individuals remained in the analysis. LD analysis indicated that the average of r2 between adjacent markers for the blocks of 5, 10, and 15 SNPs (S1 Fig) were 0.37, 0.32, and 0.30, respectively. Thus, the block sizes of 5 SNPs were chosen to perform association studies.
The haplotyping procedure generated 507,384 HapBlocks and 2,238,795 HapAlleles with allele frequency higher than 3%. The mean number of alleles per block ranged from 4.37 (BTA 1) to 1.19 (BTA 25). The distribution of the number of haplotype blocks per chromosome is shown in S1 Fig.
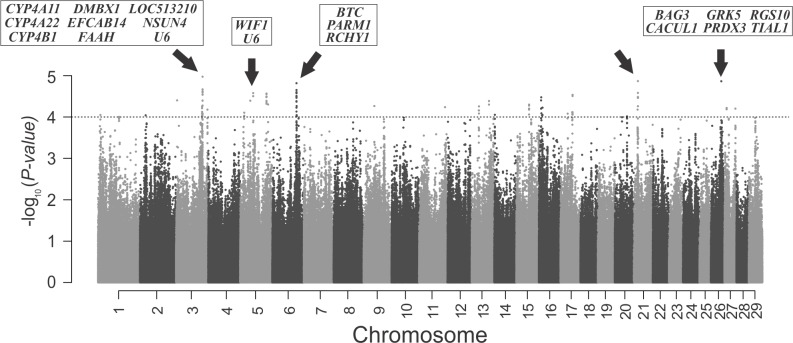
The results of GWAS between each Hapalleles and AFC in terms of – log10 (P value) are shown in Fig 1, where 68 haplotype alleles with P value < 5 × 10−5 were obtained (more details included in S1 Table). The five most significant regions selected for genomic exploration were in the detected peaks of BTA3 (BTA for Bos taurus chromosome), BTA5, BTA6, BTA21 and BTA26 (Table 1).
Fig 1. Manhattan plot of the genomic-wide association analysis of the –log10 (p-value) of haplotype alleles and AFC trait.
Each point in the graph represents a haplotype allele, arrows indicates de top five significant genomic regions and respective genes. The dashed line represent–log10(5 x 10−5) threshold.
Table 1. Top five regions of the GWAS with haplotypic blocks, indicating the chromosomal localization (Chr), start and end positions, the respective SNPs, size and p-values.
| BTA | Start (bp) | End (bp) | SNP start | SNP end | Size (bp) | P-value |
|---|---|---|---|---|---|---|
| 3 | 99,936,110 | 99,959,169 | rs133273360 | rs133299434 | 23,059 | 1.07 x10-5 |
| 5 | 49,183,430 | 49,201,226 | rs136746086 | rs136437853 | 17,796 | 2.65 x10-5 |
| 6 | 91,649,597 | 91,663,333 | rs134333101 | rs135909163 | 13,736 | 1.53x10-5 |
| 21 | 15,189,159 | 15,193,691 | rs42466473 | rs42465819 | 4,532 | 1.36 x10-5 |
| 26 | 39,765,858 | 39,773,044 | rs110416149 | rs41648222 | 7,186 | 1.37 x10-5 |
In total, 90 QTL (S2 Table) and 65 candidate genes (S3 Table) were present in these top five regions. Among these, 19 genes were related to previously described reproductive traits (Table 2). Some of the genes that were identified were associated with embryonic implantation in the uterus, development of reproductive organs in females and regulation of progesterone secretion.
Table 2. Summary of the genes present in 1-MB windows centered on the haplotypes that were top five significant haplotype alleles.
| Gene | Ensembl ID | Position (BTA:start–end [bp]) |
Description |
|---|---|---|---|
| U6 | ENSBTAG00000043170 ENSBTAG00000042928 |
3:99,562,453–99,562,559 5:48,996,775–48,996,881 |
U6 spliceosomal RNA |
| CYP4A11 | ENSBTAG00000037890 | 3:99,806,666–99,820,784 | Cytochrome P450, family 4, subfamily A, polypeptide 22 |
| CYP4A22 | ENSBTAG00000013481 | 3:99,918,428–99,933,505 | Cytochrome P450, family 4, subfamily A, polypeptide 22 |
| CYP4B1 | ENSBTAG00000011976 | 3:99,937,185–99,957,408 | Cytochrome P450 family 4 subfamily B member 1 |
| EFCAB14 | ENSBTAG00000000097 | 3:100,020,902–100,054,984 | EF-hand calcium binding domain 14 |
| DMBX1 | ENSBTAG00000009625 | 3:100,217,682–100,223,303 | Diencephalon/mesencephalon homeobox 1 |
| LOC513210 | ENSBTAG00000037858 | 3:100,285,863–100,300,972 | |
| FAAH | ENSBTAG00000007507 | 3:100,317,761–100,338,715 | Fatty acid amide hydrolase |
| NSUN4 | ENSBTAG00000015891 | 3:100,351,395–100,374,287 | NOP2/Sun RNA methyltransferase family member 4 |
| WIF1 | ENSBTAG00000014758 | 5:48,917,722–49,009,466 | WNT inhibitory factor 1 |
| BTC | ENSBTAG00000004237 | 6:91,430,305–91,480,129 | Betacellulin |
| PARM1 | ENSBTAG00000015919 | 6:91,597,122–91,723,390 | Prostate androgen-regulated mucin-like protein 1 |
| RCHY1 | ENSBTAG00000007189 | 6:92,061,508–92,076,962 | Ring finger and CHY zinc finger domain containing 1 |
| CACUL1 | ENSBTAG00000022808 | 26:39,292,197–39,356,979 | CDK2 associated cullin domain 1 |
| PRDX3 | ENSBTAG00000008731 | 26:39,672,044–39,681,392 | Peroxiredoxin 3 |
| GRK5 | ENSBTAG00000007981 | 26:39,702,550–39,930,993 | G protein-coupled receptor kinase 5 |
| RGS10 | ENSBTAG00000002647 | 26:39,976,325–40,017,296 | Regulator of G protein signaling 10 |
| TIAL1 | ENSBTAG00000004080 | 26:40,041,404–40,060,834 | TIA1 cytotoxic granule associated RNA binding protein like 1 |
| BAG3 | ENSBTAG00000013641 | 26:40,118,556–40,141,989 | BCL2 associated athanogene 3 |
Within the top five regions, we found several QTL that were related to reproductive traits (Table 3), and considering the QTL with AFC linkage, we can highlight the age at puberty, daughter pregnancy rate, and conception rate.
Table 3. QTL related to reproductive traits that were located in 1-Mb windows near haplotypes.
| QTL description | Position (BTA:start–end [bp]) |
QTLdb ID | PubMed ID |
|---|---|---|---|
| Calving ease | 3:84,669,155–102,513,835 | 15173 | 21183059 |
| Calving index | 3:84,669,155–102,513,835 | 15172 | 21183059 |
| Stillbirth | 3:84,669,155–102,513,835 | 15174 | 21183059 |
| Stillbirth | 3:99,853,456–101,636,723 | 30488 | 22888914 |
| Calving ease (maternal) | 5:49,553,247–51,015,255 | 106488 | 27328805 |
| Calving ease | 5:48,080,259–48,993,294 | 24565 | 24906442 |
| Age at puberty | 5:49,485,934–49,485,974 | 29989 | 22100599 |
| Interval to first estrus after calving | 5:49,399,050–49,399,090 | 30162 | 22100599 |
| Stillbirth | 5: 49,350,626–51,029,985 | 30495 | 22888914 |
| Calving to conception interval | 6:87,658,297–92,845,663 | 126853 | 28109604 |
| Interval to first estrus after calving | 6:87,658,297–92,845,663 | 126854 | 28109604 |
| Conception rate | 6:91,677,962–91,678,002 | 57143 | 23759029 |
| Daughter pregnancy rate | 6:91,677,962–91,678,002 | 57130 | 23759029 |
| Early embryonic survival | 6:91,677,962–91,678,002 | 57089 | 23904513 |
| Calving ease (maternal) | 26: 39,288,039–39,288,079 | 52924 | 21831322 |
| Daughter pregnancy rate | 26:39,288,039–39,288,079 | 52925 | 21831322 |
| Stillbirth (maternal) | 26:39,288,039–39,288,079 | 52926 | 21831322 |
| Calving ease | 26:39,288,039–39,288,079 | 52942 | 21831322 |
| Stillbirth | 26: 39,288,039–39,288,079 | 52944 | 21831322 |
| Calving ease | 26: 39,601,498–41,709,612 | 15227 | 21183059 |
5. Discussion
Most of the studies involving haplotypes generated based on the sliding windows have been applied to map alleles associated with human diseases [39]. The use of haplotypes instead of single markers gives putative advantages, the effects of individual SNPs being too small to overcome the stringent significance threshold [14].
The size of the haplotypes might interfere with the success of the analysis, because long blocks lead to the inclusion of non-informative SNPs and increase in the number of rare alleles, whereas short blocks might ignore the informative markers and reduce the power of the association analysis [40, 41]. The use of blocks with 5 SNPs was shown to be appropriate, with regards to the size and average of LD between the adjacent markers. Moreover, the detection of significant signals in the regions of BTA3, BTA5, BTA6 and BTA26, where known candidate genes and QTL of reproductive traits are located, reinforces the suitability of this block size.
The BAG3, DMBX1, and PRDX3 genes were annotated in the MGI database [35] with biological function related to abnormalities in fat deposition. Knockout of the BAG3 genes (cell death suppressor interaction Bcl-2 (Bis)) and DMBX1 (transcription factor) gave rise to mice with low fat deposition and severe thinness [42, 43]. The PRDX3 gene is highly expressed in the adipocytes and the suppression or knockout of this gene resulted in obese phenotype in rats and humans [44]. These genes can be pointed out by GWAS because of the positive or negative genetic correlation between body weight and reproductive traits, such as AFC and heifer pregnancies [45], in addition to the proximity of BAG3 (830 kb) and PRDX3 (383 kb) with the QTL affecting daughter pregnancy rate. Nelore heifers with early fat deposition can have decreased growth rate, with the nutritional resources directed for reproduction, thus presenting better reproductive performance than that of the heifers of greater weight.
The eIF gene family acts at several stages of translation during protein synthesis. Although there were no reports of association between eIFs and reproductive traits, functional search in String v10.5 identified the interaction of this family with the genes TIAL1, CACUL1, and NSUN4. The TIAL1 gene (also known as TIAR) encodes protein member of the RNA binding family, acting on post-transcriptional regulation. In rodents, changes in the expression of TIAL1 severely affects the development of primordial germ cells of gametes (cells that differentiate into sperm and oocytes), being orthologous in cattle [46]. CACUL1 (or CAC1) acts on the activation of the CDK2 gene involved in cell proliferation processes [47]. In the experiments using mice, inactivation of CDK2 gave rise to sterile animals, thereby proving to be essential for the development of germ cells in males and females [48]. It is worth mentioning that these two genes are close to the QTL associated with daughter pregnancy rate in BTA26, wherein TIAL1 is located at 753 kb and CACUL1 is located at only 4kb, corroborating the idea that these genes have roles in AFC variation. In this context, the NSUN4 gene has been reported to be differentially expressed in mouse oocytes. NSUN4 is a candidate gene involved in the association with cumulus cells, responsible for oocyte viability and embryonic developmental competence [38], which could possibly explain their association with AFC in the current study.
Further interaction between the LOC513210 and FAAH genes was found. LOC513210 belongs to the olfactory gene family that was associated with precocity in Nelore cattle in a previous study[49]. The physiological explanation is that these genes with olfactory function also act on germ cells [49, 50]. The expression of the FAAH gene occurs in the uterine epithelial cells and in the myometrium during the estrous cycle in rats. Adjustments in the expression of the FAAH gene are carried out by sex hormones and determine the success of uterine receptivity for the implantation of the embryo [51]. In heifers, sheep, and women, the decrease in FAAH gene expression has been associated with abortions [52–54]. In cows, the inactivation of FAAH can lead to infertility [54].
The CYP4A22 gene was found to be associated with precocity in Brahman heifers in previous studies [55, 56]. CYP4A22 belongs to the cytochrome P450 family and is involved in fat metabolism. Nguyen et al. [56] have suggested that this gene acts in response to the progesterone hormone. This hypothesis explains the involvement of CYP4A22 (having function in the pituitary gland) in the mechanisms of ovarian feedback established with during puberty. Using String, it was possible to detect a relationship between CYP4A22 and CYP4A11, which was the last one associated with perinatal modifications in target tissues that is necessary for the progression of pregnancy [57].
In addition to CYP4A22, the PARM1 gene has also been previously reported to be associated with precocity in heifers [58, 59]. PARM1 is expressed in the ovaries and encodes proteins involved in cell proliferation the, is regulated by androgens [60]. The PARM1 gene was also associated with daughter pregnancy rate of Holstein sires [61], and is located within two QTL that are essential for AFC, conception rate, and daughter pregnancy rate. This gene also participates in the regulation of luteinizing hormone and controls progesterone levels in rats [62, 63].
The genes RGS10, WIF1 and CYP4B1 were reported to be essential for the embryo implantation process in the uterus and for pregnancy viability [64–67]. RGS10, located 688 kb close to the daughter pregnancy rate QTL, had its expression identified in the pig endometrium. RGS10, which is orthologous in cattle and pigs, can lead to changes in the endometrium during the estrous cycle that can lead to successful embryo implantation [65]. WIF1 is located 476 kb away from the age at puberty QTL. This gene encodes the regulatory protein of canonical WNT molecules, involved in cell proliferation, differentiation, and migration [66, 68]. Expression studies in the endometrial tissue of heifers suggested the activity of the WIF1 gene as a molecule moderator, important for the implantation and development of the embryo [66, 67].
The BTC gene, localized on the BTA6, 197 kb away from the QTL affecting conception rate and daughter pregnancy rate, is essential for oocyte maturation and development, and fertilization [69]. BTC acts as the mediator of luteinizing hormone effects [70], which regulates functions related to puberty, such as oocyte maturation and ovulation [71].
Gurgan et al. [72], aiming to identify markers capable of predicting the quality of human oocytes detected the presence of the RCHY1 gene as a possible hormone regulator involved in follicular development, further emphasizing that this gene is close (383 kb) to the conception rate and daughter pregnancy rate QTL.
In a transcriptomic study using granulosa cells from the ovarian follicles of heifers, the GRK5 gene was detected in association with the maturation of granulosa cells [73]. These cells are important for the hormonal regulation occurring in the gonads and for viability of the female gamete [73, 74]. It is noteworthy that this gene is found 414 kb of daughter pregnancy rate QTL.
The EFCAB14 gene encodes a calcium ion binding protein and belongs to the EF-hand calcium binding gene family. There are still no reports available in the literature on the biological pathways in which genes from this domain (domain 14) are involved. However, this gene family has been associated with reproductive traits in women and cattle [75, 76].
Another candidate gene, located in the BTA3 and BTA5, encodes the U6 spliceosomal RNA, and had paralogous genes associated with AFC in a different population of Nellore cattle, as reported in a previous study [77]. In BTA5, this gene is close (498 kb) to an important QTL related to AFC, which is the age at puberty QTL.
There were no known genes or QTL related to the reproductive traits among the eight QTL and four genes annotated in the vicinities of the significant haplotypes of BTA21 (S2 and S3 Tables). This may be due either to the lack of knowledge regarding the functions of these genes or the existence of non-annotated genes in this region. It is noteworthy that the present study detected association in regions different from that reported by previous studies on the Nelore breed [77–79]. This can be because of herd particularities, such as the extent of LD, allelic frequencies, sample size, and statistical approaches [79], in addition to the use of haplotypes in the current study.
The number of significant peaks dispersed across the genome (Fig 1) confirms the polygenic nature of the trait AFC. Therefore, the current study was able to reveal genomic regions putatively associated with the reproductive performance of cattle.
The proximity of the QTL with the association regions and the involvement of QTL in reproductive traits (e.g., calving ease, daughter pregnancy rate, calving rate, embryonic survival, stillbirth, and conception rate) support the results obtained for the examined genes. Moreover, our results support the involvement of the identified regions in AFC.
6. Conclusion
The use of haplotypes allowed the detection of chromosomal regions associated with AFC. In these regions, genes and QTL related to reproduction were found. These results provide the basis for further studies that aim to elucidate the mechanisms underlying the roles of the examined genes during AFC expression.
Thus, a better understanding of this mechanism will allow the use of specific genotypes as a guide in animal genetics improvement programs, and will enable the construction of cheaper, low-density panels for the evaluation of specific genotypes that are advantageous to selection.
Supporting information
(TIF)
(TIF)
(CSV)
(CSV)
(CSV)
(RAR)
(GZ)
Acknowledgments
The Zebu Genome Consortia for providing the genotypes used in this paper. This research was supported by: Foundation Support to Development of Education, Science and Technology of the State of Mato Grosso do Sul (FUNDECT) and Coordination for the Improvement of Higher Education Personnel (CAPES). The DeltaGen and GenSys for the technical support and supply of the pseudo-phenotypes.
Data Availability
All relevant data are within the paper and its Supporting Information files (included raw genotypes and phenotypes).
Funding Statement
This research was supported by: Foundation Support to Development of Education, Science and Technology of the State of Mato Grosso do Sul (FUNDECT) and Coordination for the Improvement of Higher Education Personnel (CAPES). The funder (GenSys Consultores Associados S/S Ltda) provided support in the form of salaries for authors HHRN, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Abeygunawardena H, Dematawewa CMB. Pre-pubertal and postpartum anestrus in tropical Zebu cattle. Anim Reprod Sci. 2004;82:373–387. 10.1016/j.anireprosci.2004.05.006 [DOI] [PubMed] [Google Scholar]
- 2.Van Melis MH, Eler JP, Rosa GJM, Ferraz JBS, Figueiredo LGG, Mattos EC, et al. Additive genetic relationships between scrotal circumference, heifer pregnancy, and stayability in Nellore cattle. J Anim Sci. 2010;88(12):3809–3813. 10.2527/jas.2009-2127 [DOI] [PubMed] [Google Scholar]
- 3.Monteiro FM, Mercadante MEZ, Barros CM, Satrapa RA, Silva JAV, Oliveira LZ, et al. Reproductive tract development and puberty in two lines of Nellore heifers selected for postweaning weight. Theriogenology. 2013;80(1):10–17. 10.1016/j.theriogenology.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 4.Carvalheiro R. Genomic selection in Nelore cattle in Brazil. Proceedings of the 10th World Congress on Genetics Applied to Livestock Production; 2014. p. 17–22.
- 5.Baruselli PS, Gimenes LU, SALES JNS. Fisiologia reprodutiva de fêmeas taurinas e zebuínas. Revista Brasileira de Reprodução Animal. 2007;31(2):205–211. [Google Scholar]
- 6.Milazzotto MP, Visintin JA. Biotecnologias da reprodução animal-biologia molecular aplicada à biotecnologia. Ciência Veterinária nos Trópicos. 2008;11:145–148. [Google Scholar]
- 7.Boligon AA, Albuquerque LG. Genetic parameters and relationships of heifer pregnancy and age at first calving with weight gain, yearling and mature weight in Nelore cattle. Livest Sci. 2011;141(1):12–16. [Google Scholar]
- 8.Dias LT, El Faro L, Albuquerque LG. Estimativas de herdabilidade para idade ao primeiro parto de novilhas da raça Nelore. Rev Bras Zootec. 2004:97–102. [Google Scholar]
- 9.Mercadante MEZ, Lôbo RB, Oliveira HN. Estimativas de (co) variâncias entre características de reprodução e de crescimento em fêmeas de um rebanho Nelore. Rev Bras Zootec. 2000:997–1004. [Google Scholar]
- 10.Pereira E, Eler JP, Ferraz JBS. Correlação genética entre perímetro escrotal e algumas características reprodutivas na raça Nelore. Rev Bras Zootec. 2000;29(6):1676–1683. [Google Scholar]
- 11.Mota RR, Guimarães SEF, Fortes MRS, Hayes B, Silva FF, Verardo LL, et al. Genome‐wide association study and annotating candidate gene networks affecting age at first calving in Nellore cattle. J Anim Breed Genet. 2017;134(6):484–492. 10.1111/jbg.12299 [DOI] [PubMed] [Google Scholar]
- 12.Meuwissen THE, Hayes BJ, Goddard ME. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics. 2001;157(4):1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21(8):1213–1222. 10.1101/gr.111310.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Fan H, Wang Y, Zhang L, Gao X, Chen Y, et al. Genome-Wide Association Studies Using Haplotypes and Individual SNPs in Simmental Cattle. PloS ONE. 2014;9(10):e109330 10.1371/journal.pone.0109330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calus MPL, De Roos APW, Veerkamp RF. Accuracy of genomic selection using different methods to define haplotypes. Genetics. 2008;178(1):553–561. 10.1534/genetics.107.080838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grapes L, Dekkers JCM, Rothschild MF, Fernando RL. Comparing linkage disequilibrium-based methods for fine mapping quantitative trait loci. Genetics. 2004;166(3):1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. Invited review: Genomic selection in dairy cattle: Progress and challenges. J Dairy Sci. 2009;92(2):433–443. 10.3168/jds.2008-1646 [DOI] [PubMed] [Google Scholar]
- 18.Garrick DJ, Taylor JF, Fernando RL. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol. 2009;41(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donoghue KA, Rekaya R, Bertrand JK. Comparison of methods for handling censored records in beef fertility data: Field data. J Anim Sci. 2004;82(2):357–361. 10.2527/2004.822357x [DOI] [PubMed] [Google Scholar]
- 20.Minick Bormann J, Wilson DE. Calving day and age at first calving in Angus heifers. J Anim Sci. 2010;88(6):1947–1956. 10.2527/jas.2009-2249 [DOI] [PubMed] [Google Scholar]
- 21.Notter DR. Evaluating and reporting reproductive traits. Proc of the Beef Improv Fed 20th Ann Meet; Albuquerque, NM: Beef Improvement Federation; 1988. p. 21–42.
- 22.Madsen P, Jensen J. A User’s Guide to DMU: A Package for Analyzing Multivariate Mixed Models. Version 6. Center for Quantitative Genetics and Genomics, Dept. of Molecular Biology and Genetics, Research Centre Foulum, Denmark: 2006. [Google Scholar]
- 23.Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177(1):577–585. 10.1534/genetics.107.075614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 25.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 26.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genetics. 2002;3(4):299–309. 10.1038/nrg777 [DOI] [PubMed] [Google Scholar]
- 27.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38(6):226–231. 10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- 28.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Utsunomiya YT, Milanesi M, Utsunomiya AT, Ajmone-Marsan P, Garcia JF. GHap: An R package for Genome-wide Haplotyping. Bioinformatics. 2016;32(18):2861–2862. 10.1093/bioinformatics/btw356 [DOI] [PubMed] [Google Scholar]
- 31.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. [DOI] [PubMed] [Google Scholar]
- 32.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–247. 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 33.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Group MGD. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43(D1):D726–D736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016:gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Zhou T, Shao L, Zhang B, Liu K, Gao C, et al. Gene expression profiles in mouse cumulus cells derived from in vitro matured oocytes with and without blastocyst formation. Gene Expr Patterns. 2017;25–26:46–58. 10.1016/j.gep.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 39.Hamblin MT, Jannink J-L. Factors affecting the power of haplotype markers in association studies. The Plant Genome. 2011;4(2):145–153. [Google Scholar]
- 40.Mathias RA, Gao P, Goldstein JL, Wilson AF, Pugh EW, Furbert-Harris P, et al. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006;7(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.N’Diaye A, Haile JK, Cory AT, Clarke FR, Clarke JM, Knox RE, et al. Single Marker and Haplotype-Based Association Analysis of Semolina and Pasta Colour in Elite Durum Wheat Breeding Lines Using a High-Density Consensus Map. PloS ONE. 2017;12(1):e0170941 10.1371/journal.pone.0170941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto W, Shiuchi T, Miki T, Minokoshi Y, Takahashi Y, Takeuchi A, et al. Dmbx1 is essential in agouti-related protein action. P Natl Acad Sci USA. 2007;104(39):15514–15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Judge LM, Perez-Bermejo JA, Truong A, Ribeiro AJS, Yoo JC, Jensen CL, et al. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017;2(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huh JY, Kim Y, Jeong J, Park J, Kim I, Huh KH, et al. Peroxiredoxin 3 is a key molecule regulating adipocyte oxidative stress, mitochondrial biogenesis, and adipokine expression. Antioxid Redox Sign. 2012;16(3):229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paterno FM, Buzanskas ME, Koury Filho W, Lobo RB, Queiroz SA. Genetic analysis of visual assessment and body weight traits and their relationships with reproductive traits in Nellore cattle. J Agr Sci. 2017;155(4):679–687. [Google Scholar]
- 46.Beck ARP, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. P Natl Acad Sci USA. 1998;95(5):2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong Y, Kejun N, Yin Y. Identification and characterization of CAC1 as a novel CDK2-associated cullin. Cell Cycle. 2009;8(21):3552–3561. 10.4161/cc.8.21.9955 [DOI] [PubMed] [Google Scholar]
- 48.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 Knockout Mice Are Viable. Curr Biol. 2003;13(20):1775–1785. [DOI] [PubMed] [Google Scholar]
- 49.Irano N, Camargo GMF, Costa RB, Terakado APN, Magalhães AFB, Silva RMdO, et al. Genome-Wide Association Study for Indicator Traits of Sexual Precocity in Nellore Cattle. PloS ONE. 2016;11(8):e0159502 10.1371/journal.pone.0159502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melo TP, Camargo GMF, Albuquerque LG, Carvalheiro R. Genome-wide association study provides strong evidence of genes affecting the reproductive performance of Nellore beef cows. PlOS ONE. 2017;12(5):e0178551 10.1371/journal.pone.0178551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao AZ, Zhao YG, Duan EK. Expression and regulation of the fatty acid amide hydrolase gene in the rat uterus during the estrous cycle and peri-implantation period. Mol Hum Reprod. 2002;8(7):651–658. [DOI] [PubMed] [Google Scholar]
- 52.Schmid PC, Paria BC, Krebsbach RJ, Schmid HHO, Dey SK. Changes in anandamide levels in mouse uterus are associated with uterine receptivity for embryo implantation. P Natl Acad Sci USA. 1997;94(8):4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Xie H, Guo Y, Zhang H, Takahashi T, Kingsley PJ, et al. Fatty acid amide hydrolase deficiency limits early pregnancy events. J Clin Invest. 2006;116(8):2122 10.1172/JCI28621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weems YS, Lewis AW, Neuendorff DA, Randel RD, Weems CW. Endocannabinoid 1 and 2 (CB1; CB2) receptor agonists affect negatively cow luteal function in vitro. Prostag Oth Lipid M. 2009;90(3):89–93. [DOI] [PubMed] [Google Scholar]
- 55.Fortes MRS, Reverter A, Zhang Y, Collis E, Nagaraj SH, Jonsson NN, et al. Association weight matrix for the genetic dissection of puberty in beef cattle. P Natl Acad Sci USA. 2010;107(31):13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen LT, Reverter A, Cánovas A, Venus B, Islas-Trejo A, Porto-Neto LR, et al. Global differential gene expression in the pituitary gland and the ovaries of pre-and postpubertal Brahman heifers. J Anim Sci. 2017;95(2):599–615. 10.2527/jas.2016.0921 [DOI] [PubMed] [Google Scholar]
- 57.Loor JJ. Genomics of metabolic adaptations in the peripartal cow. Animal. 2010;4(7):1110–1139. 10.1017/S1751731110000960 [DOI] [PubMed] [Google Scholar]
- 58.Fortes MRS, Nguyen LT, Porto Neto LR, Reverter A, Moore SS, Lehnert SA, et al. Polymorphisms and genes associated with puberty in heifers. Theriogenology. 2016;86(1):333–339. 10.1016/j.theriogenology.2016.04.046 [DOI] [PubMed] [Google Scholar]
- 59.Hallé C, Goff AK, Petit HV, Blouin R, Palin M-F. Effects of different n-6: n-3 fatty acid ratios and of enterolactone on gene expression and PG secretion in bovine endometrial cells. Brit J Nutr. 2015;113(1):56–71. 10.1017/S0007114514003304 [DOI] [PubMed] [Google Scholar]
- 60.Fladeby C, Gupta SN, Barois N, Lorenzo PI, Simpson JC, Saatcioglu F, et al. Human PARM‐1 is a novel mucin‐like, androgen‐regulated gene exhibiting proliferative effects in prostate cancer cells. Int J Cancer. 2008;122(6):1229–1235. 10.1002/ijc.23185 [DOI] [PubMed] [Google Scholar]
- 61.Cochran SD, Cole JB, Null DJ, Hansen PJ. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet. 2013;14(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gratao AA, Dahlhoff M, Sinowatz F, Wolf E, Schneider MR. Betacellulin overexpression in the mouse ovary leads to MAPK3/MAPK1 hyperactivation and reduces litter size by impairing fertilization. Biol Reprod. 2008;78(1):43–52. 10.1095/biolreprod.107.062588 [DOI] [PubMed] [Google Scholar]
- 63.Park JY, Jang H, Curry TE, Sakamoto A, Jo M. Prostate Androgen-Regulated Mucin-Like protein 1: A Novel Regulator of Progesterone Metabolism. Mol Endocrinol. 2013;27(11):1871–1886. 10.1210/me.2013-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macklon NS, van der Gaast MH, Hamilton A, Fauser BCJM, Giudice LC. The impact of ovarian stimulation with recombinant FSH in combination with GnRH antagonist on the endometrial transcriptome in the window of implantation. Reprod Sci. 2008;15(4):357–365. 10.1177/1933719107311781 [DOI] [PubMed] [Google Scholar]
- 65.Kiewisz J, Krawczynski K, Lisowski P, Blitek A, Zwierzchowski L, Ziecik AJ, et al. Global gene expression profiling of porcine endometria on Days 12 and 16 of the estrous cycle and pregnancy. Theriogenology. 2014;82(6):897–909. 10.1016/j.theriogenology.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 66.Tribulo P, Moss JI, Ozawa M, Jiang Z, Tian XC, Hansen PJ. WNT regulation of embryonic development likely involves pathways independent of nuclear CTNNB1. Reproduction. 2017;153(4):405–419. 10.1530/REP-16-0610 [DOI] [PubMed] [Google Scholar]
- 67.Tríbulo P, Siqueira LGB, Oliveira LJ, Scheffler T, Hansen PJ. Identification of potential embryokines in the bovine reproductive tract. J Dairy Sci. 2018;101(1):690–704. 10.3168/jds.2017-13221 [DOI] [PubMed] [Google Scholar]
- 68.Hsieh J-C, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, et al. A new secreted protein that binds to Wnt proteins and inhibits their activites. Nature. 1999;398(6726):431 10.1038/18899 [DOI] [PubMed] [Google Scholar]
- 69.Boruszewska D, Sinderewicz E, Kowalczyk-Zieba I, Grycmacher K, Woclawek-Potocka I. The effect of lysophosphatidic acid during in vitro maturation of bovine cumulus–oocyte complexes: cumulus expansion, glucose metabolism and expression of genes involved in the ovulatory cascade, oocyte and blastocyst competence. Reprod Biol Endocrin. 2015;13(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caixeta ES, Machado MF, Ripamonte P, Price C, Buratini J. Effects of FSH on the expression of receptors for oocyte-secreted factors and members of the EGF-like family during in vitro maturation in cattle. Reprod Fert Develop. 2013;25(6):890–899. [DOI] [PubMed] [Google Scholar]
- 71.Nogueira MFG, Fernandes P, Ereno RL, Simões RAL, Buratini Júnior J, Barros CM. Luteinizing Hormone Receptor (LHR): basic concepts in cattle and other mammals. A review. Anim Reprod. 2010:51–64. [Google Scholar]
- 72.Gurgan T, Montjean D, Demirol A, Menezo YJR. Sequential (hFSH + recFSH) vs homogenous (hFSH or recFSH alone) stimulation: clinical and biochemical (cumulus cell gene expression) aspects. J Assist Reprod Gen. 2014;31(6):657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hatzirodos N, Irving-Rodgers HF, Hummitzsch K, Harland ML, Morris SE, Rodgers RJ. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics. 2014;15(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munhoz ALR, Luna HS. Morfometria e número de células da granulosa de folículos pré-antrais bovinos submetidos ao estresse calórico in vitro. Acta Vet Brasilica. 2008;2(3):85–88. [Google Scholar]
- 75.Peddinti D, Nanduri B, Kaya A, Feugang JM, Burgess SC, Memili E. Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst Biol. 2008;2(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou L, Li R, Wang R, Huang H-x, Zhong K. Local injury to the endometrium in controlled ovarian hyperstimulation cycles improves implantation rates. Fertil Steril. 2008;89(5):1166–1176. 10.1016/j.fertnstert.2007.05.064 [DOI] [PubMed] [Google Scholar]
- 77.Nascimento AV, Matos MC, Seno LO, Romero AR, Garcia JF, Grisolia AB. Genome wide association study on early puberty in Bos indicus. Genet Mol Res. 2016;15(1):1–6. [DOI] [PubMed] [Google Scholar]
- 78.Costa RB, Camargo GM, Diaz ID, Irano N, Dias MM, Carvalheiro R, et al. Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. Genet Sel Evol. 2015;47(1):47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Regatieri IC, Boligon AA, Costa RB, Souza FRP, Baldi F, Takada L, et al. Association between single nucleotide polymorphisms and sexual precocity in Nellore heifers. Anim Reprod Sci. 2017;177:88–96. 10.1016/j.anireprosci.2016.12.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(CSV)
(CSV)
(CSV)
(RAR)
(GZ)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files (included raw genotypes and phenotypes).