Abstract
Background:
There has been considerable progress in identifying inflammatory bowel disease [IBD] susceptibility genes but little progress in examining the role of genetic variation in the development of the extra-intestinal manifestations [EIMs] of IBD. This study identified clinical, serological, and genetic factors associated with ocular EIMs [O-EIMs] in IBD.
Methods:
We performed a retrospective case-control study of IBD patients, comparing those with and without O-EIMs using the Cedars-Sinai IBD Research Repository and the NIDDK IBD Genetics Consortium Repository. Genotyping was performed using Illumina whole genome platforms.
Results:
In all, 124 cases and 3328 controls with available clinical data were identified; 103 cases and 2808 controls had genetic data available. Erythema nodosum and peripheral arthritis particularly were common in patients with O-EIMs [ p = 2.77 x 10 -13 and p = 2.58 x 10 -13 , respectively] with increasing odds ratios for O-EIMs with each additional non-ocular-EIM [for ≥ 2 EIMs, odds ratio 14.72]. Nominal association with O-EIMs was observed at several known IBD susceptibility single nuclear polymorphisms. One locus, containing RBM19, achieved genome-wide level of significance for association with O-EIMs.
Conclusions:
In IBD, O-EIMs co-occur with musculoskeletal and skin manifestations and, in this study, are nominally associated with known IBD loci. Additional cohorts are needed to verify these results and identify additional genes.
Keywords: Crohn’s disease, ulcerative colitis, IBD, eye, uveitis, genetics
1. Introduction
It is widely recognised that the inflammatory bowel diseases [IBD], Crohn’s disease [CD], and ulcerative colitis [UC] occur in genetically susceptible individuals following exposure to, as yet poorly understood, environmental factors. Recently there have been considerable advances in the identification of IBD susceptibility loci, 12 , 3 but an understanding of the molecular associations with clinical sub-phenotypes has lagged behind. In addition to chronic, relapsing gastrointestinal [GI] inflammation, up to 40% of patients with IBD have extraintestinal manifestations including ocular inflammation which can cause significant morbidity including blindness. 4 , 5
Episcleritis, scleritis, and anterior uveitis are the most common ocular-extraintestinal manifestations [O-EIMs] in IBD. Other less common eye manifestations with reported associations to IBD include retinal vasculitis, papillitis, corneal infiltrates, myositis, scleromalacia perforans, and optic neuritis. After peripheral arthritis, O-EIMs are the second most common extraintestinal manifestation [EIM], occurring in 2–6% of adult IBD patients, with a higher prevalence reported in children. 4 , 6 , 7
In IBD, most patients who develop ocular inflammation do so at first presentation or with an established diagnosis of IBD, but eye disease can pre-date intestinal disease. 8 , 9 Uveitis is reported as occurring four times more commonly in women and has been strongly associated with sacroiliac joint abnormalities and arthritis. 10 Genetic susceptibility in part appears to explain the development of O-EIMs in IBD. Pheobe et al. demonstrated that a family history of IBD significantly increases the risk of ocular inflammation in subjects without IBD, suggesting shared immunological mechanisms for eye and intestinal disease. 10 Further evidence supporting shared aetiology comes from the observation that individuals with IBD and ocular disease are more likely to ‘express’ anti-outer membrane porin C [OmpC] antibodies and that patients with a family history of IBD and ocular inflammation are more likely to be anti-nuclear cytoplasmic antibody [ANCA] positive. 11 , 12
Few studies have evaluated the association between O-EIMs and genetic variation and the studies to date have been limited by power and the depth of available genotypes. Orchard et al. examined major histocompatibility complex [MHC] associations with O-EIMs and identified that eye manifestations are strongly associated with human leucocyte antigen [HLA]-B*27, B*58 and HLA-DRB1*0103. 13 Studies examining known, non-HLA, IBD susceptibility genes including ATG16L1, IL-23R , and NOD2/CARD15 , have found no association with O-EIMs and IBD. 14 , 15 We present the largest study to date investigating demographic, clinical, serological and genetic associations with O-EIMs in IBD.
2. Methods
2.1. Databases
The Cedars-Sinai IBD Research Repository [MIRIAD] and the National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium [NIDDK-IBDGC] database [Cedars-Sinai Medical Center, John Hopkins University, University of Chicago, University of Montreal, University of Pittsburgh, University of Toronto, and Yale University] were used to identify cases. We queried the two IBD databases for clinical, serological, and genetic factors associated with [+] O-EIMs and without [-] O-EIMs.
2.2. Subjects
We evaluated all IBD patients in MIRIAD and identified those with at least one documented episode of O-EIM. The paper and electronic charts for these patients were reviewed to verify the O-EIM. The diagnosis of O-EIMs was made based on a previous diagnosis of episcleritis, scleritis, or uveitis and/or after evaluation by an ophthalmologist. O-EIMs in MIRIAD were suspected in patients with ocular pain, double vision, sudden decreased visual acuity, redness of the eyes, and eye irritation or watering. Patients were referred to ophthalmology if they had any of these symptoms. We also included O-EIM positive cases from the NIDDK-IBDGC database that uses a validated phenotyping collection protocol. 16
In addition to O-EIMs, we collected information on demographics [gender, age], disease location and behaviour [as per Montreal classification], smoking status, family history, and surgical history from both databases. Data regarding the presence of non-ocular extraintestinal manifestations [non-ocular EIMs] also were collected. Non-ocular EIMs included erythema nodosum [EN], pyoderma gangrenosum [PG], peripheral arthritis, ankylosing spondylitis [AS], and primary sclerosing cholangitis [PSC]. IBD serologies [anti- Saccharomyces cerevisiae antibodies [ASCA IgG and IgA], perinuclear anti-nuclear cytoplasmic antibody [pANCA], anti-flagellin [anti-CBir1], anti-OmpC, and anti- Pseudomonas fluorescens associated sequence I2 [anti-I2]] were measured by enzyme-linked immunosorbent assay [ELISA] and expressed and assessed as previously described. 17 Only patients from MIRIAD had serology data available for evaluation.
Genotypes on the MIRIAD samples were generated at Cedars-Sinai Medical Center [CSMC] using Illumina Human610-quad, HumanCNV370-quad, and the HumanOmniExpress whole genome platforms. 13 , 18 Five samples [on 610-quad], 3 samples [on 370-quad], and 14 samples [on OmniExpress] were genotyped in duplicate, yielding concordance rates of 100%, 99.999% and 99.997%, respectively. Heritability concordance for three genotyping control trios was 99.53%. Samples which passed genotyping quality control exhibited genotyping call rate > 97%, and the average genotyping call rate across three platforms was 99.85%. The NIDDK IBDGC samples were genotyped using the HumanHap300 and HumanHap550 platforms as previously described. 19 , 20 Quality control was maintained as previously described. 17
To combine results for the two independent genome-wide association studies [GWAS] cohorts, we imputed untyped genotypes using Impute2 [http://hapmap.ncbi.nlm.nih.gov] and HapMap Phase III [www.hapmap.org] reference genotypes for the Cedars GWAS data and minimac software, and 1000 Genome Project data release 2010-08 genotypes for the NIDDK cohorts. Poorly imputed single nucleotide polymorphisms [SNPs], defined by an R-squared quality RSQR < 0.30 with MACH1/minimac or an information measure Is < 0.30 with IMPUTE2, were excluded from the analyses.
The Institutional Review Board at CSMC approved the study. All patients in both databases provided informed consent before genetic analysis.
2.3. Statistics
Univariate analysis
Logistic regression was performed to detect association of each clinical parameter with O-EIM status. The source of the sample [MIRIAD or NIDDK] was included as covariate to control for potential confounding effect. A significance threshold of 2.96 x 10 -3 was used to control for multiple testing.
Similarly, logistic regression was performed to detect the association of IBD serology as well as other EIMs with O-EIM. The significance threshold was set to be 8.33 x 10 -3 for both analyses.
2.4. Multivariate analysis
To account for the complicated correlation structure in the clinical characteristics, we also performed a multivariate logistic regression. Only variables with p < 0.05 in univariate analysis were included in the model. Serological factors were not included in this multivariate model as the serology data were only available in the MIRIAD cohort.
2.5. Genetic analysis
SNPtest and PLINK were used for genetic analyses in the Cedars and NIDDK cohorts, respectively. The DNA strand alignment was double-checked in the meta-analysis and confirmed to be on the same strand. Most of the SNPs had the same minor and major alleles, although a very low proportion of SNPs had switched alleles, with an MAF close to 0.5.
Logistic regression was performed to investigate the association between O-EIMs and genetic variations, with adjustment for the top four principal components from the population stratification analysis, to control for potential confounding. Meta-analysis was performed for SNPs that were common in both Cedars and NIDDK imputed GWAS dataset, using inverse variance weighting.
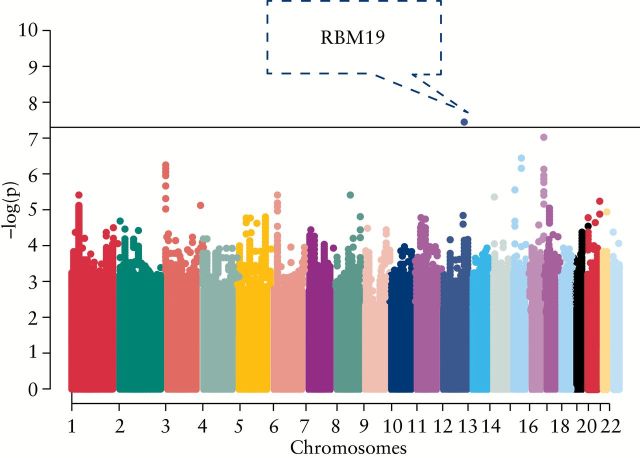
We assessed genetic associations with O-EIMs using both a genome-wide approach and also specifically at previously identified IBD susceptibility loci with an a priori nominal statistical significance defined as p < 5 x 10 -5 for O-EIMs and for any -EIM analyses. A priori significance of p < 3.1 x 10 -4 was determined for association at any known IBD loci. A Manhattan plot was completed to show chromosomal locations of associations with O-EIMs [ Figure 1 ].
Figure 1.
Manhattan plot highlighting loci associated with O-EIMs.
3. Results
3.1. Clinical factors
Among 3452 IBD patients, we identified 124 [3.6%] [+] O-EIMs [ Table 1 ]. In the MIRIAD database, 95% of [+] O-EIMs cases had been diagnosed before presentation at CSMC. Data regarding family history were available for 96.0% of cases and 97.0% of controls. Details regarding distribution of disease were available in 97.6% of cases and 98.6% of controls. Data on gender, age at diagnosis, smoking status, and surgical history were present for all patients among both cases and controls. On univariate analysis, no demographic or clinical factor was associated with O-EIMs in a statistically significant manner, though O-EIMs were more prevalent in females and CD patients [ Table 1 ]. On multivariable analysis, female association with O-EIMs approached statistical significance [ p = 8.49 x 10 -3 ] [ Table 2 ].
Table 1.
Clinical characteristics of subjects.
| Clinical characteristic | % [+] O-EIM [ n = 124] | % [-] O-EIM [ n = 3328] | p -Value | OR [95% CI] |
|---|---|---|---|---|
| Male [%] | 37.9 | 53.0 | 1.18 x 10 -3 | 0.54 [0.37–0.79] |
| CD | 72.6 | 66.7 | 0.013 | 1.79 [1.13–2.85] |
| Mean age at diagnosis | 26.7 | 26.7 | 0.931 | - |
| FH of IBD | 18.5 | 21.5 | 0.502 | 0.93 [0.61–1.41] |
| Smoking | 28.6 | 31.6 | 0.349 | 0.82 [0.55–1.24] |
| Previous surgery | 44.2 | 40.5 | 0.288 | 1.22 [0.84–1.77] |
| CD location: | ||||
| L1 | 31.0 | 28.6 | 0.945 | 1.02 [0.63–1.63] |
| L2 | 13.8 | 14.6 | 0.723 | 1.12 [0.59–2.16] |
| L3 | 55.2 | 56.8 | 0.771 | 0.93 [0.61–1.44] |
| L4 | 18.7 | 14.4 | 0.189 | 1.49 [0.82–2.72] |
| CD phenotype: | ||||
| B1 | 44.2 | 44.7 | 0.940 | 1.01 [0.66–1.57] |
| B2 | 29.1 | 29.9 | 0.958 | 0.98 [0.61–1.59] |
| B3 | 26.7 | 25.0 | 0.920 | 1.03 [0.63–4.97] |
| Perianal | 39.0 | 31.9 | 0.135 | 1.41 [0.89–2.30] |
| UC/IBD-U | ||||
| Phenotype: | ||||
| E3 | 85.3 | 69.1 | 0.061 | 2.50 [0.96–6.52] |
OR, odds ratio; CI, confidence interval; O-EIM, ocular extra-intestinal manifestation; CD, Crohn’s disease; FH, family history; IBD-U, inflammatory bowel disease unclassified.
Table 2.
Multivariable analysis of clinical factors associated with ocular-extraintestinal manifestations.
| Variable | OR [95% CI] | p -Value |
|---|---|---|
| Male gender | 0.59 [0.40–0.87] | 8.49x10 -3 |
| Crohn’s disease | 0.90 [0.58–1.40] | 0.65 |
| Peripheral arthritis | 3.09 [2.01–4.75] | 2.87x10 -7 |
| Ankylosing spondylitis | 2.73 [1.61–4.61] | 1.78x10 -4 |
| Erythema nodosum | 5.73 [2.92–11.27] | 4.13x10 -7 |
| Pyoderma gangrenosum | 5.14 [1.89–14.03] | 1.38x10 -3 |
OR, odds ratio; CI, confidence interval.
Overall, 122 O-EIM [+] [98.4%] cases and 3322 O-EIM [-] [99.8%] controls had complete data on presence of the five non-ocular EIMs. Skin and joint manifestations had clear associations with ocular inflammation in both CD and UC/IBD-U [IBD unclassified]. Among patients with CD, univariate analysis revealed peripheral arthritis was significantly associated with O-EIMs ( p = 7.72 x 10 -10 , odds ratio [OR] 4.29) [ Table 3 ]. UC/IBD-U subjects with O-EIMs had a particularly strong association with pyoderma gangrenosum [ p = 9.28 x 10 -23 ] [ Table 4 ]. On multivariate analysis, both skin and joint manifestations were significantly associated with O-EIMs [ Table 2 ].
Table 3.
Extra-intestinal manifestations in Crohn’s disease subjects
| EIM | % [+] O-EIM | % [-] O-EIM | p-Value | OR [95% CI] |
|---|---|---|---|---|
| Peripheral arthritis | 35.6 | 12.3 | 7.72x10 -10 | 4.29 [2.70–6.81] |
| Ankylosing spondylitis | 18.9 | 8.8 | 4.33x10 -3 | 2.74 [1.56–4.80] |
| Erythema nodosum | 14.9 | 2.3 | 1.99x10 -10 | 8.88 [4.53–17.39] |
| Pyoderma gangrenosum | 8.9 | 1.3 | 2.34x10 -6 | 7.14 [3.16–16.16] |
| PSC | 1.1 | 1.0 | 0.89 | 1.15 [0.15–8.63] |
| Any EIM | 54.6 | 22.9 | 7.11x10 -12 | 4.81 [3.07–7.54] |
OR, odds ratio; CI, confidence interval; O-EIM, ocular extra-intestinal manifestation; CD, Crohn’s disease; PSC, primary sclerosing cholangitis.
Table 4.
Extra-intestinal manifestations in UC/IBDU subjects.
| EIM | % [+] O-EIM | % [-] O-EIM | p -Value | OR [95% CI] |
|---|---|---|---|---|
| Peripheral arthritis | 23.5 | 6.4 | 8.92x10 -5 | 5.38 [2.32–6.52] |
| Ankylosing spondylitis | 17.6 | 1.6 | 1.14x10 -7 | 16.92 [5.95–48.14] |
| Erythema nodosum | 6.1 | 0.2 | 5.00x10 -4 | 93.0 [7.25–1194.74] |
| Pyoderma gangrenosum | 8.8 | 0 | 9.28x10 -23 | N/A |
| PSC | 8.8 | 3.2 | 0.119 | 2.66 [0.78–9.13] |
| Any EIM | 32.4 | 10.9 | 1.04x10 -4 | 4.42[2.08–9.35] |
OR, odds ratio; CI, confidence interval; O-EIM, ocular extra-intestinal manifestation; UC, ulcerative colitis; IBD-U, inflammatory bowel disease unclassified; NA, not available.
Table 5.
Extra-intestinal manifestations in inflammatory bowel disease subjects.
| EIM | % [+] O-EIM | % [-] O-EIM | p -Value | OR [95% CI] |
|---|---|---|---|---|
| Peripheral arthritis | 32.2 | 10.4 | 2.58x10 -13 | 4.58 [3.05–6.88] |
| Ankylosing spondylitis | 18.5 | 6.4 | 8.27x10 -8 | 3.88 [2.36–6.38] |
| Erythema nodosum | 12.5 | 1.6 | 2.77x10 -13 | 10.72 [5.67–20.25] |
| Pyoderma gangrenosum | 8.87 | 0.9 | 4.20x10 -11 | 11.32 [5.50–23.28] |
| PSC | 3.2 | 1.8 | 0.276 | 1.77 [0.63–4.97] |
| Any EIM | 48.4 | 18.9 | 1.18x10 -15 | 4.79 [3.26–7.03] |
| ≥ 2 any EIM | 20.5 | 0.02 | 1.62x10 -23 | 14.72 [8.69–24.94] |
OR, odds ratio; CI, confidence interval; O-EIM, ocular extra-intestinal manifestation; PSC, primary sclerosing cholangitis.
When IBD patients had at least one non-ocular-EIM, they were at increased risk for developing ocular inflammation [OR 4.79, p < 1.18x10 -15 ] [ Figure 2 ]. Additionally, the risk of O-EIMs increased with each additional non-ocular-EIM [for ≥ 2 EIMs, OR 14.72, p < 1.62 x 10 -23 ] [ Figure 2 ].
Figure 2.
Prevalence of nonocular-extraintestinal manifestations in [+] O-EIMs and [-] O-EIMs subjects.
3.2. Serology
Serological data were available for 52 MIRIAD cases and 2068 controls. There was no significant association with serology.
3.3. Genetics
Genetic analyses were performed on 103 [+] O-EIM cases and 2808 [-] O-EIM controls. No known IBD loci were significantly associated with O-EIMs. Single nucleotide polymorphisms [SNPs] at two known IBD loci had nominal association with O-EIMs; the locus containing C10ORF58 and TSPAN14 were protective for O-EIMs [OR 0.65, p = 0.035]; the TNFSF14 locus was associated with O-EIMs [OR 1.47, p = 0.010].
An unbiased approach examining all genotyped/imputed SNPs across the genome identified 14 loci with nominal significance associated with O-EIM, including one locus achieving genome-wide significance [rs4766697, p = 3.75E-8, OR = 3.31] [ Table 6 ]. The imputation score was 0.879, indicating very good imputation quality. This locus contains a long intergenic non-protein coding RNA, LINC01234 , and RBM19 , a regulator of ribosome biogenesis expressed in crypt cells of the intestinal epithelium.
Table 6.
Genetic associations with O-EIMs using a genome-wide approach.
| SNP | Chr | p -Value |
OR
[95% CI] |
Gene |
|---|---|---|---|---|
| rs4766697 | 12 | 3.75E-8 |
3.31
[1.51–5.07] |
RBM19 |
| rs11640406 | 16 | 9.50E-8 |
3.55
[0.95–5.66] |
TERF2IP, ADAT1, CHST5/6 |
| rs12050616 | 15 | 3.50E-7 |
2.21
[1.67–3.00] |
C2CD4A, VPS13C |
| rs11710167 | 3 | 5.65E-7 |
2.23
[1.72–3.05] |
CNTN4 |
| rs8031119 | 15 | 2.71E-6 |
2.69
[1.39–4.07] |
LINC00929 |
| rs645123 | 1 | 3.88E-6 |
2.49
[1.29–3.68] |
AGO1/3/4, TEKT2, COL8A2, TPAPPC3, MAP7D1, THRAP3 |
| rs16936449 | 8 | 3.91E-6 |
2.98
[1.78–4.73] |
SULF1, PRDM14, SLCO5A1 |
| rs6910165 | 6 | 3.97E-6 |
2.21
[1.79–3.10] |
LOC101928519 |
| rs35355940 | 14 | 4.57E-6 |
2.62
[1.51–3.96] |
CHD8, RAB2B, TOX4, METTL3,OR10G3, OR10G2, OR4E2,
TCRα variable region gene cluster |
| rs12624442 | 20 | 5.84E-6 |
2.65
[1.47–4.03] |
SLCO4A1, NTSR1, MRGBP, OGFR, TCFL5, DIDO1 |
| rs4401383 | 3 | 7.27E-6 | 2.15 [1.67–3.01] | MCF2L2, KLHL6/24, YEATS2 |
SNP, single nucleotide polymorphism; chr, chromosome; OR, odds ratio; CI, confidence interval.
Finally, given the co-occurrence of EIMs, we performed an analysis looking at genetic associations with the development of any extra-intestinal manifestation. We identified 504 IBD cases with any EIM and 2407 cases with no history of EIMs. A genome wide analysis revealed nominal significant associations with variants tagging, a DNA mismatch repair gene [ MSH3 ], a transmembrane protein [TMTC2], a post-transcriptional gene regulator [MIR548T], an integral membrane protein important in cell-cell recognition and adhesion [PCDH7], a tumour suppression gene [CSMD1], and a glutamate-regulated ion channel gene [GRIN3A] [ Table 7 ]. One IBD-associated SNP tagging the IL12B locus achieved our a priori level of nominal significance but did not achieve genome-wide significance [ p = 0.027, OR 1.16]. In contrast to previous studies, we found no association between HLA DRB*0103 and the presence of either ocular [ p = 0.60] or non-ocular-EIMs [ p = 0.81].
Table 7.
Genetic associations with all extra-intestinal manifestations, using a genome-wide approach.
| SNP | Chromosome | p-Value | OR [95% CI] | Gene | Reference allele | Variant allele |
|---|---|---|---|---|---|---|
| rs28678153 | 4p15 | 1.40E-07 |
0.58
[0.47–0.71] |
- | A | G |
| rs2055340 | 8p23 | 6.70E-07 |
0.71
[0.61–0.81] |
CSMD1 | A | T |
| rs16878216 | 5q14 | 1.54E-06 |
1.67
[1.35–2.06] |
MSH3, DHFR, RASGRF2 | C | G |
| rs12370878 | 12q21 | 7.72E-06 |
1.58
[1.29–1.92] |
TMTC2 | T | A |
| rs107608 | 9q31 | 8.59E-06 |
0.74
[0.65–0.85] |
- | C | G |
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
4. Discussion
We present the largest study, to date, describing characteristics associated with the development of ocular manifestations of IBD. We found that approximately 4% of IBD patients had a history of uveitis, scleritis, or episcleritis, consistent with previous findings. 4 , 21 Several previous studies have reported higher prevalence rates, mainly due to inclusion of dry eyes, conjunctivitis, and glaucoma, eye conditions not directly linked with IBD. 22 , 23 , 24 We identified higher prevalence of O-EIMs in women and Crohn’s disease, consistent with previous studies. 4 , 13 , 21 , 24
Little has been published on the relationship between O-EIMs in IBD and genetic variation. Although there was no association with known IBD loci and O-EIMs, after Bonferroni correction we observed a ‘nominal’ association between O-EIMs and the IBD loci containing TSPAN14 and TNFSF14. Further studies, in larger cohorts, will be needed to assess whether these are valid observations or not. The unbiased approach implicated genes involved in eye diseases including: COL8A2 [a major component of corneal endothelium], linked to Fuchs endothelial corneal dystrophy and posterior polymorphous corneal dystrophy type 2, and CHST6, associated with macular corneal dystrophy. Other processes potentially implicated in O-EIM development by these genetic findings include cell adhesion [ C2CD4A, CNTN4 ], golgi and endoplasmic reticulum maintenance [ TPAPPC3, GABARAPL2, RAB2B ], response to viral infection [ AGO3, SLCO5A1 ], metabolic syndromes [ ADAT1, VPS13C ], and T cell receptor alpha variable region genes. It is important to emphasise that these loci did not achieve genome-wide significance and therefore first require validation and then more detailed investigation before ‘causal’ genes and processes can be confirmed. rs4766697 tagging RBM19 achieved genome-wide significance. Little is known about this gene, but it may be implicated in regulation of ribosome biogenesis and is expressed in crypt cells of the intestinal epithelium. Despite being the largest IBD O-EIMs cohort investigated to date, it is important to stress that our study is very significantly underpowered and that the associations observed do not meet criteria for association after correction for multiple testing for known IBD loci, and only one locus achieved a genome-wide level of significance from the unbiased approach. Nevertheless these findings may provide the basis for future studies in expanded IBD cohorts with O-EIMs characterised.
Overall, our clinical and genetic data suggest that ocular manifestations are commonly associated with other EIMs, particularly those involving the skin and joints [ Figure 2 ]. Prior publications have suggested the possibility of a common antigen present in the eye, skin, joints, and gastrointestinal tract in these patients. 25 Our analysis examining genetic associations with the development of any EIMs, although not achieving genome-wide significance, are of interest given previous associations between CSMD1 and psoriasis and between MSH3 and rheumatoid arthritis. Additionally, IL12B variants have previously been associated with psoriasis, psoriatic arthritis, spondyloarthropathy, and multiple sclerosis as well as IBD. Furthermore, therapies targeted at this pathway are currently under evaluation in IBD.
Despite its size, this study has some limitations. Overall, O-EIMs occur so infrequently that our sample size may not be appropriately powered to show more subtle clinical and serological differences between those IBD subjects with and without O-EIMs. However, where data do exist, our demographic and clinical results are consistent with previous published studies. Because the databases in this study included multiple referral medical centres, and due to the retrospective nature of the study, we were limited in our ability to verify the specific ocular diagnosis. However, phenotyping of subjects from the NIDDK database has been previously validated. 18 At CSMC, experienced IBD physicians questioned patients regarding eye disease at every visit, according to standardised encounter forms. When necessary, we verified the ocular disease diagnosis of CSMC patients through an ophthalmology encounter.
In the largest study to date, we have identified several putative novel clinical and genetic parameters of O-EIMs in IBD that may help better identify those at risk in the future as well as potentially provide targets for future pharmacological intervention. Additional cohorts are needed to verify and extend our findings in order to achieve these aims.
Funding
NIDDK grants DK062431, DK62429, DK046763, and DK62422; and NIH grant DK062420. Additional resources were provided by: the F Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, Cedars-Sinai: the Leona M. and Harry B. Helmsley Charitable Trust; and the Harvey M. and Lyn P. Meyerhoff Inflammatory Bowel Disease Center. Funding sources had no role in the study design, collection or any analysis of the data, writing of the manuscript, or the decision to submit the manuscript.
Conflict of Interest
There is no conflict of interest.
Author Contributions
ST conceived the study, participated in its design, participated in data analysis, and drafted the manuscript. DL participated in data analysis and helped draft parts of the manuscript. SRT, AP, SB, JC, RD, JR, MS, EV, JR, DS, MD, and GM participated in collection of data and revised it critically for important intellectual content. TH participated in sample analysis and helped draft parts of the manuscript. DM conceived and supervised the study, participated in its design and coordination, participated in collection of data and data analysis, and helped draft the manuscript. All authors read and approved the final manuscript.
References
- 1. Jostins L, Ripke S, Weersma RK . Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease . Nature 2012. ; 491 : 119 – 24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franke A, McGovern DP, Barrett JC . Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci . Nat Genet 2010. ; 42 : 1118 – 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson CA, Boucher G, Lees CW . Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47 . Nat Genet 2011. ; 43 : 246 – 52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernstein CN, Blanchard JF, Rawsthorne P, Yu N . The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study . Am J Gastroenterol 2001. ; 96 : 1116 – 22 . [DOI] [PubMed] [Google Scholar]
- 5. Ricart E, Panaccione R, Loftus EV, Jr, et al. Autoimmune disorders and extraintestinal manifestations in first-degree familial and sporadic inflammatory bowel disease: a case-control study . Inflamm Bowel Dis 2004. ; 10 : 207 – 14 . [DOI] [PubMed] [Google Scholar]
- 6. Farhi D, Cosnes J, Zizi N, et al. Significance of erythema nodosum and pyoderma gangrenosum in inflammatory bowel diseases: a cohort study of 2402 patients . Medicine 2008. ; 87 : 281 – 93 . [DOI] [PubMed] [Google Scholar]
- 7. Manganelli C, Turco S, Balestrazzi E . Ophthalmological aspects of IBD . Eur Rev Med Pharmacol Sci 2009. ; 13 [ Suppl 1 ]: 11 – 3 . [PubMed] [Google Scholar]
- 8. Petrelli EA, McKinley M, Troncale FJ . Ocular manifestations of inflammatory bowel disease . Ann Ophthalmol 1982. ; 14 : 356 – 60 . [PubMed] [Google Scholar]
- 9. Lyons JL, Rosenbaum JT . Uveitis associated with inflammatory bowel disease compared with uveitis associated with spondyloarthropathy . Arch Ophthalmol 1997. ; 115 : 61 – 4 . [DOI] [PubMed] [Google Scholar]
- 10. Lin P, Tessler HH, Goldstein DA . Family history of inflammatory bowel disease in patients with idiopathic ocular inflammation . Am J Ophthalmol 2006. ; 141 : 1097 – 104 . [DOI] [PubMed] [Google Scholar]
- 11. Papp M, Altorjay I, Norman GL . Seroreactivity to microbial components in Crohn’s disease is associated with ileal involvement, noninflammatory disease behavior and NOD2/CARD15 genotype, but not with risk for surgery in a Hungarian cohort of IBD patients . Inflamm Bowel Dis 2007. ; 13 : 984 – 92 . [DOI] [PubMed] [Google Scholar]
- 12. Abbasian J, Martin TM, Patel S, Tessler HH, Goldstein DA . Immunologic and genetic markers in patients with idiopathic ocular inflammation and a family history of inflammatory bowel disease . Am J Ophthalmol 2012. ; 154 : 72 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orchard TR, Chua CN, Ahmad T, Cheng H, Welsh KI, Jewell DP . Uveitis and erythema nodosum in inflammatory bowel disease: clinical features and the role of HLA genes . Gastroenterology 2002. ; 123 : 714 – 8 . [DOI] [PubMed] [Google Scholar]
- 14. Cummings JR, Ahmad T, Geremia A . Contribution of the novel inflammatory bowel disease gene IL23R to disease susceptibility and phenotype . Inflamm Bowel Dis 2007. ; 13 : 1063 – 8 . [DOI] [PubMed] [Google Scholar]
- 15. Cummings JR, Cooney R, Pathan S . Confirmation of the role of ATG16L1 as a Crohn’s disease susceptibility gene . Inflamm Bowel Dis 2007. ; 13 : 941 – 6 . [DOI] [PubMed] [Google Scholar]
- 16. Dassopoulos T, Nguyen GC, Bitton A . Assessment of reliability and validity of IBD phenotyping within the National Institutes of Diabetes and Digestive and Kidney Diseases [NIDDK] IBD Genetics Consortium [IBDGC] . Inflamm Bowel Dis 2007. ; 13 : 975 – 83 . [DOI] [PubMed] [Google Scholar]
- 17. Weizman AV, Huang BL, Berel D . 26 Serologic and Genetic Profiles Suggest Distinct Immune Pathways Among Patients With Pyoderma Gangrenosum and Inflammatory Bowel Disease . Gastroenterology 2012. ; 142 : S – 7 . [Google Scholar]
- 18. McGovern DP, Gardet A, Torkvist L . Genome-wide association identifies multiple ulcerative colitis susceptibility loci . Nat Genet 2010. ; 42 : 332 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duerr RH, Taylor KD, Brant SR . A genome-wide association study identifies IL23R as an inflammatory bowel disease gene . Science 2006. ; 314 : 1461 – 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silverberg MS, Cho JH, Rioux JD . Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study . Nat Genet 2009. ; 41 : 216 – 20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vavricka SR, Brun L, Ballabeni P . Frequency and risk factors for extraintestinal manifestations in the Swiss inflammatory bowel disease cohort . Am J Gastroenterol 2011. ; 106 : 110 – 9 . [DOI] [PubMed] [Google Scholar]
- 22. Felekis T, Katsanos K, Kitsanou M, et al. Spectrum and frequency of ophthalmologic manifestations in patients with inflammatory bowel disease: a prospective single-center study . Inflamm Bowel Dis 2009. ; 15 : 29 – 34 . [DOI] [PubMed] [Google Scholar]
- 23. Yilmaz S, Aydemir E, Maden A, Unsal B . The prevalence of ocular involvement in patients with inflammatory bowel disease . Int J Colorect Dis 2007. ; 22 : 1027 – 30 . [DOI] [PubMed] [Google Scholar]
- 24. Al-Zawaideh F, Maayah J, Al-Madani M, AL-Ajlouny Y, Ba’ara B, Ghazzawi MD I . Ocular Manifestations among Jordanians with Inflammatory Bowel Disease. Hospital-Based Study in Asymptomatic Patients . Journal of the Royal Medical Services 2011. ; 18 : 17 – 21 . [Google Scholar]
- 25. Das KM . Relationship of extraintestinal involvements in inflammatory bowel disease: new insights into autoimmune pathogenesis . Dig Dis Sci 1999. ; 44 : 1 – 13 . [DOI] [PubMed] [Google Scholar]