Abstract
Plastids are critical organelles in plant cells that perform diverse functions and are central to many metabolic pathways. Beyond their major roles in primary metabolism, of which their role in photosynthesis is perhaps best known, plastids contribute to the biosynthesis of phytohormones and other secondary metabolites, store critical biomolecules, and sense a range of environmental stresses. Accordingly, plastid-derived signals coordinate a host of physiological and developmental processes, often by emitting signalling molecules that regulate the expression of nuclear genes. Several excellent recent reviews have provided broad perspectives on plastid signalling pathways. In this review, we will highlight recent advances in our understanding of chloroplast signalling pathways. Our discussion focuses on new discoveries illuminating how chloroplasts determine life and death decisions in cells and on studies elucidating tetrapyrrole biosynthesis signal transduction networks. We will also examine the role of a plastid RNA helicase, ISE2, in chloroplast signalling, and scrutinize intriguing results investigating the potential role of stromules in conducting signals from the chloroplast to other cellular locations.
Different cell, different plastid: signalling networks vary with cell and plastid type
Plastids are the organelle descendants of ancient prokaryotic endosymbionts that retain a fragment of their ancestral bacterial genomes. Approximately 3000 different proteins can be found in plastids, the vast majority of which (>95%) are encoded by the nuclear genome [1,2]. Plastid genomes have retained ~100 genes that encode parts of the machinery for plastid genome expression (i.e. ribosomal proteins, RNA polymerase subunits, rRNAs, and tRNAs), protein import from the cytosol, photosynthesis, and fatty acid biosynthesis. All plant cells contain plastids, which can differentiate into a variety of distinct types with concordant differences in plastid genome expression. Significant research efforts have focused on unravelling the complex signalling networks that coordinate nuclear and plastid genome expression and differentiation of plastid types [3–5].
Plastids are usually named for their colour or composition: green plastids are chloroplasts, translucent plastids are leucoplasts, colourfully pigmented plastids are chromoplasts, starch-storing plastids are amyloplasts, and others. This nomenclature can cause confusion, both because these names are often overlapping (e.g. amyloplasts are clear, and thus fall under the umbrella term ‘leucoplast’), and because there can be significant morphological and physiological variation within each plastid type. For example, leaves contain at least four distinct populations of plastids: (i) ground tissue chloroplasts, which are large, photosynthetic powerhouses that supply sugars to the entire plant, (ii) epidermal guard cell chloroplasts, which are smaller and primarily support only the guard cell, (iii) epidermal pavement cell leucoplasts, which are proposed to be degenerate chloroplasts and occasionally contain trace thylakoid structures and chlorophyll, and (iv) vascular leucoplasts that are small and relatively amorphous. Moreover, these plastid types can vary between species. In the ground tissue of plants with Kranz anatomy that supports C4 photosynthesis, e.g. maize, bundle sheath cells have chloroplasts with unstructured thylakoids and large starch granules, contrasted with mesophyll chloroplasts that are filled with thylakoid grana. As another example, some species, including tobacco and its relatives, have chloroplasts throughout the epidermis, including pavement cells, and thus do not have epidermal leucoplasts.
Plastid differentiation can be controlled by the nuclear genome. For example, ectopic expression of a GATA-type nuclear transcription factor, GNC, in the epidermis of Arabidopsis is sufficient to promote chloroplast identity instead of leucoplast identity in pavement cells [6]. Differences in plastid type can have key physiological consequences: epidermal pavement cell plastids in Arabidopsis are impervious to photosynthetic electron transport chain (pETC) inhibitor-induced reactive oxygen species (ROS), but epidermal pavement cell plastids in Nicotiana benthamiana are strongly oxidized under identical conditions [7]. These differences in plastid physiology can be harnessed as a sort of ‘natural variation’ to experimentally dissect signalling networks. An example of an experimentally useful difference is that during stress signalling, Nicotiana epidermal cells can all generate ROS from the pETC, but Arabidopsis epidermal pavement cells cannot. Most research on plastid signalling focuses exclusively on chloroplast signalling pathways, and among these chloroplast signalling pathways, the focus is placed on how chloroplasts can sense various environmental stresses and then promote stress responses.
Chloroplasts choosing between life and death: PAPs and EXECUTORs in oxidative stress
Plant cells undergo programmed cell death (PCD) due to developmental programs or in response to stress. Most famously, during the hypersensitive defence response in leaves, cells that recognize infection by pathogens initiate PCD to limit pathogenesis and thus form small lesions of dead cells. Lesion formation has long been known to be light-dependent, stimulating the hypothesis that chloroplasts contribute to PCD signalling. Genetic screens for mutants that produce spurious lesions (i.e. lesions in the absence of any stress or infection) in the light, but not in the dark, have been powerful tools for investigating chloroplast control of PCD. flu mutants accumulate excess protochlorophyllide (Pchlide) in the dark due to deregulation of tetrapyrrole biosynthesis [8,9]. Pchlide is a photosensitizing agent that triggers singlet oxygen (1O2) formation in the light. Subsequent studies demonstrated that when flu mutants are exposed to light at dawn the accumulated Pchlide causes 1O2 generation, which then initiates PCD in the absence of a standard environmental stress trigger [9]. A suppressor screen identified EXECUTER1 (EX1) and EX2, which both encode plastid proteins of unknown function, as necessary for transducing the flu 1O2 signal that causes PCD ([10]; Figure 1A). Despite intense investigation, little is known about the molecular functions of EX1/EX2 beyond this genetic evidence. A recent report demonstrated that EX1 is degraded in flu mutants, that this degradation is dependent on the FtsH2 protease, and that stabilization of EX1 in flu ftsh2 double mutants prevents PCD [11,12]. This is perhaps surprising, since depletion of EX1 in ex1 mutants and stabilization of EX1 in ftsh2 mutants both prevent EX1-dependent flu 1O2 PCD signalling. These results suggest that declining EX1 levels, or perhaps a specific FtsH2-dependent degradation product of EX1, are required for this signal transduction pathway.
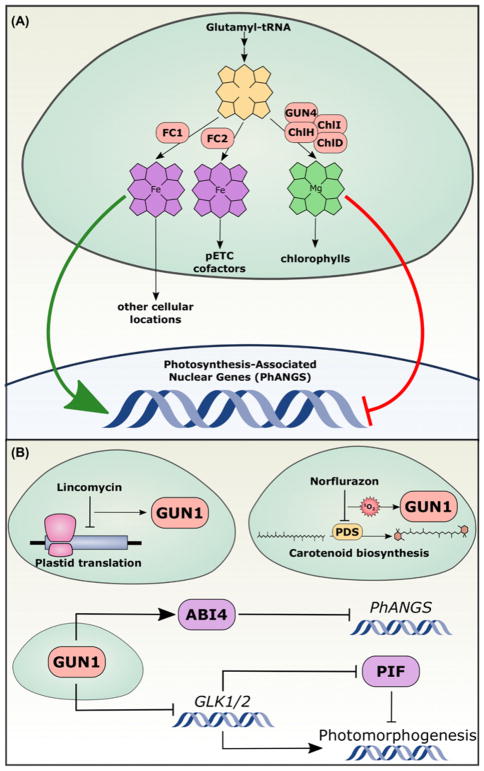
Figure 1. GENOMES UNCOUPLED (GUN) chloroplast proteins regulate the expression of nuclear genes.
(A) Tetrapyrroles are synthesized in the plastid from glutamyl-tRNA precursors. At a key branch point, PP IX is converted either into haem by the Fe chelatases or into Mg PP IX by the Mg chelatase complex (composed of ChlH = GUN5, ChlI, ChlD, and GUN4), which is then dedicated to chlorophyll biosynthesis. FC2-synthesized haem remains in the chloroplast, where it associates with pETC proteins. FC1 synthesizes haem for the rest of the cell. Nuclear genes that encode photosynthesis-associated proteins (PhANGs) are positively regulated by FC1-generated haem and/or negatively regulated by Mg PP IX accumulation. (B) GUN1 integrates diverse signals to control nuclear gene expression. Severe disruption of chloroplast function, such as inhibition of plastid translation or oxidative stress caused by inhibition of carotenoid biosynthesis, triggers a retrograde signalling pathway from the chloroplast to the nucleus that requires GUN1. GUN1 promotes the activity of ABI4, a transcription factor that repress PhANG expression, and represses transcription of GLK1/2, transcription factors that promote photomorphogenesis by antagonizing PIF transcription factors.
EX1/EX2 is not required for all chloroplast-triggered PCD pathways, however. ex1 ex2 double mutants cannot rescue the ROS-dependent necrotic phenotype of fc2 mutants (discussed at length below) nor the lesion mimic phenotype of mips1 (myoinositol phosphate synthase1) mutants. Like several other lesion mimic mutants, mips1 lesion formation is light- and chlorophyll-dependent [13], and can be suppressed in the dark or in mutant backgrounds defective in chlorophyll biosynthesis (including mutants defective in Mg chelatase activity, gun4 and gun5, and a mutant defective in divinyl protochlorophyllide 8-vinyl reductase activity, pcb2) [14]. A reverse genetic screen for suppressors of the mips1 lesion formation identified sal1/fiery1 as a strong suppressor of mips1 PCD [14]. SAL1 localizes to chloroplasts and mitochondria, where it dephosphorylates PAP (3′ phosphoadenosine 5′ phosphate) into AMP, but sal1 mutants hyperaccumulate PAP without dramatic effects on the levels of PAP precursors [15] (Figure 1C). SAL1 enzymatic activity is attenuated during oxidative stress by dimerization, by disruptive disulfide bridges that form within the SAL1 protein, and by glutathionylation, leading to accumulation of PAP in oxidative environments, e.g. chloroplasts during high light and drought stresses. PAP then moves from the chloroplast to the cytosol and nucleus, where it inhibits the activity of 5′ 3′ exoribonucleases (XRNs), raising levels of XRN-sensitive transcripts. Many of the XRN-sensitive transcripts are targets of microRNA-guided post-transcriptional cleavage, including a number of stress-related genes (Figure 1C). Through the suppression of gene silencing, and perhaps other transcripts that are sensitive to XRN activity, PAP causes increased expression of high light- and drought-inducible genes. Supporting this model, mips1 xrn2 xrn3 xrn4 quadruple mutants, which lack all Arabidopsis XRN enzymes (there is no Arabidopsis orthologue of eukaryotic XRN1), also suppresses the mips1 light-dependent cell death phenotype [14]. Thus, loss of XRN activity by either PAP inhibition in the sal1 background or genetic mutation prevents PCD in the mips1 background.
Mutual decisions: GUN1 coordinates genomes
When chloroplast function is disrupted, the expression of specific nuclear genes changes. This chloroplast-to-nucleus signalling is commonly called ‘chloroplast retrograde signalling’, and several of these signal transduction pathways have been characterized [2,16–20]. In a classic forward genetic screen to uncover mechanisms of chloroplast-to-nucleus signalling, a mutagenized population of Arabidopsis thaliana was treated with norflurazon, an inhibitor of carotenoid biosynthesis that consequently causes strong free radical photo-oxidation [21]. In wild-type plants, norflurazon treatment represses expression of both the chloroplast genome and a set of nuclear genes that encode proteins involved in photosynthesis (often called ‘photosynthesis-associated nuclear genes’, or PhANGs). Several mutants were identified that repress chloroplast genome expression after treatment with norflurazon, as expected, but do not fully inhibit PhANG expression. Since these mutants are defective in the coordinated expression of the chloroplast and nuclear genomes, they were named genomes uncoupled (gun) mutants.
The gun mutants can be divided into two classes: gun1 encodes a pentatricopeptide repeat protein (PPR) [22] and gun2 through gun6 encode proteins involved with tetrapyrrole biosynthesis ([23–26], Figure 2 ). The GUN1-dependent chloroplast-to-nucleus signalling pathway is at least partly distinct from the tetrapyrrole biosynthesis (GUN2/3/4/5/6)-dependent pathway [23,27]. For example, lincomycin specifically inhibits the prokaryotic-type plastid ribosomes, effectively inhibiting plastid translation, and when treated with lincomycin, both wild-type plants and gun2/3/4/5/6 mutants repress PhANG expression, but gun1 mutants do not [28,29]. This, and similar findings, led to the current model that GUN1 integrates a number of chloroplast signals to control downstream nuclear gene expression (reviewed in [30]). The molecular activity of GUN1 remains unresolved, however. Initially, GUN1 was suspected to promote RNA processing because almost all PPR proteins in the chloroplast are involved in highly specific recognition of nucleic acid sequences to guide RNA processing [31–33]. Curiously, GUN1 has not been found to associate with any nucleic acids [34]. While many molecular functions for GUN1 have been proposed, the current predominant hypothesis is that GUN1 interacts with other chloroplast proteins to control their stability or activity. Indeed genetic interactions between gun1 and mutants involved in organelle gene expression including mterf4/bsm/rug2 [35] and prors1 [34] have been reported, and GUN1 was found to interact with proteins with roles in chloroplast translation and protein homoeostasis including several plastid ribosomal proteins [34].
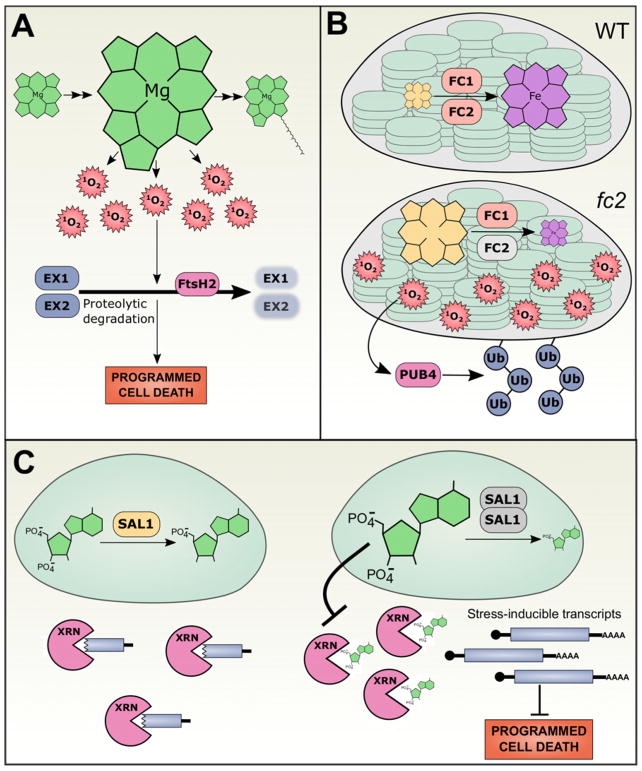
Figure 2. Chloroplast signaling networks regulate stress responses and programmed cell death.
(A) flu mutants are unable to regulate tetrapyrrole biosynthesis, leading to overaccumulation of Pchlide in the dark. In the light, Pchlide generates 1O2, which then promotes the FtsH2-dependent proteolytic degradation of EX1 and EX2. This degradation is sensed through unresolved mechanisms that trigger programmed cell death. (B) In wild-type chloroplasts, PP IX is chelated with iron by FC1 and FC2, yielding haem (top). In mutants without FC2 (grey, bottom), PP IX accumulates. PP IX generates singlet oxygen in the light, which activates PUB4-mediated polyubiquitination and degradation of the chloroplast. This pathway is invoked at low frequency in wild-type plants as a quality control measure to degrade photo-damaged chloroplasts. (C) In wild-type plants, monomeric SAL1 dephosphorylates PAP to yield AMP (left). Under oxidative stress, such as high light, SAL1 undergoes inactivating conformational changes, including dimerization, that subsequently raise PAP levels. PAP leaves the chloroplast and interferes with XRN activity, likely due to its structural similarity to the 5′ ends of uncapped RNA. Stress-inducible transcripts are stabilized by PAP-mediated inactivation of XRNs, increasing plant stress tolerance and suppressing programmed cell death.
The signal transduction pathway downstream from GUN1 is currently under investigation. Previous models suggested that a chloroplast membrane-anchored transcription factor called PTM acted downstream of GUN1 to control the activity of the nuclear transcription factor, ABI4 [36]. However, thorough efforts by multiple independent labs to reproduce the experiments arguing for a role of PTM in chloroplast-to-nucleus signalling have demonstrated that PTM is most likely not involved [37]. At least two major transcription factors are probably involved in mediating nuclear responses to the GUN1-dependent signal(s) (Figure 2B): ABI4, which is in the family of ERF/AP2 transcription factor family and represses the expression of PhANGs [22], and the GOLDEN 2-LIKE1 (GLK1) and GLK2 Myb transcription factors that promote PhANG expression [38]. abi4 mutants show a genomes uncoupled phenotype, as they do not fully repress PhANG genes after treatment with lincomycin or norflurazon (similar to gun1 mutants), genetically placing ABI4 in the genomes uncoupled pathway [22].
A recent study illustrates the relevance of the GUN1-controlled transcriptional networks to plant development and the complexity of the GUN1 signalling network. When dark-grown seedlings are transferred to the light in the presence of lincomycin, they remain etiolated, with pale yellow and unopened embryonic leaves (cotyledons). Under normal conditions, loss of the light-sensitive PHYTOCHROME-INTERACTING FACTOR (PIF) transcription factors that promote the etiolation developmental program (skotomorphogenesis) allows cotyledons to open even if seedlings are grown in complete darkness; lincomycin treatment, however, prevents cotyledon opening in pif mutants [39]. This sensitivity to lincomycin treatment is abrogated in gun1 mutants. In addition, the strong repression of GLK1 expression seen in wild-type plants treated with lincomycin is completely reversed in gun1 mutants. Moreover, overexpression of GLK1 is sufficient to confer lincomycin insensitivity during de-etiolation. Thus, GUN1 acts through unknown mechanisms to promote GLK1 expression in the nucleus, which then promotes photomorphogenesis and antagonizes the PIF-mediated skotomorphogenetic program (Figure 2B). Supporting previous studies arguing that GUN1 integrates distinct upstream signals, this process is independent of the tetrapyrrole gun2/3/4/5/6 pathway, since gun5 mutants do not rescue lincomycin sensitivity during de-etiolation. Moreover, abi4 mutants are also sensitive to lincomycin at this stage, suggesting that the signalling network downstream from GUN1 diverges, and that ABI4 is not involved in the GUN1-promoted repression of GLK1/2 during lincomycin-induced stress [39].
Tetrapyrrole biosynthesis in chloroplast signalling: new insights
Genetically disrupting tetrapyrrole biosynthesis affects the accumulation of PhANGs after norflurazon treatment [40], but the precise molecular pathway between tetrapyrrole biosynthesis and nuclear gene expression remains unresolved, and has been the topic of several recent reviews [20,1–44]. Broadly, the gun2/3/4/5/6 mutants all affect a key branch point in tetrapyrrole biosynthesis: protoporphyrin IX (PPIX) is chelated either with iron (by FER-ROCHELATASE 1, FC1, or by FC2) or with magnesium (by an enzyme with three subunits, called ChlD, ChlH, and ChlI), yielding haem or Mg-PPIX, respectively. Once chelated with iron, the tetrapyrroles may remain haems or be further processed to generate phytochromobilin, the light-sensitive cofactor of phytochromes. If instead chelated with magnesium, the tetrapyrroles are further modified to become chlorophylls. Mutants that prevent chelation with magnesium (gun4/5), that promote FC1-mediated chelation with iron (gun6-1D), or that prevent the conversion of haems into phytochromobilin (gun2/3) can all cause genomes uncoupled phenotypes [45]. This has led to the proposals that Mg-PPIX could act as a negative regulator of PhANG expression [25], that haem could act as a positive regulator of PhANG expression, or that both molecules could participate [45]. Alternatively, differential accumulation of these molecules or flux through these enzymatic pathways might act through as-yet unknown secondary messengers to affect nuclear gene expression [41].
Surprisingly, whereas overexpression of FC1 is sufficient to derepress PhANG expression in plants treated with norflurazon, overexpressing FC2 does not have the same effect [45]. This result suggested that, although the two enzymes perform the same biochemical function, they somehow generate biologically distinct pools of haem (Figure 2A). Both FC1 and FC2 are deeply conserved, implying their divergent functions. fc1 mutants are embryo-defective, typically arresting at very early stages of embryogenesis (our observations and [46]) while fc2 mutants are chlorotic but viable [45,47]. Importantly, overexpression of one of the ferrochelatases cannot complement for loss of the other. These results further support the hypothesis that FC1 and FC2 perform non-redundant functions. The inability to isolate homozygous fc1 null alleles has hindered progress on distinguishing the roles of these two enzymes, but studies with weak alleles of fc1 and both null and weak alleles of fc2 indicate that FC1 generates haem cofactors for proteins throughout the cell, whereas FC2 specifically generates haems involved in photosynthesis, such as the haem incorporated in the cytochrome b6f complex of the photosynthetic electron transport chain [46]. This finding is especially appealing because FC2 is primarily expressed in photosynthetic tissues, FC2 has a conserved hydrophobic C-terminal extension that is related to the light harvesting complex (LHC) motif that binds chlorophyll [48], and FC1 (but not FC2) generates the putative chloroplast-to-nucleus haem signal that promotes PhANG expression under normal growing conditions [45].
Recently, a distinct tetrapyrrole chloroplast signalling pathway was identified based on physiological studies of the fc2 mutant [49]. In contrast with wild-type plants, fc2 mutant seedlings are unable to de-etiolate, and exposing etiolated fc2 seedlings to light causes photo-oxidative stress, eventually leading to cell death. fc2 mutants are also unable to green under short day photoperiods (4 or 8 h light/day) due to widespread chloroplast degradation, but are nearly wild-type under longer day photoperiods. The fc2 ex1 double mutant does not rescue these phenotypes, demonstrating that this pathway is distinct from the EX1-dependent 1O2 signalling pathway. Instead, tetrapyrrole profiling suggests that fc2 specifically over-accumulates the tetrapyrrole protoporphyrin IX (PP IX) by an order of magnitude. PP IX causes generation of 1O2 in the light, and indeed, the 1O2 scavenger vitamin B6 rescued the fc2 seedling phenotypes. Moreover, genetic disruption of enzymes required to generate tetrapyrrole precursors also rescued the fc2 phenotype. These results support a model where FC2 is required to limit PP IX accumulation under specific light conditions, and in the absence of FC2, PP IX-generated 1O2 triggers chloroplast degradation. While the pathway downstream from 1O2 generation remains unknown, one recently proposed candidate is β-cyclocitral (β-CC), which can be a by-product of carotenoid exposure to 1O2, and which can induce transcriptional changes that are remarkably similar to the transcriptional changes induced by singlet oxygen [50]. Direct demonstration that β-CC accumulates in response to 1O2 in the chloroplast (e.g. in the fc2 mutants described here), and that β-CC is required for the activation of 1O2 downstream responses, are still lacking, however, and will be needed before this molecule can be considered a bona fide retrograde signal.
In an effort to uncover the molecular mechanisms downstream of fc2 signalling, a suppressor screen revealed that fc2-triggered chloroplast degradation is dependent on an ubiquitin E3 ligase, PLANT U-BOX 4 (PUB4). The screen identified several more suppressive mutations, including loss-of-function alleles of the chloroplast protein import machinery (TOC33 and TOC159), which probably broadly disrupt chloroplast biogenesis and signalling. Unexpectedly, however, recessive gun5 alleles also suppressed chloroplast degradation in fc2 mutants. GUN5 is the catalytic ChlH subunit of the magnesium chelatase that converts PP IX to Mg-PPIX [23]; under constant light conditions, mutants with the weak gun5-1 allele can accumulate nearly twice as much PP IX as can wild-type plants. Further characterization of these fc2 gun5 double mutants will clarify whether PP IX over-accumulation is sufficient to trigger PUB4-dependent chloroplast degradation, or, if PP IX also over-accumulates in this double mutant, a more complex model is required. A recent biochemical study demonstrated that, when GUN5 is oxidatively damaged (as is the case in fc2 mutants), GUN5 and PP IX synergistically interact to produce 10-fold more singlet oxygen than PP IX can on its own [51]. Thus, one appealing hypothesis is that, while the fc2 gun5 mutant might accumulate somewhat higher PP IX levels, these PP IX molecules are less photosensitizing in the double mutant, and thus do not trigger the light-dependent necrosis observed in fc2 single mutants. This hypothesis is further supported by the recent observation that gun5 can suppress expression of 1O2-responsive genes after treatment of dark-grown seedlings with either far-red light followed by white light or norflurazon in white light, and indeed that gun5 mutants do not accumulate high levels of 1O2 under these inductive conditions, both of which suggest that GUN5 is required to generate sufficient 1O2 to trigger downstream signalling events in fc2 mutants [52].
ISE2: elucidating the chloroplast-to-plasmodesmata connection
Plasmodesmata (PD) are narrow, membrane-bound pores in plant cell walls that connect the cytosol of adjacent cells, and permit molecules ranging from ions to proteins (typically up to ~80 kDa) to move from cell to cell [53,54]. Trafficking of molecules through PD is regulated both developmentally (e.g. in wild-type plants, PD transport rapidly decreases at the mid-torpedo stage of embryogenesis) and physiologically (e.g. PD trafficking decreases in response to cold stress). The Arabidopsis ise2 mutant was first described as defective in restricting plasmodesmatal (PD) transport at the mid-torpedo stage of embryogenesis [55]. In addition to increasing PD trafficking during embryogenesis, ISE2 also controls PD trafficking in adult leaves: silencing ISE2 increases PD trafficking and stimulates biogenesis of new PD [56], while overexpressing ISE2 decreases PD trafficking [57] (Figure 3 ). Unexpectedly, ISE2 was revealed to encode a conserved DEVH-box RNA helicase that is required for embryogenesis in Arabidopsis [58]. The organelle-to-PD signalling pathway (dubbed ‘ONPS’ [59]) that connects this plastid RNA helicase with PD transport remains unresolved, but several groups are investigating the mechanism of ISE2 in plastid RNA metabolism and the possible connections between ISE2 and PD transport.
Figure 3. Roles of ISE2 in chloroplast metabolism and signaling.
ISE2 is a plastid RNA helicase that contributes to many processes related to RNA metabolism within the chloroplast (pink boxes), and affects diverse processes outside of the chloroplast (blue boxes).
The first clue to the role of ISE2 in controlling PD transport came from a transcriptomic study of ise2 and ise1 mutants. ISE1 is a conserved mitochondrial RNA helicase that is also essential for embryogenesis [60]. Unexpectedly, the transcriptomes of both ise1 and ise2 are remarkably similar: nearly half of the ~3000 differentially expressed genes (DEGs) in ise1 are also differentially expressed in ise2, and of those DEGs, 93% are similarly affected (up- or down-regulated in both transcriptomes) [59]. These similarities are not simply due to embryo-lethality, because several transcriptomes of other embryo-lethal mutants at comparable developmental stages show no significant overlap with the ise1 or ise2 transcriptomes. Within this dataset of >1000 similarly regulated genes, the most striking pattern is broad repression of PhANG expression, with consistent down-regulation of genes encoding components of the photosynthetic light harvesting complexes and pETC, as well as genes encoding enzymes in the tetrapyrrole biosynthesis pathway. Thus, mitochondrial dysfunction can trigger similar changes in nuclear gene expression as plastid dysfunction, implying that either both signalling pathways converge or loss of ISE1 disrupts chloroplast functions, triggering the ise2 chloroplast-to-nucleus signalling pathway.
A complementary genetic screen for mutants defective in PD transport isolated a thioredoxin m3 mutant [61], which led to the discovery that plastid oxidative stress can also decrease PD trafficking. Mitochondrial oxidative stress, on the other hand, increases PD trafficking [62]. To test whether ISE1 and ISE2 could trigger organelle redox signalling pathways to control PD transport, a redox-sensitive GFP probe that senses the redox status of glutathione pools was targeted to plastids, mitochondria, or the cytosol in plants silencing ISE1 or ISE2 and compared with controls [62]. Silencing either RNA helicase caused a reductive shift in chloroplasts and increased PD transport, which is consonant with the finding that an oxidative shift in chloroplasts decreases PD transport. In agreement with previous results, silencing ISE1 also caused an oxidative shift in mitochondria, but silencing ISE2 did not affect the redox status of mitochondria. Importantly, loss of ISE2 also caused a significant reductive shift in the cytosolic redox status, but silencing ISE1 had no effect.
The molecular functions of ISE2 in plastid RNA metabolism have recently been illuminated. ISE2–GFP fusion proteins localize to punctae within the plastid stroma, which is consistent with its previous identification in plastid nucleoids [63]. It is now clear that ISE2 has multiple roles in plastid RNA processing (Figure 3): (i) ISE2 promotes splicing of some, but not all, group II introns [64,65], (ii) ISE2 is required for C-to-U post-transcriptional RNA editing, (iii) ISE2 controls the steady-state levels of dozens of plastid transcripts, broadly promoting accumulation of transcripts encoding pETC components but repressing accumulation of several transcripts encoding ribosomal proteins and RNA polymerase, (iv) ISE2 is similarly required for accumulation of plastid-encoded pETC proteins, and (v) ISE2 is necessary for ribosomal RNA processing and accumulation [65]. In RNA immunoprecipitation experiments, ISE2 was shown to interact with over half of plastid RNA species, including nearly all of the transcripts that require ISE2 for proper splicing and C-to-U editing, suggesting that ISE2 acts directly on a number of RNA species (in concert with other proteins) to regulate plastid RNA metabolism. Moreover, the strong effect of ISE2 on ribosomal RNA processing and accumulation probably causes extensive defects in plastid translation, triggering the canonical chloroplast-to-nucleus retrograde signalling pathways. Genetic experiments will reveal whether ise2 and the genomes uncoupled pathways are epistatic and contribute to the chloroplast-to-PD signalling pathway, in addition to the putative role of redox signalling in ise1 and ise2 mutants.
Beyond its roles in chloroplast RNA metabolism and regulation of PD transport, ISE2 also participates in disease resistance (Figure 3). Eighteen hours after infection with Tobacco mosaic virus (TMV, a tobamovirus) or Turnip mosaic virus (TuMV, a potyvirus), ISE2 transcripts are induced by as much as 30-fold compared with uninfected plants [57]. This led to the hypothesis that ISE2 induction could affect the progress of viral infections. Surprisingly, silencing or overexpressing ISE2 increases the susceptibility of Nicotiana benthamiana to TMV and TuMV. Overexpressing ISE2 in Arabidopsis thaliana similarly increased susceptibility to nematode infection. In contrast, altered expression of ISE2 had neither effect on growth of Pseudomonas syringae DC3000 nor any apparent effect on defence responses in the N. benthamiana host in the absence of pathogen infection. Further studies will be needed to clarify how modulating ISE2 expression affects disease resistance, and whether other plastid signalling pathways, such as the GUN1 or SAL1/PAP pathways, are involved.
Stromules: reaching out to communicate?
All plastids can generate narrow, membrane-bound, stroma-filled tubular extensions from the main plastid body called ‘stromules’ (Figure 4 ). Initially discovered in cytological studies over five decades ago [66], stromules were rediscovered when illuminated by studies using stroma-targeted GFP. These enigmatic structures are ubiquitous across cell types and species [67], but no clear function has yet been assigned to stromules. Stromule morphology and frequency are tightly regulated with developmental stages, physiological conditions [67–69], and cellular contexts, indicating that stromules perform conserved roles in plastid biology. Since their function(s) remain unknown, the term ‘stromule’ is used broadly to describe a variety of structures that may have different functions, including de novo extensions from the plastid surface, tubular connections that link recently divided or actively dividing plastids, and tubular extensions that ‘shed’ globular, stroma-filled vesicles (reviewed in [70]).
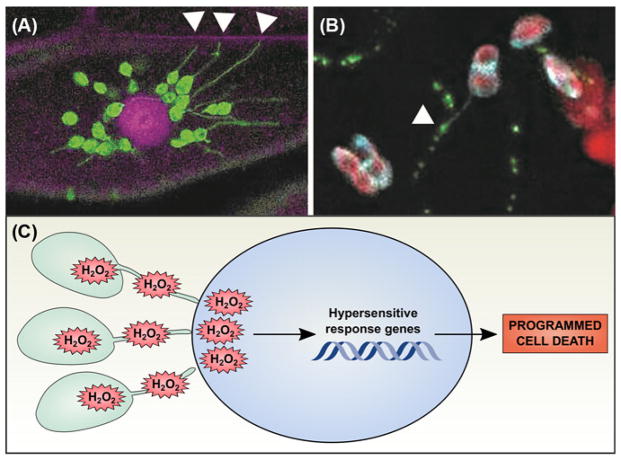
Figure 4. Stromules may participate in chloroplast signaling pathways.
(A) Chloroplast stromal GFP (green) reveals the presence of stromules, stroma-filled tubular extensions from the main body of the chloroplast. Here, chloroplasts are surrounding the nucleus (stained with propidium iodide, magenta) and extending stromules to the cell wall (also stained with propidium iodide, magenta; stromules associated with the cell wall indicated with white arrowheads). (B) Stromules are occasionally observed in close physical association with plasmodesmata. Tobacco mosaic virus movement protein P30 was fluorescently tagged with GFP (green) and transiently expressed in a transgenic N. benthamiana line expressing a chloroplast stromal Cerulean marker (cyan); chlorophyll autofluorescence is also shown (red). An example of a stromule extending from a chloroplast to associate with a PD (marked with P30-GFP) is indicated with a white arrowhead. Images were obtained by the authors with a Zeiss LSM 710 confocal scanning laser microscope. (C) During the hypersensitive response, chloroplasts generate large quantities of H2O2 that can travel through stromules to be released in the nucleus. High levels of H2O2 then promote the hypersensitive response genetic program, leading to programmed cell death.
Stromules have frequently been observed extending from chloroplasts toward other cellular locations, including the nucleus, endoplasmic reticulum, and cell wall, prompting speculation that stromules could serve as conduits for targeted signal transduction ([70,71], Figure 4A). Three recent discoveries support this hypothesis. First, chloroplasts extracted from cells and in simple buffers are able to generate stromules de novo. This suggests both that stromule formation does not necessarily rely on an external structure in the cytoplasm, and that stromules could initiate in response to signals within the chloroplast [7,72]. Second, when leaves are treated with photosynthesis inhibitors that specifically trigger ROS formation within the chloroplast, stromule frequency dramatically increases. Silencing expression of the plastid redox-signalling hub, NADPH THIOREDOXIN REDUCTASE C (NTRC), causes oxidative stress and markedly increases stromule formation, further demonstrating that redox signalling within the chloroplast controls stromule frequency [7]. A further line of evidence comes from silencing CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) [73]; in chup1 leaves, the chloroplasts are no longer able to move away from light to prevent oxidative stress, and stromule frequency increases. Thus, a likely explanation for the silenced chup1 stromule phenotype is that excess ROS are generated by the pETC, triggering stromule formation. Third, proteins have been observed moving from chloroplasts to the nucleus, and H2O2-sensitive fluorophores show that H2O2 concentrations within the nucleus are highest near stromule–nuclear contact points during chloroplast oxidative stress [73], implying that oxidative signals (perhaps H2O2 per se) move from the chloroplast to the nucleus via stromules (Figure 4C). Moreover, overexpression of cytosolic ascorbate peroxidase, which scavenges H2O2, does not have a strong impact on transfer of ROS from the chloroplast to the nucleus, suggesting that the ROS move directly from the chloroplast into the nucleus [74]. While these findings certainly do not exclude other hypotheses for stromule function, they strongly support the possibility that stromules participate in chloroplast signalling pathways.
Chloroplasts are known to associate with PD under certain circumstances to facilitate intercellular transport of metabolites, particularly in C4 plants with Kranz anatomy, stimulating speculation that stromules might also associate with PD under very specific conditions. We, and others, have observed that stromules sometimes extend from chloroplasts to the cell wall (Figure 4B), prompting us to test whether stromules could participate in chloroplast-to-plasmodesmata signalling triggered by loss of ISE2 or oxidative stress. Although we sometimes observed stromules co-localizing with PD markers, silencing ISE2 did not have any clear effect on stromule frequency or localization, and under conditions tested, chloroplast oxidative stress did not strongly increase the frequency of stromule/PD co-localization. Further studies taking advantage of the growing toolbox of fluorophore-tagged plastids across diverse plant species will be needed to determine whether stromules regulate PD-mediated cell–cell signalling and transport.
Conclusions
Plastids employ distinct signals and regulatory pathways to mediate responses to a variety of developmental and environmental cues. Not all of these pathways could be covered here, but several recent, comprehensive reviews have summarized some of the pathways we did not address [17,19,20,75]. Most research, up to now, has focused on chloroplast-generated signals and their pathways; it is likely that other plastids have their own, unique signal transduction networks that are yet to be identified and characterized. A particularly intriguing finding about chloroplast signalling is the existence and differential functioning of two haem populations; this finding suggests that plastid metabolites may sub-functionalize, and thereby increase biochemical and signalling pathway diversity. While the role of plastid gene expression in plastid signalling had long been established, new GUN1 and ISE2 findings highlight gaps in our understanding of the pathways that transduce signals from the plastid to the nucleus. Stromules were first observed over 50 years ago, and while there is copious evidence to show that they respond to changes in the cellular environment, we are only beginning to elucidate their possible roles in plastid signalling. Experiments that definitively demonstrate the function of stromule contacts with other cell compartments are eagerly awaited. Indeed, it is clear that many aspects of plastid signalling remain to be unravelled. The range of recent findings in this field herald coming years with many exciting new discoveries that will contribute to better understanding of the fundamentals of the signalling networks central to plant metabolism. As climate change progresses, we will need this understanding to predict plant responses to changes in the environment, and further, to develop plant varieties able to weather the change.
Summary.
Multiple plastid-to-nucleus retrograde signalling pathways regulate PCD.
GUN1 participates in several pathways to control nuclear gene expression.
There are many distinct pathways for tetrapyrrole-mediated retrograde signalling.
ISE2, a chloroplast RNA helicase involved in plastid gene expression, probably acts in a chloroplast-to-nucleus signalling pathway to control the flux of metabolites through plasmodesmata.
Stromules are potential routes for plastid signalling to other organelles.
Acknowledgments
Funding
J.O.B. is supported by NIH grant 5-DP5-OD023072-02. T.M.B.S. is supported by NSF award #1456761. We thank Prof. Patricia C. Zambryski and Dr Anne M. Runkel for contributions to the stromule/plasmodesmata co-localization experiment, Prof. S.P. Dinesh-Kumar for the N. benthamiana NRIP1-Cerulean line, and Dr Steve Ruzin and Dr Denise Schichnes for microscopy assistance. Research reported in this publication was supported in part by the National Institutes of Health S10 program under award number 1S10RR026866-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank Dr Claire Bendix for critical review of the manuscript.
Abbreviations
- β-CC
β-cyclocitral
- EX
EXECUTER
- FC
FERROCHELATASE
- GLK
GOLDEN 2-LIKE
- gun
genomes uncoupled
- ISE2
INCREASED SIZE EXCLUSION LIMIT2
- PAP
3′ phosphoadenosine 5′ phosphate
- PCD
programmed cell death
- Pchlide
protochlorophyllide
- PD
plasmodesmata (single, plasmodesma)
- pETC
photosynthetic electron transport chain
- PhANG
photosynthesis associated nuclear gene
- PP IX
protoporphyrin IX
- ROS
reactive oxygen species
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Leister D. Towards understanding the evolution and functional diversification of DNA-containing plant organelles. F1000 Res. 2016;5:330. doi: 10.12688/f1000research.7915.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiller N, Bock R. The translational apparatus of plastids and its role in plant development. Mol Plant. 2014;7:1105–1120. doi: 10.1093/mp/ssu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Dingenen J, Blomme J, Gonzalez N, Inze D. Plants grow with a little help from their organelle friends. J Exp Bot. 2016;67:6267–6281. doi: 10.1093/jxb/erw399. [DOI] [PubMed] [Google Scholar]
- 4.Greiner S, Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays. 2013;35:354–365. doi: 10.1002/bies.201200137. [DOI] [PubMed] [Google Scholar]
- 5.Rolland N, Curien G, Finazzi G, Kuntz M, Marechal E, Matringe M, et al. The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu Rev Genet. 2012;46:233–264. doi: 10.1146/annurev-genet-110410-132544. [DOI] [PubMed] [Google Scholar]
- 6.Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160:332–348. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunkard JO, Runkel AM, Zambryski PC. Chloroplasts extend stromules independently and in response to internal redox signals. Proc Natl Acad Sci USA. 2015;112:10044–10049. doi: 10.1073/pnas.1511570112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Kim C, Xu X, Piskurewicz U, Dogra V, Singh S, et al. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc Natl Acad Sci USA. 2016;113:E3792–E3800. doi: 10.1073/pnas.1603562113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dogra V, Duan J, Lee KP, Lv S, Liu R, Kim C. FtsH2-dependent proteolysis of EXECUTER1 is essential in mediating singlet oxygen-triggered retrograde signaling in Arabidopsis thaliana. Front Plant Sci. 2017;8:1145. doi: 10.3389/fpls.2017.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, et al. Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS One. 2009;4:e7364. doi: 10.1371/journal.pone.0007364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruggeman Q, Mazubert C, Prunier F, Lugan R, Chan KX, Phua SY, et al. Chloroplast activity and 3′phosphadenosine 5′phosphate signaling regulate programmed cell death in Arabidopsis. Plant Physiol. 2016;170:1745–1756. doi: 10.1104/pp.15.01872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foyer CH, Karpinska B, Krupinska K. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: a hypothesis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130226. doi: 10.1098/rstb.2013.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobik K, Burch-Smith TM. Chloroplast signaling within, between and beyond cells. Front Plant Sci. 2015;6:781. doi: 10.3389/fpls.2015.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gollan PJ, Tikkanen M, Aro EM. Photosynthetic light reactions: integral to chloroplast retrograde signalling. Curr Opin Plant Biol. 2015;27:180–191. doi: 10.1016/j.pbi.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Kleine T, Leister D. Retrograde signaling: organelles go networking. Biochim Biophys Acta. 2016;1857:1313–1325. doi: 10.1016/j.bbabio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 21.Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 22.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. doi: 10.1126/science.%201140516. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- 25.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 26.Chi W, Sun X, Zhang L. Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol. 2013;64:559–582. doi: 10.1146/annurev-arplant-050312-120147. [DOI] [PubMed] [Google Scholar]
- 27.Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, et al. Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 2000;24:883–894. doi: 10.1046/j.1365-313x.2000.00936.x. [DOI] [PubMed] [Google Scholar]
- 28.Gray JC, Sullivan JA, Wang JH, Jerome CA, MacLean D. Coordination of plastid and nuclear gene expression. Philos Trans R Soc Lond B Biol Sci. 2003;358:135–144. doi: 10.1098/rstb.2002.1180. discussion 144-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormac AC, Terry MJ. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 2004;40:672–685. doi: 10.1111/j.1365-313X.2004.02243.x. [DOI] [PubMed] [Google Scholar]
- 30.Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P. GUN1, a Jack-Of-All-Trades in chloroplast protein homeostasis and signaling. Front Plant Sci. 2016;7:1427. doi: 10.3389/fpls.2016.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 33.Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016;85:532–547. doi: 10.1111/tpj.13121. [DOI] [PubMed] [Google Scholar]
- 34.Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, et al. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016;170:1817–1830. doi: 10.1104/pp.15.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Xu D, Liu Z, Kleine T, Leister D. Functional relationship between mTERF4 and GUN1 in retrograde signaling. J Exp Bot. 2016;67:3909–3924. doi: 10.1093/jxb/erv525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, et al. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun. 2011;2:477. doi: 10.1038/ncomms1486. [DOI] [PubMed] [Google Scholar]
- 37.Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ. Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol. 2017;174:21–26. doi: 10.1104/pp.16.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun. 2016;7:11431. doi: 10.1038/ncomms11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voigt C, Oster U, Bornke F, Jahns P, Dietz KJ, Leister D, et al. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol Plant. 2010;138:503–519. doi: 10.1111/j.1399-3054.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 41.Terry MJ, Smith AG. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci. 2013;4:14. doi: 10.3389/fpls.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brzezowski P, Richter AS, Grimm B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta. 2015;1847:968–985. doi: 10.1016/j.bbabio.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Chi W, Feng P, Ma J, Zhang L. Metabolites and chloroplast retrograde signaling. Curr Opin Plant Biol. 2015;25:32–38. doi: 10.1016/j.pbi.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Larkin RM. Tetrapyrrole signaling in plants. Front Plant Sci. 2016;7:1586. doi: 10.3389/fpls.2016.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinas NA, Kobayashi K, Sato Y, Mochizuki N, Takahashi K, Tanaka R, et al. Allocation of heme is differentially regulated by ferrochelatase isoforms in Arabidopsis cells. Front Plant Sci. 2016;7:1326. doi: 10.3389/fpls.2016.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scharfenberg M, Mittermayr L, VON Roepenack-Lahaye E, Schlicke H, Grimm B, Leister D, et al. Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 2015;38:280–298. doi: 10.1111/pce.12248. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki T, Masuda T, Singh DP, Tan FC, Tsuchiya T, Shimada H, et al. Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: their difference in phylogeny, gene expression, and localization. J Biol Chem. 2002;277:4731–4737. doi: 10.1074/jbc.M105613200. [DOI] [PubMed] [Google Scholar]
- 49.Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salome PA, Weigel D, et al. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science. 2015;350:450–454. doi: 10.1126/science.aac7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarahi Tabrizi S, Sawicki A, Zhou S, Luo M, Willows RD. GUN4-protoporphyrin IX is a singlet oxygen generator with consequences for plastid retrograde signaling. J Biol Chem. 2016;291:8978–8984. doi: 10.1074/jbc.C116.719989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page MT, McCormac AC, Smith AG, Terry MJ. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 2017;213:1168–1180. doi: 10.1111/nph.14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brunkard JO, Zambryski PC. Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Curr Opin Plant Biol. 2017;35:76–83. doi: 10.1016/j.pbi.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 54.Paultre DS, Gustin MP, Molnar A, Oparka KJ. Lost in transit: long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell. 2016 doi: 10.1105/tpc.16.00249. [DOI] [PMC free article] [PubMed]
- 55.Kim I, Hempel FD, Sha K, Pfluger J, Zambryski PC. Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development. 2002;129:1261–1272. doi: 10.1242/dev.129.5.1261. [DOI] [PubMed] [Google Scholar]
- 56.Burch-Smith TM, Zambryski PC. Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr Biol. 2010;20:989–993. doi: 10.1016/j.cub.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganusova EE, Rice JH, Carlew TS, Patel A, Perrodin-Njoku E, Hewezi T, et al. Altered expression of a chloroplast protein affects the outcome of virus and nematode infection. Mol Plant Microbe Interact. 2017;30:478–488. doi: 10.1094/MPMI-02-17-0031-R. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi K, Otegui MS, Krishnakumar S, Mindrinos M, Zambryski P. INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell. 2007;19:1885–1897. doi: 10.1105/tpc.106.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burch-Smith TM, Brunkard JO, Choi YG, Zambryski PC. Organelle-nucleus cross-talk regulates plant intercellular communication via plasmodesmata. Proc Natl Acad Sci USA. 2011;108:E1451–E1460. doi: 10.1073/pnas.1117226108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stonebloom S, Burch-Smith T, Kim I, Meinke D, Mindrinos M, Zambryski P. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA. 2009;106:17229–17234. doi: 10.1073/pnas.0909229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, et al. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA. 2009;106:3615–3620. doi: 10.1073/pnas.0808717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stonebloom S, Brunkard JO, Cheung AC, Jiang K, Feldman L, Zambryski P. Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. 2012;158:190–199. doi: 10.1104/pp.111.186130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, et al. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlotto N, Wirth S, Furman N, Ferreyra Solari N, Ariel F, Crespi M, et al. The chloroplastic DEVH-box RNA helicase INCREASED SIZE EXCLUSION LIMIT 2 involved in plasmodesmata regulation is required for group II intron splicing. Plant Cell Environ. 2016;39:165–173. doi: 10.1111/pce.12603. [DOI] [PubMed] [Google Scholar]
- 65.Bobik K, McCray TN, Ernest B, Fernandez JC, Howell KA, Lane T, et al. The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. Plant J. 2017;91:114–131. doi: 10.1111/tpj.13550. [DOI] [PubMed] [Google Scholar]
- 66.Wildman SG, Hongladarom T, Honda SI. Chloroplasts and mitochondria in living plant cells: cinephotomicrographic studies. Science. 1962;138:434–436. doi: 10.1126/science.138.3538.434. [DOI] [PubMed] [Google Scholar]
- 67.Natesan SK, Sullivan JA, Gray JC. Stromules: a characteristic cell-specific feature of plastid morphology. J Exp Bot. 2005;56:787–797. doi: 10.1093/jxb/eri088. [DOI] [PubMed] [Google Scholar]
- 68.Schattat M, Barton K, Mathur J. Correlated behavior implicates stromules in increasing the interactive surface between plastids and ER tubules. Plant Signal Behav. 2011;6:715–718. doi: 10.4161/psb.6.5.15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buchner O, Moser T, Karadar M, Roach T, Kranner I, Holzinger A. Formation of chloroplast protrusions and catalase activity in alpine Ranunculus glacialis under elevated temperature and different CO2/O2 ratios. Protoplasma. 2015;252:1613–1619. doi: 10.1007/s00709-015-0778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gray JC, Sullivan JA, Hibberd JM, Hansen MR. Stromules: mobile protusions and interconnections between plastids. Plant Biol (Stuttg) 2001;3:223–233. doi: 10.1055/s-2001-15204. [DOI] [Google Scholar]
- 71.Schattat M, Barton K, Baudisch B, Klosgen RB, Mathur J. Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 2011;155:1667–1677. doi: 10.1104/pp.110.170480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho J, Theg SM. The formation of stromules in vitro from chloroplasts isolated from Nicotiana benthamiana. PLoS One. 2016;11:e0146489. doi: 10.1371/journal.pone.0146489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caplan JL, Kumar AS, Park E, Padmanabhan MS, Hoban K, Modla S, et al. Chloroplast stromules function during innate immunity. Dev Cell. 2015;34:45–57. doi: 10.1016/j.devcel.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat Commun. 2017;8:49. doi: 10.1038/s41467-017-00074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Souza A, Wang JZ, Dehesh K. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol. 2017;68:85–108. doi: 10.1146/annurev-arplant-042916-041007. [DOI] [PubMed] [Google Scholar]