Summary
Cysteine desulfurase plays a central role in mitochondrial iron-sulfur cluster biogenesis by generating sulfur through the conversion of L-cysteine to L-alanine and by serving as the platform for assembling other components of the biosynthetic machinery, including ISCU, frataxin, and ferredoxin. The human mitochondrial cysteine desulfurase complex consists of two copies each of NFS1, ISD11 and acyl carrier protein (ACP). We describe results from chemical crosslinking coupled with tandem mass spectrometry (XL-MS) and small angle X-ray scattering (SAXS) studies that are consistent with a closed NFS1 dimer rather than an open one for both the cysteine desulfurase-ISCU and cysteine desulfurase-ISCU-frataxin complexes. We present a structural model for the cysteine desulfurase-ISCU-frataxin complex derived from chemical crosslinking restraints in conjunction with the recent crystal structure of the cysteine desulfurase-ISCU-zinc complex (Boniecki, M.T. et al. (2017) Nat Commun 8, 1287) and distance constraints from NMR (Cai et al. (2018) J Inorg Biochem 183, 107.
Keywords: Iron-sulfur cluster biosynthesis, crosslinking coupled with tandem mass spectrometry (XL-MS), small-angle X-ray scattering (SAXS)
eTOC Blurb
Cai et al. used chemical crosslinking coupled with tandem mass spectrometry (XL-MS) and small angle X-ray scattering (SAXS) to investigate the structures of cysteine desulfurase complexes. They used these results with NMR chemical shift perturbation data and the published structure of (NIAUZ)2 to derive a structural model for (NIAUF)2.
Introduction
Iron-sulfur (Fe-S) clusters are redox-active protein cofactors that play essential roles in all kingdoms of life. Fe-S clusters are involved in numerous biological processes such as respiration, electron transfer, DNA replication and repair, cofactor biosynthesis, and gene regulation (Fontecave, 2006; Johnson et al., 2005; Lill, 2009; Pain and Dancis, 2016; Rouault, 2015). Owing to the toxicity of free iron and sulfur, the biogenesis of Fe-S clusters is a highly regulated pathway that involves the participation of many proteins. The ISC (Iron-Sulfur Cluster) machinery is conserved from yeast to man. The biosynthesis of Fe-S proteins involves two major steps: assembly of a cluster on the scaffold protein ISCU and transfer of the cluster to a recipient protein (Braymer and Lill, 2017; Lill and Muhlenhoff, 2005; Maio and Rouault, 2014; Stehling and Lill, 2013). Defects in protein components of the ISC machinery are associated with numerous diseases, underlining the essentiality of this process (Rouault, 2012; Stehling et al., 2014).
Cysteine desulfurase plays a central role in the initial steps of ISC machinery by catalyzing the conversion of cysteine to alanine and releasing sulfur for cluster assembly (Kispal et al., 1999; Pandey et al., 2012). The Escherichia coli cysteine desulfurase (IscS) is pyridoxal 5′-phosphate (PLP)-dependent homodimer, whose crystal structure was determined over a decade ago (Cupp-Vickery et al., 2003). The human homolog (NFS1), which shares 59% sequence identity with IscS, differs from IscS in that its full stability and function require two small accessory proteins, namely ISD11 and acyl carrier protein (ACP). In addition, the human cysteine desulfurase requires frataxin (FXN), which binds iron and has been proposed to be an allosteric regulator of NFS1 and control both iron and sulfur entry to the scaffold protein ISCU (Bridwell-Rabb et al., 2014; Colin et al., 2013; Fox et al., 2015; Pandey et al., 2013; Parent et al., 2015; Tsai and Barondeau, 2010). Defects in FXN are responsible for the neurodegenerative disease Friedreich ataxia (Vaubel and Isaya, 2013).
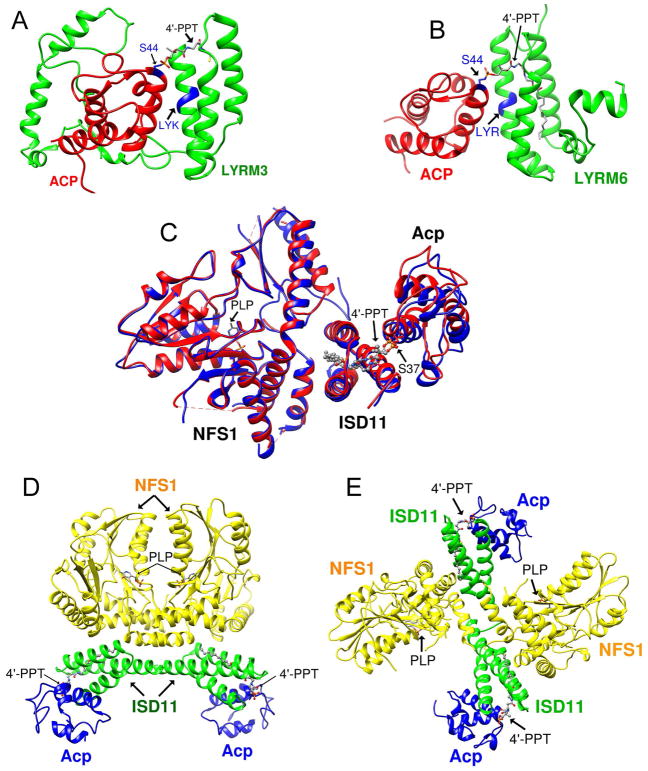
ISD11 (or LYRM4), a member of the LYRM (Leu-Tyr-Arg Motif) family proteins (Angerer, 2013, 2015), is important for both mitochondrial Fe-S cluster biogenesis and iron homeostasis (Adam et al., 2006; Shi et al., 2009; Wiedemann et al., 2006). LYRM proteins are small, basic proteins that carry a conserved Leu-Tyr-Arg sequence close to the N-terminus. The human genome contains at least ten LYRM proteins that localize predominantly to mitochondria (Angerer, 2013, 2015). ACP is a small, acidic protein known to play an essential role in mitochondrial fatty acid biosynthesis (FASII) through reactions involving its 4′-phosphopantethiene (4′-PPT) cofactor, which is conjugated to a conserved serine residue (Hiltunen et al., 2010; Kastaniotis et al., 2017). ACP has been shown to be an accessory subunit of mitochondrial respiratory chain complex I (Angerer et al., 2014; Dobrynin et al., 2010; Runswick et al., 1991), and down-regulation of ACP in mammalian cells results in compromised mitochondrial complex I and cell death (Feng et al., 2009). Recent cryo-EM structures of mammalian mitochondrial complex I have demonstrated that ACP forms a complex with two LYRM proteins (LYRM3 and LYRM6). In these ACP-LYRM protein complexes, the conserved ‘LYR’ motif interacts with ACP, and the acylated 4′-PPT cofactor of ACP is flipped-out and extends into the α-helical bundle of the LYRM proteins (Figure 1A and B) (Fiedorczuk et al., 2016; Zhu et al., 2016).
Figure 1.
Structures of LYRM-ACP and cysteine desulfurase complexes. (A) Structure of LYRM3-ACP taken from the cryo-EM structure of ovine respiratory complex I (PDB: 5LNK). (B) Structure of LYRM6-ACP taken from the cryo-EM structure of ovine respiratory complex I (PDB: 5LNK). (C) Overlay of the X-ray structures of one subunit of human cysteine desulfurase complex (NIA)1. Red, PDB entry 5WGB; cyan, PDB entry 5USR. (D) X-ray structure of the human cysteine desulfurase complex (NIA)2 by Boniecki et al. (2017) (PDB: 5WGB). (E) X-ray structure of full human cysteine desulfurase complex (NIA)2 by Cory et al. (2017) (PDB: 5USR).
The role of ACP in mitochondrial Fe-S cluster biogenesis has recently come into light. In yeast, Acp1 was found to be an essential component of cysteine desulfurase complex (Van Vranken et al., 2016). We demonstrated that Escherichia coli Acp substitutes for human mitochondrial ACP in the cysteine desulfurase complex produced by co-expressing human NFS1 and ISD11 in E. coli cells and determined the stoichiometry to be [NFS1]2:[ISD11]2:[Acp]2 (Cai et al., 2017a), henceforth abbreviated as (NIA)2. Two structures of (NIA)2 have recently been determined independently via X-ray crystallography, which confirm the stoichiometry of the complex (Boniecki et al., 2017; Cory et al., 2017). The structures of NFS1, ISD11, and Acp subunits of the (NIA)2 complex are very similar in each structure (Figure 1C), and the acyl-Acp and ISD11 interactions are similar to those in other LYRM-ACP complexes (Fiedorczuk et al., 2016; Zhu et al., 2016). However, the quaternary architectures of the two (NIA)2 complexes are strikingly different. In the (NIA)2 structure by Boniecki et al. (PDB: 5WGB), the two NFS1 subunits form a dimer similar to that of E. coli IscS (Cupp-Vickery et al., 2003; Marinoni et al., 2012; Shi et al., 2010) (Figure 1D). The structure determined by Cory et al. (PDB: 5USR) adopts a unique quaternary structure in which the two NFS1 units have little contact with each other and the substrate-binding site of NFS1 is exposed (Figure 1E).
Boniecki et al. (2017) also determined crystal structures of the cysteine desulfurase complex containing the scaffold protein ISCU (NIAU)2 (PDB: 5WKP) and ISCU plus Zn2+ (NIAUZ)2 (PDB: 5WLW). In these two structures, the NFS1 dimer architectures resembled a tightened version of 5WGB, similar to the dimeric architecture of the bacterial cysteine desulfurase in (IscS-IscU)2 (Shi et al., 2010).
We have performed chemical crosslinking of the (NIAU)2 and (NIAUF)2 complexes, where F stands for frataxin, coupled with tandem mass spectrometry (XL-MS) and small angle X-ray scattering (SAXS) to investigate the architecture of these complexes. The results are consistent with a closed NFS1 dimer as observed in the structure of (NIAU)2 published by Boniecki et al. (5WKP) but not with the open NFS1 dimer reported in the structure of (NIA)2 by Cory et al. (5USR). We present here a structural model for the complex containing frataxin (NIAUF)2 based on the crystal structure of (NIAUZ)2 (5WLW) along with structural restraints from XL-MS, SAXS, and NMR spectroscopy. This new structural model extends our knowledge of how the iron-donating protein frataxin interacts with the scaffold protein to which it delivers iron and how frataxin is positioned to activate the cysteine desulfurase reaction.
Results and Discussion
Protein Interaction Networks in Cysteine Desulfurase Complexes from XL-MS
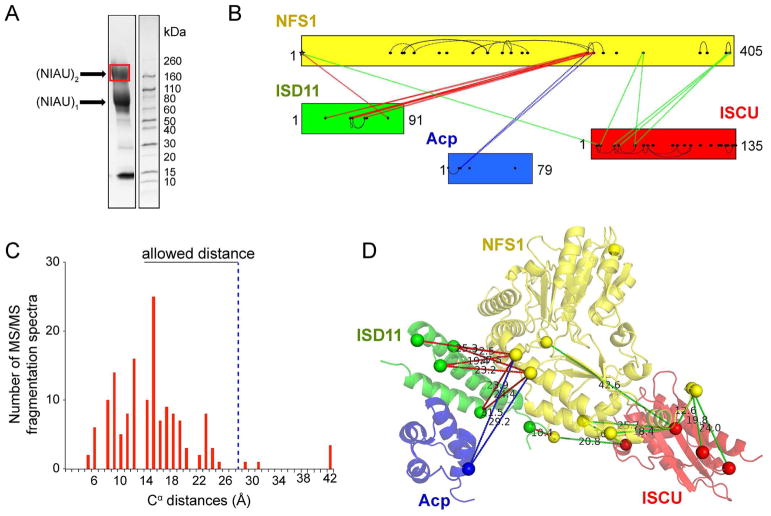
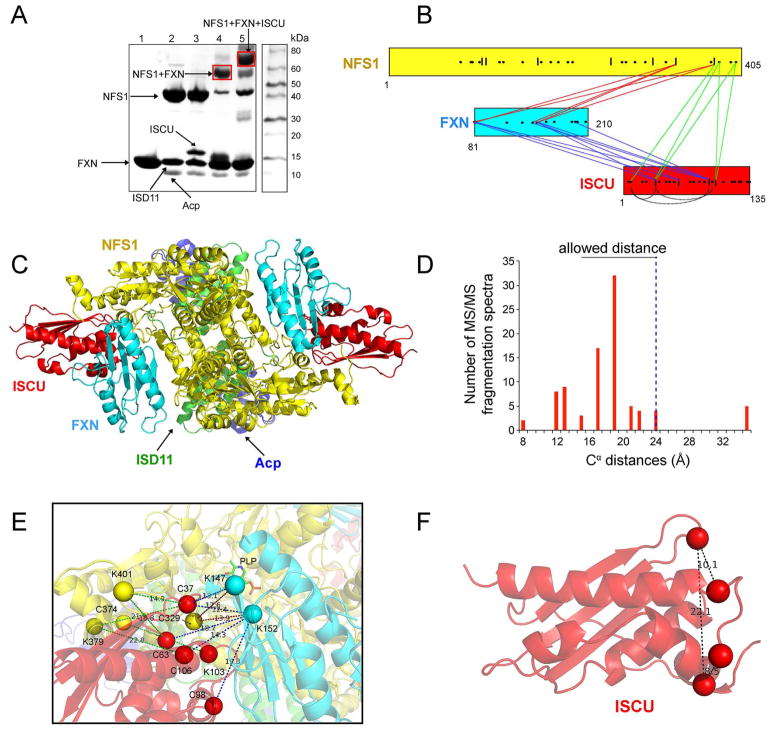
To study the protein interaction network within the (NIAU)2 complex, we subjected the sample to XL-MS. A homo-bifunctional crosslinker, bis-(sulfosuccinimidyl)suberate (BS3, Thermo Fisher Scientific), was used in the crosslinking reaction. BS3 reacts with primary amines in lysine side chains and protein N-termini. SDS-PAGE analysis showed two major bands from crosslinked species with molecular masses of about 80 and 160 kDa (Figure 2A), corresponding to the expected molecular masses of (NIAU)1 (79.7 kDa) and (NIAU)2 (159.4 kDa), respectively. The SDS-PAGE band with molecular mass 160 kDa was excised from the gel and subjected to limited proteolysis followed by and liquid chromatography-tandem mass spectrometry (LC-MS/MS). The analysis yielded 146 MS/MS fragmentation spectra corresponding to 61 high-confidence lysine-lysine or lysine-N-termini crosslinks, among which 20 are inter-subunit crosslinks and 41 are intra-subunit crosslinks (Table S1). A map of the crosslinked sites is shown in Figure 2B. The observed crosslinks were significantly different from a random selection of all possible pairs from the crystal structure of (NIAUZ)2 (5WLW) (Figure 2C and Figure S1A). The maximal Cα–Cα distance of two lysine residues linked by BS3 is 27.4 Å (Chen et al., 2010). Based on the crystal structure of (NIAUZ)1 (PDB entry: 5WLW), >95% of the NFS1, Acp, ISCU, and ISD11 inter-subunit crosslinked lysines of have predicted Cα–Cα distances below 27.4 Å (Figure 2C and D). However, three inter-subunit crosslinked lysines have predicted Cα–Cα distances above 27.4 Å (Table S1). These corresponded to NFS1(K272)–Acp(K10) (29.5 Å), NFS1(K268)–Acp(K10) (31.5 Å), and NFS1(K206)–ISCU(K42) (42.6 Å) (Figure 2D). These violations may be explained by structural flexibility of Acp and ISCU. Acp was shown to have large B-factors in the both crystal structure of (NIA)2, and solution NMR studies revealed the presence of the disordered form of ISCU in the cysteine desulfurase complex (Cai et al., 2013).
Figure 2.
XL-MS study of (NIAU)2 using bis-(sulfosuccinimidyl)suberate (BS3) as the crosslinker. (A) SDS-PAGE gel image of the crosslinked products. Red box indicates the band on the gel that was excised and digested for MS analysis. (B) Crosslink map showing the crosslinked sites in the (NIAU)2 complex. Inter-subunit crosslinks between NFS1 and ISD11 are in red, inter-subunit crosslinks between NFS1 and Acp are in blue, inter-subunit crosslinks between NFS1 and ISCU are in green, and intra-subunit crosslinks are shown by curved black lines. The lysine residues in the protein sequences are represented by dots. (C) Cα–Cα distance distribution for the experimentally observed lysine–lysine or lysine-N-termini pairs calculated from the structure of (NIAUZ)2 (PDB: 5WLW). (D) Inter-protein crosslinks between NFS1, ISD11, Acp and ISCU and their Cα–Cα distances in the structure of (NIAUZ)1 (PDB: 5WLW). The positions of the crosslinked lysine residues and N-termini are indicated by spheres.
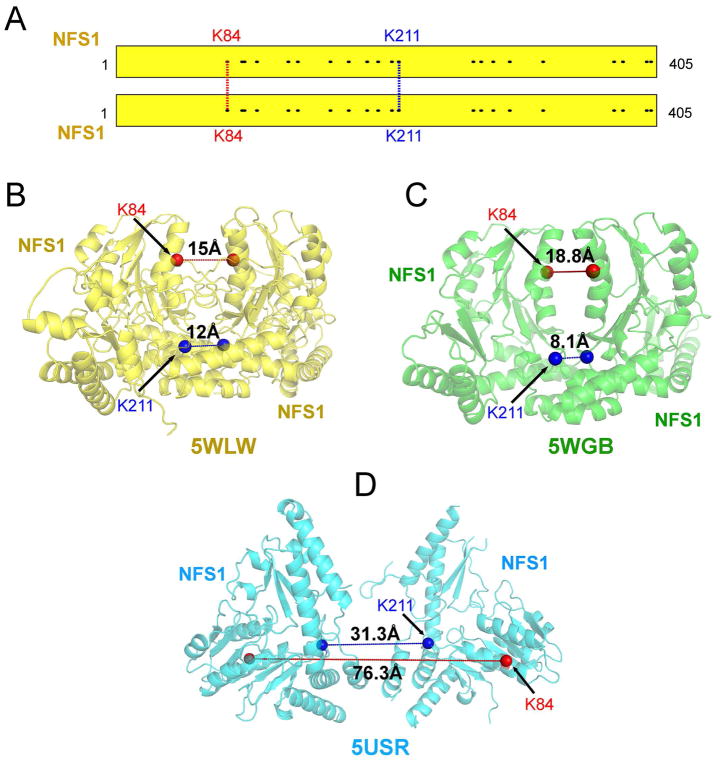
Remarkably, the analysis also identified two high-confidence NFS1-NFS1 inter-subunit crosslinks (K84–K84 and K211–K211) (Table S1 and Figure 3A). These two crosslinks were identified in a total of 21 MS/MS fragmentation spectra. Two of the MS/MS fragmentation spectra are shown in Figure S2. In particular, the K84–K84 inter-subunit crosslink was observed in two different crosslinked peptides (SNNIAIKGVAR–SNNIAIKGVAR and EIIFTSGATESNDIAIKGVAR–EIIFTSGATESNDIAIKGVAR); confidence for the match is high because these appeared in 19 MS/MS fragmentation spectra (Table S1). These two crosslinks are in agreement with distances derived from the crystal structures of (NIAUZ)2 (5WLW) and (NIA)2 (5WGB) by Boniecki et al., which show the two lysines located at the NFS1 dimer interface. In 5WLW, the Cα–Cα distances are 15.0 Å for K84–K84 and 12.0 Å for K211–K211 (Figure 3B). In 5WGB, the Cα–Cα distances are 18.8 Å for K84–K84 and 8.1Å for K211–K211 (Figure 3C). However, these crosslinks did not fit the (NIA)2 structure by Cory et al. (PDB: 5USR), which has Cα–Cα distances of 76.3 Å for K84–K84 and 31.3 Å for K211–K211 (Figure 3D).
Figure 3.
Comparison of experimental XL-MS results for (NIAU)2 with distances derived from structural models. (A) Experimental inter-subunit NFS1–NFS1 crosslinks. (B) Corresponding Cα–Cα distances in the structure of (NIAUZ)2 by Boniecki et al. (2017) (PDB: 5WLW). (C) Corresponding Cα–Cα distances the in the structure of (NIA)2 by Boniecki et al. (2017) (PDB: 5WGB). (D) Corresponding Cα–Cα distances the in the (NIA)2 structure by Cory et al. (2017) (PDB: 5USR). The positions of the crosslinked lysines are indicated by spheres. For clarity, only the two NFS1 subunits are shown.
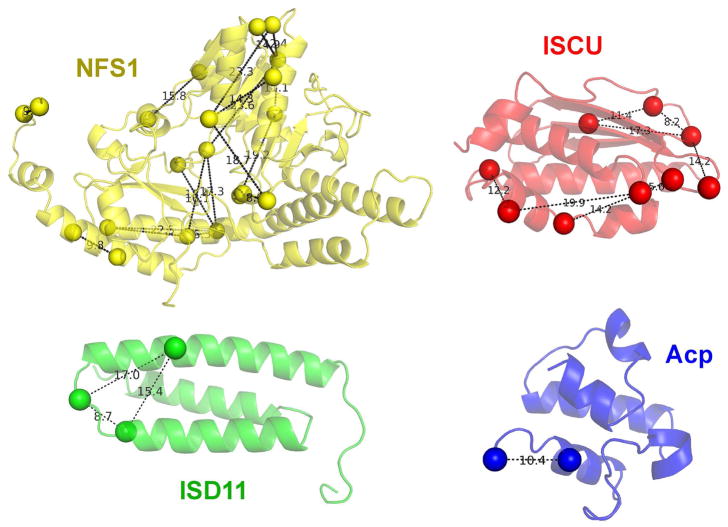
We also observed 41 intra-subunit crosslinks (Table S1 and Figure 2B, black curved traces). All these intra-subunit crosslinks could be explained by the structures of the individual proteins (Figure 4).
Figure 4.
Comparison of experimental XL-MS results for (NIAU)2 with distances derived from structural models. All the Cα distances in the intra-subunit crosslinks are below 27.4 Å. The structures shown represent NFS1, ISCU, ISD11 and Acp are all from PDB entry 5WLW. The positions of the crosslinked lysines and N-termini are indicated by spheres.
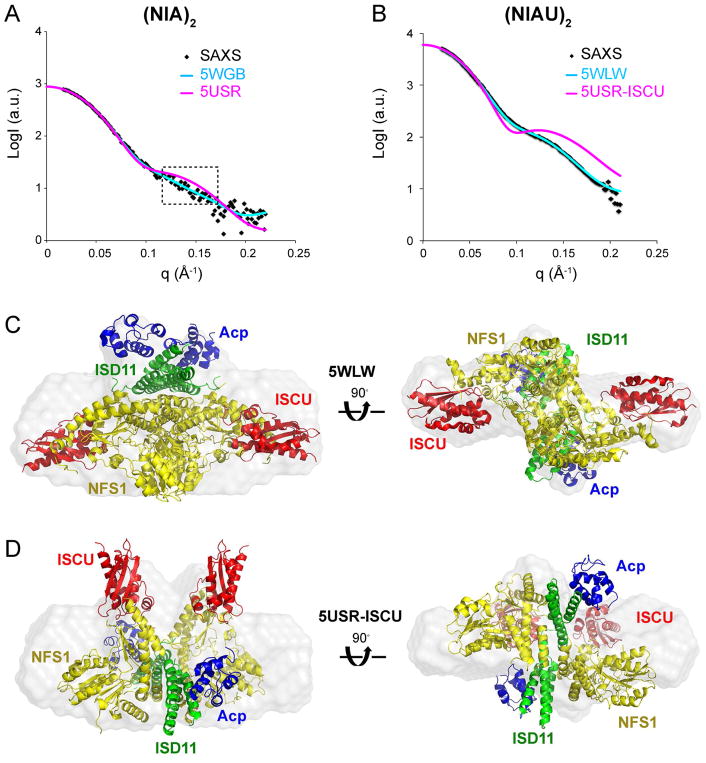
Investigation of the Structures of (NIA)2 and (NIAU)2 by SAXS
We investigated the structure of (NIA)2 by SAXS and compared the SAXS data with the two different (NIA)2 structures. The SAXS data on (NIA)2 fitted 5WGB slightly better (χ2CRYSOL = 1.15) than 5USR (χ2CRYSOL = 2.67) (Figure 5A). Visually, the theoretical scattering curve of 5WGB fitted the experimental SAXS data almost perfectly, whereas the theoretical scattering curve of 5USR showed some discrepancy with the experimental data (Figure 5A, boxed region). We next collected SAXS data on (NIAU)2. As suggested by (Cory et al., 2017), we used the highly conserved interaction between cysteine desulfurase and the scaffold protein to build a structural model for (NIAU)2 using their structure of (NIA)2 (5USR). For convenience, we named the structural model 5USR-ISCU. Our SAXS data fitted the (NIAUZ)2 structure by Boniecki et al. (5WLW) very well (χ2CRYSOL = 6.60) but fitted the 5USR-ISCU model poorly (χ2CRYSOL = 219.04) (Figure 5B). 5WLW also fitted well into the envelope generated by ab initio modeling of the SAXS data from (NIAU)2 (Figure 5C), whereas 5USR-ISCU did not (Figure 5D).
Figure 5.
SAXS studies of (NIA)2 and (NIAU)2 complexes. (A) Experimental SAXS data from (NIA)2 (black dots) overlaid with the theoretical scattering curves computed from structures of (NIA)2. Magenta trace, 5USR; cyan trace, 5WGB. The dotted boxes indicate regions of large discrepancy between the experimental SAXS data and that computed from 5USR. (B) Experimental SAXS data from (NIAU)2 (black dots) overlaid with the theoretical scattering curves computed from structure of (NIAUZ)2 (PDB: 5WLW, cyan trace) and from the model 5USR-ISCU (magenta trace). (C) Superimposition of the structure of (NIAUZ)2 (PDB: 5WLW) onto the ab initio dummy-atom envelope calculated from the SAXS data for (NIAU)2. (D) Superimposition of the structural model 5USR-ISCU onto the ab initio dummy-atom envelope calculated from the SAXS data for (NIAU)2.
Analysis of Protein Interactions in ISC Core Complexes by XL-MS
In order to investigate the protein interactions between FXN and (NIA)2 or (NIAU)2, we chose a water-soluble hetero-bifunctional chemical crosslinker, sulfo-SMCC, whose N-hydroxysuccinimide (NHS) ester reacts with primary amines (at pH 7–9) and whose maleimide group reacts with sulfhydryl groups at pH 6.5 to 7.5. The crosslinking reaction was initiated by adding sulfo-SMCC to a solution of FXN, which contains no cysteine residues, and allowing sulfo-SMCC to react with its primary amines (N-terminus and lysine side chains). After the excess reagent was removed by a desalting column, the maleimide-activated FXN was added separately to solutions containing (NIA)2 or (NIAU)2 and allowed to react with their sulfhydryl groups (cysteine residues on NFS1 and ISCU). The scheme for conjugating FXN and NFS1 with sulfo-SMCC is shown in Figure S3. Acp and ISD11, which lack cysteine residues, were not involved in the crosslinking reactions. The products were analyzed by SDS-PAGE, and bands corresponding to crosslinked species were identified by their molecular masses (Figure 6A). These bands were excised, digested by limited proteolysis, and the resulting peptides were analyzed by LC-MS/MS.
Figure 6.
XL-MS study of (NIAUF)2 using sulfo-SMCC as the crosslinker. (A) SDS-PAGE gel image of the proteins or protein complexes before and after the crosslinking reaction. Red boxes indicate bands on the gel that were excised and digested from XL-MS. (B) Crosslink map showing the crosslinked sites. Inter-subunit crosslinks between NFS1 and FXN are in red; inter-subunit crosslinks between FXN and ISCU are in blue; inter-subunit crosslinks between NFS1 and ISCU are in green; and intra-subunit crosslinks are shown as curved black lines. The cysteine residues in the protein sequences are represented by vertical lines and lysine residues are represented by dots. (C) The structural model of (NIAUF)2 built using HADDOCK server by combining NMR and XL-MS restraints with the X-ray structure of (NIAUZ)2 (5WLW). (D) Cα–Cα distance distribution for the experimentally observed lysine–cysteine pairs calculated from the structural model of (NIAUF)2. (E) Close-up view of the inter-subunit crosslinks with their Cα–Cα distances from the structural model of (NIAUF)2. (F) Intra-subunit crosslinks in the structure of ISCU with their Cα–Cα distances.
LC-MS/MS analysis of the product of crosslinking FXN with (NIA)2 revealed 31 MS/MS fragmentation spectra reporting on 5 high-confidence cysteine-lysine or cysteine-N-termini inter-subunit (Table S2). Analysis of the product of crosslinked FXN and (NIAU)2 yielded 144 MS/MS fragmentation spectra reporting on 24 inter-subunit crosslinks and 3 intra-subunit crosslinks (Table S3). A map of the crosslinked sites is shown in Figure 6B. Most of the inter-subunit crosslinks between FXN and NFS1 localized to the N-terminus and two lysine residues (K147 and K152) of FXN and the two cysteine residues (C329 and C374) of NFS1. Most inter-subunit crosslinks between FXN and ISCU localized to the N-terminus and K152 of FXN and the three cysteine residues of ISCU (C37, C63 and C98) (Figure 6B).
Interestingly, additional crosslinks were identified between K401 and K379 of NFS1 and two cysteine residues of ISCU (C37 and C106), as well as three intra-ISCU crosslinks (Figure 6B). These crosslinks likely resulted from residual chemical crosslinker not completely removed by the desalting column.
Structural Model of the (NIAUF)2 Complex
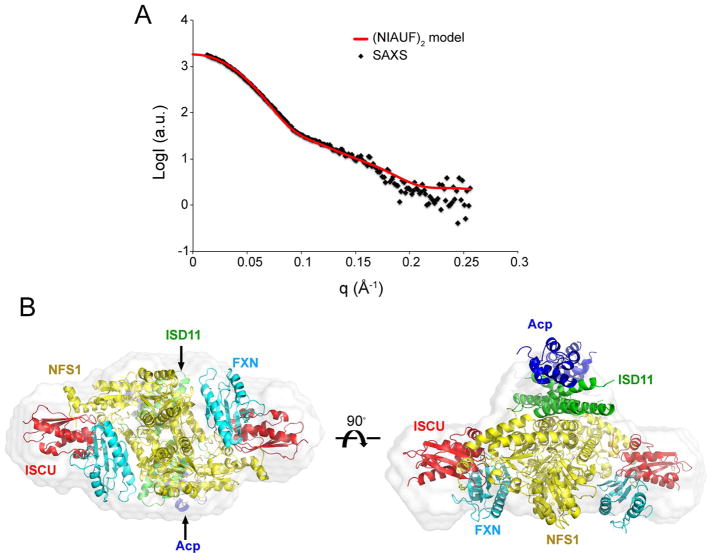
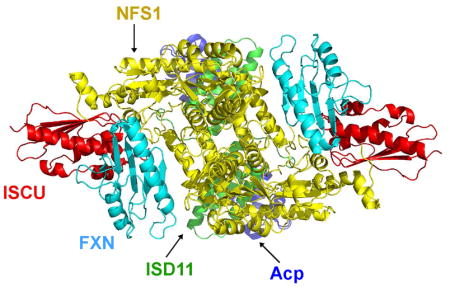
In an earlier NMR study (Cai et al., 2018b), we determined that FXN interacts with (NIA)2 through its α1 helix and β1 strand and ISCU through its β3-β5 sheet. We used the HADDOCK webserver (de Vries et al., 2010) to build a structural model of the (NIAUF)2 complex by docking the structure of FXN (PDB: 1EKG) (Dhe-Paganon et al., 2000) onto the structure of (NIAUZ)2 (5WLW) and by satisfying the restraints from NMR and XL-MS data. The resulting (NIAUF)2 structural model (Figure 6C) shows an interaction between the highly conserved arginine-rich loop of NFS1 (219RRRPRVR225) and the conserved α1-β1 acidic ridge of FXN. FXN fits into the cleft between NFS1 and ISCU. The modeled interaction between FXN and ISCU involves the β3–β5 sheet of FXN interacting with the conserved ‘98LPPVK103’ loop of ISCU (Figure 6C and Figure S4). The maximal Cα–Cα distance of cysteine-lysine pairs linked by SMCC is about 24 Å. Most of the Cα–Cα distances of NFS1–FXN and FXN–ISCU inter-subunit crosslinks (Figure 6D, Table S2 and S3) were in agreement with the structural model of (NIAUF)2. The observed crosslinks were significantly different from a random selection of all possible cysteine-lysine pairs from the structural model (Figure S1B). Some of the crosslinked residues (shown as spheres) and the distances between their Cα atoms are shown in Figure 6E. All the inter-subunit crosslinks between NFS1 and ISCU agreed with the X-ray structure of (NIAUZ)2 (5WLW). In addition, all the intra-ISCU crosslinks agreed with the structure of ISCU (Figure 6F).
We next compared experimental SAXS data from (NIAUF)2 with the structural model of that complex. The SAXS data showed good agreement with the theoretical scattering curve from the (NIAUF)2 model (χ2CRYSOL = 4.94) (Figure 7A). The structural model of (NIAUF)2 fitted well into the envelope generated by ab initio modeling of the SAXS data (Figure 7B).
Figure 7.
SAXS study of the (NIAUF)2 complex. (A) Experimental SAXS data (black dots) from (NIAUF)2 overlaid with the theoretical scattering curve (red trace) computed from the structural model of (NIAUF)2. (B) Superimposition of the structural model of (NIAUF)2 onto the dummy-atom envelope calculated from the SAXS data from (NIAUF)2.
We carried out cysteine desulfurase activity assays of non-crosslinked and crosslinked (NIAU)2 and (NIAUF)2 to determine whether the crosslinking reactions altered the activities of the protein complexes. The (NIAU)2 complex cross linked with BS3 exhibited cysteine desulfurase activity, although the activity was slightly lower than that of non-crosslinked (NIAU)2 (Figure S5, red). The decreased activity may be explained by the slight aggregation resulting from crosslinking. As reported earlier (Cai et al., 2018b), the cysteine desulfurase activity of (NIAUF)2 was much higher than that of (NIAU)2. However, (NIAUF)2 crosslinked with SMCC showed very little activity (Figure S5, blue). The low activity of SMCC-crosslinked (NIAUF)2 is expected, because the active site cysteine of NFS1 (C329) is involved in crosslinking reactions (Table S3 and Figure 6E).
Conclusions
Recent advances in instrumentation and data-analysis approaches have established SAXS and XL-MS as mainstream techniques in structural biology (Chaudhuri, 2015; Leitner et al., 2016; Tran et al., 2016; Tuukkanen et al., 2017). Here, we crosslinked the (NIAU)2 complex with the homo-bifunctional lysine-specific crosslinker BS3 and analyzed the crosslinked product by LC-MS/MS. The identified crosslinks agreed with the (NIA)2 (5WGB) and (NIAUZ)2 (5WLW) structures by Boniecki et al. (2017). In particular, we observed two high-confidence inter-subunit NFS1–NFS1 crosslinks (K84–K84 and K211–K211) that agreed with Cα–Cα distances in both 5WGB (18.8 Å for K84–K84 and 8.1 Å for K211–K211) and 5WLW (15.0 Å for K84–K84 and 12.0 Å for K211–K211). However, these NFS1–NFS1 crosslinks did not fit distances from the (NIA)2 structure by Cory et al. (2017) (5USR): 76.3 Å for K84–K84 and 31.3 Å for K211–K211. In addition, our SAXS data from (NIA)2 are in better agreement with 5WGB (χ2 CRYSOL = 1.15) than with 5USR (χ2 CRYSOL = 2.67). The differences are more pronounced in the structural models containing ISCU. The SAXS data from (NIAU)2 fit 5WLW much better than the 5USR-ISCU structural model.
The active site residues of NFS1 are better resolved in the crystal structure of (NIAUZ)2 (5WLW) than in (NIAU)2 (5WKP). Both crystal structures incorporated ISCU(M108I), a variant that we recently identified as being fully structured (Cai et al., 2018a). Our solution studies made use of complexes containing wild type ISCU, which alone in solution exists as a mixture of interconverting structured and dynamically disordered states (Cai et al., 2013). In deriving the structural model for the (NIAUF)2 complex, we chose to use the (NIAUZ)2 crystal structure along with restraints involving frataxin from our solution results.
Our only direct data on the architecture of (NIA)2 comes from SAXS. Efforts to chemically crosslink (NIA)2 with BS3 proved unsuccessful because the crosslinked product was prone to aggregation. ISCU appears to have a stabilizing effect on (NIA)2, as was suggested by a previous study (Van Vranken et al., 2016). Thus, we cannot rule out the possibility that (NIA)2 exists in solution as two different conformations, a major form with a closed NFS1 dimer (as in 5WGB) and a minor form with an open NFS1 dimer (as in 5USR). We are currently exploring this possibility by 19F NMR studies of (NIA)2 prepared from NFS1 containing 19F-labled Trp. NFS1 contains three Trp residues; however, preliminary results (Figure S6) show the presence of more than three 19F NMR peaks in spectra of the complex, as consistent with structural heterogeneity.
FXN plays an important role in mitochondrial Fe-S cluster biosynthesis. FXN has been shown to enhance sulfur transfer and control iron entry into ISCU (Bridwell-Rabb et al., 2014; Colin et al., 2013; Parent et al., 2015), however, its exact function is still a subject of debate. FXN has also been proposed as the iron donor because of its ability to bind iron and donate iron for in vitro Fe-S clsuter assembly reactions (Cook et al., 2010; Yoon and Cowan, 2003). However, the disccovery of a FXN-bypassing Isu1 mutant found in yeast (Pandey et al., 2013; Yoon et al., 2012; Yoon et al., 2014), led to speculation that FXN might not be the iron donor. Multiple lines of evidence have established that FXN forms a stable complex with (NIAU)2 (Colin et al., 2013; Parent et al., 2015; Schmucker et al., 2011; Tsai and Barondeau, 2010); however, the structure of the (NIAUF)2 complex has remained elusive. Here, we present a structural model of the (NIAUF)2 complex that is in excellent agreement with our NMR, XL-MS, and SAXS data. Our NMR data have determined that the α1 helix and β1 strand in FXN form the binding site for (NIA)2 and that the β3-β5 sheet forms the binding site for ISCU (Cai et al., 2018b). All but one of the crosslinks identified by XL-MS are in agreement with the structural model, and the SAXS data fit well with the model. The model is also supported by other lines of evidence. It is consistent with the finding that a conserved tryptophan W155 on β4 stand of FXN is involved in the interaction with ISCU (Leidgens et al., 2010). It is also in good agreement with the finding that the conserved ‘99LPPVK103’ motif of ISCU interacts with FXN (Manicki et al., 2014). In the (NIAUF)2 model, FXN fits into the cleft formed by ISCU and NFS1, which explains our finding that (NIA)2 is required for the physical contact between ISCU and FXN (Cai et al., 2018b). The structural model shows that the interaction between FXN and NFS1 involves the highly conserved arginine-rich loop ‘219RRRPRVR225’ on NFS1. Mutations of residues in this loop have been shown to compromise the interaction between E. coli IscS and CyaY (Prischi et al., 2010). Our XL-MS analysis also identified several high-confidence crosslinks between the N-terminus of FXN and the active site cysteine residues of NFS1 (C329) or ISCU (C37, C63, and C106) (Table S2 and S3). Because no structure is available for the N-terminal sequence of the mature form of human FXN (81SGTLGHPGS89), no Cα–Cα distances are available for comparison with these crosslinks. Nonetheless, based on the structure of yeast frataxin, this N-terminal peptide is likely to be disordered (He et al., 2004; Soderberg et al., 2011). In our structural model, the N-terminus of FXN is in close proximity to the active site cysteine residues of NFS1 and ISCU. Finally, it is worth noting that our model of (NIAUF)2 may not accurately show the structure of NFS1, as NFS1 may undergo a conformational change upon binding FXN (Cai et al., 2017a). It has been suggested that FXN is an allosteric activator, which exposes the otherwise buried substrate-binding site of NFS1 (Pandey et al., 2013; Tsai and Barondeau, 2010). We speculate that the N-terminal region of FXN may play a role in exposing the substrate-binding site.
STAR ★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| Rosetta 2(DE3) E. coli strain | Novagen | Cat# 71397-3 |
| Rosetta 2 (DE3) pLysS E. coli strain | Novagen | Cat# 71403 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BS3 (bis(sulfosuccinimidyl)suberate) | Thermo Fisher Scientific | Cat# 21580 |
| Sulfo-SMCC (sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate) | Thermo Fisher Scientific | Cat# 22322 |
| N,N-Dimethyl-p-phenylenediamine | Sigma Aldrich | Cat# 193992 |
| Dithiothreitol (DTT) | US Biological | Cat# D8070 |
| Sodium sulfide | Sigma Aldrich | Cat# 208043 |
| L-Cysteine | Sigma Aldrich | Cat# W326305 |
| 4-(2-Hydroxyethyl)piperazin-1-ylethanesulfonic acid (HEPES) | VWR | Cat# 97061-826 |
| Tris (2-Carboxyethyl) phosphine Hydrochloride (TCEP) | UBPBio | Cat# P1020-100 |
| Iron(III) chloride | Sigma Aldrich | Cat# 157740 |
| Trifluoroacetic acid (TFA) | Sigma Aldrich | Cat# T6508 |
| Trypsin | Sigma Aldrich | Cat# T1426 |
| Endoproteinase Glu-C | Sigma Aldrich | Cat# P6181 |
| Human NFS1 | This paper | Uniprot Q9Y697 |
| Human ISCU | This paper | Uniprot Q9H1K1 |
| Human ISD11 | This paper | Uniprot Q9HD34 |
| E. coli acyl carrier protein (Acp) | This paper | Cat# 21580 |
| Human frataxin (FXN) | This paper | Cat# 22322 |
| Oligonucleotides | ||
| Primers for
ISCU: 5′:ACTAGTGGTACCGGTCTCAAGGT CTAAAAATGTTGGAACTGGACTGG 3′:GGGCCCGCTCGAGGGATCCTCATTA TTTCTTCTCTGCCTCTCCTTTTTTG |
IDT https://www.idtdna.com/pages | N/A |
| Primers for
NFS1: 5′:ACTAGTGGTACCGGTCTCAAGGT GGGCCAGTGCTGCGACCTCTCTATATG 3′:GGGCCCGCTCGAGGGATCCTCATTA GTGTTGGGTCCACTTGATGCT |
IDT https://www.idtdna.com/pages | N/A |
| Primers for
FXN: 5′:ACTAGTGGTACCGGTCTCAAGGT TCTGGAACTTTGGGCCACCCAGGC 3′:GGGCCCGCTCGAGGGATCCTCATTA TCAAGCATCTTTTCCGGAATAGGC |
IDT https://www.idtdna.com/pages | N/A |
| Primers for
ISD11: 5′:GAAGGAGATATACC ATGGCAGCCTCCAGTCGCGCACAA 3′:GGATCCTCGAGCTC TTAATGGTGATGGTGATGGTGTTTGGTCCTGGG CATGTCTCGATTC |
IDT https://www.idtdna.com/pages | N/A |
| Recombinant DNA | ||
| cDNA of NFS1, ISD11, ISCU and frataxin | Mammalian Gene Collection | N/A |
| Plasmid: pET-SUMO, pET14b | This paper | N/A |
| Experimental Models: Constituents of Protein Complexes | ||
| Homo sapiens NFS1 | This paper | UniProt: Q9Y697 (53-457) |
| Homo sapiens ISD11 | This paper | UniProt: Q9HD34 |
| Homo sapiens frataxin | This paper | UniProt: Q16595 (81-210) |
| Escherichia coli acyl carrier protein | This paper | UniProt: P0A6A8 |
| Deposited Data | ||
| SAXS for (NIA)2 | This paper | SASDB: SASDDB3 |
| SAXS for (NIAU)2 | This paper | SASDB: SASDDC3 |
| SAXS for (NIAUF)2 | This paper | SASDB: SASDDD3 |
| NMR chemical shift data | This paper | BMRB: 27171 |
| MS data | This paper | PRIDE: PSD006938, PSD006928, PSD009079 |
| Structural model (NIAUF)2 | This paper | PDBdev: PDB_DEV00000015 |
| Software and Algorithms | ||
| ATSAS | (Franke et al., 2017) | https://www.embl-hamburg.de/biosaxs/software.html |
| DAMIF | (Franke and Svergun, 2009) | https://www.embl-hamburg.de/biosaxs/dammif.html |
| DAMAVER | (Volkov and Svergun, 2003) | https://www.embl-hamburg.de/biosaxs/damaver.html |
| CRYSOL from PRIMUS in ATSAS 2.8.1 | (Svergun et al., 1995) | https://www.embl-hamburg.de/biosaxs/crysol.html |
| SUPCOMB | (Kozin and Svergun, 2001) | https://www.embl-hamburg.de/biosaxs/supcomb.html |
| Stavrox | (Gotze et al., 2012) | https://www.stavrox.com/ |
| HADDOCK | (van Zundert et al., 2016) | https://haddock.science.uu.nl/services/HADDOCK2.2/ |
| Mascot | Matrixscience | http://www.matrixscience.com/search_form_select.html |
| FoXS | (Schneidman-Duhovny et al., 2016) | https://modbase.compbio.ucsf.edu/foxs/ |
| SAXS | Bruker | https://www.bruker.com/ |
| ProtParam | (Wilkins et al., 1999) | |
| PyMOL | Schrödinger | https://pymol.org/2/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John L. Markley (jmarkley@wisc.edu).
METHOD DETAILS
Protein expression and purification
Protein samples and complexes were prepared as described in detail in previous publications (Cai et al., 2013; Cai et al., 2017a; Cai et al., 2017b). Briefly, NFS1, ISCU and FXN81–210 (referred to as FXN) were cloned into pET-SUMO vector, and ISD11 was cloned into pET14b vector. His-tagged ISD11 (ISD11-His6) and His6-SUMO-NFS1 were co-expressed in E. coli cells, and the complex was isolated by immobilized metal affinity chromatography (IMAC) and then cleaved with SUMO protease; the released His6-SUMO was removed by size exclusion chromatography (SEC) on Superdex S200, yielding the (NIA)2 complex containing NFS1, ISD11 and E. coli Acp. His6-SUMO-ISCU and His6-SUMO-FXN were expressed in E. coli cells; ISCU and FXN were each isolated by IMAC followed by SUMO protease cleavage and subtractive IMAC to remove His6-SUMO, and then the proteins were purified by SEC. (NIAU)2 complex was prepared by mixing (NIA)2 and ISCU, and the complex was isolated by SEC. (NIAUF)2 complex was prepared by mixing (NIAU)2 and FXN, and the complex was isolated by SEC. The pH 7.5 buffer used in SEC contained 20 mM HEPES, 150 mM NaCl, and 5 mM TCEP. To produce (NIA)2 containing [19F-Trp]-NFS1, 4F-indole was added to the growth medium.
Protein crosslinking
The chemical crosslinking reaction using BS3 was started by mixing 1 mg/ml (NIAU)2 in crosslinking buffer (20 mM HEPES at pH 7.5 and 150 mM NaCl, or HN buffer) with 50-fold BS3 (Thermo Fisher Scientific). BS3 was freshly dissolved in HN buffer. The reaction was incubated at room temperature for 30 min and subsequently quenched by 1M Tris-HCl, pH 7.5. The crosslinked products were analyzed by SDS-PAGE.
The chemical crosslinking reactions using sulfo-SMCC (Thermo Fisher Scientific) was started by mixing 1 mg/mL purified FXN in PBS buffer (100mM sodium phosphate, 150mM sodium chloride, pH 7.2) with 20-fold sulfo-SMCC freshly dissolved in H2O. The reaction was incubated at room temperature for 30 min. The excess crosslinker was removed by using a Zeba Spin desalting column (Thermo Fisher Scientific). 1 mg/ml of purified (NIA)2 or (NIAU)2 in PBS buffer was added to the sulfo-SMCC labeled FXN solution. After the reactions were incubated at room temperature for 30 min, the crosslinked products were analyzed by SDS-PAGE.
Peptide preparation and mass spectrometry
Bands from the SDS–PAGE gels corresponding to crosslinked complexes were excised, and the proteins were reduced/alkylated and digested by limited proteolysis following standard protocols. The products of Sulfo-SMCC crosslinking reactions were digested by trypsin. The BS3 crosslinked products were divided into two aliquots: one was digested by trypsin alone and the other by trypsin/endoproteinase GluC. Digested peptides were extracted with 50 μL Millipore water containing 1% trifluoroacetic acide (TFA) by vortexing 10 min at room temperature. An additional extraction was performed with 80 μL of 70% acetonitrile/25% H2O/5% TFA (v/v/v). Peptides were dried in a vacuum centrifuge, reconstituted with 20 μL H2O containing 0.1% TFA for injection onto a Linear Trap Quarupole (LTQ) Orbitrap XL using an Agilent 1100 LC system at the University of Wisconsin-Madison Mass Spectrometry/Proteomics Facility. The peptides were loaded directly onto an analytical column of 75 μm inner diameter and 360 μm outer diameter house-packed with ~13 cm C18 resin (Magic-C18, 200Å, 3 μm, Bruker, Billerica, MA). MS1 spectra were collected using the Orbitrap at resolution 100,000 with preview mode enabled. The raw MS data were processed into peak lists using Mascot server. The StavroX 3.5.1 software (Gotze et al., 2012) was used to generate a list of the crosslinked peptides. The false discovery rate (FDR) cut off was set to 5%, and precursor and fragment ion tolerances were set to 10 ppm and 0.6 Da, respectively. Each crosslinked peptide was also assigned a score by StavroX 3.5.1 software based on comparison between the theoretical fragmentation and the actual MS/MS spectrum of the crosslinked peptide. Tables S1–S3 list detailed information on the generated crosslinked peptides, including the StavroX scores and predicted Cα distances.
Small angle X-ray scattering
The buffer used for the SAXS experiments (HNT buffer) contained 20 mM HEPES at pH 7.5, 150 mM NaCl, and 5 mM TCEP. Samples of (NIA)2, (NIAU)2, and (NIAUF)2 were purified with a HiLoad Superdex 200 PG size exclusion column (GE Healthcare) and dialyzed extensively against HNT buffer with three buffer changes. Protein samples were clarified by passage through a 0.2 mm filter before loading into a glass capillary cell. SAXS data for each protein or protein complex were collected at three concentrations ranging from 2 to 10 mg/mL. No significant inter-particle interactions were observed for any of the concentrations used in our SAXS studies. SAXS experiments were carried out on a Bruker Nanostar benchtop SAXS system (Bruker AXS) at the National Magnetic Resonance Facility at Madison (NMRFAM) equipped with a rotating anode (Cu) Turbo X-ray Source (1.5418 Å wavelength) and a VÅNTEC-2000 (2048X2048 pixel) detector. The sample-to-detector distance was set at ~1 m, allowing for the detection range: 0.012 > q > 0.30 Å−1. Forty microliters of protein and buffer samples were loaded separately into a capillary cell with 1 mm diameter, and scattering data were collected for 4 hr at 25 °C. The ATSAS software suite (Petoukhov et al., 2012) was used to process the SAXS data. The Rg for each protein or protein complex was determined by using the Guinier approximation in the q range (qmax Rg) < 1.3. Pairwise distance distribution functions (Pr) were obtained using GNOM software (Svergun, 1992). The output from GNOM was then used in conjunction with DAMMIF (Franke and Svergun, 2009) to generate 20 independent ab initio dummy atom models to assess the molecular shape of each sample. P2 symmetries were enforced during the calculations. Most of the models exhibited excellent agreement with experimental data and had a normalized spatial discrepancy (NSD) < 1. CRYSOL software (Svergun et al., 1995) or FoXS webserver (Schneidman-Duhovny et al., 2013, 2016) was used to compare the protein structures with experimental SAXS data. Supcomb software (Kozin and Svergun, 2001) was used to superimpose protein structures on to the SAXS ab initio dummy atom models. Vc approach was used for the molecular mass calculation from the SAXS data (Rambo and Tainer, 2013).
Homology modeling
The HADDOCK2.2 webserver (de Vries et al., 2010; van Zundert et al., 2016) (expert interface) was used to dock FXN (PDB: 1EKG) onto the structure of (NIAU)2 (PDB: 5WLW) to generate the structural model of (NIAUF)2 complex. Crosslink lengths were used to define unambiguous intermolecular distance constraints. Chemical shift perturbations observed upon complex formation were used to define ambiguous interaction constraints (AIRs) (Cai et al. 2018b). Active residues were defined as those having either chemical shifts changes larger than 0.03 ppm or severe peak broadening. Passive AIRs were defined automatically. A total of 5,000 rigid-body docking trials were carried out using the standard HADDOCK protocol. The 200 lowest-energy solutions were used for subsequent semiflexible simulated annealing and water refinement. The lowest-energy structure was used to represent the complex.
Cysteine desulfurase assay
The cysteine desulfurase assay reaction mixtures (300 μL in the buffer containing 20 mM HEPES pH 7.5 and 150 mM NaCl) contained 1 μM non-crosslinked (NIAU)2, non-crosslinked (NIAUF)2, BS3-crosslinked (NIAU)2 or SMCC-crosslinked (NIAUF)2. 500 μM DTT and 500 μM L-cysteine were added to initiate the reaction. After incubation for 30 min at room temperature, the reaction mixture was diluted to 800 μL, and 100 μL of 20 mM N,N-dimethyl-p-phenylenediamine in 7.2 M HCl and 100 μL of 30 mM FeCl3 in 1.2 M HCl was added to quench the reaction and convert sulfide to methylene blue. The quenched reaction mixture was incubated for 15 min at room temperature, and the absorbance at 670 nm was measured and used to estimate the amount of sulfide by comparison to a standard curve obtained from known concentrations of Na2S. No activity was observed with control experiments in which (NIAU)2 or L-cysteine was omitted. All experiments were performed in triplicate, and the averages and errors are reported.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analyses are given in Figure S5 (cysteine desulfurase assay), which was performed with triplicates (n=3). The statistical program was Excel.
DATA AND SOFTWARE AVAILABILITY
The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifiers PXD006928/10.6019/PXD006928, PXD006938/10.6019/PXD006938 and PXD009079/10.6019/PXD009079. SAXS data for (NIA)2, (NIAU)2 and (NIAUF)2 were deposited into Small Angle Scattering Biological Data Bank (S The mass spectrometry proteomics data were deposited into the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the dataset identifiers PXD006928/10.6019/PXD006928, PXD006938/10.6019/PXD006938 and PXD009079/10.6019/PXD009079. SAXS data for (NIA)2, (NIAU)2 and (NIAUF)2 were deposited into Small Angle Scattering Biological Data Bank (SASBDB) with entry IDs SASDDB3, SASDDC3, and SASDDD3. The structural model of (NIAUF)2 was deposited into the integrative/hybrid structure database PDB-DEV (https://pdb-dev.wwpdb.org) (Burley et al., 2017) with accession code PDBDEV_00000015.ASBDB) with entry IDs SASDDB3, SASDDC3, and SASDDD3. The structural model of (NIAUF)2 was deposited into the integrative/hybrid structure database PDB-DEV (https://pdb-dev.wwpdb.org) (Burley et al., 2017) with accession code PDBDEV_00000015.
Supplementary Material
Highlights.
We prepared cysteine desulfurase and its complexes with ISCU and ISCU-frataxin.
We identified structural restraints for these complexes from XL-MS and SAXS.
We compared these restraints with published X-ray structures.
We derived a structural model for the cysteine desulfurase-ISCU-frataxin complex.
Acknowledgments
The authors thank Brinda Vallat at the Research Collaboratory for Structural Bioinformatics-Protein Data Bank for assistance with the PDB-Dev deposition and Marco Tonelli for collecting the 19F NMR spectrum. This work received support from the Biochemistry Department, University of Wisconsin-Madison. R.O.F. is supported by NIH grant P41GM103399 and H.D. is supported by NIH grant P41 P41GM111135. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394). SAXS studies were supported by funds from NIH shared instrumentation grant S10RR027000 and the University of Wisconsin-Madison.
Footnotes
Competing Interests
The authors declare no competing interests.
Table S1, MS data on peptides from (NIAU)2 crosslinked with bis-(sulfosuccinimidyl)suberate, related to Figure 2. Table S2. MS data on peptides from NFS1-FXN crosslinked with sulfo-SMCC, related to Figure 6. Table S3, MS data on peptides from FXN-NFS1-ISCU crosslinked with sulfo-SMCC, related to Figure 6. Figures S1–S6.
Author Contributions
Conceptualization, J.L.M., K.C.; Methodology, K.C., H.D., R.O.F., J.L.M.; Investigation, K.C., H.D.; Writing – Original Draft, K.C., J.L.M; Writing – Review & Editing, K.C., H.D., R.O.F., J.L.M.; Funding Acquisition, J.L.M.; Resources, J.L.M.; Supervision, J.L.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam AC, BornhÖvd C, Prokisch H, Neupert W, Hell K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006;25:174–183. doi: 10.1038/sj.emboj.7600905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochemical Society transactions. 2013;41:1335–1341. doi: 10.1042/BST20130116. [DOI] [PubMed] [Google Scholar]
- Angerer H. Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology. 2015;4:133–150. doi: 10.3390/biology4010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer H, Radermacher M, Mankowska M, Steger M, Zwicker K, Heide H, Wittig I, Brandt U, Zickermann V. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniecki MT, Freibert SA, Muhlenhoff U, Lill R, Cygler M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nature communications. 2017;8:1287. doi: 10.1038/s41467-017-01497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. The Journal of biological chemistry. 2017;292:12754–12763. doi: 10.1074/jbc.R117.787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM, Barondeau DP. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry. 2014;53:4904–4913. doi: 10.1021/bi500532e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Kurisu G, Markley JL, Nakamura H, Velankar S, Berman HM, Sali A, Schwede T, Trewhella J. PDB-Dev: a Prototype System for Depositing Integrative/Hybrid Structural Models. Structure. 2017;25:1317–1318. doi: 10.1016/j.str.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Frederick RO, Kim JH, Reinen NM, Tonelli M, Markley JL. Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU) J Biol Chem. 2013;288:28755–28770. doi: 10.1074/jbc.M113.482042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Frederick RO, Tonelli M, Markley JL. Mitochondrial cysteine desulfurase and ISD11 coexpressed in Escherichia coli yield complex containing acyl carrier protein. ACS Chem Biol. 2017a;12:918–921. doi: 10.1021/acschembio.6b01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Tonelli M, Frederick RO, Markley JL. Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis. Biochemistry. 2017b;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Tonelli M, Frederick RO, Markley JL. Conformational and functional properties of human ISCU and the mechanism of frataxin-bypassing in iron-sulfur cluster assembly. Biochemistry. 2018a;57:1491–1500. [Google Scholar]
- Cai K, Tonelli M, Frederick RO, Markley JL. Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J Inorg Biochem. 2018b;183:107–116. doi: 10.1016/j.jinorgbio.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri BN. Emerging applications of small angle solution scattering in structural biology. Protein science: a publication of the Protein Society. 2015;24:267–276. doi: 10.1002/pro.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. Embo J. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin F, Martelli A, Clemancey M, Latour JM, Gambarelli S, Zeppieri L, Birck C, Page A, Puccio H, Ollagnier de Choudens S. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. Journal of the American Chemical Society. 2013;135:733–740. doi: 10.1021/ja308736e. [DOI] [PubMed] [Google Scholar]
- Cook JD, Kondapalli KC, Rawat S, Childs WC, Murugesan Y, Dancis A, Stemmler TL. Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe-S cluster assembly. Biochemistry. 2010;49:8756–8765. doi: 10.1021/bi1008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory SA, Van Vranken JG, Brignole EJ, Patra S, Winge DR, Drennan CL, Rutter J, Barondeau DP. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proceedings of the National Academy of Sciences of the United States of America. 2017 doi: 10.1073/pnas.1702849114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp-Vickery JR, Urbina H, Vickery LE. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. Journal of molecular biology. 2003;330:1049–1059. doi: 10.1016/s0022-2836(03)00690-9. [DOI] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE. Crystal structure of human frataxin. The Journal of biological chemistry. 2000;275:30753–30756. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- Dobrynin K, Abdrakhmanova A, Richers S, Hunte C, Kerscher S, Brandt U. Characterization of two different acyl carrier proteins in complex I from Yarrowia lipolytica. Biochimica et biophysica acta. 2010;1797:152–159. doi: 10.1016/j.bbabio.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Feng D, Witkowski A, Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. The Journal of biological chemistry. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M. Iron-sulfur clusters: ever-expanding roles. Nat Chem Biol. 2006;2:171–174. doi: 10.1038/nchembio0406-171. [DOI] [PubMed] [Google Scholar]
- Fox NG, Das D, Chakrabarti M, Lindahl PA, Barondeau DP. Frataxin Accelerates [2Fe-2S] Cluster Formation on the Human Fe-S Assembly Complex. Biochemistry. 2015;54:3880–3889. doi: 10.1021/bi5014497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Jeffries CM, Svergun DI. Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nature methods. 2015 doi: 10.1038/nmeth.3358. [DOI] [PubMed] [Google Scholar]
- Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, Sinz A. StavroX--a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- He Y, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen JK, Autio KJ, Schonauer MS, Kursu VA, Dieckmann CL, Kastaniotis AJ. Mitochondrial fatty acid synthesis and respiration. Biochim Biophys Acta. 2010;1797:1195–1202. doi: 10.1016/j.bbabio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annual review of biochemistry. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Kastaniotis AJ, Autio KJ, Keratar JM, Monteuuis G, Makela AM, Nair RR, Pietikainen LP, Shvetsova A, Chen Z, Hiltunen JK. Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology. Biochim Biophys Acta. 2017;1862:39–48. doi: 10.1016/j.bbalip.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. Embo J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- Leidgens S, De Smet S, Foury F. Frataxin interacts with Isu1 through a conserved tryptophan in its beta-sheet. Human molecular genetics. 2010;19:276–286. doi: 10.1093/hmg/ddp495. [DOI] [PubMed] [Google Scholar]
- Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends in biochemical sciences. 2016;41:20–32. doi: 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends in biochemical sciences. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicki M, Majewska J, Ciesielski S, Schilke B, Blenska A, Kominek J, Marszalek J, Craig EA, Dutkiewicz R. Overlapping binding sites of the frataxin homologue assembly factor and the heat shock protein 70 transfer factor on the Isu iron-sulfur cluster scaffold protein. J Biol Chem. 2014;289:30268–30278. doi: 10.1074/jbc.M114.596726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angewandte Chemie. 2012;51:5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- Pain D, Dancis A. Roles of Fe-S proteins: from cofactor synthesis to iron homeostasis to protein synthesis. Curr Opin Genet Dev. 2016;38:45–51. doi: 10.1016/j.gde.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Golla R, Yoon H, Dancis A, Pain D. Persulfide formation on mitochondrial cysteine desulfurase: enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem J. 2012;448:171–187. doi: 10.1042/BJ20120951. [DOI] [PubMed] [Google Scholar]
- Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A, Pain D. Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J Biol Chem. 2013;288:36773–36786. doi: 10.1074/jbc.M113.525857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A, Elduque X, Cornu D, Belot L, Le Caer JP, Grandas A, Toledano MB, D’Autreaux B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nature communications. 2015;6:5686. doi: 10.1038/ncomms6686. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nature communications. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Disease models & mechanisms. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nature reviews Molecular cell biology. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- Runswick MJ, Fearnley IM, Skehel JM, Walker JE. Presence of an acyl carrier protein in NADH:ubiquinone oxidoreductase from bovine heart mitochondria. FEBS letters. 1991;286:121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- Schmucker S, Martelli A, Colin F, Page A, Wattenhofer-Donze M, Reutenauer L, Puccio H. Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One. 2011;6:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic acids research. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang LH, Trempe JF, Matte A, Armengod ME, Cygler M. Structural Basis for Fe-S Cluster Assembly and tRNA Thiolation Mediated by IscS Protein-Protein Interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Human molecular genetics. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg CA, Shkumatov AV, Rajan S, Gakh O, Svergun DI, Isaya G, Al-Karadaghi S. Oligomerization propensity and flexibility of yeast frataxin studied by X-ray crystallography and small-angle X-ray scattering. Journal of molecular biology. 2011;414:783–797. doi: 10.1016/j.jmb.2011.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harbor perspectives in biology. 2013;5:a011312. doi: 10.1101/cshperspect.a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Tran BQ, Goodlett DR, Goo YA. Advances in protein complex analysis by chemical cross-linking coupled with mass spectrometry (CXMS) and bioinformatics. Biochimica et biophysica acta. 2016;1864:123–129. doi: 10.1016/j.bbapap.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Tsai CL, Barondeau DP. Human Frataxin Is an Allosteric Switch That Activates the Fe-S Cluster Biosynthetic Complex. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- Tuukkanen AT, Spilotros A, Svergun DI. Progress in small-angle scattering from biological solutions at high-brilliance synchrotrons. Iucrj. 2017;4:518–528. doi: 10.1107/S2052252517008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vranken JG, Jeong MY, Wei P, Chen YC, Gygi SP, Winge DR, Rutter J. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. Elife. 2016;5 doi: 10.7554/eLife.17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, Bonvin AMJJ. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. Journal of molecular biology. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Vaubel RA, Isaya G. Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Mol Cell Neurosci. 2013;55:50–61. doi: 10.1016/j.mcn.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R, et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. Embo J. 2006;25:184–195. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods in molecular biology. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Yoon H, Golla R, Lesuisse E, Pain J, Donald JE, Lyver ER, Pain D, Dancis A. Mutation in the Fe-S scaffold protein Isu bypasses frataxin deletion. Biochem J. 2012;441:473–480. doi: 10.1042/BJ20111637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Knight SA, Pandey A, Pain J, Zhang Y, Pain D, Dancis A. Frataxin-bypassing Isu1: characterization of the bypass activity in cells and mitochondria. The Biochemical journal. 2014;459:71–81. doi: 10.1042/BJ20131273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. Journal of the American Chemical Society. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.