Abstract
A specific TATA binding protein-associated factor (TAF), dTAFII110/hTAFII135, interacts with cAMP response element binding protein (CREB) through its constitutive activation domain (CAD), which recruits a polymerase complex and activates transcription. The simplest explanation is that the TAF is a coactivator, but several studies have questioned this role of TAFs. Using a reverse two-hybrid analysis in yeast, we previously mapped the interaction between dTAFII110 (amino acid 1–308) and CREB to conserved hydrophobic amino acid residues in the CAD. That mapping was possible only because CREB fails to activate transcription in yeast, where all TAFs are conserved, except for the TAF recognizing CREB. To test whether CREB fails to activate transcription in yeast because it lacks a coactivator, we fused dTAFII110 (amino acid 1–308) to the TATA binding protein domain of the yeast scaffolding TAF, yTAFII130. Transformation of yeast with this hybrid TAF conferred activation by the CAD, indicating that interaction with yTFIID is sufficient to recruit a polymerase complex and activate transcription. The hybrid TAF did not mediate activation by VP16 or vitamin D receptor, each of which interacts with TFIIB, but not with dTAFII110 (amino acid 1–308). Enhancement of transcription activation by dTAFII110 in mammalian cells required interaction with both the CAD and TFIID and was inhibited by mutation of core hydrophobic residues in the CAD. These data demonstrate that dTAFII110/hTAFII135 acts as a coactivator to recruit TFIID and polymerase and that this mechanism of activation is conserved in eukaryotes.
Transcription of a protein-coding gene requires the assembly of a large complex of general transcription factors and coactivators at the promoter to position RNA polymerase II correctly at the start site, melt the template, and initiate synthesis of an mRNA transcript (1, 2). The promoter-recognition factor, TATA binding protein (TBP), and RNA polymerase II often are isolated as components of large, preformed, multiprotein complexes in the cell, called TFIID and holoenzyme, respectively (3, 4). Activators bind to components of either or both complexes, i.e., to TBP or to one of the TBP-associated factors (TAFs) of TFIID or to the many coactivators and mediators that associate with polymerase complexes (3, 5–7), to establish a functional polymerase complex and stimulate the initiation of transcription.
The TAFs originally were proposed to serve as coactivators, molecules that bind activator and TFIID but not DNA, to mediate recruitment of TFIID to target promoters (8). However, a number of studies have questioned the role of TAFs as coactivators for recruitment of the transcription machinery. Temperature-sensitive TAFs that disrupt IID assembly are not lethal in yeast (9, 10), and TAFs were reported to serve other functions, including promoter recognition, chromatin remodeling, and covalent modification of transcription-regulating proteins (reviewed in ref. 6).
The cAMP response element (CRE) binding protein (CREB) binds constitutively to CRE-containing promoters and activates both basal and cAMP-mediated gene transcription in a variety of cell types (11–15). These functions map to separate activation domains in CREB, a constitutive activation domain (CAD) and a kinase-inducible domain (KID) (15–17). Although considerable attention has been focused on enhancement of transcription by phosphorylation of CREB on Ser-133 (18), deletion of the CAD substantially compromises kinase-induced transcription by reducing basal transcription to nearly undetectable levels (15). On the other hand, basal transcription activity of the CAD is independent of the KID and is unchanged by mutation of Ser-133 to Ala, which abolishes kinase-inducible transcription (15, 16, 18, 19).
TFIID is a large, multisubunit complex, consisting of TBP and 10–12 TAFs that are generally well conserved from yeast to humans (3). An important exception is that yeast do not contain a homologue of the metazoan TAF interacting with CREB (dTAFII110/hTAFII135) (20–22). We previously showed that CREB binds to TFIID through its CAD and that this interaction is mediated by TAFs (23). CREB and dTAFII110 interact in a yeast two-hybrid assay (20, 24), and the three subdomains in the CAD that are required for activation in vivo (25) also are required for this association (21). Mutations in any of several hydrophobic residues within a conserved motif in the CAD abrogate interaction with dTAFII110-AD in yeast (21). A similar analysis defined a hydrophobic face in hTAFII135, the human homologue of dTAFII110, that is required for interaction with Sp1 and CREB (26), suggesting that the two proteins interact through a shared hydrophobic surface.
The CREB/TAF-interaction studies depended on the inability of CREB to activate transcription in yeast. The absence of a functional homologue of dTAFII110/hTAFII135 in yeast (3) suggested to us that CREB might activate transcription in yeast if an appropriate target was provided for recruitment of yeast TFIID and a polymerase complex. To test this hypothesis, we constructed a hybrid TAF in which the domain of dTAFII110 (amino acid 1–308) that interacts with CREB was fused to the yeast-scaffolding TAF that binds directly to yeast TBP, yTAFII130 (27). We also evaluated both the coactivator potential of the TAF, which requires simultaneous binding of the activator and TFIID, and the necessity for the hydrophobic surface of the CAD. The results strongly support a role for dTAFII110/hTAFII135 as a coactivator that mediates recruitment of TFIID and RNA polymerase II by the CREB CAD to establish basal transcription, both in yeast and metazoans.
Materials and Methods
Expression and Purification of Proteins.
Proteins were synthesized in vitro by coupled transcription/translation (TNT system; Promega). Coding regions for the wild-type or mutated CADs were subcloned into the pET-3d expression vector (Novagen). TNT reactions were performed according to the manufacturer's instructions, and excess salt was removed from the reaction products in a Sephadex G-25 spin column. Recombinant CREB–Gal-4 (CRG) proteins were expressed in baculovirus and purified from Sf9 nuclear extracts by using DNA-affinity chromatography, as described (28).
Recruitment Assay.
The recruitment assay, using agarose electrophoretic mobility shift, was reported elsewhere (29). The method is an adaptation of the original agarose–electrophoretic mobility-shift assay (Ag-EMSA) method (30) for studying complex assembly of general factors in nuclear extracts. Probe DNA (150 bp), containing five Gal-4 binding sites (5XG) and a minimal TATA-containing promoter, was end-labeled with [32P]dATP by using the Klenow fragment of DNA polymerase. Five femtomoles of probe was incubated with 10 fmol of purified protein and 3 μg of rat liver nuclear extract [RLNE, prepared as described previously (28, 31)] in 1× Ag-EMSA binding buffer (12.5 mM Hepes, pH 7.9/12.5% glycerol/5 mM MgCl2/70 mM KCl/0.2 mM EDTA/10 mM 2-mercaptoethanol/0.5 mg/ml BSA/40 μg/ml poly-dIdC) for 15 min at 4°C. Reaction components were separated on a 1% Seakem agarose gel for 2 h at 100 V. The gel was dried overnight at room temperature and exposed to film to visualize labeled complexes. To determine whether specific proteins were included in the complexes, 1 μl of primary antibody was mixed with 3 μg of RLNE in 1× Ag-EMSA binding buffer for 1 h at 4°C. Ten femtomoles of purified CRG protein, 2 μl of biotinylated secondary antibody, and 3 μl of streptavidin-coated Dynabeads were added, and the incubation was continued for 1 h at 4°C. Probe (5 fmol) then was added, and the samples were incubated for an additional 15 min at 4°C. Reaction components were separated on a 1% Seakem agarose gel and visualized, as described above.
Transient Transfection Experiments.
Plasmid DNA was prepared by a standard CsCl/EtdBr density gradient-purification method. Transfection experiments were performed with JEG3 choriocarcinoma cells in culture, using a calcium phosphate protocol, as described previously (19, 32). Cells were cotransfected with 1 μg of expression plasmid, 10 μg of a 5XG-luciferase reporter plasmid, and 1 μg pRL-SV to normalize for differences in transfection efficiency. Luciferase activity of cell lysates was determined by using the Dual Luciferase Assay kit (Promega) (33).
Plasmids.
Partial coding regions encompassing the mutations within the CAD were exchanged with the wild-type CRG sequence in pCRG (15), using the SacI and HpaI sites. The dTAFII110ΔN expression plasmid was constructed from the full-length clone pTβ-dTAFII110 (R. Tjian; ref. 34) by using site-directed mutagenesis to insert a SalI site at codon 6. This site then was used in conjunction with the naturally occurring SalI site at amino acid 308 to delete the N-terminal 308 aa of the TAF. pTβ-dTAFII110ΔC was obtained from D. Wassarman (34) and contains a stop codon after amino acid 795, leading to a C-terminal truncation of the TAF protein. The TAF coding regions from these two plasmids and the full-length TAF were subcloned into the mammalian expression vector pRT by exchanging NcoI/BamHI fragments with that of pRT-CREB (19).
To prepare yeast expression plasmids for the hybrid TAF proteins, a SalI site and a stop codon were inserted at the 3′ boundary of the coding region of pET-his-myc-yTAFII130N100 (P. Weil) (27) by site-directed mutagenesis. The N-terminal 308 aa of dTAFII110 were transferred from pTβ-dTAFII110 + SalI into this plasmid at the new SalI site. The TAF coding regions of the new plasmids and control plasmids, yTAFII130N100 and the dTAFII110 1–308, were inserted as AflII/BglII fragments into the yeast expression plasmid pGAD-10 (CLONTECH).
pGN-G4-VP16 was constructed by inserting the BamHI fragment from pG4-VP16 (S. Triezenberg) into the BamHI site of the yeast expression vector pGN-1 (35). G4-VDR (vitamin D receptor) (pAS-1-VDR) and TFIIB-AD (pGAD.GH-TFIIB-WT) were described by MacDonald et al. (36).
Yeast Strain and Analyses.
The yeast strain Y-153 (gal4−, leu−, trp−, Gal:LacZ, etc.) was described by the S. Elledge laboratory (37). Yeast were transformed as described previously (21, 38). Yeast protein lysates were prepared by glass-bead lysis, as described previously (21). For the quantitative liquid assay, 2–5 mg of protein lysate was added to 1 ml of Z buffer and incubated with 200 μl of 4 mg/ml o-nitrophenyl-β-d-galactopyranoside at 30°C until a yellow color developed. The reaction was stopped with 0.5 ml of 1 M Na2CO3, and the absorbance at 420 nm was determined spectrophotometrically. To obtain specific activities, values were adjusted for incubation time and protein concentration.
Results
A Hybrid TAF Recognizing the CAD and Yeast TBP Is Sufficient to Mediate Activation in Yeast.
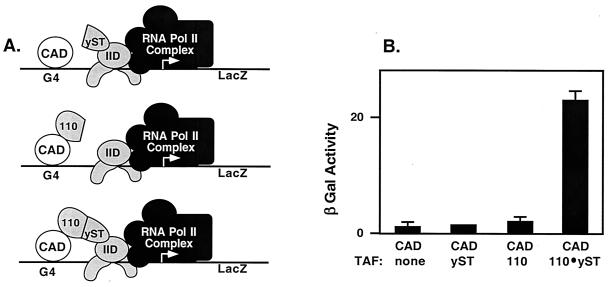
CREB does not activate transcription in Saccharomyces cerevisiae (20, 21). Yeast do not express a functional homologue of the dTAFII110/hTAFII135 subunit of TFIID that recognizes the CREB CAD (3). Other activators, such as Sp1, that interact with dTAFII110/hTAFII135 also fail to activate transcription in yeast (8, 39). Those observations suggested to us that the failure of CREB to activate transcription in yeast may result from the lack of a coactivator TAF in yeast TFIID. We predicted that integration of the CREB-interacting domain of dTAFII110 into yeast TFIID would allow CREB to activate transcription in yeast. Accordingly, we designed an expression vector encoding a hybrid TAF (labeled 110-yST in Fig. 1A), in which the CREB CAD-interacting domain of dTAFII110 (amino acid 1–308, labeled 110 in Fig. 1A) is fused to the domain of the yeast scaffolding TAF, yTAFII130 (amino acid 1–100, labeled yST in Fig. 1A), that directly binds to TBP (ref. 27; Fig. 1A). Yeast strain Y-153, bearing a Gal-4–LacZ reporter gene and lacking endogenous Gal-4 protein, was transformed with the expression plasmid for the fusion protein, or a control plasmid expressing either domain alone, together with a CAD-G4 expression plasmid. Transformants were assayed for β-galactosidase activity. Fig. 1B shows that the CAD alone did not activate the reporter, nor did either of the components of the hybrid TAF, yST or 110. In contrast, the hybrid TAF (110-yST: dTAFII110-yTAFII130) fusion protein mediated substantial activation by the CREB CAD in yeast, indicating that the provision of a coactivator for recruitment of TFIID is sufficient to allow transcription activation.
Figure 1.
A hybrid TAF recognizing the CAD and yeast TBP is sufficient to mediate activation in yeast. (A) Schematic diagram depicting the components (110 and yST) of the hybrid TAF (110-yST) and their interaction partners, CAD and TBP of IID. (B) Expression vectors for the hybrid dTAFII110/yTAFII130 fusion protein, the single TAF domain control proteins, and CAD-G4 were transformed into the yeast strain Y-153, where indicated. Colonies were grown in liquid culture, protein lysates were made, and β-galactosidase activity was determined by using the o-nitrophenyl-β-d-galactopyranoside synthetic substrate assay. Data represent three independent transformations, with three colonies assayed from each.
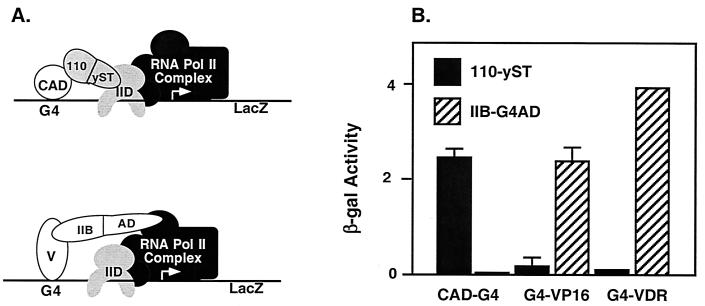
To determine whether the result obtained with the hybrid TAF was specific for the CREB CAD, we tested two other activators in this system. The viral activator VP16 does not interact with dTAFII110/hTAFII135 but does interact with TFIIB (40). The VDR interacts with dTAFII110 in a domain beyond the N-terminal 308 aa used in this experiment (41) and also interacts with TFIIB (36). To control for proper expression and activity by G4-VP16 and G4-VDR, we used a standard two-hybrid assay with a TFIIB-AD plasmid expressing the target domain of TFIIB fused to the yeast Gal-4 activation domain (IIB-AD, Fig. 2A). Yeast were transformed with expression plasmids for CAD-G4, G4-VP16, or G4-VDR, along with the hybrid TAF or TFIIB-AD. As predicted, the hybrid TAF (110-yST) mediated activation by CAD-G4 but not by G4-VP16 or G4-VDR (Fig. 2B). Conversely, IIB-AD mediated activation by G4-VP16 or G4-VDR but not by CAD-G4. Thus, all pairs of proteins had the specificity expected from their previously characterized interactions, indicating that stimulation by the hybrid TAF of transcription activation in yeast was selective. Together, these data indicate that provision of a CREB-interacting subunit in the yeast TFIID complex is sufficient to confer specific activation by the CREB CAD in yeast. The results strongly suggest that dTAFII110 serves as a coactivator of CAD-dependent transcription by acting as a direct target for the CAD to recruit TFIID and polymerase complexes.
Figure 2.
Activator-specific interactions mediate activation by hybrid TAF (110-yST) and a IIB-AD fusion protein. (A) The two hybrids tested are depicted with their interaction partners, V = VP16 or VDR. (B) Expression vectors for CAD-G4, G4-VP16, and G4-VDR were transformed into Y-153 with the hybrid TAF protein or with the TFIIB-AD two-hybrid expression vector as indicated. β-Galactosidase activity was determined by using the o-nitrophenyl-β-d-galactopyranoside synthetic substrate assay. Data represent three independent transformations, with three colonies assayed from each.
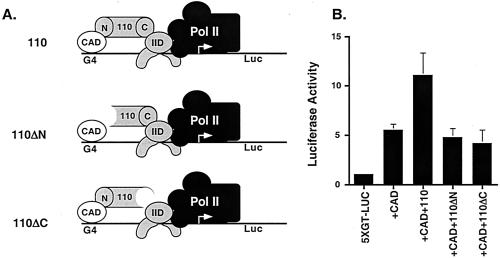
Truncated dTAFII110 Proteins That Lack Either the CREB- or IID-Interaction Domain Do Not Potentiate Transcription by the CAD in Mammalian Cells.
dTAFII110 interacts with the CREB CAD through its N terminus (amino acid 1–308) (20) and binds to the scaffolding hTAFII250 through its C terminus (amino acid 796–921), integrating it into TFIID (42) (Fig. 3A). Both domains would be required for coactivator function. To determine the contributions of the binding regions of dTAFII110 to CAD activation, we tested dTAFII110 constructs deleted of either domain for potentiation of CAD activation in mammalian cells. Full-length dTAFII110 potentiated transcription activation by the CAD in vivo (Fig. 3B). However, neither the N-terminal truncation (deleted of amino acid 1–308) nor the C-terminal truncation (deleted of amino acid 796–921) mutant of dTAFII110 produced levels of luciferase distinguishable from samples in which no dTAFII110 expression vector was added. This result demonstrates that the regions of dTAFII110 that interact with the CAD and integrate it into the TFIID complex are required for activation, as would be expected for a coactivator of the CREB CAD.
Figure 3.
Truncated dTAFII110 proteins that lack either the CREB- or IID-interaction domain do not potentiate transcription by the CAD in mammalian cells. (A) Schematic showing the regions of interaction in the dTAFII110 protein for CAD (N) and hTAFII250 (C) of TFIID. (B) Mammalian expression vectors for dTAFII110 proteins with N-terminal (Δ1–308) and C-terminal (Δ796–921) truncations, as well as full-length dTAFII110, were tested in transient transfection experiments in JEG3 cells. An expression vector for wild-type CAD and a 5XG-luciferase reporter plasmid were included as indicated. Luciferase assays were done on cell lysates to determine the relative amounts of activation. Data represent five independent experiments. Error bars depict standard error of the data.
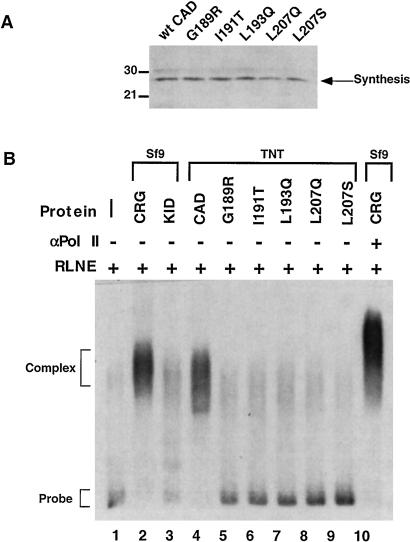
CAD Mutants That Are Defective for dTAFII110 Binding Fail to Recruit an RNA Polymerase II Complex to the Promoter in Vitro.
A prediction of the coactivator hypothesis for a TAF is that it will mediate recruitment of TFIID and a polymerase complex. We previously described an Ag-EMSA to measure activator-dependent formation of an RNA polymerase II-containing complex on a promoter DNA probe under the same conditions used to assay in vitro transcription activation (29). The activator-dependent complex that forms on the promoter contains essential components of a functional polymerase complex, including RNA polymerase II, TBP, and TFIIB (29). Here, we used this assay to assess recruitment by wild-type CAD and CAD mutants impaired for TAF binding. Western blotting with a Gal-4 antibody showed that similar amounts of CAD-G4 fusion proteins were synthesized in vitro (Fig. 4A). CRG and KID-G4 were expressed in Sf9 cells, using recombinant baculovirus, and purified by DNA affinity chromatography. CRG, KID-G4, or CAD-G4 proteins were incubated with RLNE, as a source of DNA polymerase II (pol II) and general factors, and a 32P-labeled DNA probe consisting of five Gal-4 binding sites and a minimal TATA-containing promoter (5XGT). As observed previously (29), RLNE and probe alone (Fig. 4B, lane 1) formed only a small amount of complex on the DNA. The addition of CRG produced an abundant complex that contains RNA polymerase II, as shown by the reduced mobility obtained with an antibody against RNA polymerase II cross-linked with biotinylated secondary antibody and streptavidin beads (Fig. 3B, lane 10). KID-G4 had no effect, as in a previous study (29). CAD-G4 stimulated complex recruitment, but CAD-G4 mutants defective for interaction with dTAFII110 produced only background amounts of complex. This result indicates that recruitment of a polymerase complex to the promoter requires the hydrophobic surface of the CAD that interacts with the dTAFII110 subunit of TFIID. Thus, mutation of the CAD that prevents interaction with the TAF results in failure to recruit a polymerase complex.
Figure 4.
CAD mutants that are defective for dTAFII110 binding fail to recruit an RNA polymerase II complex to the promoter in vitro. (A) Wild-type CAD and the five mutant CAD proteins were synthesized without 35S label by using the TNT system. Protein samples were run through a sephadex G-25 spin column to remove excess salts from the reactions. An aliquot of each purified TNT sample (15 μl) was tested by Western blotting with a primary antibody specific for the Gal-4 DNA binding domain to determine the amount synthesized. (B) Wild-type and mutant CAD proteins were tested for complex recruitment in an Ag-EMSA assay (see Materials and Methods). 32P-labeled probe (5 fmol) was incubated with 3 μg of RLNE and 10 fmol of the appropriate purified proteins as indicated. The sample in lane 10 included primary antibody specific for the large subunit of RNA polymerase II, biotinylated secondary antibody, and streptavidin-coated beads to supershift the complex. Reactions were incubated at 4°C for 15 min and then separated on a 1% Seakem agarose gel. The gel was dried and exposed to film to visualize the complexes.
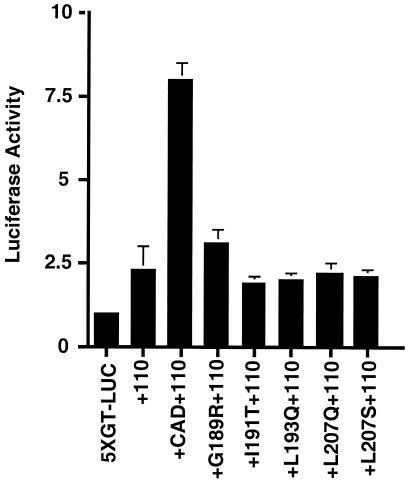
CAD Mutants That Are Defective for dTAFII110 Binding and Recruitment also Are Impaired for Stimulation of Transcriptional Activation in Mammalian Cells.
Another prediction of the coactivator hypothesis for a TAF is that it will mediate transcription activation on recruitment of TFIID and a polymerase complex. To determine the functional significance of CAD/TAF-mediated recruitment of RNA polymerase II observed in vitro, we tested TAF interaction-defective CAD mutants for activation of transcription in transiently transfected mammalian cells. The five CAD mutants that were most impaired for dTAFII110/AD interaction in the yeast two-hybrid assay were subcloned into mammalian expression vectors and cotransfected into JEG3 cells with a 5XGT-luciferase reporter plasmid and an expression vector for the full-length dTAFII110 (Fig. 5). None of the interaction-defective mutants stimulated transcription above the level seen in cells with no CAD expression plasmid. This result indicates that the CAD/TAF interaction, which promotes recruitment in vitro, also is necessary for transcription activation in mammalian cells. Thus, the coactivator function of the TAF requires interaction with the CREB CAD to mediate polymerase recruitment and transcription activation.
Figure 5.
CAD mutants that are defective for dTAFII110 binding and recruitment are also impaired for stimulation of transcriptional activation in mammalian cells. Mammalian expression vectors for the CAD mutants, as well as for wild-type CAD, were cotransfected into JEG3 cells in culture along with a 5XGT-luciferase reporter plasmid and an expression vector for full-length dTAFII110, where indicated. Luciferase assays were performed as described on all cell lysates to determine the level of transcription activation. Data are representative of three independent experiments. Error bars represent the standard error of the data.
Discussion
The work presented here provides a genetic link between the interaction of a specific TAF in TFIID with CREB, recruitment of a polymerase complex, and stimulation of transcription. The coactivator, dTAFII110/hTAFII135, must interact with hydrophobic amino acids in the CREB CAD and with the scaffolding TAF of TFIID to recruit a polymerase complex and activate transcription. Loss-of-function and gain-of-function experiments indicate that the interaction between the CREB CAD and the TAF in TFIID is necessary and sufficient to mediate recruitment of a polymerase complex and to activate transcription, both in yeast and in mammalian cells. Recruitment is an essential step for any subsequent activities of CREB. Therefore, the CAD not only mediates basal transcription but also contributes to kinase-mediated transcription activation by CREB, which occurs through a concerted mechanism involving stimulation of subsequent steps in the transcription-initiation reaction (28).
The demonstration that a hybrid TAF, which establishes communication between the promoter-recognition factor, TBP, and the CREB CAD, is sufficient to confer transcription activation in yeast provides strong evidence for an essential role of dTAFII110/hTAFII135 as a coactivator. Neither of the hybrid TAF's interaction domains had any effect alone. Therefore, they must be tethered to serve as a coactivator, rather than simply inducing allosteric changes in the proteins they bind. The recently characterized yeast homologue of dTAFII110/hTAFII135, yTAFII48 (43), lacks the amino-terminal domain of metazoan TAFs that is required for interaction with CREB and Sp1. As a result, one would not expect yTAFII48 to mediate activation by the mammalian transcription factors.
In contrast to the result with the 110-yST hybrid TAF reported here, when the N-terminal domain of dTAFII110 was fused directly to yeast TBP (110-TBP), that hybrid did not confer activation by G4-Sp1 in yeast (44). The different results obtained in these studies could be due to a difference between Sp1 and CREB CAD in the strength or type of interaction with dTAFII110 or from different activities of the two hybrid TAFs, 110-TBP and 110-yST. In the CREB CAD, the main concentration of glutamine residues is distinct from the hydrophobic cluster of amino acids (21, 25), whereas in the Sp1 activation domain, the glutamines are interspersed with hydrophobic residues (24). Thus, despite the importance of the hydrophobic cluster of amino acids to both activators, they may interact differently with dTAFII110. The other possibility is that the 110-TBP and 110-yST fusion proteins have significantly different activities. In this regard, Bai et al. (27) showed that the binding of yST to yTBP actually destabilized TBP binding to the TATA box. Thus, it is possible that the interaction between activator and TAF coactivator changes the nature of the interactions between the TAFs, TBP, and DNA in a way that would allow the 110-yST molecule to provide a function missing in the 110-TBP molecule. Regardless, our experiment clearly demonstrates that provision of a recognition surface for CREB within the yeast TFIID complex is sufficient for the activation of transcription.
Additional evidence that dTAFII110 serves as a coactivator for CREB was provided by showing that deletion of the domains of dTAFII110 binding the CAD or the scaffolding TAF that integrates it into TFIID complexes prevented augmentation of CAD-G4 activity. The potentiation of CAD-mediated transcription by full-length dTAFII110 suggests that it is functionally homologous with its mammalian counterpart hTAFII135, as predicted from the conservation of sequence in the domains that interact with activators and scaffolding TAFs (22). This result also suggests that a fraction of the TBP in cells is exchangeable and free to bind dTAFII110, consistent with reports that the TFIID pool in mammalian cells is heterogeneous (3, 45, 46). As a result, overexpression of dTAFII110 may increase the proportion of the TFIID pool available to mediate transcription activation by the CAD. A similar effect was observed with augmentation of activation by steroid hormone receptors when cells were cotransfected with hTAFII135, which serves as a coactivator for these regulators through a distinct domain in the TAF (41).
The demonstration that mutations disrupting the CAD/TAF interaction impair polymerase recruitment and transcription activation provides additional evidence for the role of dTAFII110/hTAFII135 as a coactivator. Although we cannot rule out the possibility that the CREB CAD interacts with other components of the polymerase complex, no such interaction has been demonstrated, and unphosphorylated CREB was incapable of binding holoenzyme in HeLa cell nuclear extracts (47, 48). Thus, the available data are consistent with a sequential mechanism of polymerase recruitment, involving the recruitment of TFIID, followed by recruitment of the polymerase, as described in the original studies of template commitment (49, 50). Our results are also consistent with experiments demonstrating that tethering of TFIID to a promoter can bypass the requirement for activator in establishing transcription (51–54), whereas tethering of holoenzyme cannot (54).
The recruitment activity of CREB CAD, or other transcription factors binding TFIID, plays an important role in activation of kinase-induced transcription by CREB, because later steps in transcription activation are dependent on an assembled polymerase complex. The CAD was as effective as CRG in recruiting a functional polymerase complex to the promoter in single-round transcription assays (28). A key finding of our previous studies was that phosphorylated KID-G4 could not stimulate recruitment or transcription effectively in any of the functional assays (28, 29). In light of the present results, it seems clear that the lack of effect of phospho-KID is a consequence of the lack of recruitment of a polymerase complex. In some genes, the CREB CAD may play a primary role in this recruitment process, whereas in others, different factors, working alone or in concert with the CAD, may mediate recruitment. For example, mutation of the PEPCK CRE to a Gal-4 site in G4-PEPCK diminishes but does not eliminate basal activity of the gene (33). Thus, CRG stimulates both basal and kinase-inducible activities of the G4-PEPCK promoter, whereas KID-G4 does not affect basal activity but stimulates induction of G4-PEPCK by PKA, because other factors have recruited a polymerase complex (33). Thus, the CAD or another factor is required to assemble a polymerase complex so that phosphorylation of KID can stimulate its activity (28).
The coactivator function of dTAFII110/hTAFII135 documented here is an integral part of a concerted mechanism for maximal transcription activation by phosphorylated CREB and is consistent with observations in many different biological systems. Mutation of the CREs of many genes results in decreased basal transcription (11, 14, 55, 56). Similarly, deletion of the CAD by an alternative splice in the CREB-related factor, CREMα, inhibits transcription because CREMα competes for binding with full-length CREMτ, which retains the CAD (16, 57, 58). Finally, trinucleotide expansions encoding polyglutamine regions inhibit the CAD/TAF interaction with deleterious consequences for neurons (59). Overexpression of polyglutamine repeats causes a dose-dependent inhibition of transcription and stimulates apoptosis, both of which are alleviated by overexpression of the coactivator, hTAFII135 (59). Thus, even though antiapoptotic pathways rely on kinase signaling to CREB to activate survival genes (60, 61), the TAF-dependent recruitment activity of the CAD is an essential prerequisite to this activation. It will be interesting to determine which other factors or combinations of factors activate transcription in a similar manner, using TAF coactivators to establish a polymerase complex and distinct interactions to modify the activity of the polymerase complex to promote maximal transcription activation.
Acknowledgments
We thank Jingfang Lu for excellent technical assistance, Robert Tjian for dTAFII110 constructs, Tony Weil for the yTAF130 construct, Paul MacDonald for the G4-VDR and IIB-AD constructs, and David Spector for insightful discussions of the work and critical reading of the manuscript. This work was supported by National Institutes of Health Grant DK 43871 (to P.G.Q.).
Abbreviations
- TBP
TATA binding protein
- TAF
TBP-associated factor
- CREB
cAMP response element binding protein
- CAD
constitutive activation domain
- KID
kinase-inducible domain
- CRG
CREB–Gal-4
- RLNE
rat liver nuclear extract
- VDR
vitamin D receptor
- Ag-EMSA
agarose–electrophoretic mobility-shift assay
- pol II
DNA polymerase II
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Conaway R, Conaway J. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 2.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 3.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 4.Meyer V E, Young R A. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 5.Sauer F, Tjian R. Curr Opin Genet Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- 6.Albright S R, Tjian R. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 7.Lemon B, Tjian R. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 8.Pugh B, Tjian R. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 9.Moqtaderi Z, Bai Y, Poon D, Weil P, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 10.Walker S, Reese J, Apone L, Green M. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 11.Delegeane A M, Ferland L H, Mellon P L. Mol Cell Biol. 1987;7:3994–4002. doi: 10.1128/mcb.7.11.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesler W J, Vandenbark G R, Hanson R W. J Biol Chem. 1988;263:9063–9066. [PubMed] [Google Scholar]
- 13.Montminy M R, Bilezikjian L M. Nature (London) 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 14.Quinn P G, Wong T W, Magnuson M A, Shabb J B, Granner D K. Mol Cell Biol. 1988;8:3467–3475. doi: 10.1128/mcb.8.8.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn P G. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 16.Brindle P, Linke S, Montminy M. Nature (London) 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 17.De Cesare D, Fimia G M, Sassone-Corsi P. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 19.Xing L P, Quinn P G. Mol Endocrinol. 1993;7:1484–1494. doi: 10.1210/mend.7.11.8114762. [DOI] [PubMed] [Google Scholar]
- 20.Ferreri K, Gill G, Montminy M. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felinski E A, Quinn P G. J Biol Chem. 1999;274:11672–11678. doi: 10.1074/jbc.274.17.11672. [DOI] [PubMed] [Google Scholar]
- 22.Saluja D, Vassallo M, Tanese N. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing L, Gopal V K, Quinn P G. J Biol Chem. 1995;270:17488–17493. doi: 10.1074/jbc.270.29.17488. [DOI] [PubMed] [Google Scholar]
- 24.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing L P, Quinn P G. J Biol Chem. 1994;269:28732–28736. [PubMed] [Google Scholar]
- 26.Rojo-Niersbach E, Furukawa T, Tanese N. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- 27.Bai Y, Perez G M, Beechem J M, Weil P A. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Lu J-F, Quinn P. Proc Natl Acad Sci USA. 2000;97:11292–11296. doi: 10.1073/pnas.97.21.11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felinski E A, Kim J, Lu J, Quinn P G. Mol Cell Biol. 2001;21:1001–1010. doi: 10.1128/MCB.21.4.1001-1010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman P, Berk A. Genes Dev. 1991;5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 31.Gorski K, Caniero M, Schibler U. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 32.Quinn P G. J Biol Chem. 1994;269:14375–14378. [PubMed] [Google Scholar]
- 33.Yeagley D, Agati J, Quinn P. J Biol Chem. 1998;273:18743–18750. doi: 10.1074/jbc.273.30.18743. [DOI] [PubMed] [Google Scholar]
- 34.Sauer F, Wassarman D, Rubin G, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 35.Schena M, Yamamoto K R. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 36.MacDonald P R, Sherman D R, Dowd D R, Jefcoat S C J, DeLisle R K. J Biol Chem. 1995;270:4748–4752. doi: 10.1074/jbc.270.9.4748. [DOI] [PubMed] [Google Scholar]
- 37.Durfee T, Becherer K, Chen P, Yeh S, Yang Y, Kilburu A, Lee W, Elledge S. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 38.Shultz L D, Hofmann K J, Mylin L M, Montgomery D L, Ellis R W, Hopper J E. Gene. 1987;61:123–133. doi: 10.1016/0378-1119(87)90107-7. [DOI] [PubMed] [Google Scholar]
- 39.Ponticelli A S, Pardee T S, Struhl K. Mol Cell Biol. 1995;15:983–988. doi: 10.1128/mcb.15.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 41.Mengus G, May M, Carre L, Chambon P, Davidson I. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 42.Yokomori K, Chen J L, Admon A, Zhou S, Tjian R. Genes Dev. 1993;7:2587–2597. doi: 10.1101/gad.7.12b.2587. [DOI] [PubMed] [Google Scholar]
- 43.Sanders S L, Weil P A. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 44.Keaveney M, Struhl K. Genes Cells. 1999;4:197–203. doi: 10.1046/j.1365-2443.1999.00255.x. [DOI] [PubMed] [Google Scholar]
- 45.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 46.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 47.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 49.Van Dyke M W, Sawadogo M, Roeder R G. Mol Cell Biol. 1989;9:342–344. doi: 10.1128/mcb.9.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dyke M W, Roeder R G, Sawadogo M. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 51.Majello B, Napolitano G, De Luca P, Lania L. J Biol Chem. 1998;273:16509–16516. doi: 10.1074/jbc.273.26.16509. [DOI] [PubMed] [Google Scholar]
- 52.He S, Weintraub S J. Mol Cell Biol. 1998;18:2876–2883. doi: 10.1128/mcb.18.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nevado J, Gaudreau L, Adam M, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dorris D R, Struhl K. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim K S, Lee M K, Carroll J, Joh T H. J Biol Chem. 1993;268:15689–15695. [PubMed] [Google Scholar]
- 56.Lazaroff M, Patankar S, Yoon S O, Chikaraishi D M. J Biol Chem. 1995;270:21579–21589. doi: 10.1074/jbc.270.37.21579. [DOI] [PubMed] [Google Scholar]
- 57.Foulkes N S, Sassone-Corsi P. Cell. 1992;68:411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- 58.Foulkes N S, Laoide B M, Schlotter F, Sassone C P. Proc Natl Acad Sci USA. 1991;88:5448–5452. doi: 10.1073/pnas.88.12.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, et al. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 60.Riccio A, Ahn S, Davenport C M, Blendy J A, Ginty D D. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 61.Finkbeiner S. Neuron. 2000;25:11–14. doi: 10.1016/s0896-6273(00)80866-1. [DOI] [PubMed] [Google Scholar]