Abstract
In developmental biology, localization is everything. The same stimulus –a cell signaling event or expression of a gene – can have dramatically different effects depending on the time, spatial position, and cell types in which it is applied. Yet the field has long lacked the ability to deliver localized perturbations with high specificity in vivo. The advent of optogenetic tools, capable of delivering highly localized stimuli, is thus poised to profoundly expand our understanding of development. We describe the current state-of-the-art in cellular optogenetic tools, review the first wave of major studies showcasing their application in vivo, and discuss major obstacles that must be overcome if the promise of developmental optogenetics is to be fully realized.
One of the great unsolved problems in biology is that of developmental self-organization: how cells grow, move and differentiate into the structures that robustly and reproducibly carry out an organism’s essential functions. Our ability to observe and describe development has advanced rapidly in recent years. Light sheet microscopy has made it possible to collect whole embryo data at micron-scale spatial resolution over hours or days, and a growing arsenal of approaches (e.g. live-cell fluorescent biosensors, lineage tracers, and single-cell sequencing) can be used to track cells’ molecular and phenotypic changes over time. Together these advances suggest that comprehensive maps of cell lineage, signaling pathway activity and gene expression might be achievable, at least for those species and time periods of development that can be recapitulated on a microscope.
Yet compared to these advances in measurement, our ability to perturb developmental processes has lagged far behind. Closing this gap could easily be as transformative as the current revolution in live imaging and single-cell sequencing. This review will elucidate how optogenetic tools could overcome major challenges in perturbing development, survey recent advances in their development, describe key initial successes in developmental optogenetics and outline some experimental limitations that still need to be overcome.
Recent advances in optogenetic tools for probing development
These are heady days for users of intracellular optogenetic tools. Nearly a decade ago, a series of landmark papers described how light-sensitive protein domains could be repurposed as optogenetic tools [1–5]. These initial studies introduced cryptochrome (Cry), phytochrome (Phy), light-oxygen-voltage sensing (LOV) and blue light using flavin (BLUF) domains to a broader community of cell and developmental biologists, and extended rhodopsin-based GPCRs to many additional classes of G protein-coupled signaling processes. Yet despite some early successes, many of these approaches had substantial limitations: weak expression and the requirement of adding a small molecule chromophore (in the case of plant phytochromes) [1]; competition between multiple light-dependent reactions (Cry2 clustering versus heterodimerization) [3,6,7]; and “leakiness” leading to residual activity in the dark (early LOV-based tools) [8]. Over the past ten years, multiple cycles of refinement and application have expanded the reach of optogenetic tools. We refer the interested reader to thorough, extensive reviews on this growing suite of approaches [9,10] and highlight below a few recent developments that are likely to have an outsized and immediate impact on developmental biology.
LOV domains enable the controlled presentation of a wide range of linear amino acid motifs
One major innovation for expanding the optogenetic toolbox has been the ability to reversibly uncage a huge range of distinct linear amino acid motifs upon light stimulation. All of these approaches rely on a conserved feature of many LOV domains: upon blue light absorption, a conformational change leads to the undocking and unwinding of a ~20 amino acid C-terminal alpha helix (the Jα helix; Figure 1a). The Jα helix can thus be endowed with a dual function: as long as its dark-state helical structure is left intact, its amino acid sequence can be re-engineered to expose novel linear motifs upon light stimulation. This approach has led to an explosion of tools including light-inducible degrons [11], nuclear localization and export sequences [12–15], and control over protein-protein interaction [8,16].
Figure 1. Developmental optogenetics: from tools to application.
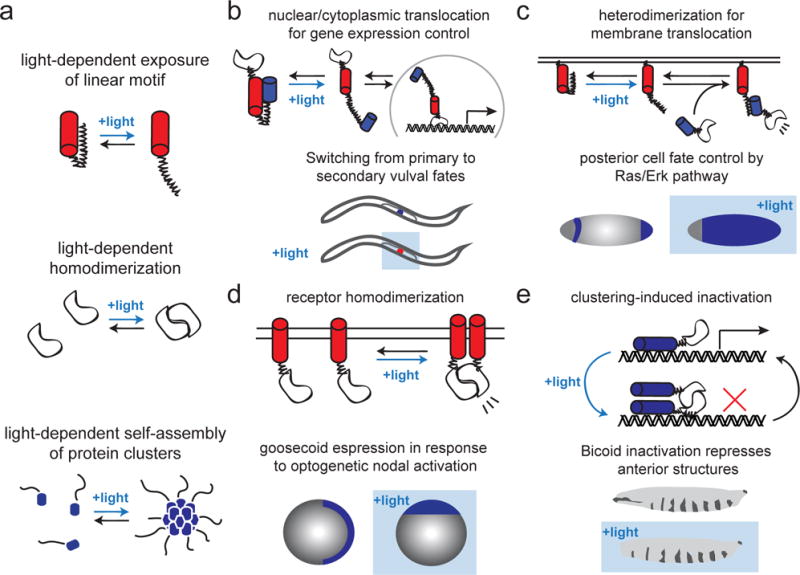
(a) Recent strategies for optogenetic control of intracellular protein activity: light-dependent display of a linear motif (e.g. degron, NLS, NES, or binding peptide); light-dependent homodimerization, and light-dependent assembly of higher-order clusters. (b–c) Classic examples of linear motif display and their applications in development, including membrane recruitment of a Ras activator, SOScat to pattern posterior fates in the early Drosophila embryo (b) and light-controlled nuclear import of the lin-1 transcription factor leading to a switch from primary to secondary vulval fates in C. elegans (c). (d–e) Classic examples of protein homo-oligomerization and their developmental applications, including homodimerization of cell surface receptors to activate Nodal signaling in zebrafish (d), clustering-based deactivation of the Bicoid transcription factor in Drosophila (e).
These light-switchable linear motifs have already begun to see application in developmental contexts. A recent study placed the nuclear import of the ETS transcription factor lin-1 under light control, leading to partial control of vulval fate (Figure 1b) [12]. This general strategy of reversibly controlling transcription factor localization is likely drive further insights into developmental gene regulatory networks. We took advantage of a second linear motif-based system, the iLID/SspB protein heterodimerization pair [16], to port the Ras-activating OptoSOS system [17] into the early Drosophila embryo, where it was shown to drive potent, localized control over MAP kinase signaling and cell fate (Figure 1c) [18].
In both studies, LOV domains proved to be highly attractive substrates for developmental control because the low blue light intensities required for photoactivation did not lead to cytotoxicity and precise spatial control could be achieved using inexpensive single-photon LED light sources. In principle, each of these tools could also be combined with mutations that are known to alter the lifetime of the illuminated, active state from seconds to hours [8,19,20]. Such photocycle mutants could enable the experimentalist to tune the tradeoff between the illumination intensity required for continuous activation (better for long-lived mutants) and the switching speed or spatial precision (both of which are better for short-lived mutants).
Light control of homo-oligomerization for activating or inhibiting intracellular processes
Light-switchable heterodimerization between two protein domains has long been achievable with a variety of optogenetic tools [1,3,16], but other forms of protein-protein interaction and assembly have only more recently been placed under light control (Figure 1a). For instance, the development of optical homo-dimerizers spurred the development of exciting new tools for gene expression control that will be discussed in detail below [21,22]. Optical dimerization also enables control over many classes of cell surface receptors whose dimerization leads to activation. Taking advantage of the homodimerizing LOV domain from Aureochrome 1, the Janovjak laboratory was able to establish light control over receptor tyrosine kinase (RTK) activity in mammalian cells and, in joint work with the Heisenberg lab, TGFβ receptor signaling during zebrafish embryogenesis (Figure 1d) [23,24].
What about molecular assemblies beyond dimers? From “signalosomes” in the Wnt pathway [25] to the RNA/protein granules that segregate asymmetrically during embryogenesis [26,27], higher-order assemblies are at the core of many developmental processes. Oligomerization can also sequester proteins away from interaction partners and inhibit signaling. Optogenetic control over protein clustering was first initiated by the observation that Cry2 weakly aggregates upon light stimulation [28] and that a point mutation termed “Cry2olig” dramatically enhanced this ability [6]. Cry2 clustering has proven useful to block Bicoid and Wnt signaling during Drosophila embryogenesis [29,30] (Figure 1e) and activate RTKs in mammalian cell culture and tissue explants [31,32].
More recently, novel Cry2 variants have been developed with an even wider range of clustering properties. An elegant mutagenesis study revealed that the relative balance of positively and negatively charged residues in the C terminal tail controls the extent of oligomerization [7]. We and the Brangwynne group further extended the Cry2 system by fusing it to various intrinsically disordered peptides with innate tendencies to phase separate, leading to light-dependent coalescence into liquid droplets or solid aggregates [33]. Such approaches will likely be useful for inducing functional signaling or receptor clusters, as well as for controlling the sequestration of intracellular factors into inactive aggregates [6].
Optogenetic control over developmental gene expression
One of the most intuitive and powerful applications of developmental optogenetics is the ability to control gene expression in illuminated cells. Befitting its central role, spatiotemporal control of gene expression has long been targeted by existing tools including the Gal/UAS system, heat-inducible promoters, and chemical inducers [34–37]. However, each of these systems suffers from substantial limitations: lineage-specific Gal drivers may not be available with sufficient precision, heat induction can have pleiotropic effects, and chemical inducers can be difficult to deliver uniformly and lack spatial control. An optogenetic system for controlling gene expression should have very low leakiness in the dark and be able to drive high levels of expression in a desired tissue. Ideally, it would also be “backwards compatible” with the wealth of genetic lines that already exist, so that strains harboring UAS-driven or LoxP-flanked genes could be simply be crossed with an “opto-Gal4” or “opto-Cre” driver line to enable light-dependent control.
Excitingly, robust tools satisfying all of these requirements are now beginning to emerge. In 2012, the Yang laboratory engineered a tripartite transcription factor – comprising the Gal4 DNA binding domain, the VVD light-sensitive dimerization domain, and the p65 transcriptional activation domain – to create the LightON system, which is capable of inducing high levels of UAS-driven gene expression upon light stimulation [20,22]. This system has already proven useful for probing the differentiation of cultured murine neural progenitor and embryonic stem cells [38,39], but its application in other developmental systems has thus far been limited. Some care is also warranted: high expression of the LightON system has been shown to be toxic during zebrafish embryogenesis, possibly due to the use of the strong p65 transactivation domain [40]. In cases where “backward compatibility” to UAS lines is not needed, the EL222 light-sensitive transcription factor has been used to drive expression from an optimized cognate promoter (termed C120) in a wide range of cellular contexts without toxicity [21,41].
Two recent reports demonstrate that a high quality, photoswitchable Cre recombinase is also within reach [42,43]. In both studies, the two halves of a split Cre recombinase were each fused to a component of a heterodimerizing pair of optogenetic proteins. This design strategy resulted in minimal leaky recombination in the dark, reconstitution of Cre function by even a brief pulse of blue light, and high levels of recombination in vivo in deep tissues (e.g. the mouse liver) using only external illumination of freely moving animals.
Emerging experimental paradigms in developmental optogenetics
Developmental optogenetics is still in its infancy, and new, unpredictable twists and turns should be expected. Nevertheless, the first wave of developmental optogenetics studies has already converged on what will likely be classic experimental design strategies for future work. One example takes advantage of the temporal control afforded by light to reveal critical periods for cell fate determination by the Erk, Nodal, Bicoid and Wnt pathways [18,24,29,30] (Figure 2a). Precisely establishing the boundaries of these critical periods can be difficult to achieve by standard genetic or pharmacological perturbations because, once they are applied, these stimuli are typically impossible to remove. Long-term perturbations can also exert distinct effects at different developmental stages. They may even trigger compensatory feedback loops that paradoxically induce gain-of-function effects in some tissues and loss-of-function effects in others, as in the case of Ras/Erk signaling during fly and zebrafish development [44]. The ability to limit light exposure to particular cells or developmental time windows may thus enable developmental biologists to mine new insights even from pathways with classic, well-characterized mutant phenotypes [18].
Figure 2. Experimental paradigms for probing development using light.
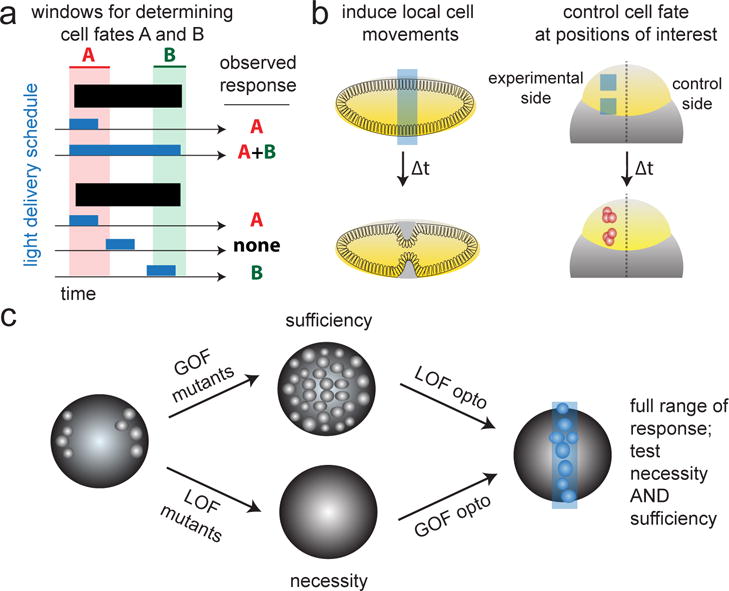
(a) Light can be applied and removed over time to reveal the temporal windows during which cells are responsive to a differentiating stimulus. Schematic shows a model developmental process where cell responses A and B are determined during distinct developmental time periods. By systematically providing a pulse of light at different points in development one can infer the windows in which response can be programmed. (b) Spatial light control opens the door to directly applying perturbations even when lineage-specific promoters are unavailable. In a straightforward application, stimuli can be restricted to one half of a symmetric axis to establish robust internal controls (left). Spatial activation can also be used to control the migration of single or groups of cells, or tissue-scale morphogenesis (right). (c) In order to achieve control over the full range of a developmental signal, optogenetics can be combined with a converse genetic perturbations (e.g. a GOF optogenetic tool + a LOF mutant), enabling the researcher to test both necessity and sufficiency of a developmental signal for controlling a desired response.
Another classic study design focuses on spatial cell-cell interactions by illuminating a subset of cells and monitoring the effect on neighboring cells and tissues. Precise spatial control was used to interrogate the morphogenesis of the ventral furrow in Drosophila [45] and the role of Rac signaling in collective cell migration in zebrafish [46]. Spatial control could also be invaluable for studying processes where lineage-specific promoters are unavailable or do not uniquely label a cell population of interest, such as symmetry-breaking processes where an initially-uniform group of cells self-organizes into distinct tissue types [47,48]. One might also take advantage of spatial perturbation to perform internal controls, such as stimulating only one side of a developmentally-symmetric axis (Figure 2b).
Beyond reinventing classical experiments, optogenetics opens the door to new classes of studies that aim to place developmental processes on a firm quantitative footing. By systematically varying the level, duration, slope, or spatial range of a light stimulus while keeping other parameters constant, it may soon be possible to systematically dissect how a morphogen is read out with quantitative precision to program specific cell fates [49], even in complex cases where the decoding function may vary between cells or change over time. Through such a systematic application of inputs, the developmental biologist may envision constructing a complete map of how a differentiation event depends on the spatiotemporal inputs received by individual cells.
Challenges facing developmental optogenetics
Despite these early successes, establishing light-based perturbation as a “plug-and-play” tool for developmental studies still faces a number of challenges. For instance, the field still awaits the union of optogenetic perturbation with classical genetic approaches. Such a union is crucial because any individual optogenetic technique typically provides spatiotemporal control over some gain-of-function (GOF) or loss-of-function (LOF) activity, but not both. For GOF optogenetic tools, light can “add” protein activity on top of its wild-type pattern, which can be used to test for its sufficiency to induce a response in illuminated tissues or probe the upper limits of activity that can support proper development [18]. On the flip side, LOF optogenetics allow us to test a signal’s necessity and its lower allowable limits. One can thus envision combining a GOF optogenetic system with a LOF genetic background (or vice versa) to achieve a full range of outcomes, from activity levels that are below those in wild-type tissues to those that are above (Figure 2c). This kind of combination will no doubt constitute one of the major next steps in developmental optogenetics.
A second roadblock for the field involves fully integrating state-of-the-art developmental imaging, such as light sheet microscopy, with precise optogenetic stimulation. We can envision an experiment that is still out of reach: to obtain real-time information about cells’ positions and activity states in a live embryo, precisely target a light stimulus to cells of interest within their 3-dimensional context, and then track them and their neighbors over time to assess the response. Making this kind of experiment a reality means overcoming at least two major problems: the ability to precisely target excitation light in 3-dimensional space, and the ability to “multiplex” fluorescent biosensors and optogenetic tools in the same cells.
Fortunately, dramatic advances in spatial light targeting have already been developed within the neuronal optogenetics field. Techniques like holographic excitation and wavefront shaping enable the experimentalist to focus coherent laser light at single or multiple 3D locations within a sample, even when these focal planes are distinct from the imaging plane [50]. A recent report even applied wavefront shaping to non-neuronal optogenetics for the first time, demonstrating Cry2-based receptor clustering in individual cells through a 300 μm thick section of mouse skull [32]. Such approaches could dramatically augment the capabilities of commonly-used existing hardware, such as digital micromirror devices, that are easy to use but more suitable for two-dimensional culture conditions [1,51], and we eagerly await their integration into modern light-sheet microscopes.
What about the spectral overlap between optogenetic photoexcitation and fluorescence imaging? The broad blue light sensitivity of all flavin chromophore-containing proteins (e.g. LOV, BLUF and CRY domains) essentially prevents non-invasive imaging of blue, cyan, green and possibly yellow fluorescent proteins. This is not a fatal flaw, as high quality red and infrared fluorescent proteins can still be used [52,53], but it can pose significant challenges when crossing to existing GFP-tagged lines. A second solution lies in the phytochromes; these photochromic proteins can be both rapidly switched on and off with two different wavelengths of light. One can thus simply apply a pulse of inactivating light after imaging any set of fluorophores to “reset” these proteins and quickly reverse any unwanted photoactivation [54]. Phytochromes have been disfavored for in vivo applications because they historically required an exogenously-supplied chromophore, but recent studies have reconstituted efficient chromophore biosynthesis in mammalian cells [55], and alternative bacterial phytochrome optogenetic tools have been developed that rely on biliverdin, a widely-available endogenous metabolite [56].
In summary, developmental optogenetics provides unprecedented control over the timescale of a perturbation, its spatial range, and its quantitative parameters. This possibility could shed new light on myriad developmental processes with complex spatiotemporal dynamics (e.g. the oscillating somite clock, symmetry-breaking cell fate choices, tissue morphogenesis) or quantitative signal interpretation (e.g. classical morphogens such as Bicoid). As light-controlled proteins continue to be refined and applied to an ever-increasing palette of intracellular processes, we envision a future where optogenetics could take a place alongside genetic screens, cell transplantation and CRISPR-based genome editing as workhorse tool of developmental biology.
Highlights.
Optogenetic techniques are ripe for application to developmental biology
Light-activated receptors, gene expression, and clustering work robustly in vivo
New experimental paradigms are emerging for optogenetic studies of development
3-D light delivery and multiplexed fluorescence imaging remain challenging
Acknowledgments
The authors acknowledge the optogenetics and developmental biology communities, whose support and openness to collaboration makes work at this interface so productive and exciting. We also apologize for any omissions in citations due to space limitations. This work was supported by NIH grant DP2EB024247 (to J.E.T.) and an NIH Kirschstein postdoctoral fellowship F32GM119297 (to H.E.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 6.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan L, Hope J, Ong Q, Lou HY, Kim N, McCarthy C, Acero V, Lin MZ, Cui B. Understanding CRY2 interactions for optical control of intracellular signaling. Nat Commun. 2017;8:547. doi: 10.1038/s41467-017-00648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. The first study where a LOV domain’s Jα helix is re-designed to display a linear motif, in this case a peptide that binds to a cognate PDZ binding domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, Kane RS. At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu Rev Chem Biomol Eng. 2017;8:13–39. doi: 10.1146/annurev-chembioeng-060816-101254. A thorough and current review of the growing optogenetic toolbox. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Yumerefendi H, Dickinson DJ, Wang H, Zimmerman SP, Bear JE, Goldstein B, Hahn K, Kuhlman B. Control of Protein Activity and Cell Fate Specification via Light-Mediated Nuclear Translocation. PLoS One. 2015;10:e0128443. doi: 10.1371/journal.pone.0128443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, Di Ventura B. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun. 2014;5:4404. doi: 10.1038/ncomms5404. This is the first study placing nuclear/cytoplasmic translocation under optogenetic control; through two rounds of design and dozens of constructs, the authors refine their system to exhibit approximately 10-fold changes in gene expression when fused to mammalian transcription factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B. Optogenetic control of nuclear protein export. Nat Commun. 2016;7:10624. doi: 10.1038/ncomms10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yumerefendi H, Lerner AM, Zimmerman SP, Hahn K, Bear JE, Strahl BD, Kuhlman B. Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Nat Chem Biol. 2016;12:399–401. doi: 10.1038/nchembio.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A. 2015;112:112–117. doi: 10.1073/pnas.1417910112. This study describes the design of the iLID/SspB light-induced heterodimerization pair, a system with substantial advantages over many of its alternatives: both proteins are small, highly expressed, tolerate both N- and C-terminal fusion, and exhibit more than a 30-fold change in affinity between the lit and dark states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Johnson HE, Goyal Y, Pannucci NL, Schupbach T, Shvartsman SY, Toettcher JE. The Spatiotemporal Limits of Developmental Erk Signaling. Dev Cell. 2017;40:185–192. doi: 10.1016/j.devcel.2016.12.002. This study establishes optogenetic control over Erk signaling during Drosophila embryogenesis. At the technical level, it demonstrates potent, tunable, and spatially-precise Erk control using single-photon excitation. At the biological level, it begins to map the spatial and temporal windows where increased Erk activity drives strong phenotypic consequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. An excellent and comprehensive review that may be especially useful to the reader for its description of the critical residues and parameters controlling the LOV domain photocycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Wang X, Du Z, Ma Z, Yang Y. Spatiotemporal control of gene expression in mammalian cells and in mice using the LightOn system. Curr Protoc Chem Biol. 2013;5:111–129. doi: 10.1002/9780470559277.ch120267. [DOI] [PubMed] [Google Scholar]
- 21*.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. This study describes a powerful optogenetic transcriptional system based on the light-induced dimerization of the EL222 helix-turn-helix transcription factor and its cognate C120 promoter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. This study establishes the LightON (or GAVPO) system, an engineered homodimeric transcription factor that is “backwards compatible” with the Gal/UAS system to induce expression of UAS-driven genes. It is likely to be highly useful for developmental studies. [DOI] [PubMed] [Google Scholar]
- 23*.Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Ingles-Prieto A, Janovjak H. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 2014;33:1713–1726. doi: 10.15252/embj.201387695. This study was the first to establish receptor activation by light-induced homodimerization, focusing on receptor tyrosine kinases (RTKs), including an “orphan” RTK whose natural ligand is unknown. To do so, the authors screened a panel of LOV domains, identifying a novel homodimerizing LOV domain as part of the study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sako K, Pradhan SJ, Barone V, Ingles-Prieto A, Muller P, Ruprecht V, Capek D, Galande S, Janovjak H, Heisenberg CP. Optogenetic Control of Nodal Signaling Reveals a Temporal Pattern of Nodal Signaling Regulating Cell Fate Specification during Gastrulation. Cell Rep. 2016;16:866–877. doi: 10.1016/j.celrep.2016.06.036. [DOI] [PubMed] [Google Scholar]
- 25.Pronobis MI, Deuitch N, Posham V, Mimori-Kiyosue Y, Peifer M. Reconstituting regulation of the canonical Wnt pathway by engineering a minimal beta-catenin destruction machine. Mol Biol Cell. 2017;28:41–53. doi: 10.1091/mbc.E16-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 27.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 28*.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. This study was the first to describe light-induced Cry2 clustering and went on to apply this effect to the light-gated activation of Wnt signaling. [DOI] [PubMed] [Google Scholar]
- 29.Huang A, Amourda C, Zhang S, Tolwinski NS, Saunders TE. Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. Elife. 2017;6 doi: 10.7554/eLife.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur P, Saunders TE, Tolwinski NS. Coupling optogenetics and light-sheet microscopy, a method to study Wnt signaling during embryogenesis. Sci Rep. 2017;7:16636. doi: 10.1038/s41598-017-16879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem Biol. 2014;21:903–912. doi: 10.1016/j.chembiol.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 32*.Yoon J, Lee M, Lee K, Kim N, Kim JM, Park J, Yu H, Choi C, Heo WD, Park Y. Optogenetic control of cell signaling pathway through scattering skull using wavefront shaping. Sci Rep. 2015;5:13289. doi: 10.1038/srep13289. This paper is an exciting proof-of-principle demonstration of subcellular Cry2 photoexcitation in live cells through a thick opaque barrier (the skull). To do so, the authors implement wavefront shaping of a 488 nm single-photon laser light source, demonstrating the potential for spatially addressing cellular optogenetic tools in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. 2017;168:159–171 e114. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Struhl G. Near-reciprocal phenotypes caused by inactivation or indiscriminate expression of the Drosophila segmentation gene ftz. Nature. 1985;318:677–680. doi: 10.1038/318677a0. [DOI] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 36.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin MK, Levorse JM, Ingram RS, Tilghman SM. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 38**.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. An elegant study directly demonstrating that oscillating and sustained expression of the Ascl1 transcription factor can induce distinct cell fate responses in neural progenitor cells. To directly test if dynamics were causally connected to cell fate choice, the authors controlled ascl1 expression using the LightON system. [DOI] [PubMed] [Google Scholar]
- 39.Sokolik C, Liu Y, Bauer D, McPherson J, Broeker M, Heimberg G, Qi LS, Sivak DA, Thomson M. Transcription factor competition allows embryonic stem cells to distinguish authentic signals from noise. Cell Syst. 2015;1:117–129. doi: 10.1016/j.cels.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci U S A. 2009;106:13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reade A, Motta-Mena LB, Gardner KH, Stainier DY, Weiner OD, Woo S. TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development. 2017;144:345–355. doi: 10.1242/dev.139238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol. 2016;12:1059–1064. doi: 10.1038/nchembio.2205. This excellent study describes an optimized light-controlled Cre recombinase based on the heterodimerization between two split Cre fragments. The optimization of a split site and use of LOV domain variants with slow photocycle kinetics (with photoinactivation occurring over hours) enabled recombination in vivo even within deep tissues. [DOI] [PubMed] [Google Scholar]
- 43.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goyal Y, Jindal GA, Pelliccia JL, Yamaya K, Yeung E, Futran AS, Burdine RD, Schupbach T, Shvartsman SY. Divergent effects of intrinsically active MEK variants on developmental Ras signaling. Nat Genet. 2017;49:465–469. doi: 10.1038/ng.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Guglielmi G, Barry JD, Huber W, De Renzis S. An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Dev Cell. 2015;35:646–660. doi: 10.1016/j.devcel.2015.10.020. A pioneering study in which optogenetic control over Rho contractility was used to probe cell-and tissue-level control of morphogenesis in the developing Drosophila embryo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- 48.Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NNM, Campbell A, Devito L, Ilic D, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Shemesh OA, Tanese D, Zampini V, Linghu C, Piatkevich K, Ronzitti E, Papagiakoumou E, Boyden ES, Emiliani V. Temporally precise single-cell-resolution optogenetics. Nat Neurosci. 2017;20:1796–1806. doi: 10.1038/s41593-017-0018-8. One of a series of excellent papers from the same laboratories, this recent work demonstrates the rapid progress toward fast, 3-D spatial addressing of individual cells within intact tissue using two-photon excitation and computer-generated holography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shemiakina II, Ermakova GV, Cranfill PJ, Baird MA, Evans RA, Souslova EA, Staroverov DB, Gorokhovatsky AY, Putintseva EV, Gorodnicheva TV, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 53.Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE. Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell. 2017;67:757–769 e755. doi: 10.1016/j.molcel.2017.07.016. This study pushes the limits of multicolor fluorescence imaging and optogenetic activation, demonstrating simultaneous recording from blue, green, red and infrared fluorescent proteins while controlling ERK activity with the PhyB/PIF6 optogenetic heterodimerization system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uda Y, Goto Y, Oda S, Kohchi T, Matsuda M, Aoki K. Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc Natl Acad Sci U S A. 2017;114:11962–11967. doi: 10.1073/pnas.1707190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Redchuk TA, Omelina ES, Chernov KG, Verkhusha VV. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat Chem Biol. 2017;13:633–639. doi: 10.1038/nchembio.2343. A long-sought development in the cellular optogenetics community: an optogenetic tool with all the benefits of the phytochromes (photoreversibility in response to two different wavelengths; red/infrared spectral sensitivity that is complementary to the blue optogenetic tools and fluorescent proteins), and which does not require the addition of an exogenous chromophore. [DOI] [PMC free article] [PubMed] [Google Scholar]


