Abstract
Purpose
The causative agent of most cases of Merkel cell carcinoma (MCC) has been identified as the Merkel cell polyomavirus (MCV). MCV-encoded T-antigens (Tags) are essential not only for virus-mediated tumorigenesis but also for maintaining MCC cell lines in vitro. MCV Tags are thus an appealing target for viral oncoprotein-directed T cell therapy for MCC. With this study, we aimed to isolate and characterize Tag-specific T cell receptors (TCR) for potential use in gene therapy clinical trials.
Experimental Design
T cell responses against MCV Tag epitopes were investigated by immunizing transgenic mice that express a diverse human TCR repertoire restricted to HLA-A2. Human lymphocytes genetically engineered to express Tag-specific TCRs were tested for specific reactivity against MCC cell lines. The therapeutic potential of Tag-specific TCR gene therapy was tested in a syngeneic cancer model.
Results
We identified naturally processed epitopes of MCV Tags and isolated Tag-specific TCRs. T cells expressing these TCRs were activated by HLA-A2-positive cells loaded with cognate peptide or cells that stably expressed MCV Tags. We showed cytotoxic potential of T cells engineered to express these TCRs in vitro and demonstrated regression of established tumors in a mouse model upon TCR gene therapy.
Conclusion
Our findings demonstrate that MCC cells can be targeted by MCV Tag-specific TCRs. Although recent findings suggest that approximately half of MCC patients benefit from PD1 pathway blockade, additional patients may benefit if their endogenous T cell response can be augmented by infusion of transgenic MCV-specific T cells such as those described here.
Keywords: Merkel cell carcinoma, Merkel cell polyomavirus, T cell receptor gene therapy
INTRODUCTION
Merkel cell carcinoma (MCC) is a rare but highly aggressive primary cutaneous neuroendocrine neoplasm. With a 33% mortality rate, MCC is deadlier than other common forms of skin cancer (1). MCC most often occurs in immunosuppressed or elderly individuals, suggesting a role for immune surveillance in blocking the development of MCC.
A clonally integrated polyomavirus (Merkel cell polyomavirus, MCV) has been found in association with at least 80% of MCCs (2,3). Similar to other polyomaviruses, MCV encodes alternatively spliced large (LT) and small (sT) tumor antigen transcripts that share exon 1 of the T antigen (Tag) gene (4,5). Sequence analysis demonstrated that LT protein is prematurely truncated in all MCV-positive MCC cases which results in deletion of the helicase domain (6). Integration of the virus before clonal expansion of the tumor cells as well as addiction of MCV-positive MCC cells to the expression of viral oncoproteins suggest that MCV is a major driver of MCC development and progression (7,8). However, details of the function of MCV in the development of MCC are still being investigated.
Programmed death 1 (PD-1) pathway blocking agents have greatly improved the options for MCC patients, and demonstrated that augmenting responses of the existing tumor-specific T cell repertoire is efficient in about half of cases (9,10). One plausible explanation for the disappointing results in the other half of patients is that the number and or avidity of tumor-specific T cells were insufficient. These patients could benefit from T cell based immunotherapy targeting MCV Tags.
To determine the immunogenic epitopes on MCV Tags and isolate specific T cell receptors (TCR), we used the previously described ABabDII mouse model (11,12). ABabDII mice are transgenic for human TCR-αβ gene loci and HLA-A*0201 (HLA-A2 hereafter) as a chimeric HLA-A2/Db molecule fused to human β2-microglobulin (HHD). Additionally, they are deficient for mouse TCR-αβ and mouse MHC I (H2-Db and β2-microglobulin) expression. Thus, ABabDII mice express a diverse human TCR repertoire restricted to HLA-A2. We isolated HLA-A2-restricted Tag-reactive TCRs for potential use in a gene therapy clinical trial. These high-avidity TCRs mediated recognition of nanomolar levels of peptide pulsed on T2 cells as well as HLA-A2+ target cells expressing MCV Tags.
METHODS
Cell lines
The human MCC cell lines WaGa (MCV+/HLA-A2+) (7), MS-1 (MCV+/HLA-A2+) (13), MKL-2 (MCV+/HLA-A2+) (14), MKL-1 (MCV+/HLA-A2−) (15), PeTa (MCV+/HLA-A2−) (16), and MCC-13 (MCV−/HLA-A2+) (17) were kindly provided by R. Houben (University of Wurzburg) and were kept in RPMI (Gibco) supplemented with 10% fetal calf serum (FCS, Pan Biotech), 50 µM 2-mercaptoethanol, 1 mM sodium pyruvate and non-essential amino acids. The human lung cancer cell line H1299 was purchased from ATCC and maintained in RPMI 1640 supplemented with 10% FCS. The HLA-A2 expressing variants (H1299-A2, MKL-1-A2, and PeTa-A2) were generated by transducing the respective cell line with HLA-A2 cDNA. MCV LT or sT expressing H1299-A2 was generated by transducing H1299-A2 with LT or sT cDNA, respectively, linked with an internal ribosomal entry site (IRES) to the reporter constructs GFP. TCR-α and TCR-β deficient Jurkat clone 76 (Jurkat-76) (18) and T2 cells (ATCC: CRL-1992) cells were kept in RPMI supplemented with 10% FCS. The viral producer cell line HEK-GALV (19) was cultured in DMEM supplemented with 10% FCS. Human peripheral blood mononuclear cells (hPBMCs) were cultured in RPMI 1640 supplemented with 10% FCS, 50 µM 2-mercaptoethanol, 1 mM sodium pyruvate and non-essential amino acids. The fibrosarcoma MC703 was previously generated in HHD mice and maintained in RPMI supplemented with 10% FCS (12,20). The variant MC703-KLL was generated by transducing MC703 with cDNA encoding for MCV-derived epitope KLL fused to GFP. All MCC cell lines were authenticated by sequencing for the presence of characteristic LT truncating mutations, which lead to a distinct molecular weight of the protein detectable by immunoblotting.
Generation of MCV Tag-specific T cells in ABabDII mice
For priming, ABabDII mice were injected subcutaneously with 100 µg of peptide (SMFDEVDEA [SMF9], SMFDEVDEAPY [SMF11], KLLEIAPNC [KLL], KLLEIAPN{Abu} [KLL-Abu], Genscript) in a 200 µl 1:1 solution of incomplete Freund’s adjuvant and PBS supplemented with 50 µg CpG. Successive boosts were performed with 100 µg of respective peptide including the same adjuvants. The time between immunizations was at least 4 weeks. The presence of MCV-specific CD8+ T cells in the peripheral blood of immunized animals was assessed by in vitro peptide stimulation and subsequent intracellular cytokine staining 7 days after each boost.
Isolation of MCV Tag-specific TCRs
Splenocytes and lymphocytes from inguinal lymph nodes were prepared from responding animals at days 10–14 after the last boost and stained either with SMF/HLA-A2 or KLL/HLA-A2 multimers (iTAg HLA class I Tetramers from MBL International Corporation) or after in vitro peptide (107 M) stimulation for 24 h with anti-mCD137 antibody (17B5, BioLegend). Between 500 and 30,000 multimer+ (or CD137+) CD8+ T cells were sorted directly into 300 µl RLT lysis buffer (Qiagen) on a BD FACSAria III sorter. Total RNA from sorted cells was extracted using RNeasy Plus Micro kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA synthesis and 5’-RACE PCR were carried out using SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories) according to the manufacturer’s instructions. In particular, subsequent TCR-specific amplification was carried out with 1–2 µl of the reverse transcription reaction, 1 U Phusion HotStart II polymerase (Thermo Scientific), 0.1 µM of either hTRAC (5’-CGG CCA CTT TCA GGA GGA GGA TTC GGA AC-3’) or hTRBC (5’-CCG TAG AAC TGG ACT TGA CAG CGG AAG TGG-3’) primers and 0.1 µM 5’ Primer (5’-CTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA TCA ACG CAG AGT-3’). The amplified TCR genes were analyzed on an agarose gel and specific bands were cut out and cloned using a Zero Blunt® TOPO® PCR cloning kit (Life Technologies). Plasmids from individual clones were isolated and sequenced using a T3 primer (5’-AAT TAA CCC TCA CTA AAG GG-3’) at Eurofins Genomics.
Generation of TCR transgene cassettes
Pairing TCR-α/β chains were linked in silico via 2A “self-cleaving” peptide sequence from Porcine teschovirus-1 (P2A) in the fashion TCRβ-P2A-TCRα as described before (21). The human TCR constant regions were replaced by their murine counterparts to improve the pairing between the chains of the introduced TCR and avoid mispairing with endogenous TCR-α and -β chains. The transgene cassettes were codon-optimized for human expression and synthesized by GeneArt (Life Technologies). The transgenes were cloned into pMP71-PRE (22) using NotI and EcoRI restriction sites.
TCR gene transfer
TCR gene transfer was carried out as described before with minor modifications (23). Packaging 293-GALV (HEK-293 cells stably expressing GALV-env and MLV-gag/pol) cells were grown to approximately 80% confluence and transfected with 3 µg of the plasmid containing the transgene in the presence of 10 µg Lipofectamine2000 (Life Technologies). 3 ml of retrovirus containing supernatant were harvested 48 h and 72 h after transfection.
Human PBMCs were isolated from healthy donors by ficoll gradient centrifugation. 1 × 106 freshly isolated or frozen hPBMCs were stimulated with 5 µg/ml anti-CD3 (OKT3) and 1 µg/ml anti-CD28 (CD28.2) (Biolegend) coated plates in the presence of 300 U/ml recombinant human interleukin 2 (hIL-2, Peprotech). Transductions were performed 48 h and 72 h after stimulation by addition of retrovirus containing supernatant and 4 µg/ml protamine sulfate followed by spinoculation for 90 min at 800 g and 32°C (1st transduction) or preloading of virus onto retronectin (Takara)-coated plates and spinoculation for 30 min at 800 g and 32°C (2nd transduction). Transduced T cells were kept in the presence of 300 U/ml hIL-2 for a total of 2 weeks followed by at least 2 days of culture in presence of 30 U/ml hIL-2, before they were used for experiments. TCR-negative Jurkat-76 cells were transduced without prior stimulation and the second transduction was omitted. TCR gene transfer into murine T cells was done as described (20).
Functional assays
IFN-γ production was measured by ELISA after 16 h coculture of 5×104 TCR-transduced T cells with 5×104 target cells (human tumor cell lines or peptide-loaded T2 cells). Stimulation with phorbol-12-myristate-13-acetate (PMA) and ionomycin was used as a positive control. Cytotoxic activity of 2×105 of transduced human PBLs was analyzed by incubation with target cells (E:T ratio of 2:1) in 200 µl T cell medium in presence of 1 µg CD107a-specific antibody (H4A3; Alexa Fluor® 647, BioLegend), 1 µg monensin (Sigma), and 1 µg brefeldin A (BD). After 6 h incubation, cells were fixed and stained using fixation/permeabilization kit (BD) and antibodies against hCD8 (HIT8α), mTCR-β (H57-597), and hIFN-γ (4S.B3), (all from BioLegend).
Flow cytometry
The following conjugated antibodies were obtained from BioLegend: anti-hCD3 (HIT3α), anti-hCD8 (HIT8α), anti-hHLA-A2 (BB7.2), anti-hHLA-ABC (W6/32), anti-hVβ8 (JR.2), anti-mCD3 (145-2C11), anti-mCD8 (53-6.7), anti-mIFN-γ (XMG1.2), anti-mTCR-β (H57-597). Anti-hVβ3 (JOVI-3) was purchased from BD Biosciences. The peptide/HLA-A2 multimers were produced by MBL International Corporation. Samples were analyzed using a FACSCanto II (BD Biosciences). Data Analysis was performed using FlowJo (Treestar).
Immunblotting
For the preparation of whole-cell extracts, cell pellets were washed in cold PBS and resuspended in lysis buffer (Sigma-Aldrich) supplemented with protease inhibitors. Cells were lysed on ice for 45 min. Aliquots of equal amounts of protein were mixed with NuPAGE® sample buffer (Invitrogen), heated at 70°C for 10 min, and electrophoresed for western transfer using the Novex® NuPAGE® SDS-PAGE Gel system (Invitrogen) according to the manufacturer’s instructions. Monoclonal antibodies CM2B4 (Santa Cruz), recognizing LT and CM8E6 (24), recognizing both the LT and sT, were used to detect Tag expression. MAGE-A1 was detected using monoclonal antibody MA454 (Santa Cruz). β-Actin antibody (abcam) was used as loading control.
Tumor challenge and adoptive T cell transfer
In vivo testing of TCR KLL-85 was performed as previously described (12,20). Briefly, 5×106 tumor cells were subcutaneously injected in 100 µl PBS into the left flank of HHDxRag−/− mice (12–16 weeks old). Tumor growth was analyzed 2–3 times a week by determination of tumor volume by caliper measurements according to π/6 × (abc). On the day of T cell transfer, mice were ranked by tumor size and sequentially allocated to treatment groups to ensure equal average tumor sizes between groups. Mice were treated by adoptive transfer of TCR-engineered HHD T cells 4 weeks after tumor cell injection when tumors were established. HHD T cells were analyzed for expression of CD8 and transgenic TCR (KLL-85 (TCRvβ8), 1367 (TCRvβ3)) by flow cytometry and intravenously injected in 100 µl PBS (adjusted to 1×106 CD8+ TCR+ HHD T cells per mouse) 3 days after transduction. Examiners were not blinded with respect to treatment groups. Mice were sacrificed when either tumors reached the maximum permitted size or if due to tumor burden the overall health-condition was poor.
Study approval
All animal experiments were performed according to institutional and national guidelines, after approval by the responsible authority (Landesamt für Gesundheit und Soziales, Berlin). Blood collection from healthy human donors was done after prior informed consent and experiments were conducted in accordance with the ethical standards of Declaration of Helsinki.
RESULTS
Identification and evaluation of CD8 T cell epitopes on MCV LT
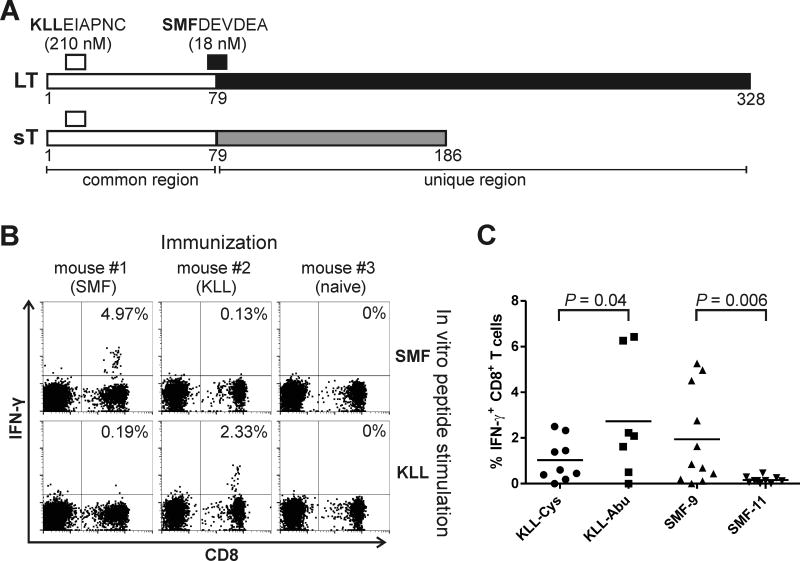
The MCV LT and sT proteins result from splice variants of a single transcript. The two oncoproteins share a common 79 amino acid N-terminal region, followed by regions unique to each isoform (Fig. 1A). Using in silico analysis of LT protein sequence with NetMHCpan algorithm (25), 4 potential HLA-A2-restricted epitopes were detected in the non-truncated region. Of these, only the epitope at position 74–82 SMFDEVDEA (hereafter SMF) with predicted IC50 of 18 nM was considered as a strong binder and was selected for further analysis. A second epitope in the ranking with the sequence KLLEIAPNC (KLL) and IC50 of 210 nM is predicted to be a weak binder (NetMHCpan). However, we selected this epitope for analysis as well, due to its presence on both, LT and sT antigens and because a T cell response against KLL was previously described (26,27). To analyze the immunogenicity of those epitopes, we immunized ABabDII mice expressing diverse human TCR repertoire in HLA-A2-restricted manner with respective peptides. Antigen-specific CD8+ T cells were detected in immunized mice but not in naïve mice by in vitro re-stimulation of peripheral blood lymphocytes (PBL) at day 7 after last immunization (Fig. 1B). Both epitopes induced specific CD8+ T cell response in the ABabDII mice. Because the presence of cysteine residues in antigenic peptides causes problems with formulation due to possible oxidative damage and dimerization (28), C-terminal cysteine in P9-anchor position of KLL was replaced by a non-natural isosteric analog of this residue 2-aminoisobutyric acid (Abu) resulting in KLL-Abu peptide. Immunization with this alternative peptide significantly increased the magnitude of the response (P = 0.04, t-test) to this particular epitope (Fig. 1C).
Figure 1.
MCV Tag epitopes SMF and KLL induce CD8+ T cell response in ABabDII mice. A. MCV LT and sT antigens: open boxes illustrate common region encoded by exon 1; black (LT) or grey (sT) filled areas show unique regions. Numbers indicate amino acid positions. Positions of the respective epitopes with amino acid sequences and predicted (by netMHC4.0) MHC affinities are also indicated. B. Representative dot plots showing intracellular IFN-γ staining as an indicator of activated CD8+ T cells after in vitro peptide stimulation of PBLs from mice immunized with SMF- or KLL-peptide or an unimmunized mouse. C. Summary of CD8+ T cell responses to immunization with indicated peptide epitopes as percentage of IFN-γ-secreting CD8+ T cells in blood after in vitro peptide stimulation. Data sets were compared using unpaired t-test.
In a peptide/MHC-based screen for MCV-specific CD8+ T cells in MCC patients’ blood and tumor-infiltrating lymphocytes (TILs), Lyngaa et al. (27) detected T cells specific to HLA-A2-multimers loaded with the peptide SMFDEVDEAPY. We sought to investigate immune response against this 11mer epitope. However, in contrast to the 9mer SMF peptide, SMF-11 didn’t induce any response in any of immunized mice (n=10) even after multiple boosts, suggesting only the 9mer variant of SMF epitope is truly immunogenic in ABabDII mice (Fig. 1C).
Cloning and characterization of TCRs directed against epitopes of MCV T antigens
To isolate specific TCRs, we sorted SMF- or KLL-reactive CD8+ T cells from splenocytes of responder mice using either CD137 as activation marker (Fig. 2A) or peptide/HLA-A2-multimers loaded with the respective peptide (Fig. 2B). By rapid amplification of cDNA end (5’RACE)-PCR, we cloned rearranged TCR-α and TCR-β genes. Matching of the correct TCR-αβ pairs was achieved by combining of the most abundant clones for each individual mouse (Supplementary table 1). Two different TCRs directed against SMF epitope (SMF-48 and SMF-72) and two TCRs specific for KLL epitope (KLL-40 and KLL-85) were isolated. The codon-optimized sequences encoding for the α- and β-chains were linked with a P2A element and inserted into retroviral expressing vector (Fig. 2C). To verify the specificity of αβ-combinations, we transduced TCR-negative Jurkat-76 cells expressing all components of the CD3 complex. Staining of transduced Jurkat-76 cells with SMF- or KLL/HLA-A2-multimers confirmed the specificity of αβ-combinations for all TCRs except for SMF-72 which was isolated through the CD137 assay (Fig. 2D). Next, we confirmed expression of the isolated TCRs in primary human T cells. Expression of all TCRs was detected by using an antibody directed against the murine TCR-β constant region (Fig. 2E, upper panel).
Figure 2.
Isolation and re-expression of MCV-specific TCRs. A. Detection of activated CD137+/CD8+ T cells after in vitro peptide (SMF) stimulation of splenocytes from a mouse immunized with SMF peptide. Unstimulated splenocytes served as negative control. Sorted cells are indicated. B. SMF/HLA-A2 multimer staining of splenocytes from a mouse immunized with SMF peptide. Staining with an irrelevant (ELA/HLA-A2) multimer served as negative control. Sorted cells are depicted. C. Schematic of MP71 vector encoding TCR-β and -α chains separated by P2A cleavage element and flanked by 5’ and 3’ long terminal repeats (LTR). D. Flow cytometry analysis of Jurkat-76 cells transduced with TCRs as indicated or untransduced (UT) and stained with SMF- or KLL-specific multimers (pMHC), respectively, and co-stained with antibody recognizing constant region of murine TCR-β. Numbers indicate percentage of double-positive cells. E. Flow cytometry analysis of human peripheral blood lymphocytes (PBLs) transduced with TCRs as indicated and stained with antibody recognizing constant region of murine TCR-β (upper panel) or with SMF or KLL/HLA-A2 multimers (lower panel). Numbers indicate percentage of CD8+ T cells positive for multimer or TCR-β staining, respectively. Images are representative of 3 independent experiments using PBLs of different donors.
Similar to Jurkat-76, all PBLs transduced with any of three TCRs except of SMF-72 were able to bind the cognate multimer CD8-independently. In contrast, SMF-72 expressing PBLs exhibited only weak binding in the CD8+ T cell compartment (Fig. 2E, lower panel).
Comparison of functional avidity of isolated TCRs
To compare the functional avidity of the isolated TCRs, we co-cultured TCR-transduced PBLs with TAP-deficient T2 cells loaded with titrated amounts of SMF or KLL peptides and measured release of IFN-γ as indicator for activation of T cells. Surprisingly, despite the poor tetramer binding, SMF-72 exhibited similar functional avidity (EC50 of 1 nM) compared to SMF-48 (Fig. 3A and C). Both TCRs induced robust IFN-γ release at peptide concentration of 10−10 M suggesting high functional avidity for these TCRs. KLL-TCRs, on the other hand, differed in their functional avidity. Cells transduced with the KLL-85 TCR recognized target cells incubated with a minimum concentration of 0.01 nM altered peptide, a 10-fold lower concentration than that required for recognition by cells transduced with the KLL-40 TCR, indicating that the KLL-85 possessed a higher functional avidity than the KLL-40 (EC50: 0.1 nM vs 1 nM) (Fig. 3B and D). When we used the native KLL peptide with cysteine residue at P9 anchor position, the functional avidities for both KLL-TCRs dropped by 2 logs (Fig. 3D, dashed lines), indicating either that the epitope analogue binds more efficiently to HLA-A2 compared to wild type epitope or due to oxidative damage of the native peptide upon formulation, the functional peptide amount was decreased. The half-maximal effective concentration of native KLL peptide for activation of T cells was comparable to that of SMF peptide, indicating that TCRs against both epitopes exhibit similar functional avidities.
Figure 3.
Functional avidity of T cells expressing MCV Tag-specific TCRs. A. & C. SMF-TCR- and B. & D. KLL-TCR-engineered human T cells generated from normal donor PBLs were assayed for response via IFN-γ production after co-culturing with T2 target cells pulsed with a range of peptide concentrations. A and B show absolute IFN-γ concentrations. In C and D data were normalized to maximum IFN-γ release. D. T2 cells were loaded either with the KLL native peptide (Cys, dashed lines) or with KLL-Abu (Abu, solid lines). Diagrams show means and standard deviations from 3 independent experiments with PBLs from different donors.
Recognition of cell lines overexpressing MCV antigens by T cells transduced with SMF- or KLL-TCRs
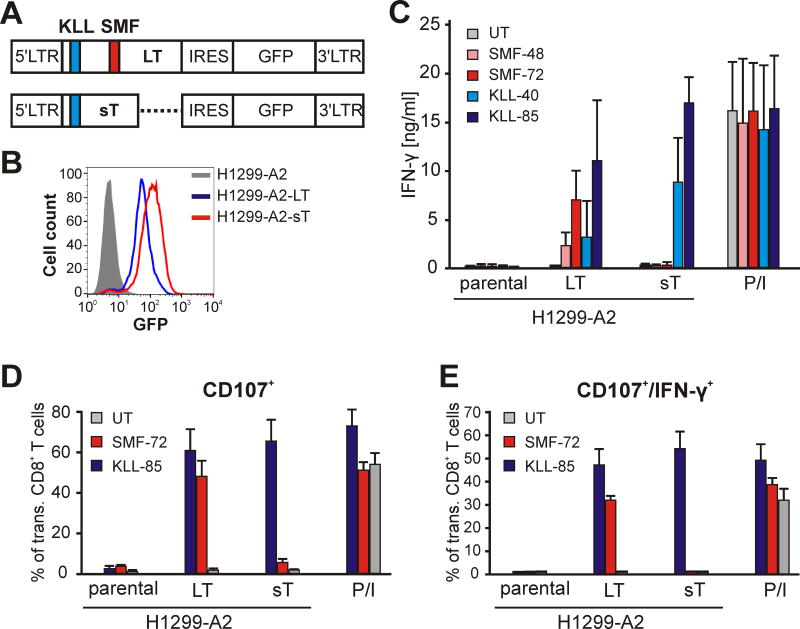
The processing and presentation of SMF- and KLL-epitopes as well as recognition by TCR-engineered PBLs were tested in vitro by culturing the PBLs with derivatives of H1299 cells that express HLA-A2 (H1299-A2) with or without concomitant expression of LT (H1299-A2-LT) or sT (H1299-A2-sT), as determined by GFP expression as reporter (Fig. 4A). Expression of MCV Tag was verified by flow cytometry of GFP-positive cells. H1299-A2-LT and H1299-A2-sT did not differ significantly in the mean fluorescence intensity (MFI) of GFP signal indicating similar expression strength for both antigens (Fig. 4B).
Figure 4.
SMF- and KLL-epitopes are naturally processed. A. Schematic of MP71 vectors encoding LT or sT antigens as bicistronic constructs with GFP-expressing sequence segregated by internal ribosomal entry site (IRES). LTR: long terminal repeats. Positions of KLL (blue) and SMF (red) epitopes are indicated. B. Expression of LT (blue) and sT (red) antigens in H1299-A2-LT or H1299-A2-sT, respectively, cells as determined by fluorescence intensity of GFP. C. TCR-engineered human T cells as indicated were assayed for response in IFN-γ production after co-culturing with MCV-negative H1299-A2 cell line or derivatives expressing LT or sT antigens. TCR-independent stimulation of T cells with PMA/Ionomycin (P/I) served as positive control. Diagrams show means and standard deviations of 3 different experiments. D and E. Proportion of CD107+ or CD107/IFN-γ-double positive cells among SMF- or KLL-TCR expressing CD8+ T cells as indicator of cytotoxic activity after co-incubation with target cells as indicated. Diagrams show means and standard deviations of two experiments with T cells from different donors.
In a co-culture with LT-transfectants, T cells transduced with SMF-72 secreted higher levels of IFN-γ compared to SMF-48 (Fig. 4C). As expected, no recognition was observed in a co-culture with H1299-A2 or H1299-A2-sT cells (the latter construct lacks a part of the SMF epitope). In contrast, KLL-85 and KLL-40 expressing T cells reacted to both, LT and sT expressing H1299-A2 cells but not to the parental H1299-A2 cells. In accordance with the functional avidity assay (Fig. 3D), KLL-85 expressing T cells secreted more IFN-γ compared to KLL-40 (Fig. 4C).
We next examined the ability of each epitope to induce the expression of CD107 on CD8+ T cells transduced with SMF- or KLL-specific TCRs. Upon TCR engagement and epitope stimulation, CD107 is exposed on the cell membrane of CD8+ CTLs and is used as assay for the detection of antigen-specific cytolytic activity (29). When co-cultured with the H1229-A2-LT cells, T cells expressing SMF-specific TCRs displayed CTL activity to the same extent as KLL-TCR expressing CD8+ T cells (Fig. 4D and E). As expected, the H1299-A2-sT cells induced CTL activity in T cells expressing KLL-TCRs only. In contrast, no significant CD107 expression was detected in untransduced T cells or in transduced T cells when co-cultured with parental H1299-A2 cells. The results of these experiments verify the natural processing and presentation of both SMF and KLL epitopes as well the specificity of the isolated TCRs.
T cells expressing KLL-specific TCR react to a Merkel cell carcinoma cell line
A panel of established MCV-positive (WaGa, MS-1, MKL-2, MKL-1, and PeTa) and an MCV-negative (MCC-13) MCC cell lines were evaluated for their ability to be recognized by the TCR-transduced T cells. Neither population of the transduced T cells recognized any of the 3 HLA-A2-positive cells lines WaGa, MS-1, or MKL-2, nor (as expected) the HLA-A2-negative MKL-1 and PeTa cells, nor the HLA-A2-positive but MCV-negative MCC-13 cell line (data not shown).
Immunoblotting with Tag-specific antibodies confirmed LT and sT protein expression in WaGa, MS-1, MKL-2, MKL-1, and PeTa cells (Fig. 5A). Different LT band size pattern reflects the different tumor-derived truncating LT mutations in these cell lines. Additionally, WaGa and MKL-1 cells exhibited higher expression of LT compared to the MS-1, MKL-2, or PeTa cells. As expected, no Tag expression was detected for the MCV-negative cell line MCC-13.
Figure 5.
Recognition of MCC cell lines by MCV Tag-specific TCRs. A. Expression of LT and sT in MCC cell lines. Cell lysates from indicated cell lines were resolved by electrophoresis, transferred to PVDF membrane, and probed with anti-LT (clone CM2B4) or anti-sT (clone CM8E6) antibodies. Immunoblot against β-actin (ACTB) was used as loading control. B. Expression of constitutive MHC class I (upper panel) or HLA-A2 (lower panel) in MCC cell lines after IFN-γ treatment. C. SMF- and D. KLL-specific TCR-transduced human T cells were co-cultured with MCC cell lines as indicated, loaded or unloaded with cognate peptide. TCR-independent stimulation of T cells with PMA/Ionomycin (P/I) served as positive control. Data are means and standard deviations of duplicates and representative of 3 independent experiments using PBLs from different donors.
Since MCV infection induces MHC-I downregulation (30), we evaluated the expression of MHC-I in MCC cell lines by flow cytometry. Indeed, all MCV-positive cell lines exhibited low expression of MHC-I in contrast to the MCV-negative MCC-13 cell line (Fig. 5B). However, after treatment with IFN-γ expression of MHC-I molecules increased on the surface of all cell lines except of MKL-2. To determine whether the levels of HLA-A2 on the cell surface were sufficient to enable recognition by T cells, we pulsed the tumor cell lines with SMF (Fig. 5C) or KLL (Fig. 5D) peptides and evaluated the recognition by the transduced T cells. The HLA-A2+ cell lines (WaGa, MS-1, and MCC-13) prepulsed with the peptide were specifically recognized in contrast to the HLA-A2− cells MKL-1 and PeTa. No recognition was observed for the HLA-A2+ cell line MKL-2, reflecting the unresponsiveness of this particular cell line to IFN-γ-inducible upregulation of HLA class I.
We then tested whether IFN-γ-treated MCC cell lines were recognized by the TCR-transduced PBLs. The HLA-A2+ cell line WaGa reproducibly induced significant IFN-γ-release by the T cells transduced with the KLL-85 TCR (Fig. 5D). No response was observed to WaGa cells by the T cells expressing KLL-40 or the SMF-TCRs. Similarly, no response was seen after co-culture of the MS-1 and MKL-2 cells with all four TCRs.
MCC cell lines bear intact antigen processing and presentation machinery
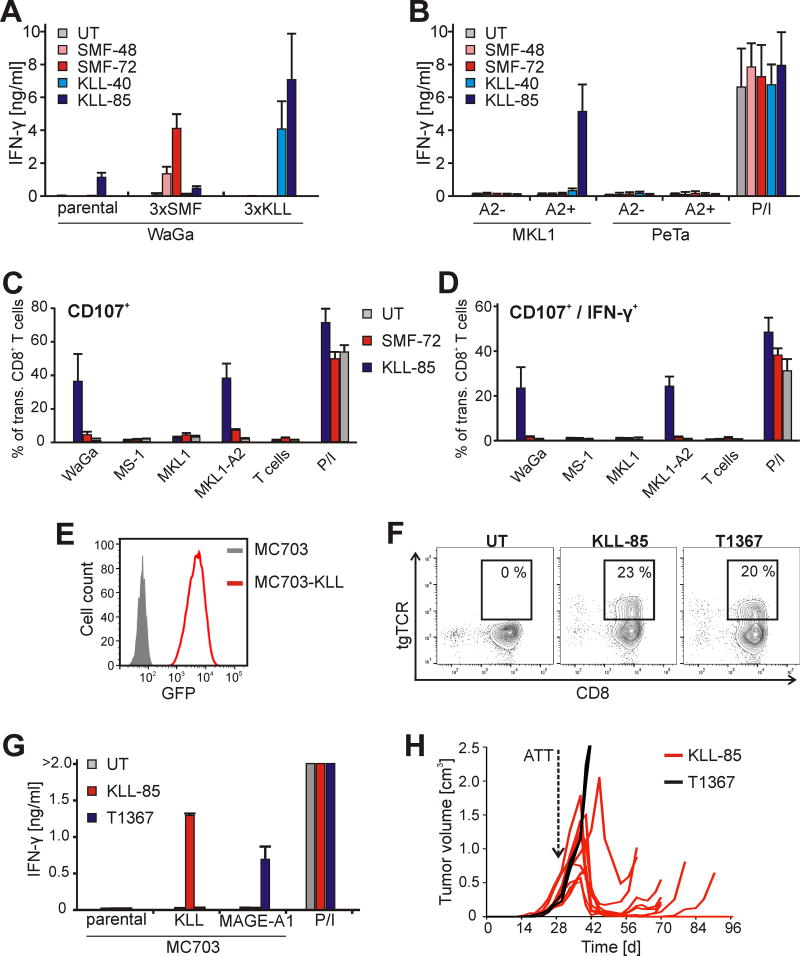
To investigate whether the expression level of MCV Tag was the limiting factor for the recognition by our TCRs, we transduced WaGa cells with trimeric minigenes encoding either SMF- or KLL-epitopes, fused to GFP, resulting in WaGa-3×SMF or WaGa-3×KLL, respectively. To enhance epitope presentation, epitope sequences were linked by an AAY motif (31). Expression of transgenes was verified by flow cytometry of GFP-positive cells (Supplementary Fig. S1). Again, parental WaGa cells were able to activate KLL-85 expressing T cells. T cells expressing KLL-40 or any of the SMF-TCRs were now activated in response to overexpression of the cognate epitopes in the respective cell lines (Fig. 6A). Interestingly, even though the expression of SMF epitope in WaGa-3×SMF was 2 logs higher than of KLL in WaGa-3×KLL as determined by MFI of GFP, T cells transduced with SMF-TCR secreted less IFN-γ than the KLL-TCR expressing T cells. This might be a consequence of a better presentation of the KLL epitope on the cell surface despite the predicted superior MHC-I binding affinity of the SMF epitope.
Figure 6.
MCV T-antigen derived KLL epitope mediates recognition of tumor cells in vitro and in vivo. A. TCR-engineered human T cells as indicated were assayed for response in IFN-γ production after co-culturing with WaGa cells and derivatives expressing either SMF- or KLL-epitopes. B. TCR-engineered human T cells as indicated were assayed for response in IFN-γ production after co-culturing with HLA-A2-negative MCC cell lines MKL-1 and PeTa and the derivatives ectopically expressing HLA-A2 (MKL1-A2 and PeTa-A2). Stimulation of T cells with PMA/Ionomycin served as positive control. In A and B diagrams show means and standard deviations of 3 different experiments. C and D. Proportion of CD107+ or CD107/IFN-γ-double positive cells among SMF- or KLL-TCR expressing CD8+ T cells as indicator of cytotoxic activity after co-incubation with target cells as indicated. Diagrams show means and standard deviations of two experiments with T cells from different donors. E. Antigen (GFP) expression in MC703-KLL cells (red). F. Flow cytometry analysis of murine T cells transduced with TCRs as indicated and stained with an antibody recognizing the variable region of the TCRs (a-hVβ8 for KLL-85 and a-hVβ3 for T1367). Gates show percentage of CD8+ T cells expressing the transgene TCR (tgTCR). G. TCR-engineered murine T cells were assayed for IFN-γ secretion after incubation with MC703 tumor cells or derivatives expressing the KLL epitope of MCV Tag or MAGE-A1. Stimulation of T cells with PMA/Ionomycin served as positive control. Data are means of duplicates ± mean deviation. H. HHDxRag−/− mice bearing established MC703-KLL tumors were treated with HHD T cells expressing KLL-85 (n=10, red lines), or T1367 (n=2, black lines).
We also tested whether the MCC cell line MS-1, which expresses the cancer-testis antigen MAGE-1 (Supplementary Fig. S2A), can process endogenously expressed antigen. We transduced T cells with the MAGE-A1-specific TCR T1367 (12) and co-cultured them with the MS-1 cells. In contrast to T cells expressing Tag-specific TCRs, T1367-transduced T cells were activated by the MS-1 target cells indicating that the processing machinery is intact in this cell line and that the expression level of MCV Tag doesn’t suffice to activate a T cell response (Supplementary Fig. S2B).
Next, we introduced cDNA encoding HLA-A2 into HLA-A2-negative cell lines PeTa and MKL-1. Whereas MKL1-A2 cells were recognized by KLL-85 and to a lesser extent by KLL-40, they didn’t induce reactivity by T cells transduced with any of the SMF-TCRs (Fig. 6B). In contrast, PeTa-A2 cells were not recognized by any of the transduced T cells, reflecting the low Tag expression in this cell line (Fig. 5A). Surface expression of the HLA-A2 transgene in the resulting cell lines PeTa-A2 and MKL1-A2 was verified by immunostaining and flow cytometry (Supplementary Fig. S3).
We also examined the ability of MCC cell lines to induce cytolytic activity in T cells expressing SMF- or KLL-specific TCRs. In accordance with the IFN-γ secretion assay, only HLA-A2 positive cells with abundant Tag expression (WaGa and MKL1-A2) induced CD107 expression (Fig. 6C–D). Again, this activity was KLL epitope-specific, since expression of CD107 in SMF-specific T cells was only slightly above the background.
Established tumors expressing the MCC-derived KLL epitope regress upon TCR gene therapy
To analyze the suitability of the KLL epitope as target for TCR gene therapy, we made use of a syngeneic cancer model of TCR gene therapy (12,20). As target cells, we generated a fibrosarcoma cell line, which was isolated from an HHD mouse expressing a chimeric HLA-A2/Db molecule (32) and was modified to express the KLL epitope (MC703-KLL, Fig. 6E). Antigen presentation was verified by co-culture with HHD-derived T cells engineered with either the KLL-85 or the T1367 TCR (Fig. 6F). TCR-transduced mouse T cells secreted IFN-γ upon co-culture with MC703-KLL tumor cells when expressing the KLL-85 TCR (Fig. 6G). Tumor cells were not recognized by untransduced mouse T cells or T cells transduced with the T1367 TCR. For in vivo analysis, HHDxRag−/− mice were challenged with MC703-KLL tumor cells and tumor-bearing animals were treated with KLL-85-engineered T cells when tumors were established (4 weeks after challenge, average tumor size: 380 mm3 ±140 mm3). Eight to eleven days after T cell transfer, tumors started to regress in all of the treated mice and tumors were rejected in 2/10 animals (Fig. 6H). In the remaining animals, tumor relapsed and escaped T cell treatment by downregulating MHC I (Supplementary Fig. S4). Tumor growth was not affected when either treating animals with T1367-engineered T cells or when left untreated.
DISCUSSION
MCV Tags are constitutively expressed in MCV-positive MCC malignancies (7). They are therefore attractive targets for TCR-based immunotherapy. The fact that immune-suppressed individuals are more likely to develop MCC indicates that host immune responses play an important role in preventing MCC tumorigenesis (33). This notion is supported by the higher rate of spontaneous regression than expected (12 in 400–700 cases or 1.7–3.0%) (34,35). Studies of regressing tumors demonstrated infiltration by CD4+ and CD8+ T cells suggesting that T-cell-mediated immunity plays a key role in tumor regression. Additionally, high intratumoral CD8+ and CD4+ lymphocyte infiltration predicts better survival (36,37). Although over 90% of the MCC patients are immune competent, they fail to clear MCC tumors that express non-self MCV antigens. These observations suggest that MCV-induced MCCs are capable of escaping immune surveillance.
Different virus species exhibit diverse immune evasion strategies (38). Downregulation of peptide presentation of intracellular proteins to CD8+ T cells is an effective mechanism for immune escape. This could be achieved either through downregulation of MHC-I surface expression and/or protein expression. Indeed, it has been shown that 84% of MCC tumors downregulate MHC-I and that MHC-I expression in MCV-positive tumors is lower than in MCV-negative tumors (39). This observation is reflected by the MCC cell lines used in our study where downregulation of MHC-I was the hallmark of all five MCV-positive cell lines but not of the MCV-negative MCC-13 cell line. However, the impaired MHC-I expression can be therapeutically overcome either by administration of interferons or by low-dose irradiation (30,39). Indeed, we observed up-regulation of MHC-I in most of the MCC cell lines after treatment with IFN-γ. However, one particular cell line, MKL-2, was unable to upregulate MHC-I after IFN-treatment. In experimental tumor models, IFN-γ-unresponsive variants have been described as a result of T cell pressure (40). Importantly, T cells transduced to express KLL-TCR reacted to WaGa and MKL1-A2 cells after treatment with IFN-γ. However, MCC cell lines MS-1 and PeTa-A2 were not recognized by any of the MCV Tag-specific TCRs, despite the up-regulation of MHC-I, suggesting there is an insufficient amount of antigen expressed in these particular cell lines. In fact, LT expression varies strongly among different MCC cell lines ((7) and Fig. 5A).
Low antigen expression might be also the reason for lack of recognition of MCC cell lines by TCRs directed to SMF epitope. We showed that this epitope is naturally processed, since T cells expressing SMF-TCRs react to H1299-A2 cells genetically modified to over-express LT cDNA. It is possible that our TCRs did not have sufficient avidity to mediate recognition of the very low levels of MCV antigens endogenously expressed in human tumor samples. In principle, TCR affinity can be enhanced by in vitro mutagenesis which however bears the risk that T cells become dysfunctional or lose their specificity. After overexpression of SMF epitope in one of the MCC cell lines, WaGa, recognition could be restored. Since the KLL epitope is present on both LT and sT, it is conceivable that the amount of processed KLL is higher than that of the SMF epitope, which is present on LT only. However, it is also possible that the KLL epitope is more efficiently processed and presented and/or has better binding affinity to MHC-I despite in the silico predictions that SMF is the epitope with highest affinity for HLA-A2. It is known, that the binding prediction of cysteine-containing peptides (such as KLL) is problematic, because algorithms do not account for potential oxidation and cysteinylation during assays (20,41). However, the most convincing explanation for the absent recognition of SMF epitope is suggested by a recent study demonstrating that MCV sT inhibits LT turnover by preventing LT proteolysis, while sT itself is subject to rapid proteasomal turnover (42). This would mean that the KLL peptides (encoded in the common region of both sT and LT oncoproteins) presented by MCC cells are preferentially derived from sT due to the rapid proteolysis of sT and the relative stability of LT. These data suggest KLL as a more suitable HLA-A2-restricted epitope to be targeted in a potential TCR gene therapy. This notion is supported by our in vivo experiments showing that a KLL-specific TCR induced regression/rejection of large established tumors in a syngeneic mouse cancer model.
A limitation of the present study is the small number of available MCC cell lines that may not adequately represent the broader population of MCC cases. There is also little information about patients from whom the tested cell lines originate. We cannot exclude that in vivo, Tag expression levels differ from that observed in MCC cell lines. Some studies suggest immunogenicity of MCC tumors. In a single patient case study, regression of metastatic MCC could be observed following transfer of autologous polyclonal T cells specific to HLA-A*2402-restricted MCV LT92–101 epitope (30).
Recent findings of a study using anti-PD-1 therapy in patients with advanced MCC showed promising results with a 56% objective response rate (43). Responses to anti-PD-1 occurred in both MCV-positive and MCV-negative MCC. Taken together with other studies of PDL-1 blockade (10), recent clinical data further support the notion that MCC is an unusually immunogenic cancer. Unfortunately, half of MCC patients do not derive persistent benefit from PD1 pathway blockade, despite the presence of prominent/abundant T cell antigens in both subsets of MCC tumors: virus-positive (viral oncoproteins) and virus-negative (UV-induced neo-antigens) (44). It is plausible that patients who do not benefit from PD1 pathway blockade therapy may have tumor-specific T cells that are too few in number or too poorly avid to support a sustained anti-tumor response in the patients. Thus, an immunotherapy approach with TCR-engineered T cells could supplement the PD1 blockade agents that have recently entered routine clinical practice (45).
In this study we describe the isolation and characterization of four HLA-A2-restricted TCRs specific to two epitopes derived from MCV Tags. Genetically modified T cells expressing these TCRs consistently recognized peptide-pulsed T2 cells and HLA-A2-positive target cells transduced with cDNA encoding MCV Tags. We also showed reactivity of one of the TCRs against HLA-A2+ MCV+ tumor cell lines in vitro and confirmed its potential to destroy large established tumors in vivo. Thus, our isolated TCR against the KLL epitope might have therapeutic potential via TCR gene therapy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
TCR gene therapy is an emerging cancer treatment modality. MCC is an aggressive skin cancer affecting an increasing number of individuals with limited treatment options for advanced disease. The majority of MCC cases are associated with MCV and expression of viral tumor antigens is essential for growth of MCC cells. Thus, MCV T-antigens could be targets for T cell-mediated immunotherapy. In this study, we describe the isolation and characterization of TCRs specific to MCV Tags using a humanized mouse model. Human T cells genetically engineered to express those TCRs recognize cell lines established from MCC tumors and display cytotoxic activity. Thus, our isolated TCRs have potential applications for TCR gene therapy, which may supplement the emerging use of PD1 pathway blocking agents that provide durable benefit for only about half of patients with advanced MCC.
Acknowledgments
Supported by Deutsche Forschungsgemeinschaft through grant SFB-TR36 (T.B.) and NIH-R01-CA162522 (P.N.).
We are grateful to Dr. Roland Houben for providing MCC cell lines and Dr. Yuan Chang for providing CM8E6 antibody. We would like to thank Melanie Manzke, Angelika Gärtner, and Kimberley Borutta for excellent technical support.
Footnotes
Conflict of interest: The Max-Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC) (T.B., I.G.) applied for a patent on the MCV-specific TCRs.
References
- 1.Becker JC. Merkel cell carcinoma. Ann Oncol. 2010;21:vii81–vii85. doi: 10.1093/annonc/mdq366. [DOI] [PubMed] [Google Scholar]
- 2.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest. 2012;122:4645–53. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, et al. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras M-T, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–9. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci USA. 2008;105:16272–7. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–72. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer. 2012;130:847–56. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 9.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med. 2016;374:2542–52. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman HL, Russell J, Hamid O, Bhatia S, Terheyden P, D’Angelo SP, et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–85. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L-P, Lampert JC, Chen X, Leitao C, Popovic J, Muller W, et al. Transgenic mice with a diverse human T cell antigen receptor repertoire. Nat Med. 2010;16:1029–34. doi: 10.1038/nm.2197. [DOI] [PubMed] [Google Scholar]
- 12.Obenaus M, Leitão C, Leisegang M, Chen X, Gavvovidis I, van der Bruggen P, et al. Identification of human T-cell receptors with optimal affinity to cancer antigens using antigen-negative humanized mice. Nat Biotechnol. 2015;33:402–7. doi: 10.1038/nbt.3147. [DOI] [PubMed] [Google Scholar]
- 13.Guastafierro A, Feng H, Thant M, Kirkwood JM, Chang Y, Moore PS, et al. Characterization of an early passage Merkel cell polyomavirus-positive Merkel cell carcinoma cell line, MS-1, and its growth in NOD scid gamma mice. J Virol Methods. 2013;187:6–14. doi: 10.1016/j.jviromet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Gele M, Leonard JH, Van Roy N, Van Limbergen H, Van Belle S, Cocquyt V, et al. Combined karyotyping, CGH and M-FISH analysis allows detailed characterization of unidentified chromosomal rearrangements in Merkel cell carcinoma. Int J Cancer. 2002;101:137–45. doi: 10.1002/ijc.10591. [DOI] [PubMed] [Google Scholar]
- 15.Rosen ST, Gould VE, Salwen HR, Herst CV, Le Beau MM, Lee I, et al. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab Invest. 1987;56:302–12. [PubMed] [Google Scholar]
- 16.Houben R, Dreher C, Angermeyer S, Borst A, Utikal J, Haferkamp S, et al. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J Invest Dermatol. 2013;133:2453–60. doi: 10.1038/jid.2013.169. [DOI] [PubMed] [Google Scholar]
- 17.Leonard JH, Bell JR, Kearsley JH. Characterization of cell lines established from merkel-cell (“small-cell”) carcinoma of the skin. Int J Cancer. 1993;55:803–10. doi: 10.1002/ijc.2910550519. [DOI] [PubMed] [Google Scholar]
- 18.Heemskerk MHM, Hoogeboom M, de Paus RA, Kester MGD, van der Hoorn MAWG, Goulmy E, et al. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102:3530–40. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 19.Hennig K, Raasch L, Kolbe C, Weidner S, Leisegang M, Uckert W, et al. HEK293-based production platform for γ-retroviral (self-inactivating) vectors: application for safe and efficient transfer of COL7A1 cDNA. Hum Gene Ther Clin Dev. 2014;25:218–28. doi: 10.1089/humc.2014.083. [DOI] [PubMed] [Google Scholar]
- 20.Leisegang M, Kammertoens T, Uckert W, Blankenstein T. Targeting human melanoma neoantigens by T cell receptor gene therapy. J Clin Invest. 2016;126:854–8. doi: 10.1172/JCI83465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leisegang M, Engels B, Meyerhuber P, Kieback E, Sommermeyer D, Xue S, et al. Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J Mol Med. 2008;86:573–83. doi: 10.1007/s00109-008-0317-3. [DOI] [PubMed] [Google Scholar]
- 22.Schambach A, Wodrich H, Hildinger M, Bohne J, Kräusslich H-G, Baum C. Context Dependence of Different Modules for Posttranscriptional Enhancement of Gene Expression from Retroviral Vectors. Mol Ther. 2000;2:435–45. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- 23.Engels B, Cam H, Schüler T, Indraccolo S, Gladow M, Baum C, et al. Retroviral vectors for high-level transgene expression in T lymphocytes. Hum Gene Ther. 2003;14:1155–68. doi: 10.1089/104303403322167993. [DOI] [PubMed] [Google Scholar]
- 24.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, et al. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118–28. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–12. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller NJ, Church CD, Dong L, Crispin D, Fitzgibbon MP, Lachance K, et al. Tumor-Infiltrating Merkel Cell Polyomavirus-Specific T Cells Are Diverse and Associated with Improved Patient Survival. Cancer Immunol Res. 2017;5:137–47. doi: 10.1158/2326-6066.CIR-16-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyngaa R, Pedersen NW, Schrama D, Thrue CA, Ibrani D, Met O, et al. T-cell responses to oncogenic Merkel cell polyomavirus proteins distinguish Merkel cell carcinoma patients from healthy donors. Clin Cancer Res. 2014;20:1768–78. doi: 10.1158/1078-0432.CCR-13-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb AI, Dunstone MA, Chen W, Aguilar M-I, Chen Q, Jackson H, et al. Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J Biol Chem. 2004;279:23438–46. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- 29.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 30.Chapuis AG, Afanasiev OK, Iyer JG, Paulson KG, Parvathaneni U, Hwang JH, et al. Regression of metastatic Merkel cell carcinoma following transfer of polyomavirus-specific T cells and therapies capable of re-inducing HLA class-I. Cancer Immunol Res. 2014;2:27–36. doi: 10.1158/2326-6066.CIR-13-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velders MP, Weijzen S, Eiben GL, Elmishad AG, Kloetzel PM, Higgins T, et al. Defined flanking spacers and enhanced proteolysis is essential for eradication of established tumors by an epitope string DNA vaccine. J Immunol. 2001;166:5366–73. doi: 10.4049/jimmunol.166.9.5366. [DOI] [PubMed] [Google Scholar]
- 32.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Pérarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185:2043–51. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J, Brewer J. Merkel Cell Carcinoma in Immunosuppressed Patients. Cancers. 2014;6:1328–50. doi: 10.3390/cancers6031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wooff JC, Trites JR, Walsh NMG, Bullock MJ. Complete spontaneous regression of metastatic merkel cell carcinoma: a case report and review of the literature. Am J Dermatopathol. 2010;32:614–7. doi: 10.1097/DAD.0b013e3181cd3158. [DOI] [PubMed] [Google Scholar]
- 35.Pang C, Sharma D, Sankar T. Spontaneous regression of Merkel cell carcinoma: A case report and review of the literature. Int. J. Surg. Case Rep. 2015;7C:104–8. doi: 10.1016/j.ijscr.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sihto H, Böhling T, Kavola H, Koljonen V, Salmi M, Jalkanen S, et al. Tumor infiltrating immune cells and outcome of Merkel cell carcinoma: a population-based study. Clin Cancer Res. 2012;18:2872–81. doi: 10.1158/1078-0432.CCR-11-3020. [DOI] [PubMed] [Google Scholar]
- 37.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-Wide Studies of Merkel Cell Carcinoma and Validation of Intratumoral CD8+ Lymphocyte Invasion As an Independent Predictor of Survival. J Clin Oncol. 2011;29:1539–46. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuren AB, Costa AI, Wiertz EJ. Recent advances in viral evasion of the MHC Class I processing pathway. Curr Opin Immunol. 2016;40:43–50. doi: 10.1016/j.coi.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Paulson KG, Tegeder A, Willmes C, Iyer JG, Afanasiev OK, Schrama D, et al. Downregulation of MHC-I expression is prevalent but reversible in Merkel cell carcinoma. Cancer Immunol Res. 2014;2:1071–9. doi: 10.1158/2326-6066.CIR-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Textor A, Schmidt K, Kloetzel P-M, Weißbrich B, Perez C, Charo J, et al. Preventing tumor escape by targeting a post-proteasomal trimming independent epitope. J Exp Med. 2016;213:2333–2348. doi: 10.1084/jem.20160636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2:522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. Merkel Cell Polyomavirus Small T Antigen Controls Viral Replication and Oncoprotein Expression by Targeting the Cellular Ubiquitin Ligase SCFFbw7. Cell Host Microbe. 2013;14:125–35. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med. 2016;374:2542–52. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2015;7:3403–15. doi: 10.18632/oncotarget.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauschild A, Schadendorf D. Checkpoint inhibitors: a new standard of care for advanced Merkel cell carcinoma? Lancet Oncol. 2016;17:1337–9. doi: 10.1016/S1470-2045(16)30441-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.