Abstract
Developments in CRISPR/Cas9-based technologies provide a new paradigm in functional screening of the genome. Conventional screening methods have focused on high-throughput perturbations of the protein-coding genome with technologies such as RNAi. However, equivalent methods for perturbing the non-coding genome have not existed until recently. CRISPR-based screening of genomic DNA has enabled the study of both genes and non-coding gene regulatory elements. Here we review recent progress in assigning function to the non-coding genome using CRISPR-based genomic and epigenomic screens, and discuss the prospects of these technologies to transforming our understanding of genome structure and regulation.
Introduction
Mammalian genomes are primarily composed of DNA that does not code for gene sequences. This non-coding sequence is thought to integrate cellular signaling into dynamic, cell type-specific patterns of gene expression. In fact, since humans share a large proportion of their protein-coding genome with much simpler organisms, it is thought that the non-coding sequence is critical in specifying diverse and complex phenotypes. Furthermore, the vast majority of genetic variation associated with complex human disease is found within non-coding regions of the genome [1]. Thus, understanding how genomic regulatory elements in the non-coding DNA encode gene expression programs could elucidate mechanisms of disease initiation and genetic contributions to drug responses, providing an avenue to develop next-generation therapies. Large-scale research projects such as the Encyclopedia of DNA Elements (ENCODE) [2] and the Roadmap Epigenomics Project [3] have mapped the epigenetic profiles of many human cell types and revealed common epigenetic signatures of regulatory elements genome-wide. However, these large datasets cannot precisely determine which elements are functional, identify the target gene(s) of a particular element, or specify which gene networks are influenced by dynamic epigenetic changes.
From the >3 billion base pairs in the human genome, millions of putative regulatory elements have been identified through projects such as ENCODE and genome-wide association studies [4]. Given the scale of the human genome and the diversity of genetic variation therein, high-throughput approaches are needed to annotate the function of non-coding regions. Tools for genome engineering based on programmable DNA-binding platforms enable the targeted perturbation of endogenous genomic sites and can be used to assess the functional role of putative regulatory elements in their native context [5]. Although the advent of these technologies initially inspired efforts to use programmable DNA-binding proteins, such as zinc finger domains, for high-throughput functional genomics [6], the considerable protein engineering required to build these systems has limited their scalability. The recently discovered and repurposed clustered regularly interspaced short palindromic repeats (CRISPR) technology for genome editing is much more compatible with high-throughput genetic and epigenetic screening [7]. The RNA-guided nature of CRISPR-based systems permits pooled screening to test thousands of unique hypotheses in parallel. Here, we review recent advances in using CRISPR screening to interrogate the function of the non-coding genome, with particular emphasis placed on epigenetic editing approaches.
CRISPR for genomic and epigenomic editing and application to pooled screening
CRISPR evolved as an adaptive immune system in prokaryotes, and has recently been repurposed for gene editing in mammalian cells [8,9]. In type II CRISPR systems, a CRISPR-associated (Cas) endonuclease is guided to a DNA target via a short guide RNA (gRNA) where it catalyzes a double-strand break [10]. Endogenous cellular pathways can repair the break by either non-homologous end joining (NHEJ) or homology-directed repair (HDR). NHEJ in particular is useful for screening as it is an error-prone process resulting in small insertions or deletions (indels) that can knockout gene function by disrupting the reading frame or alter gene regulation by mutating transcription factor binding sites. CRISPR systems have also been repurposed for programmable gene regulation by deactivating the catalytic activity of the Cas endonuclease [11]. The nuclease-deactivated version retains its ability for DNA targeting via the gRNA, and thus can serve as a scaffold for recruiting transcriptional machinery or epigenetic modifiers. The nuclease-deactivated versions have been engineered as repressors and activators by fusing transcriptional regulatory domains to the termini of the Cas protein [12].
The Cas9 endonuclease from the type II CRISPR system of Streptococcus pyogenes has been the most widely used CRISPR system for implementing high-throughput gRNA screens. The first screens with nuclease-active Cas9 designed pools of gRNAs targeting early constitutive exons of over 18,000 protein coding genes [13,14]. The indels formed in the early exons frequently result in out-of-frame mutations, leading to deactivation of the target gene. Similar screens were implemented with deactivated Cas9 (dCas9) fusion proteins to knockdown or activate genes via targeting with dCas9-based effectors [15,16]. Gene repression is typically achieved by fusing dCas9 to the Krüppel-associated box (KRAB) domain that recruits epigenetic modifiers that catalyze the formation of heterochromatin [17]. Gene activation can be achieved with any one of several distinct dCas9-based effectors, all of which rely on the recruitment of transcriptional machinery via multiple scaffold domains, such as VP64, p65 or HSF1 [15–19]. Contrary to gene knockout with nuclease-active Cas9, dCas9-based transcriptional regulation is achieved by targeting gene promoters and is highly specific in manipulating gene expression, epigenetic marks, and chromatin structure [20,21].
A high-throughput screen with either Cas9 or dCas9-based effectors applies pools of gRNAs to test thousands of hypotheses in parallel. gRNAs are typically synthesized as pools of oligonucleotides and assembled into a lentiviral vector library. The lentiviral library of gRNAs is then transduced into a cell population at a low multiplicity of infection, such that a single gRNA expression cassette integrates into each genome and serves as a genetic barcode for each cell. The cell line can either stably express the Cas9 effector, or the lentiviral library can contain both Cas9 and the gRNA. The cell population is then selected for cells that received a gRNA and cultured for a period of time to ensure adequate expression of the CRISPR components and permit the phenotypic effect resulting from the corresponding genomic perturbation.
In order to screen for genes or genomic regions corresponding to a particular phenotype, there must be selectable criteria by which cells in which that phenotype is perturbed can be identified. Common selection methods include resistance or sensitivity to drug treatment, changes to cellular proliferation rates or viability, or changes in gene expression assessed by fluorescence-activated cell sorting (FACS) or single cell RNA-seq. Following selection, the gRNA abundance in the selected population is quantified via next-generation sequencing to identify enriched or depleted gRNAs relative to the initial gRNA pool. This general approach to CRISPR-based screening was first implemented to identify essential genes in cancer, uncover mechanisms of drug resistance, and dissect cellular signaling networks [13,14,22]. In addition, data collected from such screens have helped to develop algorithms to predict optimal gRNAs for next-generation libraries [23,24].
Although the majority of CRISPR-based screens to date have targeted coding genes, several recent studies have applied a similar screening methodology to probe the function of non-coding regions distal to gene bodies. Non-coding screens have been performed with both Cas9 and dCas9-based effectors [25], each of which harbors its own advantages and disadvantages in elucidating the role of the non-coding genome in coordinating gene expression. With nuclease-active Cas9, saturation mutagenesis screens that densely tile a region with gRNAs can better resolve minimal sites within a putative regulatory element that influence function, such as transcription factor binding sites [26]. In addition, the indels produced via NHEJ can define sequences that impart differing influence on activity of a particular element. Complementing this approach, CRISPR-based screens with dCas9 fusion proteins can aid in elucidating the role of particular genomic regions and epigenetic marks in regulating gene expression. Fusions of epigenetic modifying domains to dCas9 have been shown to deposit chemical modifications to local histones and DNA and influence gene expression [12]. These tools can be applied to high-throughput screens to identify the sites that harbor regulatory function, and also to specify the epigenetic marks that guide that function. Furthermore, in some cases, a single gRNA can modulate the chromatin state of an entire enhancer [20,27], enabling the coverage of a larger region of genomic space with a fixed gRNA library size.
Nuclease Mutagenesis Screens
The largest collection of work performed thus far in screening non-coding sequence has been with CRISPR nuclease-based mutagenesis, exploiting the indel footprint of Cas9 to identify important regulatory sites (Figure 1A). One of the earliest examples probed the erythroid intronic enhancer of BCL11A which regulates fetal hemoglobin levels and is a therapeutic target for several hemoglobin disorders [26]. A gRNA library was designed consisting of ~500 gRNAs targeting the three Dnase I Hypersensitivity Sites (DHSs) in the composite enhancer, ~50 gRNAs targeting exon 2 of BCL11A as a positive control and 120 non-targeting gRNAs. The screen revealed several transcription factor motifs required for BCL11A expression including the erythroid-specific transcription factor GATA1 as well as STAT1, EHF, and EFL1. Another study used a similar approach to interrogate regulatory elements that control PD-1 expression in mouse T cells during exhaustion induced by chronic infection [28]. A later study looked to increase the number of perturbations per region by targeting putative regulatory elements found in a GWAS locus with two variants of the SpCas9 protein that have different PAM requirements [29].
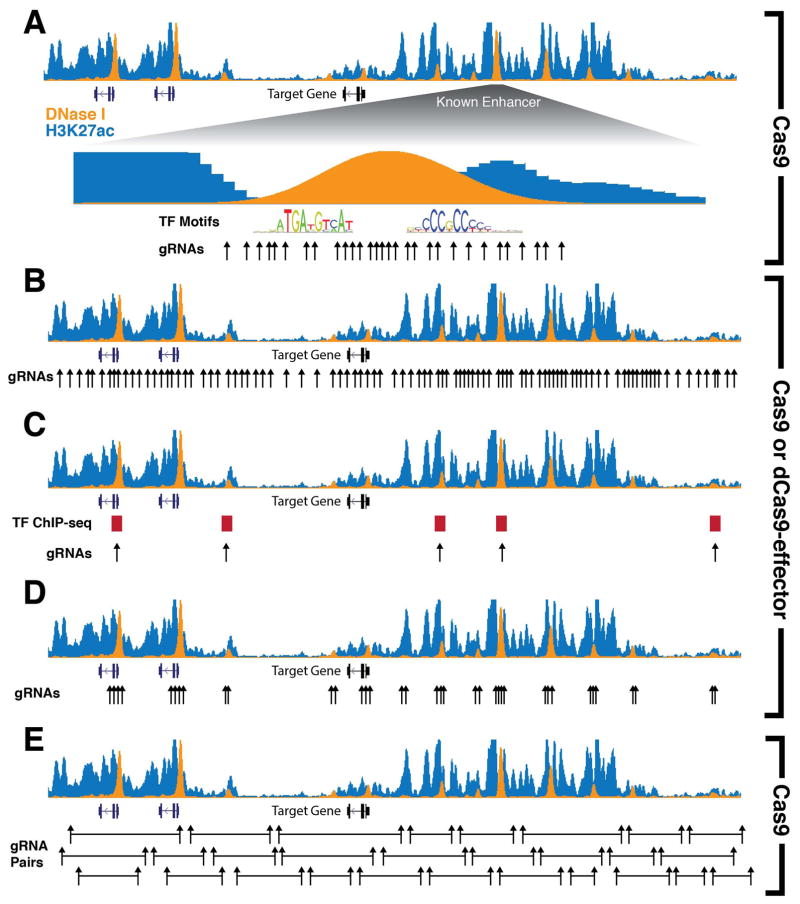
Figure 1.
An overview of library design approaches to screen the non-coding genome. (A) A saturation mutagenesis approach to dissect known or putative enhancers. Targeted regions of DNA are saturated with gRNAs to determine which sequence disruptions, generated by indels via genome editing, will alter gene expression. This method can be used to determine which transcription factor motifs are required for proper enhancer function. (B) An unbiased tiling approach to scan across a genomic region of interest. This method does not rely on any prior knowledge of transcription factor binding or other epigenetic marks, but is limited in the amount of sequence space that can be interrogated. (C) Targeting specific transcription factor binding sites via ChIP-seq signal or transcription factor motifs. This method relies on prior knowledge of important DNA-binding proteins to target and disrupt their binding sites. The focus on particular binding sites facilitates using a reduced number of gRNAs and enables the possibility of genome-wide screens. (D) A DHS-targeted approach that provides intermediate coverage between strategies in (B) and (C). gRNAs are designed to target DNase I hypersensitive sites (DHSs) in order to reduce library size and target a larger genomic region without requiring previous knowledge of important transcription factors. (B) Overlapping paired gRNAs can be used to create deletions that span across a locus and disrupt regulatory DNA. Pairs of gRNAs are delivered together such that in one cell a specific region of DNA is deleted. Signal from overlapping deletions can narrow down which region is important for gene regulation. Approaches A,B,C, and E have been used with Cas9-based nuclease screens while approaches B-D have been used with dCas9-based approaches.
An alternative approach to saturating defined putative regulatory elements is to design an unbiased library of gRNAs surrounding genes of interest (Figure 1B). One such study performed a saturation mutagenesis screen to identify cis-regulatory elements surrounding four genes specifically expressed in mouse embryonic stem cells (mESCs). Each gene had an in-frame knock-in GFP sequence to quantify gene expression via flow cytometry [30]. Additionally, the authors developed a non-viral approach to deliver the gRNA library to cells. A gRNA expression cassette containing a placeholder gRNA was integrated at the ROSA26 locus in mESCs. Subsequently, a pool of oligos encoding gRNA spacer sequences and containing homology arms was transfected into the cell line along with Cas9 and a gRNA targeting the placeholder gRNA sequence to facilitate homology-directed repair and integration of the gRNA library. Cells were sorted based on GFP expression into different bins and gRNA abundance was quantified within each bin. These screens identified promoters of nearby genes as well as unannotated regions containing no chromatin features of canonical regulatory elements (i.e. DNase signal or ChIP-seq signal for known transcription factors) as having an impact on gene expression.
Downstream validation of gRNAs enriched in the initial pooled screens is essential to sufficiently characterize the regulatory function of those defined genomic regions. For example, a recent study used chromosome conformation capture (3C) to validate putative enhancers identified with a gRNA library surrounding three genes found to be involved in vemurafenib resistance in melanoma cells [31]. The greatest enrichment of gRNAs was found in regions surrounding the CUL3 gene. In addition to 3C analysis, the authors found several correlating genomic and epigenomic features at the gRNA-enriched regions, such as increased chromatin accessibility in melanoma cells and increased DNA sequence conservation within primates, indicating evolutionarily important regulatory activity. These types of characteristics could help inform future algorithmic identification of active regulatory elements.
High density saturation mutagenesis surrounding genes of interest is effective at resolving functional regulatory elements proximal to target genes. However, this approach is limited in genomic coverage with a library of constrained size, which is typically restricted by cell number or throughput of the selection method, such as FACS. Rather than saturating regions with all possible gRNAs, a more targeted approach can be used to only focus on previously identified chromatin annotations that attempt to define putative regulatory elements, such as DNase-seq and ChIP-seq (Figure 1C,D). For instance, a recent study looked for regulators of the embryonic stem cell gene POU5F1 (OCT4) by targeting putative enhancers that were previously identified by epigenetic marks [32]. A gRNA library was designed to target 174 DHSs with ~11 gRNAs per DHS resulting in a library of ~1900 gRNAs (Figure 1D). This library was delivered with Cas9 to a POU5F1-GFP reporter stem cell line, and low reporter expressing cells were isolated with FACS. The authors identified known enhancers of POU5F1 as well as previously unknown enhancers that when perturbed, resulted in transient knockdown of reporter expression. Another study designed a gRNA library to target specific transcription factor binding motifs identified by ChIP-seq (Figure 1C). This targeted approach allowed for a reduced library size and increased number of regions targeted. The authors performed screens targeting p53 motifs with a library of 1,116 gRNAs to map enhancers involved in oncogene-induced senescence and ERα motifs with a library of 97 gRNAs to identify sites required for proliferation in breast cancer cell types [33]. These studies demonstrate that prior knowledge of transcription factor binding and chromatin accessibility can significantly reduce the size of the library required for identification of important non-coding regions.
Larger genomic regions can be analyzed with deletion screens, where a pair of gRNAs is co-expressed and mediates a targeted genomic deletion (Figure 1E). This approach was applied in two recent studies. The first delivered a library of paired gRNAs for programmed deletions around the OCT4 locus [34]. Overlap in deletions allowed for a cumulative signal to be recovered after sequencing gRNAs in cells with low GFP expression. The second study used programmed genetic deletions to identify regulatory elements of HPRT1 in HAP1 cells after selection with a chemotherapeutic drug [35]. The average deletion size in this library was ~1–2 kb in length across a 206 kb region of the HPRT1 locus. This study found a lack of distal enhancers essential for HPRT1 expression with the strongest effects of deletion occurring in the exons of the gene.
CRISPRi Screens
Rather than using Cas9 to perturb regulatory elements through DNA sequence mutation, dCas9-based CRISPR interference (CRISPRi) can be used to identify regulatory regions in similar pooled formats. One study used dCas9-KRAB to tile a total of ~1.3 megabases surrounding GATA1 and MYC in K562 cells [36]. These genes are required for proliferation in this cell line, so depletion of gRNAs from the library was measured after 14 population doublings. Two key features of the enriched regions around these genes were DNase I hypersensitivity and H3K27ac signal, supporting the use of these features for designing targeted gRNA libraries. Together with Hi-C enhancer-promoter contact frequencies, these features were used to construct a predictive model of enhancer activity and correctly ranked six of the seven identified regulatory elements as well as other known enhancers.
Another study used dCas9-KRAB to perturb DHSs in two loci, the human β-globin locus and the HER2 locus [37]. DNase I hypersensitivity was used to identify putative regulatory elements and gRNAs were designed to tile each DHS. For the β-globin locus, transcriptional activity was measured via an endogenous HBE1-mCherry reporter while HER2 levels were measured using immunofluorescence. After applying the library of gRNAs and sorting off high and low expressing cells, known enhancers in the β-globin locus control region were enriched as well as promoters of the downstream HBG1/2 genes. For the HER2 screen, a previously characterized HER2 intronic enhancer was identified as well as a downstream promoter of GRB7 and a novel intronic enhancer also located in GRB7.
CRISPRa Screens
Far fewer studies have utilized CRISPR activation (CRISPRa) to probe for enhancer activity, but next-generation dCas9-based activators have made this approach feasible [27]. An early study to use CRISPRa gain-of-function screens for regulatory element discovery utilized the dCas9-p300 fusion for activation [37]. The p300 core domain deposits acetylation marks to histones, including histone H3 lysine 27, which is found at active promoters and enhancers and is thought to open chromatin by reducing electrostatic interactions between DNA and lysine residues. This study used a library of gRNAs targeting DHSs surrounding HER2 in the A431 and HEK293T cell lines. High and low HER2 expressing cells, as measured using cell surface immunostaining, were isolated using FACS. Enriched DHSs were found in both cell lines, some of which were non-overlapping between cell types, possibly attributable to cell type-specific enhancer activity. This demonstrates the importance of choosing appropriate cell lines or systems that best model the biology being studied in order to find the most relevant hits. This study also directly compared CRISPRi and CRISPRa screens at the same locus and found only partially overlapping results, demonstrating the importance of both gain- and loss-of-function screens to map the regulatory landscape of a locus.
In another study, dCas9-VP64 was used to screen for enhancers of CD69 and IL2RA in Jurkat T cells [38]. One of the identified enhancers of IL2RA contained an autoimmunity disease variant identified through GWAS. A mouse model was generated harboring the disease variant and activated T cells showed a delayed expression of Il2ra rather than complete loss of expression. When the same enhancer was deleted, mice produced more pro-inflammatory T helper cells and decreased numbers of regulatory T cells in response to stimulus. These results demonstrate that certain regulatory elements are specific to cell type and stimulus, further supporting the importance of choosing a relevant cell type and screening approach for identifying significant hits.
Single-cell methods
CRISPR-based screens in a pooled format enable testing of thousands of hypotheses in parallel. Such a systematic and scalable approach has revolutionized the annotation of genome function. However, in the case of most CRISPR screens performed to date, the phenotypic readouts are complex cellular functions, such as cell growth and drug resistance. These phenotypes can be influenced by myriad cellular processes, yet a pooled CRISPR screen cannot inform as to the molecular mechanisms deriving from a single or multiplexed genomic or epigenomic perturbation. Such analyses are commonly performed after the initial screen during which individual gRNAs are validated in low throughput.
To address some of the limitations of traditional CRISPR screening approaches, several groups have developed technologies to combine massively parallel single-cell RNA sequencing methods [39–41] with pooled CRISPR genetic and epigenetic perturbations [42–45]. In this way, complete transcriptomic effects can be comprehensively determined for single or combinatorial perturbations. Since this approach is limited in the number of perturbations that can be tested in parallel because of constraints in sequencing depth or cell numbers, the bulk screening approaches described above can aid in narrowing down targets to choose from. For example, Adamson et al. applied their Perturb-seq technology to dissect the regulatory mechanisms of the unfolded protein response (UPR) in mammalian cells. The authors first performed a genome-wide CRISPR knockout screen to identify a set of candidate genes involved in UPR regulation. Subsequently, they implemented Perturb-seq on a subset of 82 genes and assessed the transcriptional effects resulting from the knockdown of these genes. Interestingly, they identified gene sets that modulated the UPR response through distinct signaling branches, and uncovered cellular subpopulations that respond differently to a given perturbation [42].
The initial studies combining single-cell RNA sequencing with pooled CRISPR screens have used gRNA libraries targeting protein-coding genes. However, this approach holds unique value in uncovering functions of non-coding regulatory elements. For example, the unbiased nature of profiling whole transcriptomic effects of CRISPR-based perturbations enables the identification of genes and gene networks downstream of particular regulatory elements. This is of particular importance when the target genes of candidate enhancers are not known a priori, which is often the case in the identification of disease-associated genetic variation from GWAS. In addition, the single-cell approaches facilitate combinatorial analysis of concurrent perturbations, which can help to delineate the contribution of multiple regulatory elements.
Some of these advantages were demonstrated in a recent study which targeted 71 constituent enhancers from 15 super enhancers with dCas9-KRAB and measured the transcriptional profile of 12,444 cells [46]. Cells were transduced such that each cell received an average of ~3.2 gRNAs, allowing for multiple perturbations to be studied in one cell and reducing the overall number of cells needed for the screen. An advantage to this approach is the ability to determine the cell-to-cell variability in the usage of each enhancer in a given region. This was measured by determining the number of cells that had changes to gene expression as well as the level of gene expression change in those cells. Some enhancers were found to be active in a majority of cells and also led to significant knockdown of gene expression. Other enhancers were found to be active only in a subset of cells but also led to significant knockdown of gene expression. Finally, a third class of enhancers were found to be active in the majority of cells but had only mild effects on gene expression. In addition to classification of enhancer usage, combinatorial perturbations can be measured, as was seen with cells harboring gRNAs targeting multiple enhancers within the same super enhancer. Cells containing only one gRNA targeting an enhancer near PIM1 resulted in no detectable knockdown. However, when a cell contained two gRNAs targeting enhancers nearby PIM1, significant knockdown was detected. These types of pooled screen analyses are greatly facilitated through single-cell readouts.
Perspective
High-throughput CRISPR screens have greatly improved our ability to dissect the function of the non-coding genome (Figure 2). The complement of Cas9 and dCas9-based approaches enables the identification of regulatory motifs within putative enhancers and the epigenetic signatures associated with their function. Though most screens thus far have been performed with Cas9 from Streptococcus pyogenes that is limited to target sites with an NGG PAM, recent success implementing Streptococcus aureus Cas9 into pooled screening formats will expand the sequence space for higher resolution perturbations and enable combinatorial and orthogonal screens revealing novel genetic and epigenetic interactions [47].
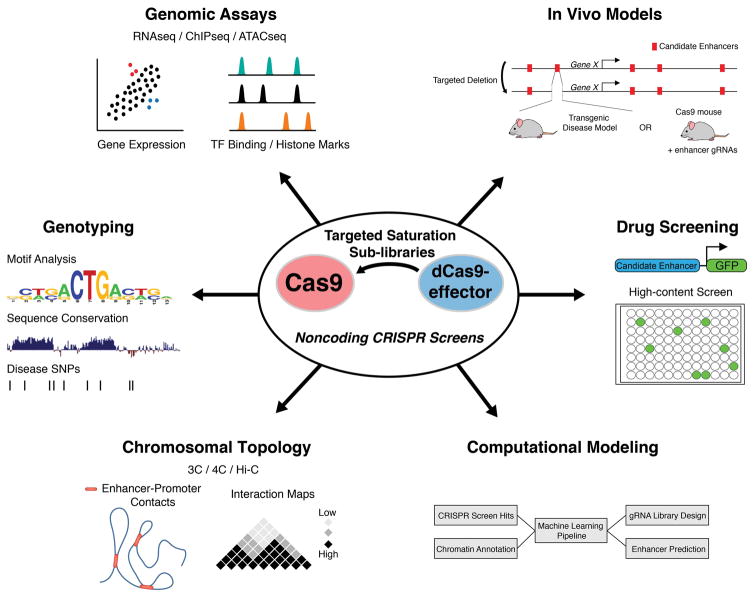
Figure 2.
Validation and characterization of hits from noncoding CRISPR screens. Pooled screens with dCas9-effectors can potentially scan a larger region of the genome since enhancer activity can be increased or decreased with a single gRNA. The results from these screens can inform target regions for more focused Cas9-based mutagenesis screens. Following a pooled screen, individual gRNAs are assayed independently to validate their function in gene regulation. Genome-wide genomic datasets (RNA-seq, ChIP-seq, ATAC-seq, Hi-C, etc…) can be used to determine the target genes of putative enhancers and the epigenetic marks and genomic topology associated with their function. With Cas9 mutagenesis, secondary screens with individual gRNAs or smaller sub-libraries can be performed to define functional genotypes resulting from Cas9-induced indels. These sequence features can then be compared to datasets of genetic variation associated with human disease, which can direct the generation of in vivo disease models or drug screening platforms. The data acquired from CRISPR-based screens of the noncoding genome can be useful in the development of algorithms to predict enhancer function or define features of highly active gRNAs.
Most CRISPR screens of the non-coding genome to date have identified regulatory elements that harbor epigenetic signatures predictive of enhancers (i.e. DNase I hypersensitivity). This finding reinforces the dogma that epigenetic marks can denote regulatory function and reduces the sequence space needed for future screens. However, one study did identify functional elements lacking any canonical epigenetic signatures [30], although it is unclear how ubiquitous this phenomenon is across the genome. It will be important to perform comparative screens using different Cas9 and dCas9-based effectors to elucidate how the deposition or removal of specific epigenetic marks influences the regulatory function of a genomic site. Data accumulated from such screens could inform computational approaches to better predict enhancer function in homeostasis or in response to perturbation. Lastly, the knowledge gained from CRISPR screens could inform the rational design of de novo enhancers, perhaps with aide from recent technologies for artificially modulating chromosomal topology [48].
Table 1.
A survey of studies using CRISPR-based screens to annotate function of the non-coding genome.
| Technology Used | Screen Phenotype Measured | Library Design | Area of Genomic Region Screened | Number of Sites Screened | Number of gRNAs in library |
|---|---|---|---|---|---|
| SpCas9 | Antibody Staining (FACS) | Saturation Mutagenesis | ~4.2 Kb | 3 DHSs, 1 Exon | 702 |
| SpCas9 | Antibody Staining (FACS) | DHS Targeting | ~60 Kb | 9 DHSs | 2,071 |
| SpCas9 | Reporter Expression (FACS) | Unbiased Tiling/Predicted Enhancer | 40 Kb – 150 Kb | 4 genes | 3,908 |
| SpCas9 | Reporter Expression (FACS) | DHS Targeting | ~1 Mb | 174 DHSs | 1,964 |
| SpCas9 | Reporter Expression (FACS) | Paired gRNA Deletions | 2 Mb | 1 gene | 11,570 |
| SpCas9 | Proliferation | TF Motif/ChIP-seq Peak Targeting | Genome wide | 685 or 73 Regions | 1,116 or 97 |
| SpCas9 | Resistance to Drug Treatment (Proliferation) | Unbiased Tiling | 200 Kb | 3 genes | 6,682, 6,934, or 4,699 |
| SpCas9 | Resistance to Drug Treatment (Proliferation) | Paired gRNA Deletions | 206 Kb | 1 gene | 8,684 |
| SpCas9, SpCas9-VQR | Antibody Staining (FACS) | Saturation Mutagenesis | ~340 Kb | 98 DHSs | 5,542 |
| dCas9-KRAB | Proliferation | Unbiased Tiling | 1.29 Mb | 2 genes | 98,000 |
| dCas9-KRAB | Single cell RNA-seq | DHS Targeting | ~2 Mb across 7 TADs | 71 constituent enhancers in 15 super enhancers | 243 |
| dCas9-KRAB, dCas9-p300 | Reporter Expression or Antibody Staining (FACS) | DHS Targeting | ~4.5 or ~4 Mb | 281 or 433 DHSs | 12,472 |
| dCas9-VP64 | Antibody Staining (FACS) | Unbiased Tiling | ~135 Kb or ~178 Kb | 2 genes | 10,780 or 20,412 |
Highlights.
Non-coding regulatory DNA determines gene expression and complex phenotypes
Technologies have not been available to annotate function of the non-coding genome
High-throughput synthesis of gRNAs facilitates CRISPR-based genomic screens
Screens include Cas9-based mutagenesis and dCas9-based gain- and loss-of-function
Diverse screening strategies can be employed with varying degrees of genomic coverage
Acknowledgments
This work was supported by an Allen Distinguished Investigator Award from the Paul G. Allen Frontiers Group, the Thorek Memorial Foundation, US National Institutes of Health (NIH) grants R01DA036865, U01HG007900, R41GM119914, and P30AR066527, and a NIH Director’s New Innovator Award (DP2OD008586). T.S.K. and J.B.B. were supported by a NIH Biotechnology Training Grant (T32GM008555), and J.B.B. was also supported by an NIH Predoctoral Fellowship (F31NS105419).
Footnotes
Conflict of Interest
T.S.K., J.B.B., and C.A.G. have filed patent applications related to genome engineering. T.S.K. and C.A.G. are co-founders and advisors to Element Genomics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hindorff LAMJ, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA. A Catalog of Published Genome-Wide Association Studies [Google Scholar]
- 5.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blancafort P, Segal DJ, Barbas CF., 3rd Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 7.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. This study characterized the function and specificity of a CRISPR/Cas9-based transcriptional repressor in modulating enhancer function. The authors demonstrate that dCas9-KRAB modulates the epigenome and gene expression from distal enhancers in a highly specific manner, establishing an epigenome editing tool sufficient for pooled screening of regulatory DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polstein L, Perez-Pinera P, Kocak D, Vockley C, Bledsoe P, Song L, Safi A, Crawford G, Reddy T, Gersbach C. Genome-wide specificity of DNA-binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015 doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016:5. doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montalbano A, Canver MC, Sanjana NE. High-Throughput Approaches to Pinpoint Function within the Noncoding Genome. Mol Cell. 2017;68:44–59. doi: 10.1016/j.molcel.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. This study demonstrates a CRISPR saturation mutagenesis screen of a previously identified enhancer of BCL11A. In addition to being the first example of a pooled CRISPR screen of a noncoding genomic region, this study identified essential sequence features defining the regulatory function of the BCL11A enhancer and defined a potential therapeutic strategy for treating globinopathies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canver MC, Lessard S, Pinello L, Wu Y, Ilboudo Y, Stern EN, Needleman AJ, Galacteros F, Brugnara C, Kutlar A, et al. Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat Genet. 2017;49:625–634. doi: 10.1038/ng.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Rajagopal N, Srinivasan S, Kooshesh K, Guo Y, Edwards MD, Banerjee B, Syed T, Emons BJ, Gifford DK, Sherwood RI. High-throughput mapping of regulatory DNA. Nat Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. This work performed Cas9 screens of the noncoding space surrounding four embryonic stem cell-specific genes. These screens identified putative regulatory elements lacking canonical chromatin features of enhancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. This study performed a Cas9 screen in noncoding regions surrounding three genes involved in BRAF inhibitor resistance in melanoma cells. The authors identified enriched gRNAs located within sites harboring chromatin marks, transcription factor binding sites, and long-range interactions associated with enhancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diao Y, Li B, Meng Z, Jung I, Lee AY, Dixon J, Maliskova L, Guan KL, Shen Y, Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. doi: 10.1038/nbt.3450. This study used ChIP-seq datasets to develop gRNA libraries targeting binding sites of p53 and ERα. A subset of enriched gRNAs was shown to significantly reduce eRNA production from the target enhancers. [DOI] [PubMed] [Google Scholar]
- 34.Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods. 2017;14:629–635. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, Cusanovich DA, Shendure J. CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. Am J Hum Genet. 2017;101:192–205. doi: 10.1016/j.ajhg.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. This study used epigenome editing with dCas9-KRAB to screen for functional noncoding regions surrounding two transcription factors MYC and GATA1. The results of these screens, in combination with previously published chromatin annotation datasets, were used to develop a prediction model of MYC enhancer activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, Gersbach CA. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35:561–568. doi: 10.1038/nbt.3853. The authors of this study used DNase-seq datasets to design gRNA libraries surrounding the β-globin and HER2 loci. This work used the dCas9-KRAB repressor and the dCas9-P300 activator in independent screens in multiple cell types, and found that enhancer function could depend on cell type and/or direction of perturbation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. This study used a tiled CRISPR activation screen to identify enhancers of CD69 and IL2RA. The authors generated mouse models based on an identified IL2RA enhancer to characterize enhancer function and define the effect on T cell phenotype in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882. e1821. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896. e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 44.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. e1817. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol Cell. 2017;66:285–299. e285. doi: 10.1016/j.molcel.2017.03.007. The authors of this study developed a method to combine pooled CRISPR screening with single cell RNA-seq to define the whole transcriptome effects of dCas9-KRAB-mediated epigenome editing. This approach enabled the identification of combinations of gRNAs that synergistically modulate enhancer activity. [DOI] [PubMed] [Google Scholar]
- 47.Najm FJ, Strand C, Donovan KF, Hegde M, Sanson KR, Vaimberg EW, Sullender ME, Hartenian E, Kalani Z, Fusi N, et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat Biotechnol. 2017 doi: 10.1038/nbt.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan SL, Mariano NC, Bermudez A, Arruda NL, Wu F, Luo Y, Shankar G, Jia L, Chen H, Hu JF, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017;8:15993. doi: 10.1038/ncomms15993. [DOI] [PMC free article] [PubMed] [Google Scholar]