Abstract
Recent advances in imaging techniques have enabled visualizations of nascent transcripts or individual protein molecules at high spatiotemporal resolution, revealing the complex nature of transcriptional regulation. Here, we highlight recent studies that have provided comprehensive insights to transcriptional dynamics using such quantitative imaging techniques. Specifically, they demonstrated that transcriptional activity is stochastic, and such transcriptional bursting is modulated by multiple components like chromatin environments, concentration of transcription factors, and enhancer-promoter interactions. Moreover, recent studies suggested that regulation of transcriptional activity is more complex than previously thought, by showing that transcription factors and RNA polymerases also move within the cell with distinct kinetics and sometimes form dynamic clusters to mediate transcriptional initiation.
Graphical Abstract
Introduction
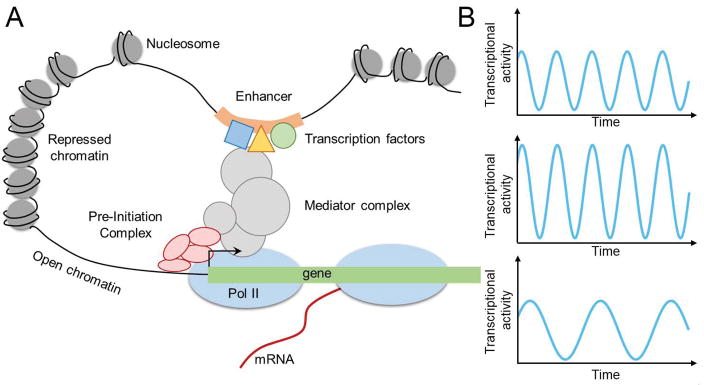
Transcriptional regulation is a critical yet complex biological process that requires precise spatiotemporal regulation to ensure normal functioning of organisms. Despite its importance, transcriptional regulation is yet to be fully understood due to its complex nature. Also, transcription is not a static, but rather a dynamic process that involves multiple layers of DNA-DNA, DNA-protein, and protein-protein interactions (Figure 1A) [1]. Pre-initiation complexes are stably formed at the promoter on the order of seconds to minutes, and RNA polymerase II (Pol II) molecules elongate and produce mRNAs at a rate of a few kb per minute [2,3]. The resulting gene expression is also dynamic in nature, where some genes exhibit oscillatory behaviors, while others show stable expressions [4]. In such dynamic environment, it is important to understand the kinetics of transcription machineries, such as transcription factors and RNA polymerases, and the effects of changes in transcription kinetics on cellular processes (Figure 1A–B).
Figure 1. Transcriptional regulation modules that affect transcriptional dynamics.
(A) Schematic of regulatory modules that control transcription.
(B) Example trajectories of transcriptional activities that can result from modulations on one of the transcriptional machineries.
Recently developed quantitative imaging methods, such as live-cell and single-molecule imaging techniques, have significantly transformed our view of transcriptional regulation [5,6]. Development of such techniques enabled visualizations of various components of transcription, which were not readily accessible previously due to technical challenges. Assays like the MS2-MCP detection system have allowed labeling of nascent transcripts in living cells, and advancement in microscopy techniques has allowed visualization of single molecules at high spatiotemporal resolution (Figure 2). This review highlights recent studies that take an advantage of such quantitative imaging to provide better insights on the dynamics of transcriptional regulation.
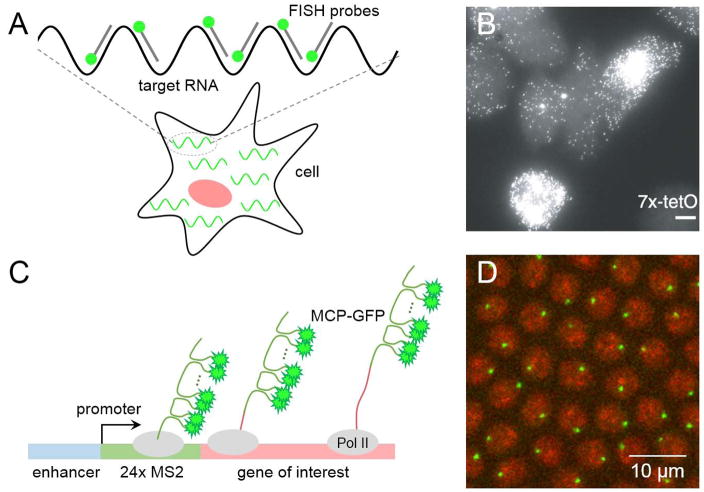
Figure 2. Imaging techniques for visualizing nascent transcription activity.
(A) Schematic of single molecule fluorescent in situ hybridization (smFISH). Several short oligo probes that are complementary to the target RNA are designed to detect individual RNA molecules. Binding of multiple fluorescently labeled probes to the target RNA provides enough sensitivity and allows detection of single nascent RNA transcripts within cells.
(B) A representative image of cells from clonal cell lines where each mRNA is hybridized to smFISH probes (image from [6]).
(C) Schematic of MS2-MCP imaging method. The system uses bacteriophage MS2 and MS2 coat proteins (MCP) that is labeled with fluorescent proteins such as GFP. Typically, 6–24 tandem repeats of MS2 sequences are being inserted to the 5′ or 3′ untranslated region (UTR) of the gene of interest. Upon transcription, MS2 forms a stem loop and dimeric MCP-GFP binds to each stem loop, allowing the detection of de novo transcripts through fluorescent imaging.
(D) A representative snapshot of an early Drosophila embryo, where nascent transcripts are visualized with the MS2-MCP system. Nuclei are shown in red and nascent transcripts are shown in green.
Visualization of transcriptional bursting in multicellular eukaryotes
There is emerging evidence that transcription is discontinuous, consisting of a series of stochastic bursts. McKnight and Miller showed the first evidence for transcriptional bursting by visualizing sequential clusters of irregularly spaced nascent RNAs, using transmission electron microscopy on chromatin spreads from Drosophila embryos [7]. Recent advances in imaging techniques have enabled further support for this concept of transcriptional bursting.
Studies with single molecule fluorescent in situ hybridization (smFISH) have revealed that only a fraction of cells in a population shows active transcription at any given time point (Figure 2A–B). Such heterogeneity in gene expression implies that transcription occurs in bursts rather than continuously. This has been inferred from the behavior of many genes across various cell types and organisms [6,8,9]. In the past three years, transcriptional bursting was exhibited by Nanog and Oct4 in mouse embryonic stem cells [10–12], the β-globin in erythroid cells [13], and the Pck1 in intact mammalian livers [14].
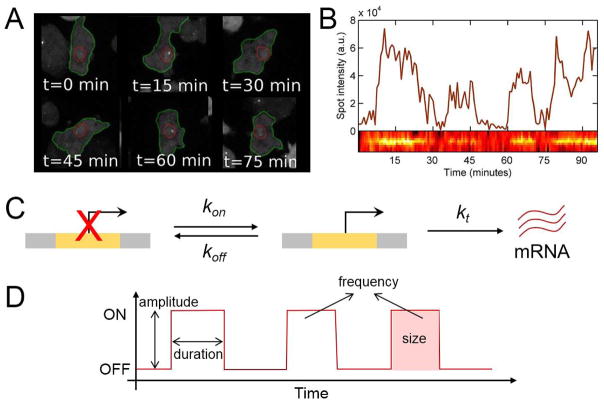
Rather than inferring from a population of fixed tissues, nascent transcripts can now be directly monitored at single-cell resolution using live imaging techniques. Using the widely used MS2-MCP system as a detection method [5], transcriptional bursting was first observed in living bacteria in 2005 [15]. Since then, bursting has been visualized in living multicellular eukaryotes as well, both in transgenic reporter genes and in endogenous genes (Figure 2C–D) [16–22]. In all cases, bursting was visualized as fluctuating fluorescent signals, with the duration of bursts ranging from a few minutes (Drosophila embryos) up to an hour (human U2OS cells) (Figure 3A–B). Such observations support the view that transcriptional bursting is a general property of transcription.
Figure 3. Visualization of transcriptional bursting and characterization of bursting parameters.
(A) Snapshots of living Dictyostelium cells where transcription activity of actin5 is detected as a fluorescent spot. (adapted from [22])
(B) Fluorescent intensity for the cells shown in (A), which reveals transcriptional bursting of actin5. (Image from [22]).
(C) Schematic of the “two-state” model of transcription. A promoter is either in an OFF state or in an ON state with the probability kon and koff,, respectively. Only when the promoter is in an ON state, it can produce mRNA at a rate of kt.
(D) Description of bursting parameters such as bursting amplitude, duration, frequency, and size.
Until recently, transcriptional bursting has been characterized in the context of a “two-state” model, wherein a promoter stochastically switches between an ON state, where transcription occurs at a fixed rate, and an OFF state where no transcription happens (Figure 3C) [23,24]. This model serves as the simplest explanation for the observed heterogeneity of transcriptional activity. However, many live-imaging studies have recently suggested that the promoter at its ON state produces mRNA at changing rates rather than at a single, fixed rate. Using live imaging and modeling, Corrigan et al. showed that endogenous actin5 in Dictyostelium exhibits continuous changes in the rates of active transcription before reaching an inactive state [22]. Varying levels of transcription activation rates were shown in pituitary tissues as well as in early Drosophila embryos [25,26]. Featherstone et al. identified multiple levels of active transcription rates across individual cells in living pituitary tissues, where each cell showed distinct duration of ON states from each other [25]. Holloway et al. used live imaging data of eve2-mediated transcription by Bothma et al. and developed a stochastic model to demonstrate that there exist at least two distinct ON rates of transcription, in agreement with studies in Dictyostelium and in pituitary cells [26,27]. Such high-resolution imaging data combined with mathematical modeling have provided better understanding on the stochastic nature of transcriptional dynamics.
Modulation of transcriptional dynamics
With the availability of single-molecule sensitivity, extensive studies have been performed to understand how different transcription machineries can modulate the property of transcriptional bursting, such as bursting frequency, size, and durations (Figure 3D). Many studies have tried to elucidate the mechanism of the regulation of bursting kinetics by the level of transcription factors. For example, using live imaging, Larson et al. controlled the level of steroids through light activation, and showed that steroids mediate the level of steroid-responsive genes by modulating bursting frequency [20]. Similarly, Senecal et al. showed that concentration of ERK or p38 can be used to regulate bursting frequency of the target gene c-Fos, while not affecting other parameters such as burst size or duration [28]. Most recently, Kafri et al. showed that it is the rate of nuclear β-catenin accumulation that shapes the transcriptional output of downstream target gene cyclin D1, by modulating both bursting frequency and size [29].
Other studies have examined the role of nucleosome occupancy, chromatin states, or mediators on transcriptional regulation [30–33]. Dey et al. used the HIV-1 promoter to show that promoters with more repressed chromatin produce fewer bursts and exhibit more noise, suggesting the effects of nucleosome occupancy on gene expression heterogeneity [30]. It has been shown that the local chromatin environment affect transcriptional dynamics as well, by changing bursting frequency or size. When reporter genes were integrated into random locations in the genome, distinct stochastic gene expressions were observed in different genetic loci [30–32]. In addition, a study with the HIV-1 promoter showed that the time scale of bursting is mediated independently by Mediator and by TBP-TATA box interaction, showing modulation of transcriptional dynamics at the level of the pre-initiation complex [33].
Lastly, two recent studies examined how enhancers control transcriptional bursting [13,34]. By forcing looping between the β-globin enhancer and the β-globin promoter in murine and human erythroid cells, Bartman et al. showed that forced tethering between the enhancer and the promoter increased burst frequency, but not burst size [13]. Using MS2-MCP-based live imaging in early Drosophila embryos, Fukaya et al. showed that different enhancers produce transcriptional bursts with similar size, but with different frequencies, such that strong enhancers produce more bursts than weak enhancers [34]. Their results also support the idea that bursting frequency is a key parameter of gene control in development.
Given all these different modes of modulating transcriptional dynamics, one would wonder why cells would exhibit such stochastic behaviors. A previous study demonstrated that heterogeneity in single-cell organisms could help overall fitness of the population [35]. Recently, it was suggested that transcriptional bursts might sensitize cells for repression. Esposito et al. showed that Snail repressor silences the target gene expression during the refractory period between bursts, implying that the repressor can act more effectively during the inactive state [36]. Similarly, Antolovic et al. showed that genes that will eventually be downregulated show more stochasticity, supporting the idea that discontinuous transcription might be beneficial for repression [37]. This indicates that transcriptional bursting can be used as a mechanism to facilitate dynamic repression of gene expression.
Kinetics of transcription factors and RNA polymerases
In addition to extensive imaging-based studies on transcription dynamics, many researchers have tried to understand the kinetics of proteins that comprise transcriptional machineries, such as transcription factors and RNA polymerases (Figure 1A). Recent advances in microscopy and fluorophores made single-molecule imaging readily available in living tissues. The Betzig lab recently developed the lattice light-sheet microscope, which has allowed quantitative imaging of single molecules with high spatiotemporal resolution [38]. Moreover, traditionally used fluorophores like rhodamine dyes were improved to obtain a few-fold increase in brightness and sensitivity [39,40].
Through these improved imaging techniques, the dynamic nature of protein-DNA interactions has been visualized. Many studies showed how different transcription factors move with different kinetics to find their target DNA. Izeddin et al. demonstrated that the transcription factor c-Myc sweeps an entire nucleus in a diffusive manner, while the elongation factor P-TEFb explores only a compact domain within a nucleus sub-diffusively [41]. Chen et al. and Xie et al. also showed that the transcription factors Sox2, Oct4, STAT3, and ESRRB, all of which bind to the same enhancer, actively search for their target DNA with distinct kinetics [42,43]. Moreover, general transcription factors bind to the promoters in a very dynamic fashion. Zhang et al. showed that general transcription factors TFIIA and TFIID bind to the promoter stably, while TFIIB-promoter interaction is transient [2]. These studies have provided valuable insights into the dynamics of the search process between transcription factors and target DNAs.
Other studies have provided explanations for the mechanism of how DNA-bound transcription factors and RNA polymerases initiate transcription. One possible mechanism is that transcription factors and Pol II form a cluster at active transcription loci. Cisse et al. first reported the formation of Pol II clusters in live human cells by using exogenously labeled Pol II, and it was later found that endogenous Pol II also forms clusters [44,45]. Following up on these findings, Cho et al. showed that the lifetime of Pol II clusters are correlated with nascent mRNA output, suggesting a function of the clustering [46]. Moreover, protein clustering has also been observed for transcription factors. By tracking dynamics of the transcriptional activator Msn2 and the repressor Mig1 in yeast, Wollman et al. suggested that clustering of proteins facilitates the search process for target DNAs [47]. In the Drosophila embryo, the transcription factor Bicoid is reported to form a cluster, which helps increase its local density and facilitates binding to target DNAs [48]. Altogether, single-molecule live imaging has enabled the exploration of the kinetics of transcriptional machineries with a high spatiotemporal resolution, which helped reveal dynamic nature of complex transcription processes.
Future perspectives
One of the biggest advantages of live imaging techniques is that transcriptional activity of a single cell can be directly monitored over time. Utilizing these techniques, it was shown that early developmental enhancers in early Drosophila embryos drive dynamic gene expression patterns, such that transcripts were expressed very broadly across the embryo and refined gradually over time to form a final pattern [27,49]. Other examples of gene expression dynamics include anterior shift of Krüppel gene expression patterns and a gradual activation of T48 along the dorsoventral axis [50,51]. These interesting findings from in vivo studies of multicellular eukaryotes, however, were mostly limited to those using Drosophila. It will be interesting to see how dynamics of transcriptional activity can affect patterning or other key developmental processes in various other multicellular organisms.
Live imaging assays can also be used to understand long-range chromatin dynamics. Through chromosome conformation capture assays, it has been suggested that non-neighboring parts of a chromosome interact to form chromosomal loop domains [52]. However, kinetic information is missing from these models. For example, is loop domain formation static or dynamic? Is gene expression significantly affected if the loop formation is delayed or expedited? Recent studies have already shown that loop domain formation is indeed dynamic [53,54]. Moreover, Hansen et al. characterized the kinetics of CTCF and Cohesin, the two key molecules for the loop formation, providing better understanding on the chromatin loop dynamics [55]. More of such imaging studies in vivo will provide helpful insights into chromatin dynamics and its effects on transcriptional regulation.
Advancement in imaging technique has brought unforeseeable changes to the field of transcriptional regulation, especially through visualization of transcriptional dynamics at a remarkable spatiotemporal resolution that was unfeasible before. This enabled many interdisciplinary works, including molecular biology, imaging, and modeling, which highlighted an important aspect of gene control: the critical role of kinetics in transcriptional regulation. Further advances in imaging techniques and interdisciplinary works will provide more sophisticated and comprehensive insights to the complex world of transcriptional regulation.
Highlights.
Imaging-based studies demonstrate that transcription is a dynamic process
Quantitative imaging methods have revealed transcriptional bursting
Changes in transcription machineries can modulate transcriptional dynamics
Transcription factors search through target DNA with distinct kinetics
Acknowledgments
BL would like to thank Sang Beom Kim, Tyler Heist, Yuji Yamazaki, Takashi Fukaya, Kai Chen, and other Levine Lab members for helpful discussions. This work was supported by the National Institutes of Health [F32GM122186].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hager GL, McNally JG, Misteli T. Transcription Dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••2.Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG, et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 2016;30:2106–2118. doi: 10.1101/gad.285395.116. Using in vitro single-molecule imaging platform, the authors characterized the dynamics of general transcription factors TFIID, TFIIA, and TFIIB. They showed that TFIIB binding to the target promoter is transiet, while TFIID and TFIIA binding is stable. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukaya T, Lim B, Levine M. Rapid Rates of Pol II Elongation in the Drosophila Embryo. Curr Biol. 2017;27:1387–1391. doi: 10.1016/j.cub.2017.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA Particles in Living Yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 6.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:1707–1719. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKnight SL, Miller OL. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell. 1979;17:551–563. doi: 10.1016/0092-8674(79)90263-0. [DOI] [PubMed] [Google Scholar]
- 8.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abranches E, Guedes AMV, Moravec M, Maamar H, Svoboda P, Raj A, Henrique D. Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development. 2014;141:2770–2779. doi: 10.1242/dev.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skinner SO, Xu H, Nagarkar-Jaiswal S, Freire PR, Zwaka TP, Golding I. Single-cell analysis of transcription kinetics across the cell cycle. Elife. 2016:5. doi: 10.7554/eLife.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochiai H, Sugawara T, Sakuma T, Yamamoto T. Stochastic promoter activation affects Nanog expression variability in mouse embryonic stem cells. Sci Rep. 2015;4:7125. doi: 10.1038/srep07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •13.Bartman CR, Hsu SC, Hsiung CCS, Raj A, Blobel GA. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol Cell. 2016;62:237–247. doi: 10.1016/j.molcel.2016.03.007. This is one of the first studies that examined the impact of enhancers on transcriptional bursting. The authors characterized the looping interaction between the β-globin enhancer and the promoter in mice and human erythroid cells, and showed that the enhancer-promoter interaction affects burst fraction but not burst size. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahar Halpern K, Tanami S, Landen S, Chapal M, Szlak L, Hutzler A, Nizhberg A, Itzkovitz S. Bursty gene expression in the intact mammalian liver. Mol Cell. 2015;58:147–156. doi: 10.1016/j.molcel.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional Pulsing of a Developmental Gene. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian Genes Are Transcribed with Widely Different Bursting Kinetics. Science (80-) 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 18.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-Time Observation of Transcription Initiation and Elongation on an Endogenous Yeast Gene. Science (80-) 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muramoto T, Cannon D, Gierlinski M, Corrigan A, Barton GJ, Chubb JR, Cannona D, Gierli’nski M, Corrigan A, Barton GJ, et al. Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc Natl Acad Sci U S A. 2012;109:7350–7355. doi: 10.1073/pnas.1117603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson DR, Fritzsch C, Sun L, Meng X, Lawrence DS, Singer RH. Direct observation of frequency modulated transcription in single cells using light activation. Elife. 2013 doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell PD, Chao JA, Singer RH, Marlow FL. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development. 2015;142:1368–1374. doi: 10.1242/dev.118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Corrigan AM, Tunnacliffe E, Cannon D, Chubb JR. A continuum model of transcriptional bursting. Elife. 2016:5. doi: 10.7554/eLife.13051. The authors visualized transcription activity of endogenous actin5 in Dictyostelium, revealing transcriptional bursting. In combination with mathematical modeling, the authors suggest that each cell exhibits continuously changing rate transcriptional activity, challenging the traditional “two-state” model, where promoters switch between a rigid on and an off state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peccoud J, Ycart B. Markovian Modeling of Gene-Product Synthesis. Theor Popul Biol. 1995;48:222–234. [Google Scholar]
- 24.Kumar N, Singh A, Kulkarni RV. Transcriptional Bursting in Gene Expression: Analytical Results for General Stochastic Models. PLoS Comput Biol. 2015:11. doi: 10.1371/journal.pcbi.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Featherstone K, Hey K, Momiji H, McNamara AV, Patist AL, Woodburn J, Spiller DG, Christian HC, McNeilly AS, Mullins JJ, et al. Spatially coordinated dynamic gene transcription in living pituitary tissue. Elife. 2016:5. doi: 10.7554/eLife.08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holloway DM, Spirov AV. Transcriptional bursting in Drosophila dEvelopment: Stochastic dynamics of Eve stripe 2 expression. PLoS One. 2017:12. doi: 10.1371/journal.pone.0176228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci. 2014;111:10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senecal A, Munsky B, Proux F, Ly N, Braye FE, Zimmer C, Mueller F, Darzacq X. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014;8:75–83. doi: 10.1016/j.celrep.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Kafri P, Hasenson SE, Kanter I, Sheinberger J, Kinor N, Yunger S, Shav-Tal Y. Quantifying β-catenin subcellular dynamics and cyclin D1 mRNA transcription during Wnt signaling in single living cells. Elife. 2016:5. doi: 10.7554/eLife.16748. The authors examined transcriptional dynamics of cyclin D1 in human HEK293 cells, and showed that the kinetics of Wnt signaling, not the level, is correlated with the transcriptional output of cyclin D1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••30.SS D, JE F, PLDVS, APA Orthogonal control of expression mean and variance by epigenetic features at different genomic loci. Mol Syst Biol. 2015 May 5;:806. doi: 10.15252/msb.20145704. By integrating the long terminal repeats (LTR) of HIV-1 into random locations within the human genome, the authors demonstrated that cells exhibit very distinct transcriptional dynamics upon different local chromatin environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viñuelas J, Kaneko G, Coulon A, Vallin E, Morin V, Mejia-Pous C, Kupiec J-J, Beslon G, Gandrillon O. Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts. BMC Biol. 2013;11:15. doi: 10.1186/1741-7007-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoller B, Nicolas D, Molina N, Naef F. Structure of silent transcription intervals and noise characteristics of mammalian genes. Mol Syst Biol. 2015;11:823–823. doi: 10.15252/msb.20156257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tantale K, Mueller F, Kozulic-Pirher A, Lesne A, Victor J-M, Robert M-C, Capozi S, Chouaib R, Bäcker V, Mateos-Langerak J, et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••34.Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. Using MS2-MCP system in early Drosophila embryos, the authors showed that enhancers regulate transcriptional acitivity by modulating bursting frequency, such that a strong enhancer produces more bursts than a weak enhancer. This study also suggests the formation of dynamic chromosomal loop domains to mediate transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Losick R, Desplan C. Stochasticity and Cell Fate. Science (80-) 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposito E, Lim B, Guessous G, Falahati H, Levine M. Mitosis-associated repression in development. Genes Dev. 2016;30:1503–1508. doi: 10.1101/gad.281188.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antolović V, Miermont A, Corrigan AM, Chubb JR. Generation of Single-Cell Transcript Variability by Repression. Curr Biol. 2017;27:1811–1817. e3. doi: 10.1016/j.cub.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science (80-) 2014;346:1257998–1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017 doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izeddin I, Récamier V, Bosanac L, Cissé, Boudarene L, Dugast-Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife. 2014:3. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Xie L, Torigoe SE, Xiao J, Mai DH, Li L, Davis FP, Dong P, Marie-Nelly H, Grimm J, Lavis L, et al. A dynamic interplay of enhancer elements regulates Klf4 expression in naive pluripotency. Genes Dev. 2017;31:1795–1808. doi: 10.1101/gad.303321.117. Using genetic and single-cell imaging approaches, the authors characterized the dynamics of transcription factor assembly at the Kruppel-like factor 4 enhancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X, et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–7. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 45.Cho W-K, Jayanth N, Mullen S, Tan TH, Jung YJ, Cissé II. Super-resolution imaging of fluorescently labeled, endogenous RNA Polymerase II in living cells with CRISPR/Cas9-mediated gene editing. Sci Rep. 2016;6:35949. doi: 10.1038/srep35949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••46.Cho WK, Jayanth N, English BP, Inoue T, Andrews JO, Conway W, Grimm JB, Spille JH, Lavis LD, Lionnet T, et al. RNA Polymerase II cluster dynamics predict mRNA output in living cells. Elife. 2016:5. doi: 10.7554/eLife.13617. The authors used live super-resolution imaging to visualize Pol II clustering in living mouse embryonic fibroblasts. They showed that the dynamics of Pol II clustering is correlated with mRNA synthesis of endogenous β-actin. [DOI] [PMC free article] [PubMed]
- •47.Wollman AJ, Shashkova S, Hedlund EG, Friemann R, Hohmann S, Leake MC. Transcription factor clusters regulate genes in eukaryotic cells. Elife. 2017:6. doi: 10.7554/eLife.27451. The authors characterized the dynamics of transcription factors Mig1 and Msn2 in Saccharomyces cerevisiae, and revealed clustering of transcription factors to mediate gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mir M, Reimer A, Haines JE, Li X-Y, Stadler M, Garcia H, Eisen MB, Darzacq X. Dense Bicoid hubs accentuate binding along the morphogen gradient. Genes Dev. 2017;31:1784–1794. doi: 10.1101/gad.305078.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia HGG, Tikhonov M, Lin A, Gregor T. Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Curr Biol. 2013;23:2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Sherif E, Levine M. Shadow Enhancers Mediate Dynamic Shifts of Gap Gene Expression in the Drosophila Embryo. Curr Biol. 2016;26:1164–1169. doi: 10.1016/j.cub.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim B, Levine M, Yamakazi Y. Transcriptional Pre-patterning of Drosophila Gastrulation. Curr Biol. 2017;27:286–290. doi: 10.1016/j.cub.2016.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science (80-) 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao SSP, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320. e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707. e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •55.Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife. 2017:6. doi: 10.7554/eLife.25776. Using single-molecule imaging, the authors characterized the kinetics of CTCF and cohesin in mouse embryonic stem cells. They showed that CTCF exhibits faster binding kinetics to chromatin than cohesin, revealing dynamic chromatin loop formation. [DOI] [PMC free article] [PubMed] [Google Scholar]