Abstract
Clinical genomics promises unprecedented precision in understanding the genetic basis of disease. Understanding the impact of variation across the genome is required to realize this potential. Currently, clinical genomics analyses focus on protein-coding genes. However, the noncoding genome is substantially larger than the protein-coding counterpart, and contains structural, regulatory, and transcribed information that needs to be incorporated into genome annotations if the full extent of the opportunity to use genomic information in healthcare is to be realized. This article reviews the challenges and opportunities in unlocking the clinical significance of coding and noncoding genomic information and translating its utility in practice.
Medical genetics: The importance of the whole genome
Most of the DNA in the genome does not consist of genes that code for proteins, and understanding the function of these less examined parts of our genetic material is essential to fully understand human development and disease. Brian Gloss and Marcel Dinger at the Garvan Institute of Medical Research in Sydney, Australia, review the challenges and opportunities in unraveling the clinical significance of all parts of our DNA. Many regions of DNA that do not encode protein molecules perform crucial functions in regulating the activity and interactions of the protein-coding genes. Variations in these regions may significantly influence the risks and causes of disease. Studying all parts of the genome will be critical for ensuring that the powerful modern techniques of genetic analysis have maximal impact on healthcare.
The evolution and revolution of clinical genomics
The rise of genomics
Resolving the genetic basis of disease seemed like a certainty once the human genome sequence was completed. This new comprehensive map of the body’s operating system meant that the historically difficult task of mapping diseases with evidence of Mendelian inheritance patterns to a causative locus was now readily accessible to the scientific community.
This map also enabled the development of tools with unprecedented capacity to survey genomes at population scale. Microarray technology interrogation of common single-nucleotide polymorphisms (SNPs) revolutionized our understanding of genetic inheritance patterns through the hapmap project1 (Fig. 1) and genome-wide association studies (GWAS) seemed set to unravel the genetic basis of monogenic and complex disease2.
Fig. 1. The rise of genomics at a glance.
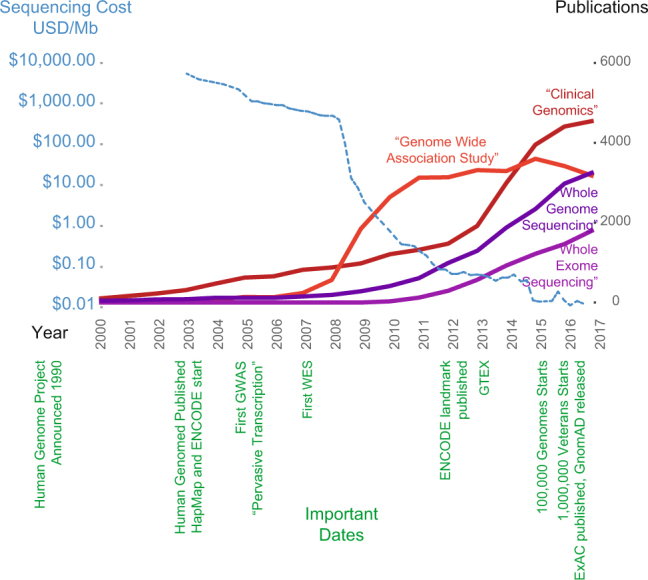
DNA sequencing costs (blue) over time compared to the number of publications containing specific phrases in PubMed. Some key events in genomics are shown in green
The increased power and precision of GWAS facilitated the mapping of monogenic traits but highlighted “missing heritability” where observed inherited traits could not be explained by observed genetic variance. In the clinical genetics field, this was considered likely due to the inability of SNP-chip technology to adequately measure rare variants, structural defects, polygenic and/or complex inheritance patterns. Furthermore, epistatic interactions, where coinheritance of two or more variants could more adequately explain heritability, continues to be difficult to estimate due to computational limitations3,4. At the same time, next-generation sequencing was poised to further revolutionize approaches to not only survey the genome, but also redefine the understanding of how the genome behaved and the diverse mechanisms through which genetic disease could manifest.
The rise of clinical genomics
Exome sequencing, where the coding regions of genes are enriched from DNA and sequenced, has allowed the direct measurement of variance at the genetic level. It has provided clinicians and researchers base-scale resolution of the coding genome, giving rise to a quantum leap in the resolving power of associating genetic variation directly to altered protein. The scalability and relatively low cost has resulted in large databases characterizing and annotating the variation in the coding genome such as ExAC5. But like many technological advances, exome sequencing has exposed its own limitations, namely technical artefacts of the DNA capture used and overdependence on extant genome annotations. This latter limitation proved significant as the emergence of exome sequencing coincided with the observation of the pervasively transcribed genome6 and the rise of lncRNAs as important functional transcripts, which for the most part were overlooked by the approach.
Importantly, the understanding that the majority of informative variants identified by GWAS occurred within the noncoding genome, and a shift in how the genome was known to encode function through transcribed noncoding regulatory RNAs gave rise to many theories regarding the genetic basis of disease, particularly in providing an explanation for the source of missing heritability. Broadly, theories explaining missing heritability fell into two areas—(1) that variants in regulatory DNA sequences such as promoters, enhancers, and structural elements and regions encoding regulatory RNAs were responsible or (2) that large numbers of individual genetic features, potentially with complex interactions, contributed collectively to inherited traits.
Increasingly inexpensive whole-genome sequencing, particularly with PCR-free library preparations, has made it possible to overcome many of the technical artefacts of exome sequencing—thus yielding high-quality surveys of coding gene variants including SNP, copy-number, and structural variations, and insertion/deletion events. These technical advantages in analyzing coding regions alone enabled improved diagnostic yield of genome sequencing and has led to growing numbers of whole-genome clinical sequencing services worldwide.
Whole-genome sequencing consortia producing large databases of genomic variation, such as GnomAD5, 100,000 genomes7, and the Million Veterans Project8 (Fig. 1) are making publicly available their data for interrogation. As well as assisting in the distinction between rare pathogenic variants and those common in the population, this abundance of data provides measurements of the variation in the 98% of the genome that is non-protein-coding. Therefore, observed noncoding variants as well as protein-coding variants of unknown significance are increasing and there is potential now to advance the use of the noncoding genome to improve clinical diagnostic rates of genetic disease9.
The challenge
When the human genome project was completed, the implications of the complexity evident in the noncoding genome were staggering10. After more than a decade of research, considerable advances have been made in understanding how the genome instructs the development and function of organisms and it is increasingly pertinent that this knowledge is harnessed to maximize diagnoses in clinical genomics practice.
In essence, clinical genomics seeks to causatively associate a clinical feature (disease, drug response, risk) with one of the ~5 million variants (relative to a reference genome) present in every individual. This poses the challenge of effectively developing variant filtering algorithms that narrow the search space for variants to regions where pathogenicity can be most clearly determined, i.e., protein-coding regions related to well-described biological function.
Typical approaches for interpreting clinical genomes involve reducing a genome down to rare coding variants with the appropriate inheritance patterns in a gene list of interest. This approach typically yields a handful of variants for consideration. Various annotations of variant impact are then added including predicting the impact on protein structure (SIFT11, Polyphen12, and VEP13) and observation in disease (COSMIC14, ClinVar15, and HGMD16). The proliferation of these tools have led to aggregator services such as VarCards17 that allow multiple scores for a given variant to be interrogated in one place. A clinical molecular geneticist, molecular genetic pathologist, or other certified professional can interpret these data to assign a likely causal variant (Fig. 2). If a candidate is not apparent from these approaches even in cases where there is a strong genetic component, a diagnosis becomes difficult since biochemical testing of variants of unknown significance is not feasible in a typical pathology laboratory setting and may not be considered to be cost-effective. Furthermore, although expanding the search space to include more variants increases the number of candidates, there is typically insufficient evidence to associate any particular variant with the phenotype.
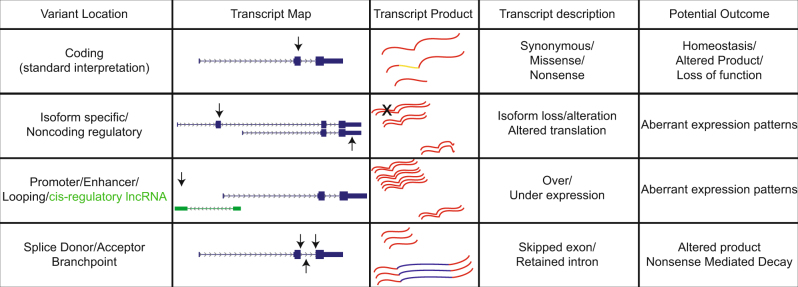
Fig. 2. How coding and noncoding variation can impact gene function.
Variants (arrows) at a hypothetical locus are shown along with potential functional impacts
Efforts worldwide are attempting to expand the annotation of the genome beyond the pure coding and to better understand how variations in these regions can have biological impact to expand the understanding of genetic basis of disease18 and to thus fully realize the clinical utility of the whole genome.
Advances in functional annotation of the genome
Resolving the annotation of gene-level variation
The interpretation of disease-associated variation at the level of the gene is undergoing a shift in understanding. Protein-coding mutations have historically be considered deletarious where they lead to truncations (nonsense/deletions), amino acid alterations (missense/in-frame in/del), frame shifts (in/del) and splicing defects (splice-site donors/acceptors). However, these kinds of mutations have been shown to be relatively common, even in healthy genomes19. Furthermore, these variants can be difficult to interpret in a clinical setting if the mutation occurs in a region not previously reported, or in a gene whose function within the context of the disease in question has not been investigated20. It is also becoming apparent that mutations that do not affect the encoded amino acid (synonymous) can affect gene products in the context of codon frequency and RNA structure21,22. Furthermore, the concept of multiplicity, where gene expression can be impacted by combinations of genetic alterations23 is only starting to be addressed. This implies that the even annotation of coding variants is far from complete.
It is also important to note that the coding proportion of a gene comprises a small percentage of the genetic information encoded by the locus and that alterations in the noncoding sequence can have impact on gene function (Fig. 2) Variation at gene promoters can impact the expression of the gene24, e.g., the TERT promoter is frequently mutated, which leads to overexpression, and in turn, can be a pathogenic basis for causing or driving cancer development25. Variation at imprinted loci can drive the deposition of epigenetic marks responsible for imprinting26, which can lead to aberrant expression. Alterations in 5′ and 3′ untranslated regions of genes can impact transcript stability and translation primarily through RNA structural alterations27,28. Introns can similarly contain important genetic information that can be influenced by mutation29, e.g., disease-associated SNPs within branch points can be associated with altered splicing patterns30. Together, these investigations show that a significant proportion of the clinically relevant genetic information elucidated by whole-genome sequencing is not typically interpreted in diagnostic laboratories.
Resolving the transcriptional and regulatory landscape of the genome
Ever since the first observation of the pervasively transcribed genome more than a decade ago, there has been an explosion in the identification and functional characterization of long noncoding RNA (lncRNA)31 and other noncoding transcript types32. The encyclopedia of DNA elements consortium (ENCODE) raised considerable controversy in 2012 by using tissue-specific transcript profiling, supported by epigenetic profiling of the genome, to suggest that 82% of the human genome was functionally important33. As the vast majority of transcribed species of the genome are noncoding, of which little is still known31, efforts are ongoing to describe the detail and regulation of noncoding RNA. LncRNAs are of particular interest to the field of clinical genomics as their exquisite tissue-specific expression and regulatory behavior34 indicate that a role in disease will become apparent as more is understood about lncRNA biology.
As a result, several large-scale efforts have been undertaken to comprehensively annotate the noncoding transcriptional landscape, particularly through the FANTOM projects6,35,36, ENCODE33 and Roadmap Epigenomics37. The large-scale GTEx project38 has set out to further understand the genetic drivers of tissue-specific gene expression via expression quantitative trait loci (eQTL) analysis. Large-scale screens for noncoding RNA function have elucidated functional annotations for thousands of lncRNAs39 and molecular tools tailored to the unique biology of lncRNA behavior are ongoing40. These efforts have enhanced the understanding of gene transcription and hint at a complexity that requires expanded resolution of functional annotation at the genetic level to inform interpretation in a clinical diagnostic setting.
Interpreting functionality at the whole-genome level
Traditional indicators of functionality (and thus of potential clinical utility), such as conservation, have thus been challenged by this expanding annotation of the genome. The volume of available data has fueled recent computational efforts to annotate functional parts of the genome without necessarily depending exclusively on the coding genome (Table 1). Early attempts used existing annotations to train computational models that could assess the potential function of a variant genome-wide (CADD41/GWAVA42). Newer approaches have used genome-wide data itself to assign functional importance, either through association with DNA binding proteins (Eigen43), or direct measures of resistance to variation (Orion44), to provide comprehensive maps of coding and noncoding regions likely to be impacted by variation. These maps are expanding the pool of potentially clinically relevant variants and continue to evolve with growing interest and innovation.
Table 1.
Genome-wide tools for estimating impact of noncoding variation
| Tool | Year | Method used to build model |
|---|---|---|
| CADD41 | 2014 | Support vector machine |
| GWAVA42 | 2014 | Random forest algorithm |
| DeepSEA73 | 2015 | Deep learning |
| FATHMM-MKL74 | 2015 | Multiple Kernel learning + SVM |
| Eigen43 | 2016 | Unsupervised partitioning |
| Basset75 | 2016 | Deep convolutional neural network |
| LINSIGHT76 | 2017 | Generalized linear model (INSIGHT and fitCons) |
| Orion44 | 2017 | Observed/expected variation |
Noncoding variation and disease
Structural alterations
The physical arrangement of the genome is also critical to homeostasis. Copy-number alterations are associated with many diseases, but can also have no pathogenic effect45,46. The study of disease-associated genomic translocations has typically focused on the generation of gene fusions, which are particularly clinically relevant in cancer47. However, studies of intergenic translocations can also perturb local gene expression, possibly by interrupting chromatin looping and by rearranging regulatory sequence48–50. Moreover, chromatin looping51 and nucleosome occupancy52 are also susceptible to alteration by DNA mutation and structural rearrangement.
Localized DNA structures have been associated with genetic disease such as Huntington’s disease mostly as recognition sites for genomic rearrangements53. However, such quaternary structures recently gained traction as important mediators of biological information in themselves with left handed helices (z-DNA54), G-quadruplexes55,56, and DNA:DNA/DNA:RNA triplexes57,58 showing evidence of regulatory function. Indeed the interplay between the physical state of the DNA appears to be intimately associated with the process of gene expression59 and transcription factor binding60. Importantly, it was recently shown that disease-associated variations that disrupt G-quadruplex formation in RNA can affect post-transcriptional regulation of genes27, suggesting that variants in structural features can directly impact cellular function.
Noncoding transcription at GWAS loci
The prevalence of intergenic, disease-associated SNPs from GWAS studies provoked diverse studies into how these variants were contributing to disease, revealing impacts on DNA conformation51, DNA-protein interactions61, and epigenetic marks62. Recent application of RNA-capture sequencing63 to haplotype blocks associated with GWAS disease-associated SNPs revealed a multitude of transcripts of which less than half were in extant transcript databases64. Combined with fine mapping of SNPs associated with breast cancer, this approach revealed enhancer alterations affecting novel transcript expression65. These studies raise the possibility of direct and indirect impacts of disease-associated SNPs on tissue-specific transcription patterns and illustrate that both the resolution of disease-associated variants and genome annotation remain incomplete. The ongoing accumulation of whole-genome data worldwide will eventually resolve the exact disease associations and a greater understanding of the noncoding transcriptome will continue to provide context for elucidating the impact of these variants34.
New classes of functional repeats in the human genome
In a similar vein, pseudogenes have classically been regarded as nonfunctional byproducts of retrotransposition66. With the observation of transcription and evidence of disease linkage67, pseudogene biology is being revisited, however, consensus as to a generic biological role has not yet been reached68,69. Indeed, the process of retrotransposition itself in shaping the genome is undergoing a renaissance through evidence of gene regulatory roles70.
A place for noncoding annotations in clinical genomics
Rules of evidence
In 2015, the American College for Medical Genetics (ACMG) described a set of evidence lines that could be used to ascribe degrees of pathogenicity to a particular variant71. Importantly, these recommendations sought to distinguish deleterious impacts on a gene from contribution to disease. Predicting the impact of coding variation is a more mature process, especially in the case of missense and nonsense mutations. Tools like PolyPhen and VEP are commonly used to estimate genic pathogenicity, although the likely impact of the variant can be open to interpretation. Evidence for disease contribution is usually achieved by cross-referencing rare variants with lists of genes with known roles in the disease of interest, reports in the literature, and clinical databases such as COSMIC and ClinVar. The point at which there is sufficient evidence of a variant causing a disease is becoming refined20. However, due to the complexities in the WGS data, interpretation, and phenotyping, associations can be subject to how the data are evaluated by genetic professionals and can still require in vitro testing. Including non-protein-coding into this framework would require extra complexity predominantly due to the lack of functional data to support impact of a particular variant with precision, given the ongoing genome annotations outlined above (Fig. 3). However, noncoding variants can clearly be clinically relevant and their inclusion into clinical genomics frameworks is necessary for realizing the full clinical utility of genomic information.
Fig. 3. The challenge of assigning variant impact in a complex genome.
Assigning variants (red arrows) at a hypothetical locus where protein-coding transcripts (blue), lncRNA (green), and regulatory regions (magenta) are incorporated
A framework for noncoding inclusion in clinical genomics
The clinical interpretation of variants typically begins strictly as an informatics exercise where variants are filtered and ranked according to likelihood of clinical trait association. One of the earliest steps is to omit variants that are noncoding, which in the light of the evidence outlined above may miss vital insights into the molecular basis of a disease. To address this limitation, existing frameworks that estimate noncoding impact such as the GTEX eQTLs and tools outlined in Table 1 should be integrated into existing variant interpretation frameworks such as GEMINI72. While less data is available for accurately calculating variant frequency in noncoding regions, growing whole-genome reference databases are now available for this purpose. These annotations can then be interpreted alongside existing lines of evidence within the context of disease.
The primary paradigm shift required by these additions to clinical genome interpretation workflows will be the expansion of the concept of what part of the genome constitutes a gene. Impacts on a specific gene function can theoretically occur anywhere within the genome. This represents a currently insurmountable computational obstacle for the same reason that epistasis remains an intractable issue in genomics. However, splicing and promoter variations are directly linked to genes and are currently well annotated. For this reason, we propose that variants occurring at splice sites and branch points as well as promoters annotated by ENCODE should be included in clinical genomics where they occur in disease relevant genes. We expect that a more inclusive approach to impacts on gene function will facilitate an improved picture of the clinical landscape, particularly in the case of disease with strong evidence of inheritance where no coding candidate can be found. For example, a promoter variant may be the second-hit in a recessive heterozygous locus leading to total loss of a gene product. Furthermore, as our knowledge of the biology of the genome grow, more interpretative power will become available in the context of clinical genomics. We contend that the potential to improve diagnostic rates using a multi-level whole-genome annotation approach will outweigh the necessary increased time for manual variant review and ruling out of false positives.
The future
Understanding the genetic basis of disease has been an aim of science since heritable traits were first observed. Technological and conceptual progress have given rise to a picture of the genome that is as complex as one would expect from a four letter code that gives rise to living multicellular organisms. Research is currently at the point of attempting to describe and unravel this complexity as discussed above. We expect that tools and knowledge of the noncoding genome will continue to expand and that continued refinement of an integrated coding and noncoding genomic landscape through comprehensive genomic, transcriptomic, and epigenomic profiling will improve the prediction of variant outcomes. The computational issues of epistasis and polygenetic impacts will be improved as more data is generated and more powerful computational frameworks emerge, such as quantum computing to enable large combinatorial calculations that are currently unfeasible. These will go hand in hand with more widespread adoption of moderate throughput screens for rapid and direct measurements of the impact of candidate variants such as CRISPR-Cas9 tools in patient-derived iPS cell lines. It will be important for clinical scientists involved in variant interpretation to remain mindful of the growing clinical significance of the whole genome and for developers of software and knowledgebases used to inform variant interpretation to consider non-protein-coding data sources and algorithms that act on noncoding genomic regions in their workflows.
Acknowledgements
We thank James Torpy for manuscript feedback, and Dr. Mark Pinese and Dr. Eric Lee for constructive criticism.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 3.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 7.The 100,000 Genomes Project Protocolv4, Genomics England 10.6084/m9.figshare.4530893.v4, https://www.genomicsengland.co.uk/100000-genomes-project-protocol/ (2017).
- 8.Gaziano JM, et al. Million veteran program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little PF. Structure and function of the human genome. Genome Res. 2005;15:1759–1766. doi: 10.1101/gr.4560905. [DOI] [PubMed] [Google Scholar]
- 11.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaren W, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes SA, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson PD, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46:D1039–D1048. doi: 10.1093/nar/gkx1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spielmann M, Mundlos S. Looking beyond the genes: the role of non-coding variants in human disease. Hum. Mol. Genet. 2016;25:R157–R165. doi: 10.1093/hmg/ddw205. [DOI] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature467, 1061–1073 (2010).. [DOI] [PMC free article] [PubMed]
- 20.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 22.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RB, Chan EK, Cowley MJ, Little PF. The influence of genetic variation on gene expression. Genome Res. 2007;17:1707–1716. doi: 10.1101/gr.6981507. [DOI] [PubMed] [Google Scholar]
- 24.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc. Natl Acad. Sci. USA. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 26.Chuang TJ, Tseng YH, Chen CY, Wang YD. Assessment of imprinting- and genetic variation-dependent monoallelic expression using reciprocal allele descendants between human family trios. Sci. Rep. 2017;7:7038. doi: 10.1038/s41598-017-07514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeraati M, et al. Cancer-associated noncoding mutations affect RNA G-quadruplex-mediated regulation of gene expression. Sci. Rep. 2017;7:708. doi: 10.1038/s41598-017-00739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesole G, et al. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/S0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 29.Vaz-Drago R, Custodio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum. Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 30.Signal B, Gloss BS, Dinger ME, Mercer TR. Machine learning annotation of human branchpoints. Bioinformatics. 2018;34:920–927. doi: 10.1093/bioinformatics/btx688. [DOI] [PubMed] [Google Scholar]
- 31.Quek XC, et al. lncRNAdbv2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris KV, Mattick JS. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012).. [DOI] [PMC free article] [PubMed]
- 34.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim. Biophys. Acta. 2016;1859:16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 36.Kawai J, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet.45, 580–585 (2013).. [DOI] [PMC free article] [PubMed]
- 39.Liu SJ, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:35–39. doi: 10.1126/science.aah4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat. Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gussow AB, et al. Orion: detecting regions of the human non-coding genome that are intolerant to variation using population genetics. PLoS ONE. 2017;12:e0181604. doi: 10.1371/journal.pone.0181604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C, Scherer SW. The clinical context of copy number variation in the human genome. Expert Rev. Mol. Med. 2010;12:e8. doi: 10.1017/S1462399410001390. [DOI] [PubMed] [Google Scholar]
- 46.Shaikh TH, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 48.Chiang C, et al. The impact of structural variation on human gene expression. Nat. Genet. 2017;49:692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lupianez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David D, et al. Identification of OAF and PVRL1 as candidate genes for an ocular anomaly characterized by keratolenticular dysgenesis and ectopia lentis. Exp. Eye Res. 2018;168:161–170. doi: 10.1016/j.exer.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Schierding W, Cutfield WS, O’Sullivan JM. The missing story behind genome wide association studies: single nucleotide polymorphisms in gene deserts have a story to tell. Front. Genet. 2014;5:39. doi: 10.3389/fgene.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 55.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bacolla A, Wang G, Vasquez KM. New perspectives on DNA and RNA triplexes as effectors of biological activity. PLoS Genet. 2015;11:e1005696. doi: 10.1371/journal.pgen.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levens D, Baranello L, Kouzine F. Controlling gene expression by DNA mechanics: emerging insights and challenges. Biophys. Rev. 2016;8:23–32. doi: 10.1007/s12551-016-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou T, et al. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl Acad. Sci. USA. 2015;112:4654–4659. doi: 10.1073/pnas.1422023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maurano MT, et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015;47:1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pai AA, Pritchard JK, Gilad Y. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 2015;11:e1004857. doi: 10.1371/journal.pgen.1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartonicek N, et al. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 2017;18:241. doi: 10.1186/s13059-017-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Betts JA, et al. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am. J. Hum. Genet. 2017;101:255–266. doi: 10.1016/j.ajhg.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 67.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl Acad. Sci. USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grander D, Johnsson P. Pseudogene-expressed RNAs: emerging roles in gene regulation and disease. Curr. Top. Microbiol. Immunol. 2016;394:111–126. doi: 10.1007/82_2015_442. [DOI] [PubMed] [Google Scholar]
- 69.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 70.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shihab HA, et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang YF, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017;49:618–624. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]