Abstract
Several studies have examined associations between peripheral DNA methylation patterns of the serotonin transporter gene (SLC6A4) promoter and symptoms of depression and anxiety. The SLC6A4 promoter methylation has also been associated with frontal-limbic brain responses to negative stimuli. However, it is unclear how much of this association is confounded by DNA sequence variations. We utilized a monozygotic-twin within-pair discordance design, to test whether DNA methylation at specific CpG sites in the SLC6A4 promoter of peripheral cells is associated with greater frontal-limbic brain responses to negative stimuli (sadness and fear), independently of DNA sequence effects. In total 48 pairs of healthy 15-year-old monozygotic twins from the Quebec Newborn Twin Study, followed regularly since birth, underwent functional magnetic resonance imaging while conducting an emotion-processing task. The SLC6A4 promoter methylation level was assessed in saliva samples using pyrosequencing. Relative to the co-twins with lower SLC6A4 promoter methylation levels, twins with higher peripheral SLC6A4 methylation levels showed greater orbitofrontal cortical (OFC) activity and left amygdala-anterior cingulate cortex (ACC) and left amygdala-right OFC connectivity in response to sadness as well as greater ACC-left amygdala and ACC-left insula connectivity in response to fearful stimuli. By utilising a monozygotic-twin design, we provided evidence that associations between peripheral SLC6A4 promoter methylation and frontal-limbic brain responses to negative stimuli are, in part, independent of DNA sequence variations. Although causality cannot be determined here, SLC6A4 promoter methylation may be one of the mechanisms underlying how environmental factors influence the serotonin system, potentially affecting emotional processing through frontal-limbic areas.
Introduction
Animal and human studies suggest that environmental exposures can affect our epigenome and these epigenetic marks can persist through cell divisions1. Epigenetic modifications can change gene expression without altering the genetic sequence. These epigenetic modifications play an important role in setting up tissue- and cell type-specific gene expression programs during development and are crucial in maintaining normal cellular physiology2. DNA methylation—as one of these possible epigenetic mechanisms—implies a covalent modification of the DNA molecule itself through enzymatic addition of a methyl group to (mostly) cytosine bases2. DNA methylation, involved in epigenetic regulation of gene expression, at critical regulatory regions such as promoters and enhancers, can lead to permanent silencing of the gene, either directly by blocking transcriptional factors from binding to the DNA sequence or by attracting proteins to form corepressor complexes3,4.
DNA methylation patterns are shaped during development and cellular differentiation but are also responsive to environmental signals, particularly during gestation5. Indeed, even in newborns, variations in DNA methylation levels can be detected across twins, suggesting that environmental factors might be influencing the normal developmental trajectories of DNA methylation pattern6. DNA methylation alterations in response to early environmental exposure also appear to be stable, at least in some gene regions, for many years. For example, a study in humans has shown that individuals who were exposed to the Dutch famine in the perinatal period had, six decades later, altered DNA methylation in particular sites compared to their non-exposed siblings7.
One of the most widely studied genes in relation to DNA methylation and mental health is the serotonin transporter gene, SLC6A4. The serotonin transporter is of interest as it has been shown to play an important role in mood, cognition and psychophysiology, as well as brain development8. Following results of association studies showing a link between SLC6A4 methylation assessed from peripheral cells and early life environment and later behavior in human clinical and nonclinical populations9,10, a number of studies have now investigated possible associations between peripheral SLC6A4 methylation and brain processes. For instance, Wang et al.11 found that SLC6A4 promoter methylation in T lymphocytes of adults was associated with lower in vivo Positron Emission Tomography (PET) measures of brain serotonin (5-hydroxytryptamine; 5-HT) synthesis in the orbitofrontal cortex and higher childhood aggression. Furthermore, a recent study found associations between SLC6A4 methylation and in vivo PET measures of serotonin transporter availability12. We also previously reported that SCL6A4 methylation obtained from white blood cells was significantly associated with activation in response to negative emotional stimuli in the insula13 and lower hippocampal volume14 in depressed patients and controls. Using functional magnetic resonance imaging (fMRI), greater peripheral SLC6A4 methylation from saliva and whole-blood DNA samples has also been associated with an increased amygdala response to threat-related stimuli in adolescents and young adults15,16. In one (f)MRI study, we reported that greater peripheral SLC6A4 methylation from whole-blood, saliva, and buccal DNA samples have been associated with greater (superior) prefrontal cortical GM volume and parietal-frontal regional functional connectivity at rest in healthy adults17. Together, these findings support the relevance of peripheral SLC6A4 methylation measures for frontal-limbic brain processes and support the notion that SLC6A4 methylation level changes may be an underlying mechanism of how environmental factors can influence aspects of the 5-HT system and, consequently, emotion processing.
However, these associations could be confounded by the effects of variations in DNA sequence. Indeed, certain genotypic variants have been shown to have different DNA methylation patterns than others in the context of similar exposures and experiences18. For instance, lower methylation was found in association with unresolved trauma or loss in individuals with the s/s genotype of the SLC6A4 promoter polymorphism, while an inverse correlation was found in the l/l genotype group18. Furthermore, the influence of DNA sequence on methylation status may depend on genomic location, cell type, and developmental stage19.
Monozygotic (MZ) twin study designs are unique in their ability to control for DNA sequence differences because each pair of MZ twins shares essentially the same genetic sequence. Thus, differences within twin pairs in gene expression and phenotype, including brain processes, can be attributed to environmental effects rather than DNA sequence influences.
In the present study, we utilized a MZ design to test, in a sample of healthy adolescents, whether DNA methylation at the SLC6A4 promoter in peripheral cells is associated with greater frontal-limbic activation and connectivity in response to negative stimuli, irrespective of DNA sequence differences. Specifically, we focused on sad and fearful stimuli because neural responses to these negative emotional facial expressions have been relatively consistently associated with SLC6A4 genetic and epigenetic variation13,15,20.
Subjects and methods
Participants were 96 monozygotic twins (48 pairs: 21 male and 27 female pairs; age 15 years) from the Quebec Newborn Twin Study (QNTS)21,22, followed longitudinally since birth, in the area of Montreal, Canada, between April 1995 and December 1998. The number of individuals (n = 96, 48 pairs) was larger than the sample size of our previous studies in non-twins and that focused on SLC6A4 methylation and its link with brain processes (e.g., 13, 17). They were all physically healthy, free of any medication liable to affect brain function and free of current and past psychopathology. Mental health status was confirmed by the Schedule for Affective Disorders and Schizophrenia for School-Age Children- Present and Lifetime Version (K-SADS)23. Furthermore, we used the Dominic Interactive (Adolescent version), a computerized questionnaire to assess subclinical levels of internalizing symptoms (phobias, generalized anxiety and depression)24. The appropriate ethics committees (CHU Sainte-Justine and Montreal Neurological Institute) approved the research protocol. Written assent was obtained from all participants and written consent from parents of all participants.
DNA methylation protocol
Whole saliva was collected using the Oragene TM DNA self-collection kit following manufacturer instructions (DNA Genotek Inc., 2004, 2006). Participants did not eat, chew gum, or drink anything but water for 30 min prior to saliva collection. Once extracted, whole-saliva DNA was converted with EZ DNA Methylation-Gold Kit (D5006, Zymo Research, Irvine, CA, USA). Following recommendation by Tost and Gut25, 20 ng of bisulfite-treated DNA was used in the PCR amplification to reach high reproducibility of pyrosequencing, using previously published primer sets optimized for bisulfite-treated DNA template (11, Fw: 5′TTGTTAGGTTTTAGGAAGAAAGAGAGA-3′, Rev: 5′Biosg-AAAAAAACTACACAAAAAAACAAATATAC-3′) with EpiMark Hot Start Taq DNA Polymerase (New England Biolabs, Ipswich, MA, USA). Ten CpG sites (numbered as 5–14) in the 28563060–28563221 region of chromosome 17 (according to hg19) were assayed.
Site-specific methylation analyses were performed by pyrosequencing using PyroMark Q96 (Qiagen, Venlo, Limburg, Netherlands) using the CFI Imaging and Molecular Biology Platform at McGill University in the Department of Pharmacology and Therapeutics using 2 sequencing primers: 5′AAGAGAGAGTAGTTT-3′ and 5′GTAGATTTTTGTGTG-3′. The DNA methylation level at each CpG site was computed after quality control of the triplicates. If a sample did not pass quality control at a certain CpG site, the mean DNA methylation value for that CpG site was calculated from the remaining two replicates. We then computed the average methylation level from our ten CpG sites of interest (CpG sites 5–14). In line with other studies that linked brain processes with SLC6A4 methylation13,16,17,26, average methylation levels across the investigated CpG sites values were calculated and used in the analyses. The average value of promoter methylation level was used in the analyses rather than the DNA methylation level of each individual CpG site to limit the number of statistical comparisons. The choice to calculate the mean of the ten studied CpG sites was further based on our previous work in healthy and clinical samples, showing associations between average SLC6A4 methylation levels across these 10 sites and in vivo measures of brain 5-HT synthesis in the frontal-limbic brain circuits as well as frontal-limbic functional and structural processes11,13,17.
Neuroimaging
Participants were scanned on a 3 T Siemens TIM Trio Scanner located at the Montreal Neurological Institute (MNI) with a 32-channel head coil. The scan included a magnetization-prepared rapid acquisition gradient-echo (MPRAGE) 9 min sequence (176 slices, 1 mm thickness, TR = 2300 ms, TE = 2.98 ms, TI = 900 ms, FA = 9°, FOV = 256 × 256 mm, matrix size 256 × 256) to assess brain anatomy. Next, a 15-min functional scan during an event-related emotion-processing task, adapted from the task used in Canli and colleagues27, was performed. Briefly, the task consisted of a series of 120 Ekman faces with different emotions (happy, sad, angry, fearful, and neutral) from the Pictures of Facial Affect series28. Faces were presented randomly for 2 s and followed by a fixation cross (~2 s; jittered) and a question asking whether the face belonged to a man or a woman. Two hundred and 26 functional whole-brain images (multi-slice gradient echo EPI with 3.5 mm isotropic resolution and TR/TE = 2110/30 ms, 40 slices, FOV = 224 × 224 mm, matrix size 64 × 64, interleaved descending slice acquisition) were acquired.
Image processing
Task-based fMRI
Pre-processing steps (slice timing and motion correction, coregistration with the anatomical scan, normalization into an EPI stereotactic Space (MNI template) and spatial smoothing at 7 mm FWHM with a Gaussian Kernel were performed in SPM8 (Wellcome Department of Cognitive Neurology, London UK). For each emotion, activation during neutral stimuli was subtracted from the activation during the different emotions. Voxel-wise sad minus neutral and fear minus neutral contrasts were generated at the first level. We calculated within-pair discordance by subtracting the activation contrast of the twin with higher SLC6A4 methylation levels from the activation contrast of the co-twin with lower SLC6A4 methylation levels29. Discordance values were then used in the regression analyses, by regressing discordance in brain activation in response to sadness>neutral and fear>neutral independently onto discordance in methylation. Whole-brain analysis was conducted. The threshold p value was set at p < 0.001. Only clusters showing a spatial extent of at least 10 contiguous voxels were considered (except for the amygdala, in which we required at least 5 contiguous voxels). We corrected for multiple comparisons at the cluster level using the family-wise error rate (FWE)30.
Functional connectivity
Following the pre-processing steps, functional connectivity analyses were performed using the CONN Functional Connectivity toolbox v174 (http://www.nitrc.org/projects/conn). We controlled for the possible confounding effects of head motion artifacts using the ComCor strategy and default preprocessing parameters31,32. In addition, we applied the following denoising options in CONN: detrending, which removes linear/quadratic/cubic trends within each functional session, and despiking, which applies a squashing function to reduce the influence of potential outlier scans32.
We conducted ROI-to-ROI analyses to assess functional connectivity during sad and fearful conditions. The following ROIs were chosen, namely bilateral amygdala, bilateral orbitofrontal cortex (OFC), anterior cingulate cortex (ACC) and bilateral insula. Amygdala was chosen in light of the link between peripheral SLC6A4 methylation and amygdala responses to fearful stimuli in (non-twin) adolescents16. OFC was of particular interest in light of the results of our study in healthy (non-twin) individuals showed an association between lower brain serotonin synthesis in the OFC and greater peripheral SLC6A4 methylation11. ACC was chosen given the findings of activation in this region during sadness in (non-twin) individuals with depressive symptoms33,34. Insula was chosen based on our previous study showing an association between insula response to negative stimuli and peripheral SLC6A4 methylation in (non-twin) healthy and depressed adults13. Mean BOLD time series were extracted from each ROI and correlated with the BOLD time-series signal of every other ROI in the network to create a ROI-to-ROI connectivity matrix (connectome) showing connectivity between each region within the network.
In order to assess discordance between ROI-to-ROI connectivity values based on peripheral SLC6A4 promoter methylation discordance, ROI-to-ROI connectivity for twins with greater SLC6A4 methylation level was compared to their co-twins with lower methylation level. Given the sample size and the correlational nature of functional connectivity and to balance the risk for type I and type II errors, we set the statistical threshold for each voxel at p < 0.05, uncorrected.
Results
DNA methylation
The mean SLC6A4 promoter methylation level was 3.74% (SD = 2.37, range: 1.23–14.37%). This level is comparable to that found in other peripheral SLC6A4 promoter methylation studies, derived from various biological tissues including saliva, using the same CpG sites11,13,17. Within pair, the average discordance in SLC6A4 promoter methylation varied from 0.23 to 8.58%. There were no sex differences in mean SLC6A4 methylation levels (F(1,94) = 0.01, p = 0.98), nor in SLC6A4 methylation discordance levels (F(1,46) = 2.53, p = 0.12). Mean level of internalizing symptoms on the Dominic scale was 10.30 (SD = 6.16, range 1–27 out of maximum total score of 52), indicative of absent/low likelihood of internalizing symptomatology. Within pair, the average discordance in symptoms was 4.37 points (SD = 3.39, range 0–15). SLC6A4 promoter methylation did not correlate with internalizing symptoms (as assessed by the Dominic Interactive questionnaire) neither globally (r = 0.12, p = 0.25) nor within pair (r = 0.12, p = 0.42).
fMRI results
Sad condition
Relative to their co-twin with lower methylation, twins with higher SLC6A4 promoter methylation levels had greater left OFC activation (k = 206, t = 5.11, cluster pFWE = 0.03) (Fig. 1), as well as greater functional connectivity between left amygdala and ACC and between left amygdala and right OFC (Fig. 2 and Table 1). No other significant fMRI results were found in the sad condition.
Fig. 1. Greater within-pair difference in peripheral SLC6A4 promoter methylation was associated with greater responses to sad stimuli in left orbitofrontal cortex (T = 5.11, pFWE = 0.032).
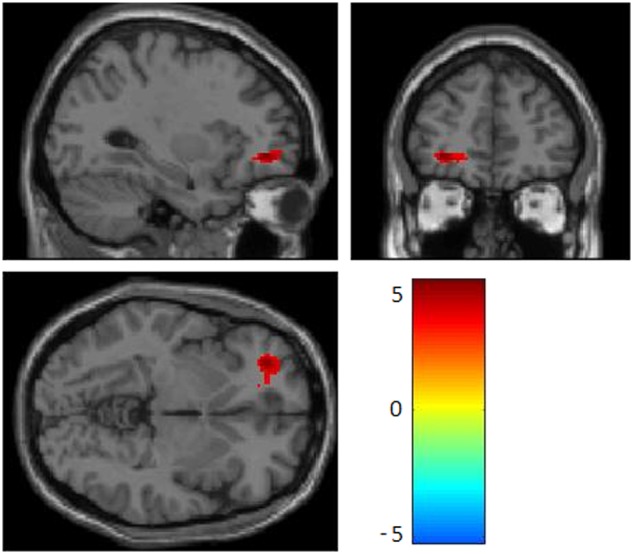
The color bar represents T values
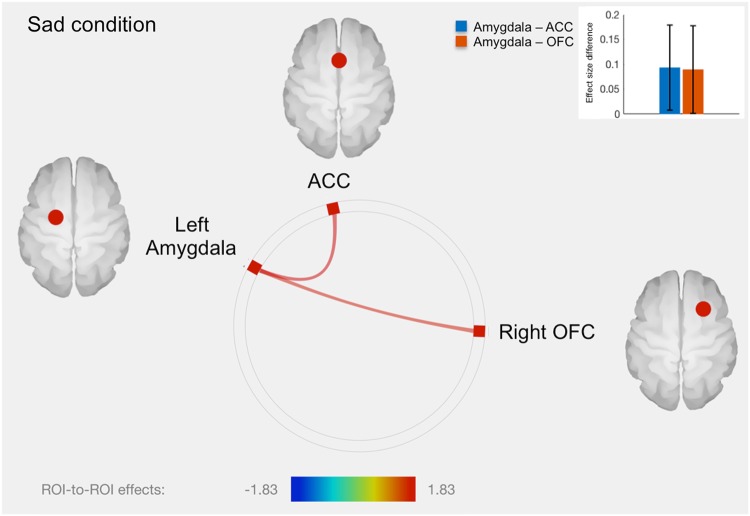
Fig. 2. Connectome ring representation of ROI-to-ROI connectivity between amygdala, anterior cingulate cortex (ACC), orbitofrontal cortex (OFC) and insula in the sad condition.
Only significant connectivity discordances are represented: greater ROI-to-ROI connectivity in the sad condition for twins with greater methylation level compared to their co-twins with lower methylation level, between left amygdala and ACC (T = 1.83, p = 0.036), as well as between left amygdala and right OFC (T = 1.71, p = 0.047). The color bar represents T values. The bars plot in the top right-hand corner represents differences in effect sizes (difference in Fisher-transformed correlation coefficients) between twins with greater methylation level and their co-twins with lower methylation level, for each connection
Table 1.
Significant within-pair fMRI associations with SLC6A4 promoter methylation
| Direction of effect: Condition | Region | t | p-value |
|---|---|---|---|
| Positive: Sad | left amygdala-ACC | 1.83 | 0.036 |
| left amygdala-right OFC | 1.71 | 0.047 | |
| Positive: Fearful | ACC-left amygdala | 2.79 | 0.004 |
| ACC-left insula | 1.85 | 0.035 |
fMRI functional Magnetic Resonance Imaging, SLC6A4 serotonin transporter gene, OFC orbitofrontal cortex, ACC anterior cingulate cortex
Fearful condition
There was no significant association with neural activity in the fearful condition. However, twins with higher SLC6A4 promoter methylation level had greater functional connectivity between ACC and left amygdala and between ACC and left insula, relative to the co-twin with lower SLC6A4 methylation (Fig. 3 and Table 1). No other significant fMRI results were found in the fearful condition.
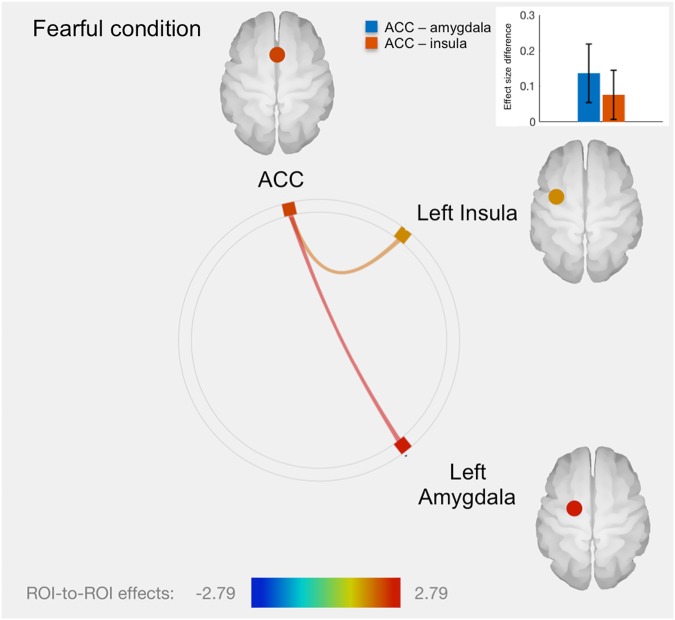
Fig. 3. Connectome ring representation of ROI-to-ROI connectivity between anterior cingulate cortex (ACC), left amygdala and insula in the fearful condition.
Only significant connectivity discordances are represented: greater ROI-to-ROI connectivity in the fearful condition for twins with greater methylation level: between ACC and left amygdala (T = 2.79, p = 0.004), as well as between ACC and left insula (T = 1.85, p = 0.035). The color bar represents T values. The bars plot in the top right-hand corner represents differences in effect sizes (difference in Fisher-transformed correlation coefficients) between twins with greater methylation level and their co-twins with lower methylation level, for each connection
Discussion
The aim of the present study was to assess potential neural correlates of peripheral SLC6A4 promoter methylation during processing of negative facial expressions, i.e., sadness and fear. While an emerging number of studies have now shown that peripheral SLC6A4 methylation correlates with frontal-limbic processing of negative emotions, it is not clear whether this association is due to the environment or to genetic factors. Here, we utilized a within-pair MZ-twin design to control for the putative role of DNA sequence in this association. We found that relative to the co-twins with lower SLC6A4 promoter methylation, twins with greater SLC6A4 methylation levels had greater left OFC activation, greater amygdala-OFC and left amygdala-ACC connectivity in the sad condition, as well as greater ACC-amygdala and ACC-insula connectivity in the fearful condition. In spite of relatively low within-pair variability, current findings are of particular interest given the implication of these brain regions in emotion regulation. Indeed, regional functioning of OFC, ACC, insula and amygdala, all part of the frontal-limbic circuitry, has previously been associated with emotion regulation35,36 and SLC6A4 methylation16. Our findings do not only support the previously suggested hypothesis that peripheral SLC6A4 promoter methylation shares potentially functional relevant information about frontal-limbic functioning and emotion-processing, but, most importantly, demonstrate for the first time that these associations occur, at least in part, independently of DNA sequence.
The finding of within-pair differences in neural processing of sad stimuli in the OFC regions fits well with the results of our previous study in which we compared neural activation to sad stimuli between 8-year-old monozygotic (MZ) and dizygotic (DZ) twins, allowing separation of genetic factors from environmental factors shared between twin pairs and non-shared environmental factors that are unique for each twin member37. Among other brain areas, neural activation during processing of sad stimuli in Broadmann Area (BA) 47, a key region in the OFC, was found to be fully driven by non-shared/unique environmental factors37. Interestingly, results of a positron emission tomography (PET) study, investigating the neural basis of various emotions (sadness, happiness, anger and fear) in healthy individuals, showed that the orbitofrontal regional activity was found to peak solely during the sadness condition38, emphasizing the role of the OFC in the detecting and processing of sad stimuli. That being said, in addition to the OFC, Damasio and colleagues38 reported increases in insula and ACC activity during sad condition (and, respectively, during fearful and angry condition). The absence of these insula-ACC regional responses to sad or fearful cues in the current study may be, in part, explained by differences in methodology (e.g., use of current implicit emotion-processing task versus self-generated emotional conditions by recall).
Furthermore, we previously found—using in vivo PET measures of brain 5-HT synthesis—that serotonergic synthesis in the OFC is not only modulated by DNA sequence39 but also by early environmental factors40. The environmental influences on serotonin functioning in the OFC observed in the present study also fits well with our previous PET study showing that greater peripheral SLC6A4 methylation correlated with lower in vivo brain serotonin synthesis in the OFC region11. Current findings are also in line with the results of a study by another research group reporting positive association between peripheral SLC6A4 promoter methylation and resting-state functional connectivity between amygdala, insula and ACC in healthy adults26. Notably, OFC, ACC, insula and amygdala constitute an integral part of the salience functional network, involved in affective processes41. Interestingly, results of an fMRI study examining neural correlates of emotion regulation in healthy individuals, showed that functional connectivity between amygdala and several (pre)frontal regions, including OFC and ACC, was associated with attenuated emotional response to negative emotional stimuli41, thereby underscoring the involvement of frontal-limbic functional connectivity in processing and regulation of emotions. Interestingly, salience network has been found to show relatively low cross-twin correlation in both MZ and DZ twins42, indicative of scarce evidence for either genetic or non-shared environmental effect on this network. Results of the present study suggest that this SLC6A4-related salience network is, at least in part, independent of variation in DNA sequence. Altogether the findings suggest an important role for OFC, ACC, and amygdala in the processing of sadness and fear, which appear to be partly driven by SLC6A4-modulated non-shared/unique environmental factors.
Some laterality effects were observed as well. Notably, the finding that the association in the amygdala was specific for the left side is in line with other research showing that the left amygdala is more frequently activated in response to emotional stimuli than the right amygdala (see ref. 43 for meta-analysis). Furthermore, stronger functional connectivity between left (but not right) amygdala and bilateral ACC and OFC has been associated with decreased negative affect induced by negative emotional content in healthy individuals44. Besides, conjoint activity of ACC and left (but not right) insula has been linked to increased awareness of one’s own emotions45. Interestingly, greater SLC6A4 promoter methylation has been previously linked to greater left (but not right) amygdala and insula activation in response to negative emotional stimuli13,16. In light of these findings, we might advance that these lateralized regional activities reflect serotonin functioning-related neurobiological processing of the (sad or fearful) emotionally salient content. However, additional SLC6A4 methylation fMRI studies of the connectivity of amygdala, OFC and insula subdivisions are necessary to deepen the understanding of the laterality.
While we found significant associations between peripheral SLC6A4 promoter methylation discordance and neural processing of negative stimuli, we did not find any correlations between depressive symptoms and peripheral SLC6A4 methylation. However, the levels of symptomatology variation were very low. The observation of an association between SLC6A4 methylation and neural processing of negative stimuli in the absence of an association with anxiety- or depression-related symptoms is consistent with studies that include healthy individuals (e.g., 16). Thus, variation in frontal-limbic brain activation, as a function of variation in SLC6A4, occurs in the absence of behavioral variation. This supports the idea that SLC6A4 methylation could reflect a marker for neural regulation of negative stimuli, but additional factors may be necessary for an overt behavioral phenotype to be expressed8. Nevertheless, it would be of particular interest to test MZ twins who are highly discordant in levels of internalizing symptoms and in SLC6A4 promoter methylation and link their discordance to differences in frontal-limbic processes.
Furthermore, peripheral SLC6A4 promoter methylation levels and methylation differences within MZ twin pairs were low. However, considering the binary (either methylated or unmethylated allele at a particular CpG site) nature of the DNA methylation, the percentage change in DNA methylation indicates the fraction of cells having possibly experienced a complete change in methylation at a given site17. In other words, a small increase in SLC6A4 promoter methylation of a tissue sample may suggest that few cells are potentially turning off their expression. Indeed, using an in vitro model, higher DNA methylation levels in the selected SLC6A4 promoter region have been previously shown to lower its transcriptional activity11, thereby corroborating the regulatory role of DNA methylation of this region. In addition, peripheral DNA methylation in this particular gene region has been shown to be a potential marker of serotonin synthesis in the OFC11.
The strengths of this study include the use of a well-characterized, prospective, homogeneous longitudinal sample of adolescents. All twins were 15 years of age and carefully screened for the absence of psychiatric history. Furthermore, the investigated SLC6A4 region and primary CpG sites were chosen a-priori, based on previous work and validation in vivo and in vitro in three independent samples11,13,14,17. However, the present findings should also be considered in the context of certain limitations. First, methylation was assessed from peripheral tissue (i.e., saliva), as DNA methylation generally cannot be assessed in the living human brain. However, studies combining epignegative stimuli are detectable while controlling for DNA sequenceenetics and various fMRI measures have shown substantial correlations between methylation assessed from peripheral tissue and brain functioning13–17,46. Moreover, future studies will be necessary to investigate potential moderating roles of age, sex, and various psychosocial variables, as well as to identify individual participant characteristics that account for the within-pair variation. Furthermore, we will continue to follow these twins in the future to assess the clinical relevance of these alterations in peripheral SLC6A4 promoter methylation and processing of negative emotional stimuli. In spite of these limitations, the findings in our MZ twin study provide evidence that previously reported associations between peripheral SLC6A4 promoter methylation and frontal-limbic processing of negative stimuli are detectable while controlling for DNA sequence. These associations may have implications for the future use of non-invasive DNA methylation markers in diagnosis, treatment and risk prediction in mental health problems in which serotonin plays an important role.
Acknowledgements
We are grateful to the children and parents of the Quebec Newborn Twin Study (QNTS), and the participating teachers and schools. We thank Jocelyn Malo and Marie-Élyse Bertrand for coordinating the data collection, as well as Hélène Paradis and Nadine Forget-Dubois for managing the data bank of QNTS. The study was supported by a grant from the H.F. Guggenheim foundation awarded to Dr. Booij, and by an operating grant from the Canadian Institutes of Health Research (CIHR) awarded to Drs. Booij, Szyf, Tremblay, and Boivin. Dr. Lévesque and E. Ismaylova, MSc, were funded by doctoral awards from CHU Sainte-Justine—Foundation of Stars and from the Fonds de Recherche du Québec-Santé (FRQ-S). Dr. Linda Booij was supported by a New Investigator Award from CIHR, and Michel Boivin by the Canada Research Chair program. QNTS was supported by a series of grants from the Fonds de Recherche du Québéc -Société et Culture (FQRSC), the Fonds de Recherche du Québec-Santé (FRQ-S), the Social Science and Humanities Research Council of Canada (SSHRC), the National Health Research Development Program (NHRDP), the Canadian Institutes for Health Research (CIHR), Sainte-Justine Hospital Research Center, Université Laval and Université de Montréal. The authors thank the fMRI unit technicians of the McConnell Brain Imaging Centre of the Montreal Neurological Institute for assistance with collection of the brain-imaging data; and Dr. Farida Vaisheva, Victoria Ly, BSc, Mr. Jun Yi and Marie-Pier Verner, MSc, for other technical assistance.
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McGowan PO, Szyf M. The epigenetics of social adversity in early life: implications for mental health outcomes. Neurobiol. Dis. 2010;39:66–72. doi: 10.1016/j.nbd.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcriptional modulation and epigenetic regulation. Nat. Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razin A, Cedar H. DNA methylation and embryogenesis. EXS. 1993;64:343–357. doi: 10.1007/978-3-0348-9118-9_15. [DOI] [PubMed] [Google Scholar]
- 6.Ollikainen M, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum. Mol. Genet. 2010;19:4176–4188. doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- 7.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 8.Booij L, Tremblay RE, Szyf M, Benkelfat C. Genetic and early environmental influences on the serotonin system: consequences for brain development and risk for psychopathology. J. Psychiatry Neurosci. 2015;40:5–18. doi: 10.1503/jpn.140099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booij L, Wang D, Lévesque ML, Tremblay RE, Szyf M. Looking beyond the DNA sequence: the relevance of DNA methylation processes for the stress–diathesis model of depression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120251. doi: 10.1098/rstb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell C, Schneper LM, Notterman DA. DNA methylation, early life environment, and health outcomes. Pediatr. Res. 2015;79:212–219. doi: 10.1038/pr.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, et al. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS ONE. 2012;7:e39501. doi: 10.1371/journal.pone.0039501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drabe M, et al. Serotonin transporter gene promoter methylation status correlates with in vivo prefrontal 5-HTT availability and reward function in human obesity. Transl. Psychiatry. 2017;7:tp2017133. doi: 10.1038/tp.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frodl T, et al. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J. Psychiatry Neurosci. 2015;40:296–305. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booij L, et al. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PLoS ONE. 2015;10:e0119061. doi: 10.1371/journal.pone.0119061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swartz JR, Hariri AR, Williamson DE. An epigenetic mechanism links socioeconomic status to changes in depression-related brain function in high-risk adolescents. Mol. Psychiatry. 2016;22:209–214. doi: 10.1038/mp.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolova YS, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat. Neurosci. 2014;17:1153–1155. doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ismaylova E, et al. Serotonin transporter gene methylation in peripheral cells in healthy adults: neural correlates and tissue specificity. Eur. Neuropsychopharmacol. 2017;27:1032–1041. doi: 10.1016/j.euroneuro.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 18.van IJzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R. Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol. Psychiatry. 2010;68:405–407. doi: 10.1016/j.biopsych.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiarella J, Tremblay RE, Szyf M, Provencal N, Booij L. Impact of early environment on children’s mental health: lessons from DNA methylation studies with monozygotic twins. Twin Res. Hum. Genet. 2015;18:623–634. doi: 10.1017/thg.2015.84. [DOI] [PubMed] [Google Scholar]
- 20.Hariri AR, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 21.Boivin M, et al. The genetic-environmental etiology of parents’ perceptions and self-assessed behaviours toward their 5-month-old infants in a large twin and singleton sample. J. Child Psychol. Psychiatry. 2005;46:612–630. doi: 10.1111/j.1469-7610.2004.00375.x. [DOI] [PubMed] [Google Scholar]
- 22.Brendgen M, et al. Examining genetic and environmental effects on social aggression: a study of 6-year-old twins. Child Dev. 2005;76:930–946. doi: 10.1111/j.1467-8624.2005.00887.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaufman J, et al. Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): initial reliability and validity data. J. Am. Acad. Child Adolesc. Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- 24.Scott TJ, Short EJ, Singer LT, Russ SW, Minnes S. Psychometric properties of the Dominic 5interactive assessment: a computerized self-report for children. Assessment. 2006;13:16–26. doi: 10.1177/1073191105284843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 26.Muehlhan M, Kirshbaum C, Wittchen HU, Alexander N. Epigenetic variation in the serotonin transporter gene predicts resting state functional connectivity strength within the salience-network. Hum. Brain Mapp. 2015;36:4361–4371. doi: 10.1002/hbm.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canli T, et al. Amygdala reactivity to emotional faces predicts improvement in major depression. Neuroreport. 2005;16:1267–1270. doi: 10.1097/01.wnr.0000174407.09515.cc. [DOI] [PubMed] [Google Scholar]
- 28.Ekman P., Frisen W. V. Pictures of Facial Affect (Consulting Psychologists Press: Palo Alto, CA, 1976).
- 29.Casey KF, et al. Birth weight discordance, DNA methylation, and cortical morphology of adolescent monozygotic twins. Hum. Brain Mapp. 2017;38:2037–2050. doi: 10.1002/hbm.23503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer JP. Multiple hypothesis testing. Annu Rev. Psychol. 1995;46:561–584. doi: 10.1146/annurev.ps.46.020195.003021. [DOI] [Google Scholar]
- 31.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- 33.Lévesque ML, et al. Altered patterns of brain activity during transient sadness in children at familial risk for major depression. J. Affect Disord. 2011;135:410–413. doi: 10.1016/j.jad.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Beauregard M, Paquette V, Lévesque J. Dysfunction in the neural circuitry of emotional self-regulation in major depressive disorder. Neuroreport. 2006;17:843–846. doi: 10.1097/01.wnr.0000220132.32091.9f. [DOI] [PubMed] [Google Scholar]
- 35.Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception I: The neural basis of normal emotion perception. Biol. Psychiatry. 2003;54:504–514. doi: 10.1016/S0006-3223(03)00168-9. [DOI] [PubMed] [Google Scholar]
- 36.Mak AK, Hu ZG, Zhang JX, Xiao ZW, Lee TM. Neural correlates of regulation of positive and negative emotions: an fMRI study. Neurosci. Lett. 2009;457:101–106. doi: 10.1016/j.neulet.2009.03.094. [DOI] [PubMed] [Google Scholar]
- 37.Cote C, et al. Individual variation in neural correlates of sadness in children: a twin fMRI study. Hum. Brain Mapp. 2007;28:482–487. doi: 10.1002/hbm.20400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damasio AR, et al. Subcortical and cortical brain activity during the feeling of self-generated emotions. Nat. Neurosci. 2000;3:1049–1056. doi: 10.1038/79871. [DOI] [PubMed] [Google Scholar]
- 39.Booij L, et al. Tryptophan hydroxylase2 gene polymorphisms predict brain serotonin synthesis in the orbitofrontal cortex in humans. Mol. Psychiatry. 2012;17:809–817. doi: 10.1038/mp.2011.79. [DOI] [PubMed] [Google Scholar]
- 40.Booij L, et al. Perinatal effects on in vivo measures of human brain serotonin synthesis in adulthood: a 27-year longitudinal study. Eur. Neuropsychopharmacol. 2012;22:419–423. doi: 10.1016/j.euroneuro.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Northoff G. Spatiotemporal psychopathology I: no rest for the brain’s resting state activity in depression? Spatiotemporal psychopathology of depressive symptoms. J. Affect Disord.10.1016/j.jad.2015.05.007 (2015). [DOI] [PubMed]
- 42.Yang Z, et al. Genetic and environmental contributions to functional connectivity architecture of the human brain. Cereb. Cortex. 2016;26:2341–2352. doi: 10.1093/cercor/bhw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baas D, Aleman A, Kahn RS. Lateralization of amygdala activation: a systematic review of functional neuroimaging studies. Brain Res. Rev. 2004;45:96–103. doi: 10.1016/j.brainresrev.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Banks SJ, Eddy KT, Angstadt M, Nathan PJ, Phan KL. Amygdala–frontal connectivity during emotion regulation. Soc. Cogn. Affect Neurosci. 2007;2:303–312. doi: 10.1093/scan/nsm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips ML, et al. Depersonalization disorder: thinking without feeling. Psychiatry Res. 2010;108:145–160. doi: 10.1016/S0925-4927(01)00119-6. [DOI] [PubMed] [Google Scholar]
- 46.Won E, et al. Association between reduced white matter integrity in the corpus callosum and serotonin transporter gene DNA methylation in medication-naïve patients with major depressive disorder. Transl. Psychiatry. 2016;6:e866. doi: 10.1038/tp.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]