Abstract
Despite the remarkable number of scientific breakthroughs of the last 100 years, the treatment of neurodevelopmental disorders (e.g., autism spectrum disorder, intellectual disability) remains a great challenge. Recent advancements in genomics, such as whole-exome or whole-genome sequencing, have enabled scientists to identify numerous mutations underlying neurodevelopmental disorders. Given the few hundred risk genes that have been discovered, the etiological variability and the heterogeneous clinical presentation, the need for genotype—along with phenotype-based diagnosis of individual patients has become a requisite. In this review we look at recent advancements in genomic analysis and their translation into clinical practice.
Neurodevelopmental disorders: Translating genomic advances into clinical practice
The identification of genetic mutations associated with neurodevelopmental disorders (NDDs) along with routine diagnosis based on patients’ characteristics is aiding the delivery of personalized therapies. Dora Tarlungeanu and Gaia Novarino at the Institute of Science and Technology in Klosterneuburg, Austria, review recent advances in genetic technologies, such as whole exome sequencing, that can lead to early intervention, guide choice of treatment and prompt genetic counseling. Introducing the mutations associated with NDDs into model organisms or stem cells is revealing some of the mechanisms underlying NDDs and enabling the evaluation of novel therapeutic strategies that target core symptoms of the disorders. To accelerate the implementation of individualized treatments for NDD the authors highlight the need to adopt interdisciplinary research approaches and to keep clinical staff updated on the latest findings in NDD genetics.
Introduction
The past decade has seen a rapid development of precise technological and methodological advancements in genetics and genomics, thus allowing an unprecedented identification of mutations that are involved in complex neurodevelopmental conditions. Neurodevelopmental disorders (NDDs) affect more than 3% of children worldwide and can be attributed to mutations at over 1000 loci1.
Understanding the etiology of NDDs faces many challenges that range from delineating the heritable genetic components to defining individual factors that predispose to NDD risk and identifying the precise mechanisms through which these factors together lead to the disorder2. In addition, the clinical heterogeneity of NDDs make diagnosing a lengthy and costly process, complicating the quest for personalized medicine. However, the identification of bona fide genetic risk factors and the use of functional genomics to progress from mutation to phenotype represent a solid foundation for the development of individualized therapeutic approaches. In this review, we begin by mentioning some features of these disorders and continue by emphasizing the importance of genomics in determining the etiology of NDDs. We then describe advantages and limitations in the use of animal or stem cell models to study patient-specific genetic mutations. Finally, we discuss successful examples of translational research creating an evidence-based framework of how personalized medicine can advance the treatment of NDDs.
Neurodevelopmental disorders
NDDs are a group of early onset neurological disorders, including autism spectrum disorders (ASD), intellectual disability (ID) and language disorders among others. ASDs are characterized by early dysfunction in social interactions, communication deficits, and the presence of repetitive and restricted behaviors3. ASDs, with an estimated prevalence of 1 in 68 births3, represent an issue of public concern. Typically, ASDs have been classified into syndromic—Rett syndrome (RS)4, Fragile X syndrome (FXS)5, and tuberous sclerosis (TSC)6—and nonsyndromic. Evidence suggests that the causes involve both genetic and environmental factors7. Patients diagnosed with ASD often present with other comorbidities such as intellectual disability (ID)8, epilepsy9, and motor abnormalities10. Intellectual disability affects ~1.5–2% of the Western population11. The severe forms of ID are thought to have a genetic origin, but in at least 50% of cases, the cause remains elusive. Over the past years many autosomal or X-linked mental retardation genes have been identified12, with FMR1 (FXS) being one of the most common inherited monogenic causes of ID and ASD in male patients13. The core features of ASD and ID often coexist with recurrent seizures or epilepsy. Epileptic seizures are due to abnormal neuronal activity such as excessive excitation or hypersynchronization, which can occur as a result of developmental defects or due to brain insults (e.g., trauma, stress, etc.) later on in life. With over 65 million people affected worldwide, epilepsy is the most common, chronic neurological disorder14. Although in many cases seizures can be controlled by existing anti-epileptic drugs, the treatment gap is still large15. Genetic underpinnings for epilepsies have been long recognized and over the past 20 years a significant number of epilepsy-risk genes have been identified16,17.
The genetics of NDDs
On average, a newborn acquires between 50 and 100 new genetic variants, resulting in 0.86 new amino acid-altering mutations (i.e., de novo mutations) per individual18. Given such a high individual variability, a plethora of variants associated with NDDs have been found in hundreds of different genes, ranging from single nucleotide changes (single nucleotide variants (SNV)) to loss or gain of up to thousands of nucleotides (copy number variants (CNV)).
Sequencing of the human and other mammalian genomes has provided an important set of tools to start understanding the human genetic variation. The first steps to elucidate the genetic heterogeneity of NDDs were done by using karyotyping or fluorescence in situ hybridization (FISH). As the need for more accurate detection of nucleotide variations in the context of developmental disabilities grew, chromosome microarray (CMA) technology was developed and rapidly implemented as part of first-line evaluation for children with a NDD19,20. CMA set the stage for genetic variation detection, but the advent of whole-genome and whole-exome sequencing (WGS and WES) led to the identification of many inherited and de novo germline variants that significantly contribute to total NDD risk21–24 (Fig. 1a, b). In the case of ASD for instance, it is estimated that rare genetic mutations, both de novo and inherited, are causal in ~11% of simplex cases25. Similarly, common inherited genetic variants contribute substantially to ASD risk (49%); however, the individual common genetic variant liability is lower than that for rare genetic mutations26. Likewise, in the case of epilepsy, rare CNVs have been shown to explain ~3% of individuals suffering from idiopathic generalized epilepsy27. In addition, ~300 de novo mutations were identified in patients suffering from epileptic encephalopathies. These mutations emphasize the convergence on specific biological pathways due to their enrichment in certain gene sets including genes regulated by the fragile X protein28. Finally, germline mutations do not explain all NDD cases, indicating that other genetic defects also come into play. For example, postzygotic (i.e., somatic) mutations explain a significant proportion of NDD cases29–31. Along with the previously identified CNVs, such mutations have meaningful implications for risk prediction, diagnosis and patient management32.
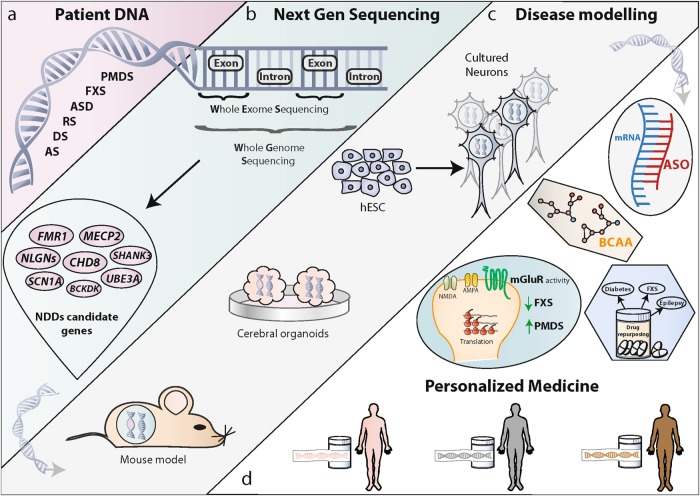
Fig. 1. Genomic sequencing guides the way from patient DNA to personalized medicine.
a DNA from patients diagnosed with NDDs used for sequencing; FXS Fragile X Syndrome, RS Rett Syndrome, PMDS Phelan McDermid Syndrome, DS Dravet Syndrome, AS Angelman Syndrome, ASD autism spectrum disorder. b Next-generation sequencing can be used to decipher the genetic code within exons (dark blue section—whole-exon sequencing) or throughout the entire genome (dark and light blue section—whole-genome sequencing). Mutations are identified in a series of genes with predisposition to NDDs (pink ovals). c The mutations are regenerated in models (mice, organoids, or hESC-derived neurons) in order to understand their underlying mechanism. d Disease modeling reveals targets that enable the implementation of personalized medicine. ASO (antisense oligonucleotides—gray panel) and BCAA (branched chain amino acids—beige panel) are two examples of personalized therapies probed in mouse models. mGLUR (metabotropic glutamate receptor) activity (green panel) needs to be decreased in FXS and increased in PMDS. Drug repurposing (blue panel) enables the usage of the same drug for different diseases due to novel mechanisms identified
Thus, the abovementioned technologies represent powerful tools for the molecular genetic dissection of patients affected by NDDs. Their introduction into clinical practice and association with routine phenotype-driven diagnosis holds promise for personalized diagnosis and therapy of NDDs.
The promise of genetics
The early occurrence of genetic glitches and the relatively late onset of symptoms that enable the diagnosis of NDDs, represent a major pitfall in identifying the cause and delivering the right kind of therapy. To complicate things further, for most NDDs, therapies hinge largely on behavioral or educational interventions33 and on treating associated rather than core symptoms of the disorder. Thus, for the majority of people with NDDs, the outcomes are poor or very poor in adulthood34. Given such challenges, we must ask how genetics may contribute to their improvement.
First and foremost, genetic testing can lead to active monitoring and early intervention, even before the onset of the disorder. Furthermore, knowing the genetic cause of a disorder may reveal the role of a specific biological pathway in its onset. Thus, targeted pharmacological interventions could be made with already existing drugs. Studies reported that 55% of 187 genetic findings led to changes in clinical management35 and 10 out of 118 probands undergoing WES benefited from a revised diagnosis and clinical assessment36. Similarly, WES with targeted gene analysis (e.g., SCN1A) influenced decisions on antiepileptic drug selection and reconsideration of surgical interventions37. Lastly, since NDDs are associated with cognitive and behavioral abnormalities, genetic information can guide the choice of behavioral treatment38. However, despite these advantages, the multiple guidelines proposing the use of genetic testing for individuals with NDDs are not implemented routinely in clinical practice20. This lack of use is either due to scarcity of resources or due to a lack of medical staff prepared to analyze and interpret genetic results. To circumvent this issue, a proper dissemination of up-to-date findings about NDD genetics to clinical staff is desirable. In addition, genetic counseling should inform parents about recurrence risk assessment.
Modeling NDDs: potentials and limitations
An ideal model of a human disorder is characterized by construct validity (model mimics the genetic insult that causes the disease), face validity (the model’s phenotype resembles that of the human disease), and predictive validity (the model and the patients respond similarly to certain treatments). Several systems (cells, rodents, primates) have been used to generate models of NDDs that can partially reproduce disease features and can be of interest for understanding underlying mechanisms (Fig. 1c).
The most favored model organism, the mouse, has been extensively employed for modeling neurological disorders with a known genetic cause, such as FXS (Fmr1 KO) (FXS)39, Dravet syndrome (DS-Scn1a KO)40, ASD Nrxn1a KO41, Nlgn3 KO42, Phelan-McDermid syndrome (PMDS-Shank 3 KO)43 or RS (Mecp2 KO)44. Mice share 95–98% of their genomic information with humans, have a relatively rapid reproduction time, are cost-effective and allow scientists to precisely manipulate their genome in a temporal/spatial specific manner. However, mice also present with important limitations. For example, assessment of higher brain functions, such as language and facial recognition, is difficult in large screens. To overcome some of these limitations, non-human primates can be employed to model complex behavior and higher cortical functions45, whereas zebrafish and invertebrates can be used for high-throughput genetic screens46.
Alongside animal models, in vitro reprograming of stem cells has enabled the generation and analysis of human neurons. Employing either human embryonic stem cell (hESC)-derived or human induced pluripotent stem cell (hiPSC)-derived neurons, researchers have recapitulated several neuronal synaptic defects for monogenic forms of NDDs such as RS47, FXS48, Prader-Willi and Angelman syndromes (AS)49, PMDS50, DS51 and Timothy syndrome (TS)52,53. The experimental tractability, the ability to model diseases directly from affected individuals and the unlimited source of cells are just some of the advantages of stem cell-based models. Conversely, the high heterogeneity among iPSC clones, the immature identity of neurons differentiated in vitro, the lack of high-order connectivity and the difficulty to model lamination in a 2D system are some of the obvious shortcomings of iPSC-derived disease models. Fortunately, recently several researchers have developed protocols for the generation of 3D cortical organoids (mini-brains/spheroids), providing avenues to study features of cortical lamination and brain development in vitro54,55, thus contributing additional tools for studying the mechanisms underlying NDDs52,56.
Bridging the gap between research and the clinic—personalized therapeutic approaches for NDDs
Axiomatically, the biggest advantage of genetic studies is to provide clues about the underlying neurobiology of NDDs and to transition those clues into clinical practice (Fig. 1d).
At present, the available treatments for NDDs consist of a combination of behavioral therapies57 and drugs approved for ameliorating comorbidities such as irritability and anxiety, while in many cases the core symptoms of NDDs remain unsolved.
However, the combination of genetics and functional analysis led to the discovery of several molecular pathways involved in NDDs that were targeted to evaluate novel therapeutic strategies. Particularly, inhibiting the mechanistic target of rapamycin (mTOR) rescues physiological, morphological and behavioral abnormalities in mice modeling diseases associated with protein translation defects such as TSC58, PTEN- associated macrocephaly59 or 15q11-13 duplication syndrome60. Multiple clinical trials are investigating the pharmacokinetics and pharmacodynamics of rapamaycin and its analogs (sirolimus, everolimus) for treating TSC with associated ASD61,62. Likewise, increasing levels of (IGF1) -like growth factor 1 and brain-derived neurotrophic factor via transcriptional modulation improves physiological and behavioral anomalies in RS mouse models63,64 and IGF1 administration leads to a higher endurance to social and cognitive testing in patients with RS65 or PMDS66.
Modulation of the excitation/inhibition ratio by employing antagonists of mGluRs or agonists of GABA A and GABA B receptors, has also been considered as a potential strategy to treat NDDs67. However, contrary to what was predicted by a decade of studies in FXS animal models, administration of mavoglurant, an mGluR5 antagonist68, or arbaclofen, a GABA B receptor agonist69,70, to adolescents and adults with FXS showed no significant improvement in behavioral traits in a randomized, double-blind, placebo-controlled phase 2 trial. Conversely, in a mouse model of PMDS with complete deletion of Shank3, researchers reported decreased mGluR5 signaling in the striatum and cortex. Administration of a benzamide derivative resulted in augmentation of mGluR5 activity and rescue of functional and behavioral defects in mice. Thus, pharmacological treatments aimed at increasing mGluR5 activity may represent an option for patients with SHANK3 mutations71,72. The contrasts between mGluR5 activity in FXS and PMDS suggest that the dysfunction leads to distinct phenotypes in different brain regions/genetic backgrounds. Hence, genetically discriminating between different forms of NDDs and identifying the convergence of the common molecular pathways underlying NDD pathophysiology are important goals (Fig. 1d).
Recently, new therapeutic strategies have been designed based on genetic findings. For example, AS is mostly caused by loss-of-function mutations in the maternal allele of the imprinted UB3A gene, while the paternal allele is silenced by a long noncoding RNA (UBE3A antisense transcript). Using antisense oligonucleotides (ASOs) (Fig. 1d) the paternal allele was unsilenced, thereby restoring the UB3A protein levels and leading to improvement in cognitive deficits in an AS mouse model73. Following a similar rationale, ASOs are used for restoring normal levels of MeCP2 and rescuing neurological deficits in mice carrying an extra copy of Mecp274. Replacing a defective gene may also be achieved by gene therapy using adeno-associated virus (AAV) vectors75. However, making ASOs and AAV amenable to translation into clinical trials is challenging due to their safety, pharmacokinetics and distribution in the brain76. In a similar vein, WES of consanguineous families with ASD, ID and epilepsy led to the identification of mutations in the gene BCKDK (Branched Chain Ketoacid Dehydrogenase Kinase), encoding an enzyme regulating the catabolism of the branched-chain amino acids (BCAAs). The Bckdk mouse model displays an abnormal brain amino acid profile and dietary supplementation with the missing BCAAs reverses certain neurological phenotypes (Fig. 1d). Furthermore, BCAA dietary supplementation in patients led to normalization of plasma BCAA levels, demonstrating the potential of BCAA supplementation as a therapy in patients with BCKDK mutations77,78.
In addition to identifying new targets for therapy, genetic findings are useful for personalizing existing pharmacotherapy or behavioral interventions. In this sense, WES with targeted gene analysis (e.g., SCN8A, KCNQ2) is an effective diagnostic tool for patients with epilepsies as it can influence anti-epileptic drug selection, adverse effect minimization and consideration for surgery, based on each patient’s genetic script37. In the case of people with SHANK3 deletions, they tend to have more advanced receptive communication skills than verbal language ability79 and therefore could benefit from assistive communication strategies that may not have been in mind unless the genetic cause of their ASD was known.
Recently, a very common trend uses genetic findings for the application of targeted drug repurposing based on single gene defects (Fig. 1d). Such an approach already shows promise for personalizing therapies for epilepsy cases arising from gain-of-function mutations in ion-channel subunit genes (e.g., GRIN2A, GRIN2B, SCN8A). Nonetheless, important barriers remain in order to translate these approaches to non-ion channel epilepsy genes and loss-of-function mutations80,81. Likewise, recent observations indicate that metformin, a worldwide first-line therapy for type 2 diabetes, rescues core phenotypes in adult FXS mice due to normalization of ERK signaling, eIF4E phosphorylation and matrix metalloproteinase 9 expression (MMP-9)82. Given that the previously mentioned clinical trials with mGluR5 antagonists have failed, metformin represents a new therapeutic avenue for clinical studies involving FXS patients. Administration of oxytocin, which is a peptide usually administered to initiate uterine contractions that also appears to be involved in modulating social behavior, improves ASD-like social deficits in several mouse models83 and in a Shank3-deficient rat84, but the clinical effectiveness of oxytocin on ASD should still be considered tentative due to mixed findings85.
Conclusion
The quick development of novel and efficient sequencing technologies made the identification of genetic causes for a number of NDDs possible. Using these techniques, an underlying genetic cause of many NDD cases can be identified. This progress allows the design of personalized therapeutic strategies and the implementation of genetic counseling. Furthermore, studies employing animal and human cell models carrying specific genetic glitches are underscoring potential novel therapeutic approaches.
In the past few years, potential treatments derived from genetic and functional analysis made it to clinical trials. Although several clinical trials have failed, the treatment of some NDDs seems much closer. Due to the very complex nature of NDDs, interdisciplinary approaches combining genetics, functional genomics, robust biological models and objective measures of response, such as biomarkers86, as well as the capability of researchers and clinicians to work side by side, will be essential.
Data for this review was collected by typing the following keywords into PubMed: genomics, genetics, personalized therapy, neurodevelopmental disorders (all in combination with NDDs, ASD, ID).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shashi V, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet. Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 2.de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elsabbagh M, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 5.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- 6.Kandt RS, et al. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat. Genet. 1992;2:37–41. doi: 10.1038/ng0992-37. [DOI] [PubMed] [Google Scholar]
- 7.Kim YS, Leventhal BL. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol. Psychiatry. 2015;77:66–74. doi: 10.1016/j.biopsych.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson JL, Shoemaker M. Intellectual disability and its relationship to autism spectrum disorders. Res. Dev. Disabil. 2009;30:1107–1114. doi: 10.1016/j.ridd.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Canitano R. Epilepsy in autism spectrum disorders. Eur. Child Adolesc. Psychiatry. 2007;16:61–66. doi: 10.1007/s00787-006-0563-2. [DOI] [PubMed] [Google Scholar]
- 10.Esposito G, Pasca SP. Motor abnormalities as a putative endophenotype for autism spectrum disorders. Front. Integr. Neurosci. 2013;7:43. doi: 10.3389/fnint.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mefford HC, Batshaw ML, Hoffman EP. Genomics, intellectual disability, and autism. N. Engl. J. Med. 2012;366:733–743. doi: 10.1056/NEJMra1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topper S, Ober C, Das S. Exome sequencing and the genetics of intellectual disability. Clin. Genet. 2011;80:117–126. doi: 10.1111/j.1399-0004.2011.01720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thurman DJ, et al. Standards for epidemiologic studies and surveillance of epilepsy. Epilepsia. 2011;52(Suppl 7):2–26. doi: 10.1111/j.1528-1167.2011.03121.x. [DOI] [PubMed] [Google Scholar]
- 15.Cameron A, et al. Mapping the availability, price, and affordability of antiepileptic drugs in 46 countries. Epilepsia. 2012;53:962–969. doi: 10.1111/j.1528-1167.2012.03446.x. [DOI] [PubMed] [Google Scholar]
- 16.Noebels J. Pathway-driven discovery of epilepsy genes. Nat. Neurosci. 2015;18:344–350. doi: 10.1038/nn.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl Acad. Sci. USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning M, Hudgins L, Professional P, Guidelines C. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet. Med. 2010;12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volkmar F, et al. Practice parameter for the assessment and treatment of children and adolescents with autism spectrum disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2014;53:237–257. doi: 10.1016/j.jaac.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen RK, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 25.Sanders SJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaugler T, et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Need AC, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am. J. Human. Gen. 2012;91:293–302. doi: 10.1016/j.ajhg.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epi4K Consortium, Epilepsy Phenome/GenomeProject. Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupp DR, et al. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am. J. Hum. Genet. 2017;101:369–390. doi: 10.1016/j.ajhg.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim ET, et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 2017;20:1217–1224. doi: 10.1038/nn.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riviere JB, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Gen. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soden SE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren Z, et al. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics. 2011;127:e1303–e1311. doi: 10.1542/peds.2011-0426. [DOI] [PubMed] [Google Scholar]
- 34.Fountain C, Winter AS, Bearman PS. Six developmental trajectories characterize children with autism. Pediatrics. 2012;129:e1112–e1120. doi: 10.1542/peds.2011-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson LB, et al. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet. Med. 2014;16:657–664. doi: 10.1038/gim.2014.18. [DOI] [PubMed] [Google Scholar]
- 36.Dixon-Salazar TJ, et al. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perucca P, et al. Real- world utility of whole exome sequencing with targeted gene analysis for focal epilepsy. Epilepsy Res. 2017;131:1–8. doi: 10.1016/j.eplepsyres.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Bruining H, et al. Behavioral signatures related to genetic disorders in autism. Mol. Autism. 2014;5:11. doi: 10.1186/2040-2392-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernardet M, Crusio WE. Fmr1 KO mice as a possible model of autistic features. Sci. World J. 2006;6:1164–1176. doi: 10.1100/tsw.2006.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han S, et al. Autistic- like behaviour in Scn1a+/- mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 2013;8:e67114. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudouin SJ, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulbert SW, Jiang YH. Monogenic mouse models of autism spectrum disorders: common mechanisms and missing links. Neuroscience. 2016;321:3–23. doi: 10.1016/j.neuroscience.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson KK, Platt ML. Of mice and monkeys: using non-human primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J. Neurodev. Disord. 2012;4:21. doi: 10.1186/1866-1955-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCammon JM, Sive H. Addressing the Genetics of Human Mental Health Disorders in Model Organisms. Annu. Rev. Genom. Hum. Genet. 2015;16:173–197. doi: 10.1146/annurev-genom-090314-050048. [DOI] [PubMed] [Google Scholar]
- 47.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell. Stem. Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc. Natl. Acad. Sci. USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shcheglovitov A, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiao J, et al. Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum. Mol. Genet. 2013;22:4241–4252. doi: 10.1093/hmg/ddt275. [DOI] [PubMed] [Google Scholar]
- 52.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hattori N. Cerebral organoids model human brain development and microcephaly. Mov. Disord. 2014;29:185. doi: 10.1002/mds.25740. [DOI] [PubMed] [Google Scholar]
- 56.Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 2016;22:1220–1228. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- 57.Zarafshan H, Salmanian M, Aghamohammadi S, Mohammadi MR, Mostafavi SA. Effectiveness of non-pharmacological interventions on stereotyped and repetitive behaviors of pre-school children with autism: a systematic review. Basic Clin. Neurosci. 2017;8:95–103. doi: 10.18869/nirp.bcn.8.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, et al. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J. Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oguro-Ando A, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol. Psychiatry. 2015;20:1069–1078. doi: 10.1038/mp.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Budde K, et al. Pharmacokinetics and pharmacodynamics of everolimus in patients with renal angiomyolipoma and tuberous sclerosis complex or lymphangioleiomyomatosis. Br. J. Clin. Pharmacol. 2016;81:958–970. doi: 10.1111/bcp.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohrman MH. Emerging treatments in the management of tuberous sclerosis complex. Pediatr. Neurol. 2012;46:267–275. doi: 10.1016/j.pediatrneurol.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 63.Kline DD, Ogier M, Kunze DL, Katz DM. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J. Neurosci. 2010;30:5303–5310. doi: 10.1523/JNEUROSCI.5503-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castro J, et al. Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc. Natl Acad. Sci. USA. 2014;111:9941–9946. doi: 10.1073/pnas.1311685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pini G, et al. Illness severity, social and cognitive ability, and EEG analysis of ten patients with Rett syndrome treated with mecasermin (recombinant human IGF-1) Autism Res. Treat. 2016;2016:5073078. doi: 10.1155/2016/5073078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolevzon A, et al. A pilot controlled trial of insulin-like growth factor-1 in children with Phelan- McDermid syndrome. Mol. Autism. 2014;5:54. doi: 10.1186/2040-2392-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat. Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berry-Kravis E, et al. Mavoglurant in fragile X syndrome: Results of two randomized, double-blind, placebo-controlled trials. Sci. Transl. Med. 2016;8:321ra5. doi: 10.1126/scitranslmed.aab4109. [DOI] [PubMed] [Google Scholar]
- 69.Lozano R, Martinez-Cerdeno V, Hagerman RJ. Advances in the understanding of the gabaergic neurobiology of FMR1 expanded alleles leading to targeted treatments for Fragile X spectrum disorder. Curr. Pharm. Des. 2015;21:4972–4979. doi: 10.2174/1381612821666150914121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veenstra-VanderWeele J, et al. Arbaclofen in children and adolescents with autism spectrum disorder: a randomized, controlled, phase 2 trial. Neuropsychopharmacology. 2017;42:1390–1398. doi: 10.1038/npp.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vicidomini C, et al. Pharmacological enhancement of mGlu5 receptors rescues behavioral deficits in SHANK3 knock-out mice. Mol. Psychiatry. 2017;22:689–702. doi: 10.1038/mp.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 2016;7:11459. doi: 10.1038/ncomms11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sztainberg Y, et al. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature. 2015;528:123–126. doi: 10.1038/528S123a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gadalla KKE, et al. Development of a novel AAV gene therapy cassette with improved safety features and efficacy in a mouse model of Rett syndrome. Mol. Ther. Methods Clin. Dev. 2017;5:180–190. doi: 10.1016/j.omtm.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beaudet AL, Meng L. Gene-targeting pharmaceuticals for single-gene disorders. Hum. Mol. Genet. 2016;25(R1):R18–R26. doi: 10.1093/hmg/ddv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia-Cazorla A, et al. Two novel mutations in the BCKDK (branched-chain keto-acid dehydrogenase kinase) gene are responsible for a neurobehavioral deficit in two pediatric unrelated patients. Hum. Mutat. 2014;35:470–477. doi: 10.1002/humu.22513. [DOI] [PubMed] [Google Scholar]
- 78.Novarino G, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phelan K, McDermid HE. The 22q13.3 Deletion Syndrome (Phelan- McDermid Syndrome) Mol. Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Symonds JD, Zuberi SM, Johnson MR. Advances in epilepsy gene discovery and implications for epilepsy diagnosis and treatment. Curr. Opin. Neurol. 2017;30:193–199. doi: 10.1097/WCO.0000000000000433. [DOI] [PubMed] [Google Scholar]
- 81.Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. Human GRIN2B variants in neurodevelopmental disorders. J. Pharmacol. Sci. 2016;132:115–121. doi: 10.1016/j.jphs.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gantois I, et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat. Med. 2017;23:674–677. doi: 10.1038/nm.4335. [DOI] [PubMed] [Google Scholar]
- 83.Insel TR. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron. 2010;65:768–779. doi: 10.1016/j.neuron.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harony-Nicolas H, et al. Oxytocin improves behavioral and electrophysiological deficits in a novel Shank3-deficient rat. eLife. 2017;6:e18904. doi: 10.7554/eLife.18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ooi YP, Weng SJ, Kossowsky J, Gerger H, Sung M. Oxytocin and autism spectrum disorders: a systematic review and meta-analysis of randomized controlled trials. Pharmacopsychiatry. 2017;50:5–13. doi: 10.1055/s-0042-109400. [DOI] [PubMed] [Google Scholar]
- 86.Loth E, et al. Identification and validation of biomarkers for autism spectrum disorders. Nat. Rev. Drug. Discov. 2016;15:70–73. doi: 10.1038/nrd.2015.7. [DOI] [PubMed] [Google Scholar]