Graphical abstract
Keywords: AFB1, Hepatocellular carcinoma, Cirrhosis, Markers
Highlights
-
•
The first study of hepatic aflatoxin residues in the Brazilian population.
-
•
AFB1 levels ranged from 10 to 418 pg/g in 27.1% of diseased liver samples (N = 48).
-
•
Residual AFB1 in liver negatively affected the p53 and protein Rb pathways in HCC.
-
•
AFB1 exposure can contribute for the progression of cirrhotic liver diseases.
Abstract
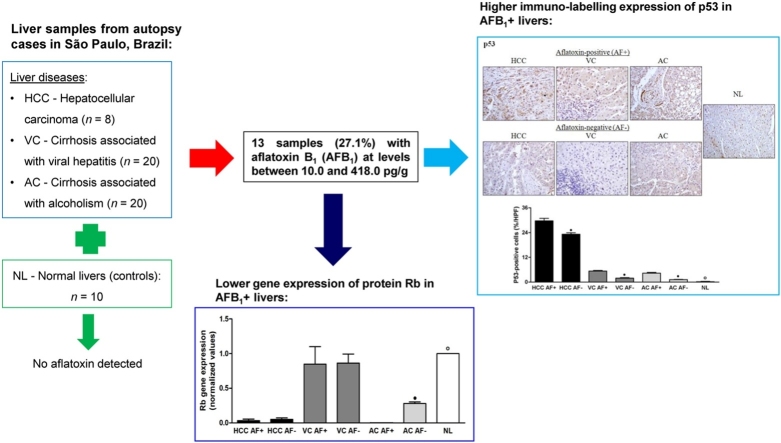
In this study, hepatic biopsies from autopsy cases in São Paulo, Brazil, showing hepatocellular carcinoma (HCC, n = 8), cirrhosis associated with viral hepatitis (VC, n = 20), cirrhosis associated with alcoholism (AC, n = 20), and normal livers (NL or controls, n = 10) were subjected to determination of aflatoxin B1 (AFB1) and its main metabolites, and of markers of hepatic carcinogenesis Only non-metabolized AFB1 was detected in 13 samples (27.1%, N = 48) of liver disorders (HCC, VC and AC), at levels between 10.0 and 418.0 pg/g (mean: 76.6 ± 107.7 pg/g). Immuno-labeling of p53, cyclin D1, p21, β-catenin, and Prohibitin (PB) increased mainly in HCC patients, in relation to the controls. AFB1+ samples of HCC presented higher expressions of p53, cyclin D1, p21, and β-catenin compared with AFB1-livers. In contrast, p27, p16, and Rb immuno-labeling decreased in HCC, VC, and AC samples, compared with NL, with lowest values in AFB1+ samples for all liver disorders. Compared with NL, gene expression of cyclin D1 and PB in AFB1+ samples of HCC and AC were also higher, along with higher gene expression of p21 in VC and AC AFB1+ livers. Results indicated that patients with liver disorders were exposed to dietary aflatoxins, and that residual AFB1 in liver negatively affected the p53 and protein Rb pathways in HCC. Moreover, the presence of AFB1 in cirrhotic livers warrants concern about the potential contribution of dietary aflatoxin to disease progression during VC and AC.
1. Introduction
Aflatoxins are carcinogenic secondary metabolites produced by fungi in the genus Aspergillus, especially the species A. flavus, A. parasiticus, and A. nomius, which naturally grow in food products (Murphy et al.). There are 20 known similar compounds called aflatoxins, although the main types of interest in terms of health are B1, B2, G1, and G2 [1]. Aflatoxin B1 (AFB1), besides being the most frequently one found in plant substrates, is the one with the highest toxigenic power [2]. The toxin is ingested with contaminated food, mainly foodstuffs that undergo storage, such as peanuts, corn, beans, and rice. AFB1 is genotoxic, and is considered one of the most potent natural mutagenic agents. Involvement in liver carcinogenesis is the most important effect of aflatoxin chronic toxicity [3]. Brazilian regulations determine maximum levels of aflatoxins (sum of AFB1, AFB2, AFG1, and AFG2) ranging from 1 to 20 μg/kg [4] in several foodstuffs.
Aflatoxins are absorbed in the gastrointestinal tract and primarily biotransformed in the liver by means of microsomal enzymes from the cytochrome P450 mixed function oxidase system [5]. AFB1 requires metabolic activation to show its toxic effects., and its carcinogenic form is the highly active electrophilic metabolite AFB1 8,9-oxide or AFB1-epoxide, which originates from the epoxidation of the vinyl-ether double bond found in the di-furan structure of AFB1 molecule [6,7]. AFB1 activated form quickly reacts with covalent bonds of macromolecules, such as cellular deoxyribonucleic acid (DNA), ribonucleic acid (RNA), as well as proteins [6]. The bond with DNA leads to the production of adducts with guanine in the N7 position at codon 249 of p53 tumor suppressor gene. Besides epoxidation, AFB1 biotransformation includes hydroxylation originating aflatoxins M1 (AFM1), Q1 (AFQ1), and B2a (AFB2α), as well as O-demethylation, originating aflatoxin P1 (AFP1), which may undergo conjugation with glucuronic acid or sulfates, and is excreted in urine, bile, and feces [8]. Aflatoxicol (AFL) can also be formed by the reduction of AFB1 by means of an NADPH-dependent cytoplasmic enzyme found in the soluble fraction of liver homogenates [9]. All these biotransformation products of AFB1 may remain in the form of residues in the liver, as observed in several species [10]. These residues are considered markers of toxin exposure in the diet.
Hepatocellular carcinoma (HCC) is the most frequent malignant neoplasm found in the liver, and the third most important cause of cancer deaths in the world. Greater occurrence of HCC is associated with the presence of chronic liver disease in up to 90% of the cases [11]. Africa and China have the greatest incidence of HCC compared with the rest of the world due to aflatoxin exposure, besides the known risk factors of western countries, such as viral hepatitis and alcoholism. In Brazil, HCC is now the 7th cause of cancer death. However, different from the pattern in the rest of the world, 42% of the HCC cases in Brazil do not show positive serology to viral hepatitis, and there are regional differences [12]. These differences may correspond to AFB1 exposure, taking into account the association between AFB1, HCC the induction of a specific mutation in p53 gene [13]. However, this relationship has not been investigated in Brazil, and there are no previous studies on the occurrence of AFB1 residues in human livers in this country.
Hepatocarcinogenesis seems to be a multifactorial process in which extrinsic stimuli induce gene changes in mature hepatocytes, leading to successive proliferation and death, and culminating in the production of monoclonal populations. Evidence suggests that hepatocarcinogenesis may begin with pre-neoplastic lesions, such as regenerative macronodules and hepatic nodules with low or high-grade dysplasia. Therefore, the accumulation of genetic changes and new mutations in pre-neoplastic lesions may probably cause HCC [14]. Mutations were identified in several critical genes involved in hepatocarcinogenesis, such as p73, p53, Rb, APC, DLC-1 (deleted in liver cancer), p16, GSTP1, PTEN, IGF-2, BRCA2, SOCS2, Smad2 and Smad4, ß-catenin, c-my + c, and cyclin D1 [15,16]. The p53 may also be related with other tumor suppressing gene, Prohibitin (PB). PB may be found in the cytoplasm or nucleus, according to the pathological characteristics of the cell [17]. In the nucleus, PB seems to be essential in the regulation of cell processes such as apoptosis, proliferation, and gene transcription [18].
Besides p53, other pathogenic mechanisms that may cause HCC after aflatoxin exposure still need to be studied. Cyclins are a family of proteins that are directly involved in the regulation of the cell cycle [19]. Changes in cyclin activities in transformed cells contribute to the acceleration of the cell cycle, which may be due to the lack of control of pathways that regulate cyclin synthesis, or to mutations that lead to the loss of function in inhibiting proteins [20]. The pathway p16/cyclin D1/Rb (retinoblastoma) may be considered the major cell cycle regulator. There are also three important inhibitors of cell cycle progression belonging to the Cip/Kip family: p27KIP1, p21WAF1, and p57KIP2. However, little is known about the interference of major chronic liver diseases, such as cirrhosis associated with viral hepatitis or alcoholism, in AFB1 biotransformation in the liver. The objective of the present study was to investigate the occurrence of residual AFB1 or its metabolites in hepatic biopsies from autopsy cases, and the association of these residues with mechanisms related to hepatocarcinogenesis.
2. Material and methods
2.1. Sampling procedures
A total of 58 human liver samples were collected from patients (46 males and 12 females) with HCC (n = 8), cirrhosis associated with viral hepatitis (VC, n = 20), cirrhosis associated with alcoholism (AC, n = 20), and normal livers (NL or controls, n = 10). These livers came from autopsies performed at the Division of Pathology of a University Hospital in Ribeirão Preto, Brazil, from 2011 to 2014. Post-mortem time for sample collection was around 6 h. Clinical data were obtained from patients’ records. Patients with evidence of human immunodeficiency virus infection, auto-immune or drug-induced hepatitis, endocrine disorders, or any other cause of liver disease were excluded from the study. The ethical guidelines from the Helsinki Declaration (1975) were followed in the study, and it also was approved by the Ethics Committee of the School of Medicine of the University of São Paulo at Ribeirão Preto (protocol no. 1611/2011). Each liver sample was subdivided in two parts: one was fixed in 10% buffered formalin for 48 h and then embedded in paraffin; and the other was placed in RNA stabilizing solution (RNAlater RNA Stabilization Reagent, Qiagen, Turnberry Lane, CA, EUA), frozen in liquid nitrogen, and stored at −80 °C. Paraffin-embedded sections were stained with hematoxylin and eosin (H&E) for confirmation of the original diagnosis.
2.2. Determination of aflatoxin B1 and metabolites
Liver samples stored at −80 °C were thawed at room temperature. Aflatoxin was extracted according to Chiavaro et al. [21] and purified through immunoaffinity columns (Aflatest WB®, Vicam, Watertown, MA, USA). Briefly, 1.0 g of liver sample was ground in a mortar and the resulting slurry was transferred to a 5-mL cryotube containing 4.0 mL of methanol/water (80:20, v/v) and 10 μL of internal standard (IS) working solution (described in the end of this section). The mixture was sonicated for 10 min, vortexed for 30 s and allowed to stand overnight at 7–10 °C. After that period, vortexing and sonication were repeated, and samples centrifuged (Sorvall® RC 3B Plus) at 3600 × G for 10 min. An aliquot (4 mL) of the supernatant was diluted with 25 mL of PBS and passed through the immunoaffinity cleanup column. The column was washed with 20 mL of PBS and then 2 mL of deionized water. After the washing steps, aflatoxins were eluted with 2 mL of methanol, dried, and reconstituted in 500 μL of acetonitrile/water (1:1, v/v). This solution was filtered (PTFE, 0.22 mm, Millex, Millipore Corp) and transferred to an amber glass vial.
Residual levels of AFB1, AFM1, AFP1, AFQ1, AFB2α, and AFL were determined by ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS using [13C17]-AFB1 (Sigma, St. Louis, MO, USA) and [13C17]-AFM1 (Biopure, Romer Labs, Tulln, Austria) as internal standards. To prepare the calibration lines for the quantification assay, a standard working solution with a mixture of all standard aflatoxins (AFB1, AFM1, AFP1, AFQ1, AFB2α, and AFL at 10 μg L−1 each) were dissolved in acetonitrile/water (1:1, v/v) and diluted to the final concentrations of 1.0, 0.750, 0.5, 0.1, 0.05, and 0.01 ng/mL. An additional calibration line was constructed for 10, 8, 4, and 2 ng/mL AFB1 by means of dilution of an individual AFB1 standard solution (10 ng/mL). The IS working solution (25 ng/mL [13C17]-AFB1 and 100 ng/mL [13C17]-AFM1) was prepared in acetonitrile/water (1:1, v/v) and added to all composite standards at a final concentration of 0.5 ng/mL and 2.0 ng/mL, respectively. The curves were determined by plotting the ratio of the standard peak area to the IS peak area against the concentration of the calibration standards. For aflatoxin quantification, the peak area ratios of the analytes to the IS were calculated, and concentration was determined by the corresponding calibration line. LOD and LOQ values were calculated based on signal-to-noise ratios of 3:1 and 10:1, respectively, of peaks corresponding to the confirmatory Multi Reaction Monitoring (MRM) transitions.
Final extracts were injected into a Waters Acquity I-Class UPLC system (Waters, Milford, MA, USA) coupled to a Xevo TQ-S mass spectrometer (Waters, Milford, MA, USA). The column (BEH C18, 2.1 × 50 mm, 1.7 μm) was kept at 40 °C during the analyses, and samples were maintained at 15 °C. The injection volume of extracted samples and standards was 10 μL. The mobile phase was composed by water (eluent A) and acetonitrile (eluent B), both containing 0.1% of formic acid. After an initial period of 0.5 min at 95% eluent A, the percentage of eluent B was linearly raised to 25% over 4.5 min (5.0 min). Then, eluent B was increased to 90% over 0.5 min, followed by a hold time of 0.25 min (5.5 min). After that, the percentage of eluent B was reduced to 5% over 0.5 min (6.0 min), and the column re-equilibrated to the initial conditions for 0.5 min. Total chromatographic run time was 6.5 min, and the flow rate was maintained at 0.5 mL/min. Retention times for AFB2α, AFQ1, AFM1, AFB1, AFP1, and AFL were 1.14, 1.16, 1.17, 1.24, 1.28 and 1.29 min, respectively. The mass spectrometer was operated in MRM mode using electrospray ionization in positive ion mode and the following conditions: capillary voltage: 0.75 kV; source temperature: 150 °C; desolvation temperature; 500 °C; desolvation gas flow: 800 L/h; and cone gas flow: 150 L/h. Quantification and confirmatory MRM transitions for aflatoxins and corresponding optimal mass spectrometric parameters used in the LC–MS/MS analysis are presented in Table 1. Data collection and processing was performed using software MassLynx version 4.1.
Table 1.
Mass spectrometry conditions used for aflatoxin analyses in human liver samples by Multi Reaction Monitoring.
| Aflatoxin | Precursor ion (m/z) | Fragment ion (m/z) | Cone voltage (V) | Collision energy (V) |
|---|---|---|---|---|
| [13C17]-AFB1 | 330.34 | 301.48 | 94 | 20 |
| [13C17]-AFM1 | 346.11 | 288.12 | 52 | 25 |
| AFB1 | 312.71 | 241.07a | 94 | 36 |
| AFB1 | 312.71 | 284.89b | 94 | 22 |
| AFB2α | 331.29 | 241.23a | 6 | 38 |
| AFB2α | 331.29 | 285.03b | 6 | 24 |
| AFM1 | 328.9 | 229.04a | 52 | 38 |
| AFM1 | 328.9 | 273.06b | 52 | 24 |
| AFP1 | 299.27 | 215.12a | 56 | 30 |
| AFP1 | 299.27 | 271.01b | 56 | 22 |
| AFQ1 | 329.28 | 177.22a | 60 | 34 |
| AFQ1 | 329.28 | 206.08b | 60 | 24 |
| Aflatoxicol | 296.91 | 114.99a | 40 | 58 |
| Aflatoxicol | 296.91 | 268.65b | 40 | 20 |
Transitions used for quantification.
Transitions used for confirmation.
2.3. Determination of aflatoxin B1–N7-guanine
The analysis of AFB1-N7-guanine in the liver samples requires DNA isolation, extraction, and hydrolysis. These procedures were carried out according to Kensler et al. [22], and Egner et al. [23]. Samples (1.0 g) were homogenized with a solution (4.0 mL) made up by sucrose 0.25 M, calcium chloride 2.0 mM, and 1.0 mM tris(hydroxymethyl)aminomethane buffer (TRIS), pH 7.4. Ten microliters of the [13C17]-AFB1 IS working solution (prepared as described in Section 2.2) were added in each mixture. The homogenate was filtered and a Triton X-100 25% solution in homogenization buffer was slowly added to a volume 9 times the weight of the liver. The homogenate was centrifuged at 1000g for 10 min, the supernatant was removed, and raw nucleus material was re-suspended in 1.0 mL of homogenization buffer and stored at −80 °C. After this step, nucleuses were frozen and suspended in 500 μL of 0.5 mol/L Tris-HCl, solution, pH 6.5. An adequate volume of sodium dodecyl sulfate (SDS) and sodium chloride was added to a final solution of SDS 1% and sodium chloride 1.0 M. The DNA concentration was measured using NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA), reaching 0.2–0.3 mg/mL, and the final volume was about 1.0 mL. An equal volume of a chloroform/isoamyl alcohol solution (24:1, v/v) was added, and the two phases were vigorously shaken for 20 min at room temperature. The organic and aqueous phases were separated by centrifugation at 7000g for 10 min. Then, the aqueous phase was separated and subjected to extraction again using the same procedure. Nucleic acids were precipitated in the aqueous phase by the addition of 3 volumes of ice-cold ethanol, removed from the solution with the aid of a glass rod, and washed with ice-cold ethanol. Nucleic acids were dissolved with 0.8–0.9 mg/mL of potassium acetate 50 mM and zinc chloride 0.1 mM, pH 5.2. DNA was denatured by heating at 95 °C for 15 min and quickly cooled in ice. Nuclease P1 (100 μL), at a concentration of 1 mg/200 mg in nucleic acid, was added and the solution was incubated at 40 °C for 12 h. The solution was deproteinized by agitation with an equal volume or chloroform, and the two phases were separated by centrifugation at 700g for 10 min. Nucleic acids were hydrolyzed with 100 μL of chloric acid 0.1 M for 16 h. The hydrolysate was filtered in a 0.45 μm filter, and AFB1-N7-guanine was extracted with an equal volume of ethyl acetate. After agitation, the organic phase was transferred to an amber glass vial, dried, and re-suspended in 500 μL methanol/water (1:1, v/v) for LC–MS/MS analysis.
The AFB1-N7-guanine standard (Toronto Research Canada, Toronto, ON, Canada) was diluted in methanol:1% acetic acid in water (50:50, v/v) and stored at −20 °C. A 5-point calibration curve was prepared in methanol/water (1:1, v/v) at the range from 0.2 to 2.5 ng/mL by means of serial dilutions of the AFB1-N7-guanine standard solution (10 ng/mL). The [13C17]-AFB1 IS peak area was used to correct the concentration of the analyte determined by means of the corresponding calibration line. The final extracts were injected in the same UPLC system coupled to a Xevo TQ-S mass spectrometer (Waters, Milford, MA, USA) as described in Section 2.2, keeping the same temperature conditions, injection volumes, elution gradients and flow rate. Total chromatographic run time was 3.0 min, and the retention time for AFB1-N7-guanine was 1.42 min. The mass spectrometer parameters were also set as described in Section 2.2. Quantification and confirmatory MRM transition for AFB1-N7-guanine were m/z 480.33 > 152.07 and 480.33 > 135.06, respectively.
2.4. Immuno-histochemical analyses
Liver sections were incubated with monoclonal primary antibodies specific for Rb, p16INK4D, β-catenin, cyclin D1, and p53 (Santa Cruz Biotechnology, Santa Cruz, CA, USA, dilution 1:100); p21WAF1/Cip1 and p27Kip1(DAKO, Carpinteria, CA, USA, dilution 1:100); and PB (Thermo Fisher Scientific Waltham, MA, USA, dilution 1:100). After that, a secondary antibody (Vectastain Elite ABC Kit, Universal, Vector Laboratories Inc.) was applied. Next, the slides were incubated with avidin-biotin-peroxidase complex (Vectastain Elite ABC Kit) and developed with 3,3-diamino-benzidinetetrahydrochloride (DAB) kit (Vector Laboratories Inc.) for 5 min. Slides were counterstained with Harris hematoxylin and mounted with Permount (Biomeda, Foster City, CA, USA). As negative controls, all specimens were incubated without the primary antibodies under identical conditions. Percentages of Rb, p16INK4D, β-catenin, cyclin D1, p53, p21WAF1/Cip1, p27Kip1, and PB-positive cells in each case were obtained blindly and independently by two of the authors (LDP and LNR), in at least 10 representative high-power fields (40×). Concordance between the authors ranged 95%.
2.5. Gene expression analyses
Gene expression was analyzed by real time PCR (RT-PCR). Frozen liver samples were used to quantify gene expression of p53, p21WAF1/Cip1, Rb, cyclin D1, and PB. Tissue fragments were homogenized and subjected to analysis of total RNA expression using a specific kit (RNeasy Mini Kit, QIAGEN, CA, USA). During the procedure, complementary DNA was obtained by reverse transcription using a retro-transcription kit (Ready-To-Go You-Prime First-Strand Beads and pd(N)6 Random Hexamer; Amersham Biosciences), beginning with 5 μg total RNA. Gene amplification with concurrent quantification was conducted by RT-PCR StepOnePlusTM (Applied Biosystems Inc., Foster City, CA, USA) using specific primers for p53, p21WAF1/Cip1, Rb, cyclin D1, PB, and GAPDH genes (Assays-on-Demand Gene Expression Products, Applied Biosystems), and a specific Taq Polymerase enzyme (TaqMan Universal PCR Master Mix, No AmpErase UNG - 2X, Applied Biosystems). All experiments were performed in duplicate, and the results were normalized to GAPDH mRNA expression.
2.6. Statistical analyses
The results on aflatoxin residues found in liver samples were used to distribute immuno-histochemical and gene expression data in two groups: AFB1 positive (AFB1+) and AFB1 negative (AFB1−). Data were analyzed using the GraphPad Prism software 4.0 (GraphPad Software, San Diego, CA, U.S.A.). All data are reported as means ± standard deviations. Statistical comparisons of the groups were performed by Kruskal-Wallis nonparametric one-way analysis of variance followed by Dunn’s posttest or Mann-Whitney test. Probability values less than 0.05 were considered statistically significant [24].
3. Results
3.1. Overview of clinical-pathological data
Table 2 shows the distribution of autopsied individuals according to the hepatic disorder, to gender and age. Most the patients were men (79.3%), older than 50 years of age (72.4%), and this proportion was generally similar for all types of liver disease, including controls (normal liver). Mean age of patients at the time of death was 59 years, ranging from 48 to 63 years. Only HCC patients had serum alpha-fetoprotein levels above 20 μg/L. Patients with HCC showed tumors grade I (n = 3), II (n = 4), or III (n = 1). Lymphovascular invasion was observed in the livers of three individuals with HCC.
Table 2.
Gender and age of the individuals autopsied in the Division of Pathology of an University Hospital in Ribeirão Preto, Brazil, from 2011 to 2014.
| Type of liver disorder | Gender |
Age (years) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Male |
Female |
≤50 |
>50 |
||||||
| N | n | % | n | % | n | % | n | % | |
| HCC | 8 | 5 | 62.5 | 3 | 37.5 | 2 | 25.0 | 6 | 75.0 |
| VC | 20 | 16 | 80.0 | 4 | 20.0 | 5 | 25.0 | 15 | 75.0 |
| AC | 20 | 18 | 90.0 | 2 | 10.0 | 7 | 35.0 | 13 | 65.0 |
| NL (control) | 10 | 7 | 70.0 | 3 | 30.0 | 2 | 20.0 | 8 | 80.0 |
| Total | 58 | 46 | 79.3 | 12 | 20.7 | 16 | 27.6 | 42 | 72.4 |
N: Number of individuals autopsied for each type of liver disorder.
HCC: Hepatocellular carcinoma.
VC: Cirrhosis associated with viral hepatitis.
AC: Cirrhosis associated with alcoholism.
NL: Normal liver.
3.2. Aflatoxin B1 residues
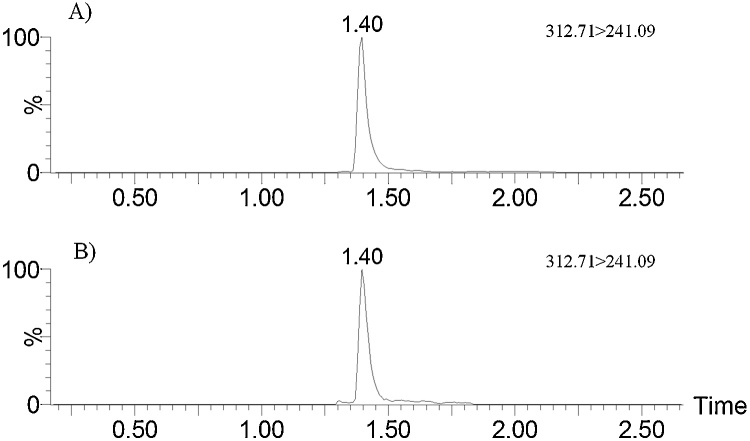
Thirteen liver samples (22.4%, N = 58) were positive for residual AFB1, with levels ranging from 10.0 and 418.0 pg/g (mean: 76.6 ± 107.7 pg/g), as shown in Table 3. The percentage of liver samples showing AFB1 in individuals with HCC (n = 8), VC (n = 20) and AC (n = 20) was 37.5%, 15%, and 35.0%, respectively. Mean levels of AFB1 in the livers with HCC and VC were similar (63.0–64.0 pg/g); however, the concentration of the toxin in the livers with AC was greater (87.9 pg/g), with large standard deviation (158.1 pg/g) due to a sample that showed much higher levels than the others (418.0 pg/g) (Fig. 1). Control patients (NL, n = 10) without history of liver disease did not show residues of AFB1 or its metabolites in their livers. Furthermore, aflatoxin metabolites (AFM1, AFP1, AFQ1, AFB2α, AFL) or AFB1-N7-guanine were not found in any of the samples analyzed. LOQ values were 10 pg/g (AFB1 and AFL), 25 pg/g (AFB2α), and 50 pg/g (AFP1, AFQ1 and AFM1). Determined LOD values were 4 pg/g (AFB1 and AFL), 8 pg/g (AFB2α), and 16 pg/g (AFP1, AFQ1, and AFM1). For AFB1-N7-guanine, LOQ and LOD values were 90 and 30 pg/g, respectively.
Table 3.
Aflatoxin B1 residues in liver samples from autopsied individuals in São Paulo, Brazil.
| Positive samplesa | Levels (pg/g) | ||||
|---|---|---|---|---|---|
| Type of liver disorder | N | n | (%) | Mean b | Variation |
| HCC | 8 | 3 | 37.5 | 63.0 ± 7.9 | 54.0–69.0 |
| VC | 20 | 3 | 15.0 | 64.0 ± 30.5 | 39.0–98.0 |
| AC | 20 | 7 | 35.0 | 87.9 ± 158.1 | 10.0–418.0 |
| NL (control) | 10 | 0 | 0 | ND | – |
| Total | 58 | 13 | 22.4 | 76.6 ± 107.7 | 10.0– 418.0 |
N: Number of samples analyzed for each type of liver disorder.
HCC: Hepatocellular carcinoma.
VC: Cirrhosis associated with viral hepatitis.
AC: Cirrhosis associated with alcoholism.
NL: Normal liver.
Samples with concentrations above the limit of quantification (LOQ) (10.0 pg/g).
Results are expressed as mean ± standard deviation of samples with concentrations above the LOQ.
Fig. 1.
Chromatograms (quantification transitions) of AFB1 standard at 1000 pg/mL (A) and a liver sample containing 418.0 pg/g (B).
3.3. Immuno-histochemical parameters and aflatoxin B1 residues
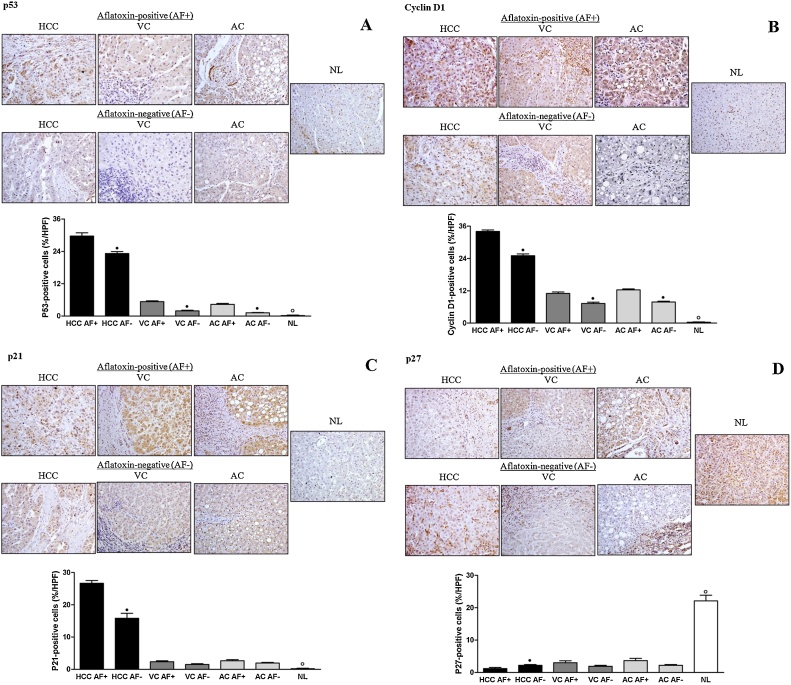
Comparative immuno-labeling of p53, cyclin D1, p21, and p27 in AFB1+ and AFB1− liver samples is presented in Fig. 2. HCC, VC and AC AFB1+ groups presented higher labeling for p53 than HCC, VC and AC AFB1- groups. Only the AC AFB1− group showed p53 expression similar to the NL group; in all the other groups, p53 expression increased significantly. HCC, VC, and AC AFB1+ groups showed increased labeling for cyclin D1 compared with HCC, VC, and AC AFB1− groups, as well as with the NL group. HCC AFB1+ group presented higher labeling for p21 than the HCC AFB1− group. There was no significant difference between VC and AC AFB1+ groups, and VC and AC AFB1− groups. However, all the other groups showed higher p21 expression than normal livers. HCC AFB1- group presented higher labeling for p27 than the HCC AFB1+ group. There was no significant difference between VC and AC AFB1+ groups, and VC and AC AFB1− groups. Additionally, the expression of p27 was decreased in all groups in relation to the NL group.
Fig. 2.
Comparative immune-histochemical expressions of p53 (A), cyclin D1 (B), p21 (C) and p27 (D) in aflatoxin-positive (AF+) and aflatoxin-negative (AF−) liver samples. HCC: hepatocellular carcinoma; VC: cirrhosis associated with viral hepatitis; AC: cirrhosis associated with alcoholism; NL: normal liver. n = 3 (HCC AF+), 5 (HCC AF−), 3 (VC AF+), 17 (VC AF−, 7 (AC AF+), 13 (AC AF−), 10 (NL). • Significant (P < 0.05) for comparisons between AF+ and AF− samples in each liver disorder. ° Significant (P < 0.05) for comparisons between NL and AF+ or AF- samples in all liver disorders.
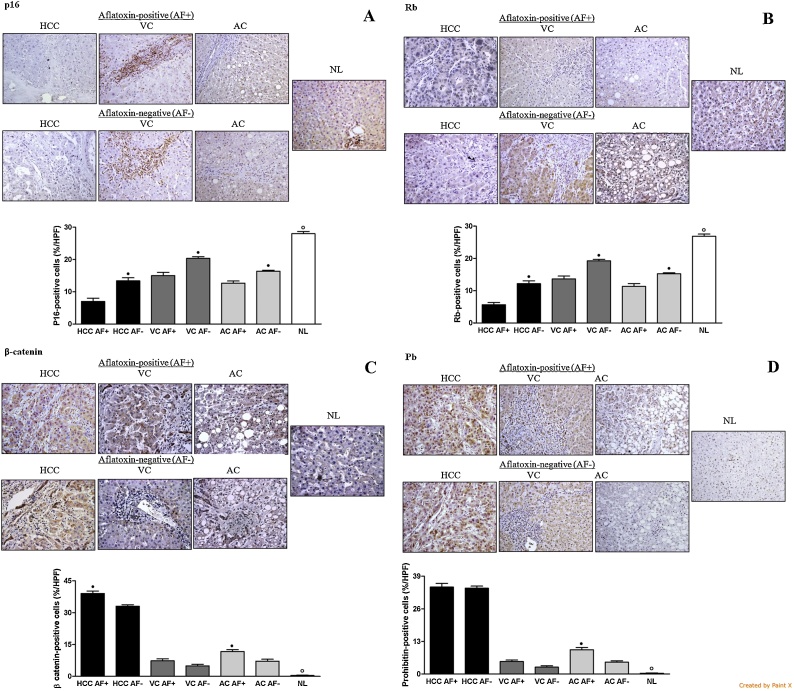
Fig. 3 presents the immuno-labeling of p16, Rb, β-catenin, and PB in the liver samples. Immuno-expression for p16 was decreased in all groups with liver disease compared with normal livers. HCC, VC, and AC AFB1+ groups showed lower labeling for p16 than HCC, VC, and AC AFB1− groups. HCC, VC, and AC AFB1+ groups showed lower labeling for Rb than HCC, VC and AC AFB1− groups. Rb expression was lower than the NL group in all HCC, VC, and AC groups. HCC and AC AFB1+ groups presented higher labeling for β-catenin than HCC and AC AFB1− groups. There was no significant difference between the VC AFB1+ groups and VC AFB1− groups. The β-catenin labeling was increased in all groups compared with normal livers. AC AFB1+ group had higher PB marking than AC AFB1− group. HCC and VC AFB1+ groups did not show higher PB expression in relation to HCC and VC AFB1− groups. PB expression was higher than the NL group in all the other groups.
Fig. 3.
Comparative immune-histochemical expressions of p16 (A), Rb (B), β-catenin (C) and Pb (D) in aflatoxin-positive (AF+) and aflatoxin-negative (AF−) liver samples. HCC: hepatocellular carcinoma; VC: cirrhosis associated with viral hepatitis; AC: cirrhosis associated with alcoholism; NL: normal liver. n = 3 (HCC AF+), 5 (HCC AF-), 3 (VC AF+), 17 (VC AF−), 7 (AC AF+), 13 (AC AF−), 10 (NL). • Significant (P < 0.05) for comparisons between AF+ and AF− samples in each liver disorder. ° Significant (P < 0.05) for comparisons between NL and AF+ or AF− samples in all liver disorders.
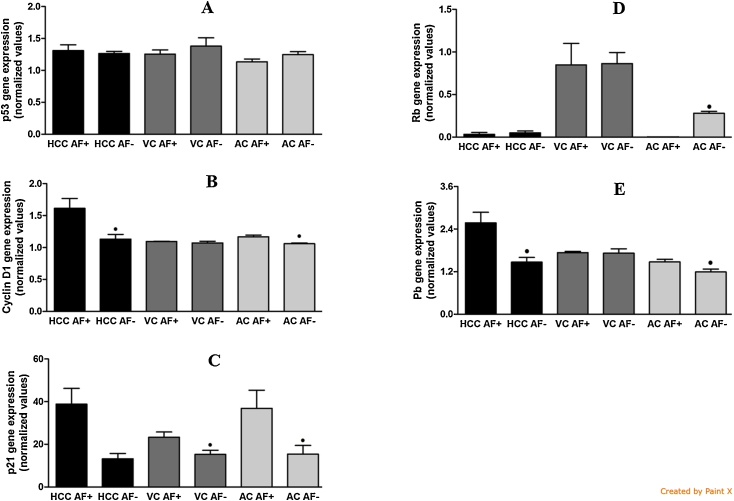
3.4. Gene expression and aflatoxin B1 residues
There was no difference in p53 expression (Fig. 4) between HCC, VC and AC AFB1+ groups, and HCC, VC and AC AFB1− groups. HCC and AC AFB1+ groups presented higher expression of cyclin D1 than HCC and AC AFB1- groups. There was no significant difference between VC AFB1+ and VC AFB1− groups. VC and AC AFB1+ groups showed higher p21 expression than VC and AC AFB1− groups. There was no significant difference between the groups HCC AFB1+ and HCC AFB1−. AC AFB1+ presented lower Rb expression than AC AFB1− group. There was no difference in Rb expression between HCC and VC AFB1+ groups, and HCC and VC AFB1− groups. HCC and AC AFB1+ groups showed higher PB expression than HCC and AC AFB1− groups. There was no significant difference between the groups VC AFB1+ and VC AFB1−.
Fig. 4.
Comparative gene expressions of p53 (A), cyclin D1 (B), p21 (C), RB (D), and Pb (E) in aflatoxin-positive (AF+) and aflatoxin-negative (AF−) liver samples. HCC: hepatocellular carcinoma; VC: cirrhosis associated with viral hepatitis; AC: cirrhosis associated with alcoholism; NL: normal liver n = 3 (HCC AF+), 5 (HCC AF−), 3 (VC AF+), 17 (VC AF−), 7 (AC AF+), 13 (AC AF−), 10 (NL). • Significant (P < 0.05) for comparisons between AF+ and AF− samples in each liver disorder.
4. Discussion
In the present study, HCC, VC, and AC liver specimens from patients autopsied in São Paulo showed detectable levels of AFB1 residues. Previous studies demonstrated that, in general, levels of aflatoxin reported in foodstuffs commercialized in Brazil [25,26] complied with the limits determined by local regulations [4]. Studies on biomarkers of AFB1 exposure in urine [27,28] and serum [29] in individuals from the state of São Paulo corroborate these findings, as they indicated low exposure to the toxin in foodstuffs. However, data obtained in the present study suggest that exposure to AFB1 may be high in some population groups in Brazil. In this study, it was not possible to analyze the eating habits of the patients in order to assess the level of previous aflatoxin exposure. Experiments conducted with human subjects indicated that 95% of AFB1 and its metabolites are eliminated in urine within 24 h [30]. However, there are no estimates on the conversion ratios of AFB1 in the diet into AFB1 and/or other residual metabolites in human livers. This ratio varies widely in experimental studies carried out with laboratory and production animals. In pigs, the estimated value was 800:1 [10]. If this ratio is directly extrapolated to humans with the mean residual AFB1 level obtained in the present study (76.6 pg/g), mean levels of AFB1 in the diet of the patients would be about 60 μg/kg, corresponding to three times the maximum limit for aflatoxins (20 μg/kg) determined in Brazil [4].
The results for HCC are similar to previous data on the incidence of aflatoxins in livers of HCC patients in Czechoslovakia [31]. Furthermore, our data demonstrated that the patients were exposed to dietary aflatoxins, providing evidence that support the association between aflatoxin exposure and the occurrence of HCC, for the first time in Brazil. However, the presence of AFB1 in cirrhotic livers (VC and AC) warrants concern about the potential contribution of dietary aflatoxin for the liver disease progression. These findings are in agreement with those described by [32], who observed that exposure to AFB1 increased the risk of cirrhosis and HCC in a dose-response manner among chronic carriers of hepatitis B virus. The absence of detectable metabolites of AFB1 (AFM1, AFP1, AFQ1, AFB2α, AFL) in liver samples was surprising, as several studies demonstrated that the accumulation of biotransformation products of AFB1 in animal tissues preferentially takes place in the liver [23,33]. The AFB1-N7-guanine adduct was not detected in any liver sample, indicating that liver DNA was not affected at the time of sample collection, although there are no available studies on the rate of AFB1-DNA repair in the liver in humans or animal models. However, taking into account the genotoxic changes caused by AFB1 [6], it can be hypothesized that its residues in the liver would play an additional role in the pathogenic pathway linking cirrhosis and HCC.
Regarding the mechanisms involved in hepatocarcinogenesis, genes Rb and p16 act as tumor suppressing genes when cyclin D1 is activated as an oncogene. However, cyclin D1 high expression in HCC is variable, ranging from 6 to 76% [34,35]. The analysis of cyclin D1 aberrant expression, its biological significance, and its relationship with p53 mutations in HCC demonstrated that mRNA of cyclin D1 is associated with larger and less differentiated tumors [36,37]. The p27KIP1 is strongly expressed in non-proliferating cells and has an important role in the regulation of both quiescence and progression of G1 phase by means of cyclin release. Loss of p27KIP1 acts together with mutations in several oncogenes and suppressing genes, making tumor growth easier. The reduction in protein synthetized by p27KIP1 has significant effect on the stage and volume of primary tumors. Therefore, p27KIP1 has been described as a crucial negative regulator of HCC progression, and its increased expression is considered an independent variable in favorable prognosis in HCC [38,39]. Reduced expression of p21WAF1 is mainly related with the mutation of gene p53 in HCC, also contributing to hepatocarcinogenesis. However, loss of p21WAF1 was not identified as an independent factor for poor prognosis in HCC [38]. In terms of frequency, the most common molecular changes in HCC are: p53 (20–70%), cyclin D (11%), p16Ink4 (0–50%), Rb (15%), and ß-catenin (16–26%). Only the increased expression of p53 was reported in association with gene interaction in hepatitis B virus and exposure to aflatoxin [40]. However, the involvement of this toxin with p53 changes in the genesis of HCC in Brazil, as observed in the present study, had not been described until now. Although the p53 pathway has an important role in HCC pathogenesis when cirrhosis is present, changes in the cell cycle regulator genes p21WAF1/CIP1 e p27KIP1 are more common in HCC cases that do not involve cirrhosis [41]. These changes may be, at least in part, attributed to aflatoxin exposure, hence confirming the molecular changes in genes p21WAF1/CIP1 and p27KIP1, as identified in the present study.
PB has been detected in the nucleus, where it is associated with retinoblastoma (Rb) and p53 proteins. PB induces changes in transcription factors, resulting in the inhibition of the cell cycle and apoptosis induction [42]. PB tumor suppressing action was associated with nuclear expression in neoplastic cells of several tumors [17,43]. In addition, during normal tumor suppressing function, PB reduces the nuclear translocation of NFkB mediated by TNF-α [44]. On the other hand, PB is involved in the stabilization of mitochondrial proteins and reduction of oxidative stress in the chronic phase of several diseases [44]. As for HCC, PB is followed by anti-neoplastic effects such as blockage of proliferation and induction of apoptosis [42], and increased PB levels were observed in HCC cells in humans [45]. Thus, changes in PB expression related to p53 were observed in human HCC samples from patients exposed to aflatoxin, with PB overexpression even in non-neoplastic livers, corroborating aflatoxin participation in oxidative stress pathways.
In our study, HCC AFB1+ cases had increased expression of p53, cyclin D1, p21, and β-catenin in relation to HCC AFB1+ livers. This pattern was also maintained in CV and AC cases, and was more important in the latter group. The opposite was observed in relation to p27, p16, and Rb expression. These data are in accordance with a previous study that showed that the Rb and p53 pathways are somewhat affected by the development of HCC of different etiologies, because of common mechanisms during the progression from chronic hepatitis lesions to cirrhosis. However, HCC cases that were more related to alcoholism showed greater frequency of changes in the Rb and p53 pathways than HCC linked to viral hepatitis. Suppression of Rb expression was inversely proportional to the suppression of p16 expression. The suppression of one or the other, though, culminates with the activation of the cyclin pathway, stimulating cell proliferation [46]. Additionally, a close relationship was observed between aflatoxin-induced depletion of gene Rb, development of HCC, and suppression of the Rb pathway promoting the activation of the cyclin pathway and tumor progression [47]. It was also reported that exposure to aflatoxin potentiates carcinogenesis associated with hepatitis B, provoking mutations and increased expression in gene p53 thus leading to synthesis of p53 anomalous proteins that do not perform their tumor-suppressing role adequately [48]. Increased expression of β-catenin was also reported in HCC [49].
In the present experiment, frozen samples of liver were not available in quantities as much as paraffin-embedded liver samples, which are commonly collected from autopsies and biopsies. Given that AFB1 residues could only be determined in frozen samples, our sampling number resulted greatly restricted especially within the liver diseases containing residual AFB1. Therefore, further studies are necessary to provide dose-response relationships between AFB1 residues in human liver samples and the abnormalities described in cell cycle or tumor suppressor pathway for the development of HCC.
5. Conclusion
Results of this study demonstrated that autopsied patients with liver disorders in São Paulo, Brazil, were exposed to dietary aflatoxins, and that residual AFB1 in liver negatively affected the p53 and protein Rb pathways in HCC. Moreover, the presence of AFB1 in cirrhotic livers warrants concern about the potential contribution of dietary aflatoxin for the disease progression during VC and AC.
Funding
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP – grant number 2010/20895-4).
Transparency document
References
- 1.Diaz D.E. Nottingham University Press; Nottingham, UK: 2005. The Mycotoxin Blue Book. [Google Scholar]
- 2.Murphy P.A., Hendrich S., Landgren C., Bryant C.M. Food mycotoxins: an update. J. Food Sci. 2006;71:51–65. [Google Scholar]
- 3.International Agency for Research on Cancer, Aflatoxin In: IARC Monograph on the Evaluation of Carcinogenic Risk to Humans, Vol. 82, Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene. IARC, Lyon, (2002), pp. 171–175. [PMC free article] [PubMed]
- 4.Agência Nacional de Vigilância Sanitária . ANVISA; Brasil: 2001. Resolução RDC n 7, de 18 de fevereiro de 2011.http://portal.anvisa.gov.br/wps/wcm/connect/bc17db804f45fe2cbd41fdd785749fbd/Resolu%C3%A7%C3%A3o+0-2011-GGALI.pdf?MOD=AJPERES 2001, Available from: (Accessed August 2014) [Google Scholar]
- 5.Gross-Steinmeyer K., Eaton D.L. Dietary modulation of the biotransformation and genotoxicity of aflatoxin B(1) Toxicology. 2012;299:69–79. doi: 10.1016/j.tox.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh D.P., Atkinson D.N. Bisfuranoid mycotoxins: their genotoxicity and carcinogenicity. Adv. Exp. Med. Biol. 1991;283:525–532. doi: 10.1007/978-1-4684-5877-0_69. [DOI] [PubMed] [Google Scholar]
- 7.Essigmann J.M., Croy R.G., Nadzan A.M., Busby Jr W.F., Reinhold V.N., Buchi G., Wogan G.N. Structural identification of the major DNA adduct formed by aflatoxin B1in vitro. Proc. Natl. Acad. Sci. U. S. A. 1977;74:1870–1874. doi: 10.1073/pnas.74.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groopman J.D., Zhu J.Q., Donahue P.R., Pikul A., Zhang L.S., Chen J.S., Wogan G.N. Molecular dosimetry of urinary aflatoxin-DNA adducts in people living in Guangxi autonomous region, People’s Republic of China. Cancer Res. 1992;52:45–52. [PubMed] [Google Scholar]
- 9.Biehl M.L., Buck W.B. Chemical contaminants: their metabolism and their residues. J. Food Prot. 1987;50:1058–1073. doi: 10.4315/0362-028X-50.12.1058. [DOI] [PubMed] [Google Scholar]
- 10.Park D.L., Pohland A.E. A rationale for the control of aflatoxin in animal feeds. In: Steyn P.S., Vleggaar R., editors. Mycotoxins and Phycotoxins. Elsevier Applied Science; Amsterdam: 1986. pp. 473–482. [Google Scholar]
- 11.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat. Ver. Gastroenterol. Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonçalves C.S., Pereira F.E.I., Gayotto L.C.C. Hepatocellular carcinoma in Brazil: report of a national survey (Florianopolis, SC, 1995) Rev. Inst. Med. Trop. S. Paulo. 1997;39:165–170. doi: 10.1590/s0036-46651997000300008. [DOI] [PubMed] [Google Scholar]
- 13.Bressac B., Kew M., Wands L., Ozturk M. Select G to T mutations of p53 gene in hepatocellular carcinoma in Southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 14.Theise N.D., Park Y.N., Kojiro M. Dysplastic nodules and hepatocarcinogenesis. Clin. Liver Dis. 2002;6:497–512. doi: 10.1016/s1089-3261(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 15.Fujimori M., Tokino T., Hino O., Kitagawa T., Imamura T., Okamoto E., Mitsunobo M., Ishikawa T., Nakagama H., Harada H. Allelotype study of primary hepatocellular carcinoma. Cancer Res. 1991;51:89–93. [PubMed] [Google Scholar]
- 16.Tsuda H., Oda T., Sakamoto M., Hirohashi S. Different pattern of chromosomal allele loss in multiple hepatocellular carcinomas as evidence of their multifocal origin. Cancer Res. 1992;52:1504–1509. [PubMed] [Google Scholar]
- 17.Fusaro G., Dasgupta P., Rastogi S., Joshi B., Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem. 2003;278:47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 18.Theiss A.L., Sitaraman S.V. The role and therapeutic potential of prohibitin in disease. Biochim. Biophys. Acta. 2011;6:1137–1143. doi: 10.1016/j.bbamcr.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Chenivesse X., Henglein B., Brechot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]
- 20.Sherr C.J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 21.Chiavaro E., Cacchioli C., Berni E., Spotti E. Immunoaffinity clean-up and direct fluorescence measurement of aflatoxin B1 and M1 in pig liver: comparison with high-performance liquid chromatography determination. Food Addit. Contam. 2005;22:1154–1161. doi: 10.1080/02652030500307115. [DOI] [PubMed] [Google Scholar]
- 22.Kensler T.W., Egner P.A., Davidson N.E., Roebuck B.D., Pikul A., Groopman J.D. Modulation of aflatoxin metabolism, aflatoxin-N7-guanine formation, and hepatic tumorigenesis in rats fed ethoxyquin: role of induction of glutathione S-transferases. Cancer Res. 1986;46:3924–3931. [PubMed] [Google Scholar]
- 23.Egner P.A., Groopman J.D., Wang J.S., Kensler T.W., Friesen M.D. Quantification of aflatoxin-B1-N7-guanine in human urine by high-performance liquid chromatography and isotope dilution tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:1191–1195. doi: 10.1021/tx060108d. [DOI] [PubMed] [Google Scholar]
- 24.Snedecor G.W., Cochran W.G. 6th ed. Iowa State Univ. Press; Ames, IA: 1967. Statistical Methods. [Google Scholar]
- 25.Magrine I.C.O., Ferrari S.C., Souza G.F., Minamihara L., Kemmelmeier C., Bando E., Machinski M. Intake of aflatoxins through the consumption of peanut products in Brazil. Food Addit. Contam. Part B. 2011;4:99–105. doi: 10.1080/19393210.2011.561931. [DOI] [PubMed] [Google Scholar]
- 26.Jager A.V., Tedesco M.P., Souto P.C.M.C., Oliveira C.A.F. Assessment of aflatoxin intake in São Paulo, Brazil. Food Control. 2013;33:87–92. [Google Scholar]
- 27.Romero A.C., Ferreira T.R.B., Dias C.T.S., Calori-Domingues M.A., Gloria E.M. Occurrence of AFM1 in urine samples of a Brazilian population and association with food consumption. Food Control. 2010;21:554–558. [Google Scholar]
- 28.Jager A.V., Tonin F.G., Souto P.C.M.C., Privatti R.T., Oliveira C.A.F. Determination of urinary biomarkers for assessment of short-term human exposure to aflatoxins in São Paulo, Brazil. Toxins. 2014;6:1996–2007. doi: 10.3390/toxins6071996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jager A.V., Tonin F.G., Baptista G.Z., Souto P.C.M.C., Oliveira C.A.F. Assessment of aflatoxin exposure using serum and urinary biomarkers in São Paulo, Brazil: a pilot study. Int. J. Hyg. Environ. Health. 2016;219:294–300. doi: 10.1016/j.ijheh.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Jubert C., Mata J., Bench G., Dashwood R., Pereira C., Tracewell W., Turteltaub K., Williams D., Bailey G. Effects of chlorophyll and chlorophyllin on low-dose aflatoxin B1 pharmacokinetics in human volunteers. Cancer Prev. Res. (Phila.) 2001;2:1015–1022. doi: 10.1158/1940-6207.CAPR-09-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garner R.C., Dvorackova I., Tursi F. Immunoassay procedures to detect exposure to aflatoxin B1 and benzo(a)pyrene in animals and man at the DNA level. Int. Arch. Occup. Environ. Health. 1998;60:145–150. doi: 10.1007/BF00378689. [DOI] [PubMed] [Google Scholar]
- 32.Chu Y.J., Yang H.I., Wu H.C., Liu J., Wang L.Y., Lu S.N., Lee M.H., Jen C.L., You S.L., Santella R.M., Chen C.J. Aflatoxin B1 exposure increases the risk of cirrhosis and hepatocellular carcinoma in chronic hepatitis B virus carriers. Int. J. Cancer. 2017;141:711–720. doi: 10.1002/ijc.30782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neeff D.V., Ledoux D.R., Rottinghaus G.E., Bermudez A.J., Dakovic A., Murarolli R.A., Oliveira C.A.F. In vitro and in vivo efficacy of a hydrated sodium calcium aluminosilicate to bind and reduce aflatoxin residues in tissues of broiler chicks fed aflatoxin B1. Poultry Sci. 2012;92:131–137. doi: 10.3382/ps.2012-02510. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg R.A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Ito Y., Takeda T., Sakon M., Tsujimoto M., Monden M., Matsuura N. Expression of p57/Kip2 protein in hepatocellular carcinoma. Oncology. 2001;61:221–225. doi: 10.1159/000055378. [DOI] [PubMed] [Google Scholar]
- 36.Peng S.Y., Chou S.P., Hsu H.C. Association of downregulation of cyclin D1 and of overexpression of cyclin E with p53 mutation, high tumor grade and poor prognosis in hepatocellular carcinoma. J. Hepatol. 1998;29:281–289. doi: 10.1016/s0168-8278(98)80014-7. [DOI] [PubMed] [Google Scholar]
- 37.Jung Y.J., Lee K.H., Choi D.W., Han C.J., Jeong S.H., Kim K.C., Oh J.W., Park T.K., Kim C.M. Reciprocal expressions of cyclin E and cyclin D1 in hepatocellular carcinoma. Cancer Lett. 2001;168:57–63. doi: 10.1016/s0304-3835(01)00403-7. [DOI] [PubMed] [Google Scholar]
- 38.Ito Y., Matsuura N., Sakon M., Miyoshi E., Noda K., Takeda T., Umeshita K., Nagano H., Nakamori S., Dono K., Tsujimoto M., Nakahara M., Nakao K., Taniguchi N., Monden M. Expression and prognostic roles of the G1-S modulators in hepatocellular carcinoma: P27 independently predicts the recurrence. Hepatology. 1999;30:90–99. doi: 10.1002/hep.510300114. [DOI] [PubMed] [Google Scholar]
- 39.Fiorentino M., Altimari A., D’Errico A., Cukor B., Barozzi C., Loda M., Grigioni W.F. Acquired expression of p27 is a favorable prognostic indicator in patients with hepatocellular carcinoma. Clin. Cancer Res. 2000;6:3966–3972. [PubMed] [Google Scholar]
- 40.Ozturk M. Genetic aspects of hepatocellular carcinogenesis. Semin. Liver Dis. 1999;19:235–242. doi: 10.1055/s-2007-1007113. [DOI] [PubMed] [Google Scholar]
- 41.Tretiakova M.S., Shabani-Rad M.T., Guggisberg K., Hart J., Anders R.A., Gao Z.H. Genomic and immunophenotypical differences between hepatocellular carcinoma with and without cirrhosis. Histopathology. 2010;56:683–693. doi: 10.1111/j.1365-2559.2010.03554.x. [DOI] [PubMed] [Google Scholar]
- 42.Sánches-Quiles V., Santamaria E., Segura V., Sesma L., Rieto J., Corrales F.J. Prohibitin deficiency blocks proliferation and induces apoptosis in human hepatoma cells: molecular mechanisms and functional implications. Proteomics. 2010;10:1609–1620. doi: 10.1002/pmic.200900757. [DOI] [PubMed] [Google Scholar]
- 43.Wang S., Fusaro G., Padmanabhan J., Chellappan S.P. Prohibitin colocalizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 44.Theiss A.L., Vijay-Kumar M., Obertone T.S., Jones D.P., Hansen J.M., Gewirtz A.T., Merlin D., Sitamaran S.V. Prohibitin is a novel regulator of antioxidant response that attenuates colonic inflammation in mice. Gastroenterology. 2009;137:199–208. doi: 10.1053/j.gastro.2009.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porta L.M.D., Ramalho F.S., Oliveira C.A.F., Silva D.M., Augusto M.J., Ramalho L.N.Z. Comparison of p53 and prohibitin expression in the spectrum of hepatitis, cirrhosis, and hepatocellular carcinoma. Hepatoma Res. 2017;3:34–42. [Google Scholar]
- 46.Edamoto Y., Hara A., Biernat W., Terracciano L., Cathomas G., Riehle H.M., Matsuda M., Fujii H., Scoazec J.Y., Ohgaki H. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int. J. Cancer. 2003;106:334–341. doi: 10.1002/ijc.11254. [DOI] [PubMed] [Google Scholar]
- 47.Reed C.A., Mayhew C.N., McClendon A.K., Yang X., Witkiewicz A., Knudsen E.S. Rb has a critical role in mediating the in vivo checkpoint response, mitigating secondary DNA damage and suppressing liver tumorigenesis initiated by aflatoxin B1. Oncogene. 2009;28:4434–4443. doi: 10.1038/onc.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi L.N., Bai T., Chen Z.S., Wu F.X., Chen Y.Y., Xiang B., Peng T., Han Z.G., Li L.Q. The p53 mutation spectrum in hepatocellular carcinoma from Guangxi, China: role of chronic hepatitis B virus infection and aflatoxin B1 exposure. Liver Int. 2015;35:999–1009. doi: 10.1111/liv.12460. [DOI] [PubMed] [Google Scholar]
- 49.Li P., Cao Y., Li Y., Zhou L., Liu X., Geng M. Expression of Wnt-5a and β-catenin in primary hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:3190–3195. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.