Abstract
Liver cells express a cytosolic α-tocopherol transfer protein (αTTP) with high binding affinity for α-tocopherol (αT) and much lower affinities for the non-αT congeners. The role of αTTP in the intracellular distribution of the different vitamin E forms is currently unknown. We therefore investigated the intracellular localization of αT, γ-tocopherol (γT), α-tocotrienol (αT3), and γ-tocotrienol (γT3) in cultured hepatic cells with and without stable expression of αTTP. We first determined cellular uptake of the four congeners and found the methylation of the chromanol ring and saturation of the sidechain to be important factors, with tocotrienols being taken up more efficiently than tocopherols and the γ-congeners more than the α-congeners, irrespective of the expression of αTTP. This, however, could perhaps also be due to an observed higher stability of tocotrienols, compared to tocopherols, in culture media rather than a higher absorption. We then incubated HepG2 cells and αTTP-expressing HepG2 cells with αT, γT, αT3, or γT3, isolated organelle fractions by density gradient centrifugation, and determined the concentrations of the congeners in the subcellular fractions. All four congeners were primarily associated with the lysosomes, endoplasmic reticulum, and plasma membrane, whereas only αT correlated with mitochondria. Neither the chromanol ring methylation or sidechain saturation, nor the expression of αTTP were important factors for the intracellular distribution of vitamin E. In conclusion, αTTP does not appear to regulate the uptake and intracellular localization of different vitamin E congeners in cultured liver cells.
Abbreviations: αT, α-tocopherol; αT3, α-tocotrienol; γT, γ-tocopherol; γT3, γ-tocotrienol; αTTP, α-tocopherol transfer protein; AUC, are under the concentration-time curve; HepG2, human hepatoma cultured cell; HepG2-TTP, HepG2 cell line expressing the human TTP cDNA; HepG2-EV, HepG2 cell line transfected with the antibiotic resistance gene
Keywords: Endoplasmic reticulum, HepG2 liver cells, Intracellular localization, Lysosomes, Mitochondria, Plasma membrane, α-Tocopherol transfer protein, Tocotrienols, Trafficking, Vitamin E
Graphical abstract
Highlights
-
•
We studied how αTTP affects intracellular distribution of αT, γT, αT3, γT3 in HepG2 cells.
-
•
All congeners associated with lysosomes, endoplasmic reticulum and the plasma membrane.
-
•
Only αT significantly correlated with mitochondria.
-
•
Neither the chemical structure, nor αTTP were important for intracellular localization.
1. Introduction
Vitamin E, discovered in 1922 as an “unknown factor X” required for fertility in female rats [1], is now known to comprise eight structurally related lipid-soluble compounds composed of a saturated (tocopherols (T)) or threefold unsaturated (tocotrienols (T3)) 16-carbon sidechain bound to a chromanol ring; the Greek letters α, β, γ, or δ are used as prefixes to designate the number and positions of methyl groups attached to the chromanol ring [2], [3].
Upon oral intake, the lipid-soluble tocopherols and tocotrienols are incorporated into mixed micelles and absorbed in the small intestine, following the general path of dietary lipids. The extent of absorption and transport to the liver is nearly similar for all eight vitamin E congeners, but the liver then selectively releases α-tocopherol (αT) into the systemic circulation, while the non-αT congeners are preferentially metabolized to the sidechain shortened carboxyethyl hydroxychromanols via a cytochrome P450-dependent pathway [4], [5]. The selective retention of αT in the organism appears to be the result of an interaction of this catabolic pathway with the hepatic α-tocopherol transfer protein (αTTP) [6], a cytosolic protein that specifically binds αT [7], [8] and has much lower affinities for β-tocopherol (38%), γ-tocopherol (9%), and δ-tocopherol (2%) [9].
Our current understanding of the involvement of αTTP in intracellular trafficking of αT in the liver is as follows: αT enters hepatocytes by endocytosis and reaches the late endosomal compartment, from where it is transported to the plasma membrane and secreted with lipoproteins into the circulation. αTTP binds αT (and probably to a lesser extent the non-αT congeners) in the outer leaflet of the endosomal membrane and facilitates its transport to the plasma membrane, where the binding of resident phosphatidylinositol 4,5-bisphosphate induces a conformational change and results in the release of αT and its incorporation into the membrane [10], [11], [12]. αT then exits the cell involving the ATP-binding cassette transporter A1, is incorporated into lipoproteins and delivered to extrahepatic tissues. αTTP then translocates to the endosomal compartment to repeat the cycle [10].
In addition to lysosomal and plasma membranes, αT is also present in the endoplasmic reticulum, mitochondria, and peroxisomes [13], [14], [15], [16], [17]; organelles that are involved in its metabolism [14]. Although the role of αTTP in intracellular trafficking of αT is partly understood, its importance for the intracellular localization of the non-αT congeners in liver cells has not yet been studied. We therefore investigated the uptake and intracellular distribution of αT, γT, αT3, and γT3 in cultured hepatic cells as a function of the expression of αTTP. These four congeners were specifically chosen to allow conclusions regarding the importance of the methylation pattern and sidechain saturation for intracellular trafficking of vitamin E.
2. Materials and methods
2.1. Test compounds
RRR-α-tocopherol and RRR-γ-tocopherol (αT, ≥ 95%, CAS number 59-02-9, cat#KP5101; γT, ≥ 95%, CAS number 54–28-4, cat#KP5103) were from Calbiochem/Merck Millipore (Darmstadt, Germany), R-α-tocotrienol (αT3; ≥ 97% pure, CAS number 58864-81-6, #07205) was from Sigma-Aldrich (Taufkirchen, Germany) and R-γ-tocotrienol (γT3; ≥ 98% pure, extracted from vitamin E capsules as previously described [18]) was a kind gift from Professor Walter Vetter (Institute of Food Chemistry, University of Hohenheim, Germany).
αT (100 mmol/L), αT3 (50 mmol/L), γT (50 mmol/L) and γT3 (20 mmol/L) stock solutions were prepared in ethanol (Carl Roth, Karlsruhe, Germany). Substances were diluted in growth medium (see below) prior to experiments and ethanol concentrations did not exceed 0.1% (v/v).
2.2. HepG2 cell lines and cultivation
The authenticity of the human hepatoma cell line HepG2 and all transfected cell lines (see below) was confirmed by Multiplex Human Cell Line Authentication Test (Multiplexion; Immenstaad; Germany). Cells were cultivated in Dulbecco's Modified Eagle Medium (DMEM; Sigma-Aldrich, Taufkirchen, Germany) with 10% (v/v) foetal calf serum (FCS; Life Technologies Corporation, Darmstadt, Germany) and 1% (w/v) penicillin/streptomycin (P/S; Biochrom AG, Berlin, Germany). HepG2 cells expressing the human TTP cDNA (HepG2-TTP) were generated by transfection with a pcDNA3 vector containing the TTP cDNA using the FuGENE HD Transfection Reagent (Roche, Grenzach-Wyhlen, Germany) according to the manufacturer's protocol. The empty vector control HepG2 cell line (HepG2-EV) not expressing the human TTP cDNA, but transfected with the antibiotic resistance gene, was generated using the same method as for HepG2-TTP. Stable transfectants were selected with geneticin 1% (w/v) (G418; Biochrom AG, Berlin, Germany) and cells were cultivated in DMEM with 10% (v/v) FCS and 0.5% (w/v) G418. All cell lines were cultivated at 5% CO2, 37 °C, and used between passages 8 and 43.
2.3. Cell viability
Cell viability was measured using the neutral red uptake [19] and MTT assays [20] to determine the maximum concentrations of the test compounds to be used in the subsequent experiments. Cells were treated with the test substances (10–100 μmol/L), solvent control (ethanol 0.1% v/v), positive control (Triton X-100, 0.1%, v/v; Merck, Darmstadt, Germany), and culture medium controls. Incubation with up to 50 μmol/L αT or αT3, and up to 30 μmol/L γT or γT3 resulted in ≥ 90% viable cells (data not shown) and these concentrations were therefore used for the subsequent experiments.
2.4. Time dependent-cellular uptake experiment
Cells were seeded in 12-well plates at a density of 3 × 105 cells and incubated for 24 h to reach 50–60% confluence. Cells were then treated with the respective test compound and control substances for 0.5, 1, 2, 4, 6, 24, 48 and 72 h. αT and αT3 were tested at 50 μmol/L, and γT and γT3 at 30 μmol/L in cultured medium. Cells incubated with culture medium alone and culture medium containing the test compounds incubated without cells were used as negative controls and stability controls, respectively.
After each incubation time, supernatants were collected and cells detached, washed, and resuspended in 20 μL lysis buffer (150 mmol/L NaCl; 50 mmol/L Tris(hydroxymethyl)-aminomethan hydrochloride (Tris-HCl; Carl Roth, Karlsruhe, Germany), pH 8.0; 1% (v/v) Nonidet P-40 (NP-40; Roche, Mannheim, Germany); 4% protease inhibitor cocktail (Roche, Mannheim, Germany)). After 20 min incubation at 4 °C, lysed cells were sonicated for 1 min and centrifuged. Four microliter lysed cell suspension were used for protein determination via Bradford assay [21]. The remaining cell lysis suspension and supernatants were stored at − 80 °C for vitamin E determination.
To determine the stability of the test compounds in the two culture media used in the experiments, 50 μmol/L of each of the four compounds were added to DMEM with penicillium/streptomycin 25 and DMEM with geneticin, respectively, and incubated at 25 and 37 °C, respectively, for up to 0.5 h in the absence of cells. Samples (n = 6) for vitamin E quantification were collected at 0 and 0.5 h.
2.5. Subcellular fractionation by density gradient centrifugation
An optimized density gradient centrifugation method was developed based on previously described protocols [22], [23]. Confluent cells were sub-cultivated in T75 flasks at a ratio of 1:2 and incubated to reach confluence. Cells were then incubated with culture medium containing the test compounds (αT and αT3, 50 μmol/L; γT and γT3, 30 μmol/L) during 24 h. Afterwards cells were washed twice with 10 mL PBS, detached with 1 mL trypsin/EDTA after 10 min incubation at 37 °C and re-suspended in 10 mL culture medium. Cells were counted with a CASY cell counter (Innovatis, Darmstadt, Germany) and 1 × 108 cells were centrifuged (163×g, 4 °C, 5 min). Cells were washed with 5 mL PBS and 5 mL homogenization buffer (0.25 mol/L sucrose (Carl Roth, Karlsruhe, Germany), 1 mmol/L EDTA, 0.1% (v/v) ethanol, 10 mmol/L morpholinopropane sulfonic acid (Mops; Serva Biochemica, Heidelberg, Germany) in double distilled water (H2Odd), pH adjusted to 7.4 with 1 mol/L sodium hydroxide (Carl Roth, Karlsruhe, Germany); 4% protease inhibitor cocktail (Roche, Mannheim, Germany)), with centrifugation in between. Cells were re-suspended in 3 mL homogenization buffer, transferred to Miltenyi tubes and disrupted with a tissue dissociator for 1 min (Miltenyi, Bergisch Gladbach, Germany). Cell breakage of 90% was monitored via trypan blue staining (50 μL PBS, 10 μL trypan blue 0.4% (v/v)) (Serva Biochemica, Heidelberg, Germany). The cell suspension was then centrifuged (1000×g, 4 °C, 10 min, without brake), the supernatant transferred, cells re-suspended in 4 mL homogenization buffer, centrifuged, and supernatants were combined. The cell pellet was discarded and post-nuclear supernatants homogenized with a dounce homogenizer using three strokes with a loose-fitting pestle. Post-nuclear supernatants were mixed with Optiprep™ working solution 50% (v/v), iodixanol (83.3% (v/v) OptiPrep™ (Axis-Shield PoC AS, Oslo, Norway), 16.7% (v/v) dilution medium (0.25 mol/L sucrose, 6 mmol/L EDTA, 0.6% (v/v) ethanol, 60 mmol/L Mops in H2Odd, pH adjusted to 7.4 with 1 mol/L NaOH)) for a final concentration of 24% iodixanol in 11 mL total volume. The solution was transferred to a centrifugation tube (OptiSeal™ tubes; Beckman Coulter Inc., Fullerton, CA, USA), overlaid with homogenization buffer and ultra-centrifuged (318,600×g, 4 °C, 2 h, without brake from 773×g on; Optima L-80 XP Ultracentrifuge with VTi 65.1 rotor, Beckman Coulter Inc., Fullerton, CA, USA). The generated gradient was unloaded carefully in 0.95 mL fractions, dense-end first. Three aliquots of 30 μL of each fraction were stored at − 80 °C for further Western blot analyses. The density of each fraction was measured in °Brix with a refractometer. Samples were stored at − 80 °C until vitamin E determination.
2.6. Western blot analysis
Fractions (30 μL) were mixed with 10 μL loading buffer (4×SDS protein sample buffer: 250 mmol/L Tris-HCl (pH 6.8), 8% (w/v) SDS, 40% (v/v) glycerol (Sigma-Aldrich), 0.03% (v/v) bromphenol blue (Serva Biochemica, Heidelberg, Germany), 20% (v/v) beta-mercaptoethanol (Merck)), and proteins were separated by 8% and 15% SDS gel electrophoresis and transferred to polyvinylidenefluoride membranes, blocked for 1 h at room temperature in blocking buffer (5% bovine serum albumin (BSA; Sigma-Aldrich) in tris-buffered saline Tween-20 (TBST: 0.8% (w/v) NaCl, 0.24% (w/v) Tris-HCl (pH 7.6), 0.05% (v/v) Tween 20 in H2Odd; Sigma-Aldrich)) and incubated with the primary antibodies (Abcam, Cambridge, UK). LAMP1 (lysosomes-associated membrane glycoprotein 1, 1:1000, ab24170), Na+-K+-ATPase (1:20000, ab76020) and calnexin (1:1000, ab22595) were incubated with the membrane from the 8% SDS gel, and catalase (1:10000, ab76024) and COX IV (cyclooxygenase IV, 1:1000, ab33985) with the membrane from the 15% SDS gel. The primary antibodies were diluted in 5% BSA in TBST and incubated for 1 h at room temperature or overnight at 4 °C (for catalase). Membranes were washed in TBST and incubated for 1 h at room temperature with the secondary antibodies (rabbit anti-mouse peroxidase conjugated for COX IV (1:10000, cat#402335); and goat anti-rabbit peroxidase conjugated for the others (1:10000, cat#401353); Calbiochem/Merck Millipore). Membranes were washed with TBST and bands were visualized using AceGlow™ Essential chemiluminescence solutions A and B (Peqlab Biotechnologie GmbH, Erlangen, Germany) and WesternBright™ Sirius; WesternBright™ Peroxide (Advansta, Menlo Park, CA, USA). Intensities were recorded on a Fusion FX and processed using the FusionCapt Advance software (Vilber Lourmat, Eberhardzell, Germany).
To verify the αTTP expression, 20 μL aliquots of cell suspensions of HepG2, HepG2-TTP and HepG2-EV, adjusted to 40 μg protein, were separated by 10% SDS gel, blotted and imaged as described using α-TTP (1:1000, ab155323) and goat anti-rabbit peroxidase conjugated as primary and secondary antibodies, respectively. The housekeeping protein β-actin was used as loading control, using β-actin (1:1000, Cell signaling Technology, Danvers, MA, USA) and goat anti-rabbit peroxidase conjugated as primary and secondary antibodies, respectively.
To correlate the association between the αTTP expression and the organelle markers in the subcellular fractions, HepG2-TTP cells incubated for 72 h were treated as described in Section 2.5 and the expression of αTTP and the different organelle markers was analyzed in the isolated subcellular fractions by Western blotting, as described above.
2.7. HPLC analysis of vitamin E congeners
All chemicals used were of highest purity and purchased from Sigma-Aldrich or Merck. Methanol was HPLC grade and all water used was deionized and filtered (Millipore, Billerica, MA, USA). αT, αT3, γT and γT3 in lysed cells and supernatants from uptake assays, and in subcellular fractions were extracted and quantified as previously described [24]. Briefly, for lysed cells and fractions, the complete cells and 800 μL samples were used, respectively, and the saponification was performed, followed by a liquid-liquid extraction with hexane. Supernatants were processed using 600 μL sample without saponification using the same extraction procedure. For all cases, the hexane extraction was performed twice, first with 2 mL n-hexane and transfer of 1 mL organic layer, and then with 1 mL n-hexane and a transfer of 1 mL organic layer. Prior to HPLC analysis, extracts were re-suspended in 100 μL methanol/water (85:15, v/v) and transferred to amber HPLC vials.
Twenty microliter sample was injected into a Jasco HPLC (system controller LC-Net II/ADC, two pumps X-LC™ 3185PU, mixing unit X-LC™ 3180MX, degasser X-LC™ 3080DG, autoinjector X-LC™ 3159AS, column oven X-LC™ 3067CO, fluorescence detector FP-2020 Plus; Jasco, Germany). Test compounds were separated on a Phenomenex Kinetex™ PFP column (2.6 µm particle size, 150 × 4.6 mm) maintained at 40 °C, using methanol/water (85:15, v/v) at a flow rate of 1.7 mL/min, for a total run time of 15 min. The fluorescence detector was operated at excitation/emission wavelengths of 296/325 nm, peaks were recorded and integrated using Chrompass software (version 1.9. 302.1124, Jasco), and quantified against external standard curves using the authentic compounds (Sigma-Aldrich, minimum ≥ 95.5% pure).
2.8. Statistical analyses
Cellular uptake experiments were performed in biological triplicates (n = 3), each consisting of three technical replicates, and subcellular fractionation experiments in biological triplicates (n = 3), each consisting of one experiment. Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Areas under the concentration-time curve (AUC) were calculated for the cellular uptake experiments and two-way ANOVA was used to test for effects of congener, cell type and congener × cell type interactions. A Bonferroni post-hoc test was used to calculate significant differences between groups.
Stability tests for αT, γT, αT3, and γT3 in culture media were performed in six replicates (n = 6) and significant differences assessed by two-way ANOVA with congener and time as factors.
Non-parametric Spearman's rank order correlation tests were performed to determine significant correlation between the percentage of test compounds or band intensity of the αTTP expression and band intensity of cell organelle markers in the subcellular fractions. Differences and correlations were considered significant at P < 0.05.
3. Results and discussion
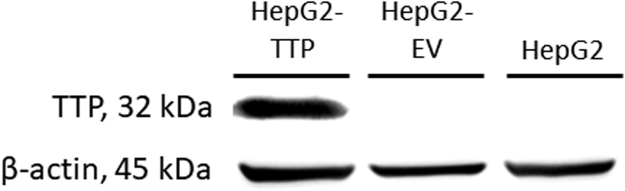
Although the human hepatoma cell line HepG2 is commonly used to investigate vitamin E uptake, trafficking and metabolism, the fact that it does not express αTTP brings about certain limitations to its predictive power for these processes in vivo. We therefore transfected HepG2 cells with a pcDNA3 vector containing the αTTP cDNA to generate HepG2 clones stably expressing αTTP (HepG2-TTP) or an empty vector (HepG2-EV; Fig. 1) [25], which allowed us to study the potential role of αTTP in the cellular uptake and intracellular distribution of αT, γT, αT3, and γT3 and to determine potential structure-dependent differences between these four congeners.
Fig. 1.
Representative Western blots of α-tocopherol transfer protein expression in HepG2, empty vector-control HepG2-EV, and α-tocopherol transfer protein-transfected HepG2-TTP cells.
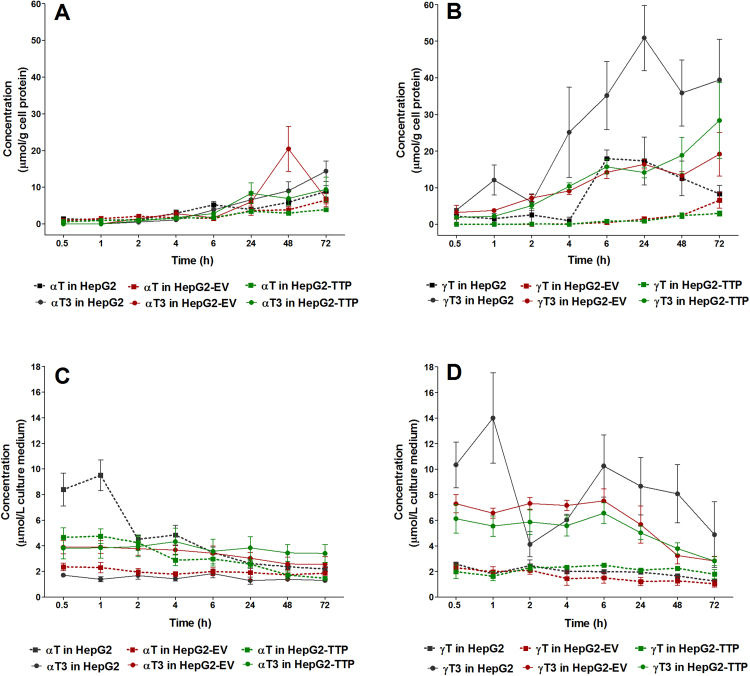
We first investigated the time-dependency of the uptake of αT, γT, αT3, and γT3 into the three hepatic cell lines by monitoring their concentrations in the culture medium and cell pellets for 72 h. We found a steady uptake into the cells over time (Fig. 2A and B), which was mirrored by decreasing concentrations in the incubation medium (Fig. 2C and D), albeit to somewhat different extents between the four compounds (see also Supplementary Tables 1 and 2). Sidechain saturation apparently affected cellular uptake, with tocotrienol (T3) concentrations being somewhat higher than those of the corresponding tocopherols (T). The differences were small within the first 4–6 h and became more apparent from 24 to 72 h.
Fig. 2.
Time course of the mean concentrations (error bars represent standard error of the mean; n = 3) of αT, γT (squares, dotted lines), αT3, and γT3 (circles, solid lines) in cell lysates (A, B) and cell culture medium (C, D) of HepG2, HepG2-EV (empty vector control) and αTTP-expressing HepG2-TTP cells incubated with 50 μmol/L of αT or αT3 or 30 μmol/L γT or γT3 for up to 72 h.
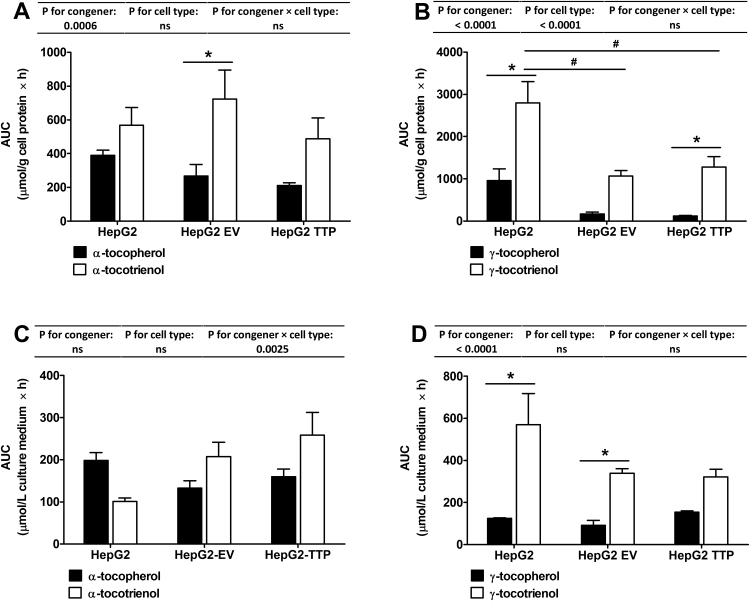
The observed higher uptake of T3 compared to T is in agreement with previous observations in hepatocytes [4], in human T-leukemia cells [16], in primary cortical neuron cells [26] and in human lung carcinoma cells [27], and has been attributed to the higher intermembrane mobility of the T3 [16]. The absence (HepG2-EV) or presence (HepG2-TTP) of αTTP in the cells did not significantly affect intracellular or medium concentrations of the congeners (Fig. 3A–D).
Fig. 3.
Area under the concentration-time curve (AUC) (error bars represent standard error of the mean; n = 3) of αT, γT, αT3 and γT3 in cell lysates (A, B) and cell culture medium (C, D) of HepG2, HepG2-EV (empty vector control) and αTTP-expressing HepG2-TTP cells incubated with 50 μmol/L of αT or αT3 or 30 μmol/L γT or γT3 for up to 72 h. AUC was calculated with GraphPad Prism 5. Two-way ANOVA with Bonferroni post-hoc test were calculated to detect significant differences (P < 0.05). P-values for the effects of congener, cell type and congener x cell type interactions are reported above each graph. Significant differences between congeners are marked with asterisks (*) and between cell types with the number symbol (#).
The better cellular uptake of the tocotrienols compared to their corresponding tocopherol congeners in all cell types was also confirmed by comparing the area under the concentration-time curves (AUC), which were higher for T3 than T (Fig. 3A and B). This was, however, not accompanied by a faster decrease in tocotrienol concentrations in the culture media. On the contrary, AUC in culture media were also higher for tocotrienols (Fig. 3C and D), which may suggest potentially lower stability of tocopherols in culture media rather than a better cellular uptake.
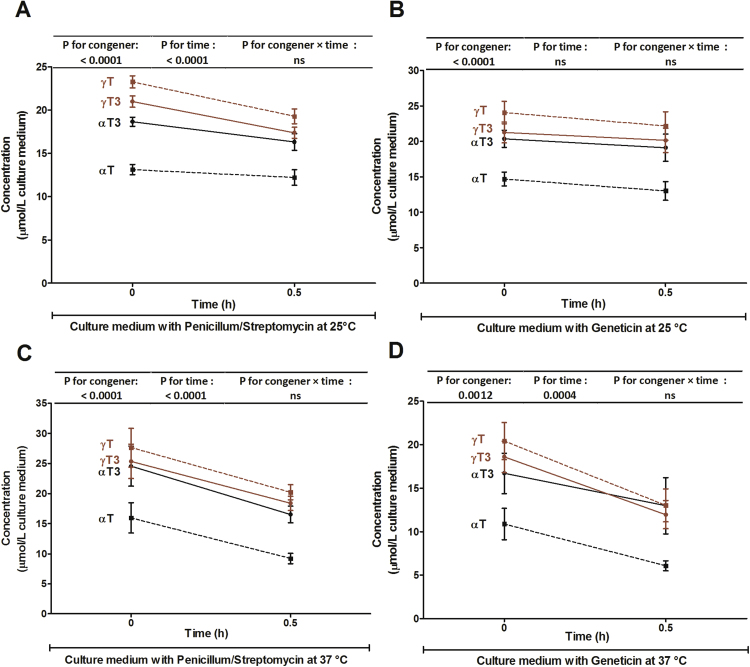
We therefore tested the recovery of all four congeners from cell culture media in the absence of cells at 0 and 0.5 h. Directly upon addition of all four congeners to cell culture media, significant losses in the order of αT > αT3 > γT3 > γT were evident and further declines, which were of similar extent for all congeners, were observed after 30 min (Fig. 4). This raises the question if the higher cellular uptake of tocotrienols observed here (Fig. 2) and previously [4], [16], [26], [27] may in fact be a result of the lower stability and resulting greater loss of tocopherols, in particular αT, which often served as the reference tocopherol, in the culture media. In agreement, considerable losses of αT and γT in cultured macrophages and culture medium were reported and had been attributed to catabolism and/or in vitro oxidation during incubation [28]. The notion of lower oxidative stability of αT is consistent with the reduction-oxidation potentials and antioxidant activities of tocopherols and tocotrienols, which are higher for αT than for γT [29], [30]. αT3, however, depending on the test system, is a similarly potent (in homogenous solutions) or even more potent (in membrane systems) antioxidant than αT and oxidative losses would therefore be expected to be similar. In support of the hypothesis that oxidative instability may at least partly explain the differences in the recovery from culture media, the recovery of the more reactive α-congeners was lower than that of the less reactive γ-congeners (Fig. 4). Another explanation could be that the congeners bind to serum proteins in culture media, perhaps with different affinity, and thereby escape detection. Although the reason for the observed loss of vitamin E upon addition to culture medium is not known and warrants further investigation, the subsequent intracellular distribution experiments were based on a qualitative, rather than a quantitative comparison between the congeners and are thus not affected by differences in the concentrations of the congeners in culture medium.
Fig. 4.
Time course of the mean concentrations (error bars represent standard error of the mean; n = 6) of αT (black dotted lines), γT (brown dotted lines), αT3 (black solid lines) and γT3 (brown solid lines) in two cell culture media and two temperatures [DMEM/ 10% FCS/1% penicillum/streptomycin at 25 °C (A) and 37 °C (C) and DMEM/ 10% FCS/0.5% geneticin at 25 °C (B) and 37 °C (D)] incubated with 50 μmol/L of αT, γT, αT3 and γT3 for up to 0.5 h. Two-way ANOVA were calculated to detect significant differences (P < 0.05). P-values for the effects of congener, time and congener x time interactions are reported above each graph.
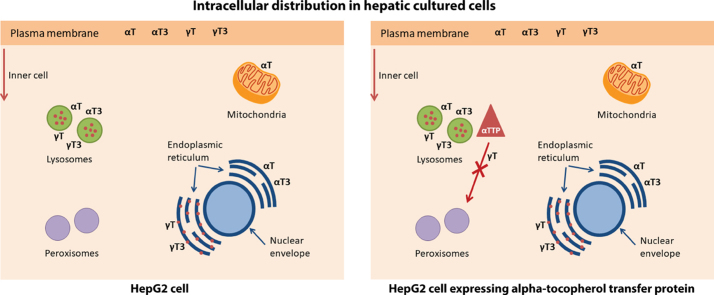
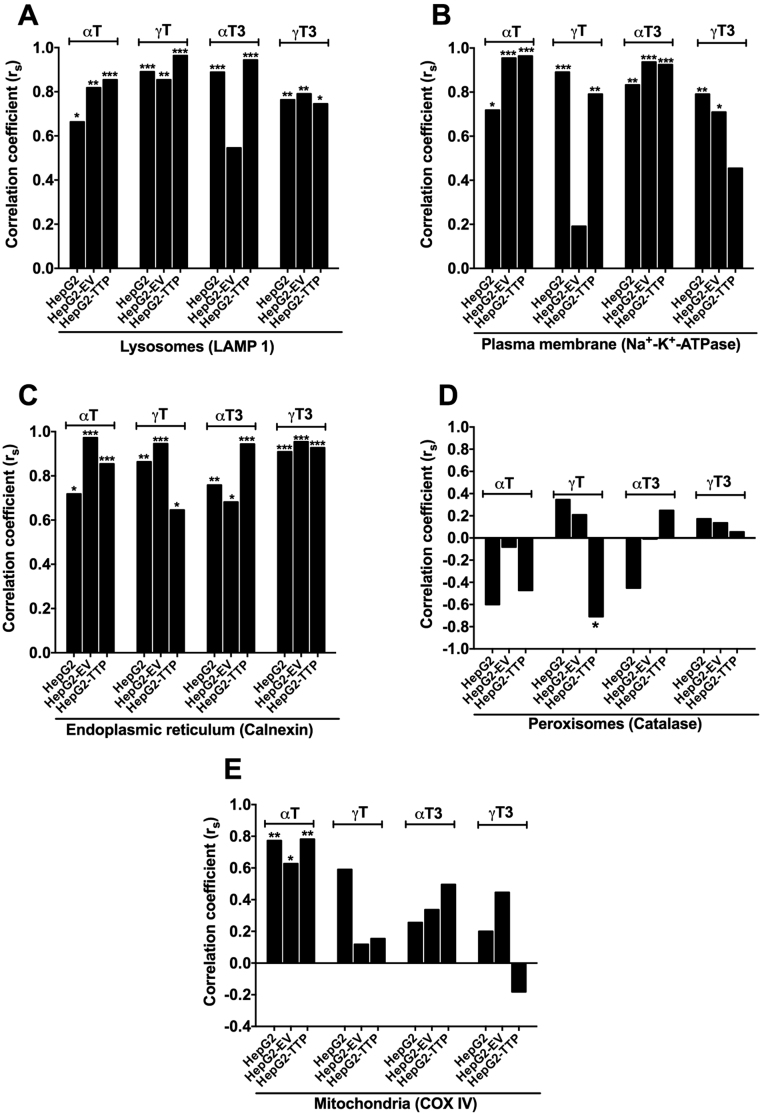
Based on the uptake experiments, an incubation time of 24 h was chosen for the intracellular distribution experiments to ensure sufficient intracellular accumulation of the test compounds, while limiting their enzymatic degradation to their respective short-chain metabolites, which becomes more pronounced after 48 h (data not shown). We employed a density gradient centrifugation technique for the separation into eleven fractions representing five different organelles, which were identified using organelle-specific marker proteins (Supplementary Fig. 1). All four congeners accumulated mainly in fractions 9–11 (Fig. 5). Because organelles overlapped over several fractions, that is each marker protein was present in more than one fraction (Supplementary Fig. 1 and Supplementary Tables 3–8), a non-parametric Spearman's correlation test was computed to identify the organelles associated with high concentrations of αT, γT, αT3, or γT3. The concentrations of all four vitamin E forms correlated mainly with the organelle markers for the endoplasmic reticulum, plasma membrane, and lysosomes, but not with those for the peroxisomes (Fig. 6, Supplementary Tables 9 and 10), which is in line with the current understanding of the intracellular trafficking of αT (see introduction and literature cited there) and data from αT-injected rats, for which ca. 75% of hepatic αT was found in the microsomal (endoplasmic reticulum) fraction [14], wild type and NPC1 mice, for which the highest concentrations of αT in the liver were found in lysosomal membranes [17], as well as data from rat pheochromocytoma PC12 and human T-leukemia cells, which also had most αT present in the microsomal fractions [15], [16]. Only αT, but not γT, αT3, or γT3 concentrations significantly correlated with the mitochondria marker intensity in all cell lines (Fig. 6E), irrespective of αTTP expression, suggesting that αTTP is not involved in mitochondrial targeting. The mitochondrial localization of αT observed here is in agreement with observations in rats fed respectively injected with αT [13], [14], in wild type and NPC1 mice fed with αT [17], and PC12 cells [15]. In human T-leukemia cells, on the other hand, both αT3 and αT were present in mitochondria [16]. The lack of significant correlations of γT, αT3, and γT3 concentrations with the mitochondria and of all four congeners with peroxisomes (Fig. 6D and E) agrees with the function of these organelles in the metabolism of vitamin E in general and the preferential metabolism of the non-αT congeners in particular. The initial ω-hydroxylation of the parent vitamin E occurs in the endoplasmic reticulum, this alcohol metabolite is then ω-oxidized in the peroxisomes to yield the 13′-carboxychromanol forms, which are then β-oxidized in the mitochondria to ultimately yield the sidechain-shortened carboxyethyl hydroxychromanol metabolites (reviewed in [3], [31]). Hence, the metabolites rather than the parent compounds are expected to localize in these organelles.
Fig. 5.
Mean percentage (n = 3) of αT (A), γT (B), αT3 (C), and γT3 (D) in eleven fractions separated by density gradient centrifugation prepared from HepG2, HepG2-EV (empty vector control) and αTTP-expressing HepG2-TTP cells incubated with 50 μmol/L αT or αT3 or 30 μmol/L γT or γT3 for 24 h.
Fig. 6.
Spearman's correlation coefficients (rs) for the correlation between the percentage of αT, γT, αT3, and γT3 and the band intensities of the cell organelle markers for the lysosomes (LAMP1, A), plasma membrane (Na+-K+-ATPase, B), endoplasmic reticulum (calnexin, C), peroxisomes (catalase, D) and mitochondria (COX IV, E) in HepG2, HepG2-EV (empty vector control) and αTTP-expressing HepG2-TTP cells incubated with 50 μmol/L of αT or αT3 or 30 μmol/L γT or γT3 for 24 h. Significant correlations are indicated by asterisks; *P < 0.05; **P < 0.01; ***P < 0.001.
The comparison of αTTP-expressing (HepG2-TTP) with the non-expressing HepG2 and HepG2-EV cells revealed only a minor impact of the protein on the intracellular localization of αT, γT, αT3, and γT3 (Fig. 6). The most pronounced and significant difference was observed for the localization of γT in peroxisomes. In the absence of αTTP (HepG2 and HepG2-EV cells), γT was not correlated with the peroxisomal marker, when αTTP was expressed (HepG2-TTP), a significant negative correlation was observed (Fig. 6D), suggesting that the protein prevented the transport of γT to peroxisomes, where it is metabolized. This observation gives further support to our previous findings in these cells that γT metabolism to γ-carboxyethyl hydroxychromanol is reduced in cells with a moderate expression of αTTP and even more strongly in cells with a high expression of αTTP [6].
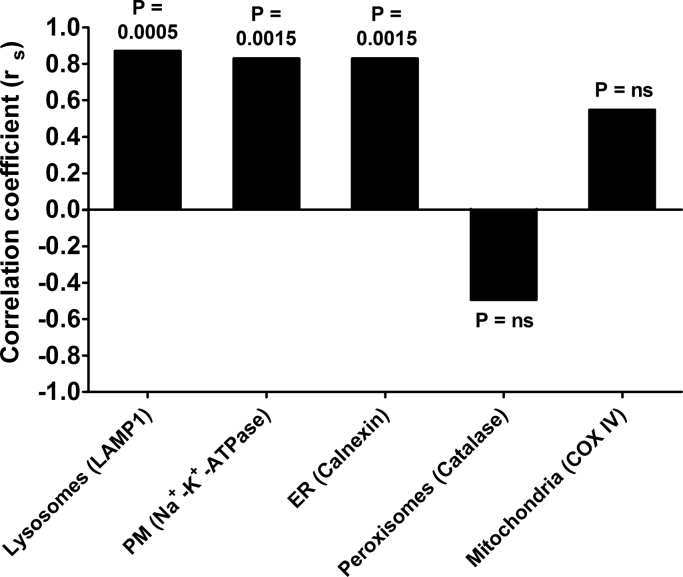
In order to better understand if αTTP may indeed interact with the organelles involved in vitamin E catabolism, we computed a non-parametric Spearman's correlation test to identify the organelles associated with the expression of αTTP. αTTP expression correlated mainly with the organelle markers for the endoplasmic reticulum, plasma membrane, and lysosomes (Fig. 7), The association with the lysosomes and plasma membrane is in line with the current understanding of the intracellular localization and trafficking of αTTP in hepatocytes (see introduction and literature cited there). The correlation of αTTP with the endoplasmic reticulum, the organelle in which the first and rate-limiting step in vitamin E catabolism takes place [3], is in agreement with our previous findings that αTTP expression reduces the metabolism of γT [6].
Fig. 7.
Spearman's correlation coefficients (rs) for the correlation between the band intensity of the αTTP expression and the band intensities of the cell organelle markers for the lysosomes (LAMP1), plasma membrane (Na+-K+-ATPase), endoplasmic reticulum (calnexin), peroxisomes (catalase) and mitochondria (COX IV) in αTTP-expressing HepG2-TTP cells incubated for 72 h. Correlations are significant at P < 0.05. P-values for the correlations are reported above each bar.
To the best of our knowledge, we are the first to directly compare the intracellular localization of the four vitamin E congeners αT, γT, αT3 and γT3. Our observation that all four congeners, irrespective of their methylation pattern and sidechain saturation, are primarily associated with the lysosomal compartment, endoplasmic reticulum, and the plasma membrane (Figs. 6C- 4E), suggests that only small, if any, differences in intracellular trafficking exist between αT, γT, αT3 and γT3 in cultured liver cells. What is more, the expression of αTTP in the liver cells did not bring about any major shifts in the intracellular distribution of the four T and T3, indicating that the protein, despite its preferential binding of αT [9], does not determine the intracellular localization of vitamin E. Because all four congeners were mainly present in lysosomes, endoplasmic reticulum, and the plasma membrane and because αTTP did not significantly affect this localization, there must be other processes, perhaps simple passive diffusion, involved in the trafficking of vitamin E within liver cells. This is further supported by previous observations that vitamin E accumulates mainly in organelles with a high lipid content [15], [16].
4. Conclusions
While in the present experiment and in agreement with published literature (see discussion above) γ-configuration of the chromanol ring and unsaturation of the sidechain promoted the uptake of tocochromanols into cultured hepatocytes, it cannot be ruled out that this is caused by the observed instability of tocopherols in cell culture media rather than differences in cellular uptake. The expression of αTTP did not affect the incorporation of T and T3 into HepG2 cells. Based on our data, ring methylation and sidechain saturation of vitamin E also do not appear to be major determinants of its intracellular localization, with the exception of mitochondria, for which a significant correlation was only found for αT. αTTP expression in liver cells did not substantially influence the intracellular distribution of αT, γT, αT3 and γT3.
In summary, our findings suggest that neither the methylation pattern of the chromanol ring or the sidechain saturation of vitamin E congeners, nor the cytosolic αTTP are major factors controlling the intracellular localization of vitamin E in cultured liver cells. This may be indicative of passive processes, such as diffusion, as main driving forces of the distribution of vitamin E forms within the cell.
Acknowledgments
This work was partially funded by the foundation Fiat Panis through a Dr. Hermann Eiselen Ph.D. grant within the framework of the Ph.D. Program “Global Food Security”, which is organized by the Food Security Center of University of Hohenheim.
Andrea Irías-Mata acknowledges a scholarship provided by the Food Security Center of the University of Hohenheim, which is supported by the German Academic Exchange Service (DAAD) with funds of the Federal Ministry of Economic Cooperation and Development (BMZ) of Germany.
Acknowledgments
Conflict of interest
The authors declare no competing financial interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.07.027.
Appendix A. Supplementary material
Supplementary material
References
- 1.Evans H.M., Bishop K.S. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science. 1922;56:650–651. doi: 10.1126/science.56.1458.650. [DOI] [PubMed] [Google Scholar]
- 2.Frank J., Chin X.W.D., Schrader C., Eckert G.P., Rimbach G. Do tocotrienols have potential as neuroprotective dietary factors? Ageing Res. Rev. 2012;11(1):163–180. doi: 10.1016/j.arr.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Galli F., Azzi A., Birringer M., Cook-Mills J.M., Eggersdorfer M., Frank J., Cruciani G., Lorkowski S., Özer N.K. Vitamin E: emerging aspects and new directions. Free Radic. Biol. Med. 2017;102:16–36. doi: 10.1016/j.freeradbiomed.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Sontag T.J., Parker R.S. Influence of major structural features of tocopherols and tocotrienols on their ω-oxidation by tocopherol ω-hydroxylase. J. Lipid Res. 2007;48:1090–1098. doi: 10.1194/jlr.M600514-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Sontag T.J., Parker R.S. Cytochrome P450 -hydroxylase pathway of tocopherol catabolism. J. Biol. Chem. 2002;277(28):25290–25296. doi: 10.1074/jbc.M201466200. [DOI] [PubMed] [Google Scholar]
- 6.Grebenstein N., Schumacher M., Graeve L., Frank J. α-Tocopherol transfer protein is not required for the discrimination against γ-tocopherol in vivo but protects it from side-chain degradation in vitro. Mol. Nutr. Food Res. 2014;58:1052–1060. doi: 10.1002/mnfr.201300756. [DOI] [PubMed] [Google Scholar]
- 7.Meier R., Tomizaki T., Schulze-Briese C., Baumann U., Stocker A. The molecular basis of Vitamin E retention: structure of human α-tocopherol transfer protein. J. Mol. Biol. 2003;331:725–734. doi: 10.1016/s0022-2836(03)00724-1. [DOI] [PubMed] [Google Scholar]
- 8.Stocker A. Molecular mechanisms of Vitamin E transport. Annu. N.Y. Acad. Sci. 2004;1031:44–59. doi: 10.1196/annals.1331.005. [DOI] [PubMed] [Google Scholar]
- 9.Hosomi A., Arita M., Sato Y., Kiyose C., Ueda T., Igarashi O., Arai H., Inoue K. Affinity for alpha-tocopherol transfer protein as a determinant of the biological activities of vitamin E analogs. FEBS Lett. 1997;409:105–108. doi: 10.1016/s0014-5793(97)00499-7. [DOI] [PubMed] [Google Scholar]
- 10.Chung S., Ghelfi M., Atkinson J., Parker R., Qian J., Carlin C., Manor D. Vitamin E and phosphoinositides regulate the intracellular localization of the hepatic α-tocopherol transfer protein. J. Biol. Chem. 2016;291(33):17028–17039. doi: 10.1074/jbc.M116.734210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian J., Morley S., Wilson K., Nava P., Atkinson J., Manor D. Intracellular trafficking of vitamin E in hepatocytes: role of tocopherol transfer protein. J. Lipid Res. 2005;46:2072–2082. doi: 10.1194/jlr.M500143-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Horiguchi M., Arita M., Kaempf-Rotzol D.E., Tsujimoto M., Inoue K., Arai H. pH-dependent translocation of α-tocopherol transfer protein (α-TTP) between hepatic cytosol and late endosomes. Genes Cells. 2003;8:789–800. doi: 10.1046/j.1365-2443.2003.00676.x. [DOI] [PubMed] [Google Scholar]
- 13.Buttriss J.L., Diplock A.T. The alpha-tocopherol and phospholipid fatty acid content of rat liver subcellular membranes in vitamin E and selenium deficiency. Biochim. Biophys. Acta. 1988;963(1):61–69. doi: 10.1016/0005-2760(88)90338-4. [DOI] [PubMed] [Google Scholar]
- 14.Mustacich D.J., Leonard S.W., Patel N.K., Traber M.G. α-tocopherol β-oxidation localized to rat liver mitochondria. Free Radic. Biol. Med. 2010;48(1):73–81. doi: 10.1016/j.freeradbiomed.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito Y., Fukuhara A., Nishio K., Hayakawa M., Ogawa Y., Sakamoto H., Fujii K., Yoshida Y., Niki E. Characterization of cellular uptake and distribution of coenzyme Q10 and vitamin E in PC12 cells. J. Nutr. Biochem. 2009;20:350–357. doi: 10.1016/j.jnutbio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Saito Y., Yoshida Y., Nishio K., Hayakawa M., Niki E. Characterization of cellular uptake and distribution of vitamin E. Annu. N.Y. Acad. Sci. 2004;1031:368–375. doi: 10.1196/annals.1331.047. [DOI] [PubMed] [Google Scholar]
- 17.Yévenes L.F., Klein A., Castro J.F., Marín T., Leal N., Leighton F., Alvarez A.R., Zanlungo S. Lysosomal vitamin E accumulation in Niemann–Pick type C disease. Biochim. Biophys. Acta. 2012;1822:150–160. doi: 10.1016/j.bbadis.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Vetter W., Hammann S., Müller M., Englert M., Huang Y. The use of countercurrent chromatography in the separation ofnonpolar lipid compounds. J. Chromatogr. A. 2017;1501:51–60. doi: 10.1016/j.chroma.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 19.Repetto G., del Peso A., Zurita J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008;3(7):1125–1131. doi: 10.1038/nprot.2008.75. [DOI] [PubMed] [Google Scholar]
- 20.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein– dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Axis-Shield, OptiPrep TM Application Sheet S13. 〈http://www.axis-shield-density-gradient-media.com/S13.pdf〉. (Accessed 5 September 2017).
- 23.Axis-Shield, OptiPrep TM Application Sheet S16. 〈http://www.axis-shield-density-gradient-media.com/S16.pdf〉. (Accessed 5 September 2017).
- 24.Grebenstein N., Frank J. Rapid baseline-separation of all eight tocopherols and tocotrienols by reversed-phase liquid-chromatography with a solid-core pentafluorophenyl column and their sensitive quantification in plasma and liver. J. Chromatogr. A. 2012;1243:39–46. doi: 10.1016/j.chroma.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 25.Qu Y.-H., Fu J.-C., Liu K., Zuo Z.-Y., Jia H.-N., Ma Y., Luo H.-L. Screening of α-tocopherol transfer protein sensitive genes in human hepatoma cells (HepG2) Int. J. Mol. Sci. 2016;17(1016):1–12. doi: 10.3390/ijms17071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito Y., Nishio K., Akazama Y.O., Yamanaka K., Miyama A., Yoshida Y., Noguchi N., Niki E. Cytoprotective effects of vitamin E homologues against glutamate-induced cell death in immature primary cortical neuron cultures: tocopherols and tocotrienols exert similar effects by antioxidant function. Free Radic. Biol. Med. 2010;49:1542–1549. doi: 10.1016/j.freeradbiomed.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Nishio K., Horie M., Akazawa Y., Shichiri M., Iwahashi H., Hagihara Y., Yoshida Y., Niki E. Attenuation of lipopolysaccharide (LPS)-induced cytotoxicity by tocopherols and tocotrienols. Redox Biol. 2013;1:97–103. doi: 10.1016/j.redox.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao R., Stone W.L., Huang T., Papas A.M., Qui M. The uptake of tocopherols by RAW 264.7 macrophages. Nutr. J. 2012;1(2) doi: 10.1186/1475-2891-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamal-Eldin A., Appelqvist L. The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids. 1996;31(7):671–701. doi: 10.1007/BF02522884. [DOI] [PubMed] [Google Scholar]
- 30.Müller L., Theile K., Böhm V. In vitro antioxidant activity of tocopherols and tocotrienols and comparison of vitamin E concentration and lipophilic antioxidant capacity in human plasma. Mol. Nutr. Food Res. 2010;54(5):731–742. doi: 10.1002/mnfr.200900399. [DOI] [PubMed] [Google Scholar]
- 31.Schmölz L., Birringer M., Lorkowski S., Wallert M. Complexity of vitamin E metabolism. World J. Biol. Chem. 2016;7(1):14–43. doi: 10.4331/wjbc.v7.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material