Abstract
Delivery of genes to mouse liver is routinely accomplished by tail-vein injections of viral vectors or naked plasmid DNA. While viral vectors are typically injected in a low-pressure and -volume fashion, uptake of naked plasmid DNA to hepatocytes is facilitated by high pressure and volumes, also known as hydrodynamic delivery. In this study, we compare the efficacy and specificity of delivery of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped lentiviral vectors to mouse liver by a number of injection schemes. Exploiting in vivo bioluminescence imaging as a readout after lentiviral gene transfer, we compare delivery by (1) “conventional” tail-vein injections, (2) “primed” injections, (3) “hydrodynamic” injections, or (4) direct “intrahepatic” injections into exposed livers. Reporter gene activity demonstrate potent and targeted delivery to liver by hydrodynamic injections. Enhanced efficacy is confirmed by analysis of liver sections from mice treated with GFP-encoding vectors, demonstrating 10-fold higher transduction rates and gene delivery to ∼80% of hepatocytes after hydrodynamic vector delivery. In summary, lentiviral vector transfer to mouse liver can be strongly augmented by hydrodynamic tail-vein injections, resulting in both reduced off-target delivery and transduction of the majority of hepatocytes. Our findings pave the way for more effective use of lentiviral gene delivery in the mouse.
Keywords: lentivirus, high-pressure tail-vein injection, hydrodynamic delivery, in vivo, liver gene therapy, gene transfer, hepatocytes
Introduction
The liver is the affected organ in many genetic and metabolic disorders involved in lysosomal storage and turnover of carbohydrates, amino acids, and organic acid, hence making the liver an attractive organ for gene therapy. Delivery of genetic material to hepatocytes and efforts to enhance gene therapy of liver tissue often begin with murine models. Experimental studies of in vivo delivery also include genome-editing methodologies, and early reports indeed were carried out in mice.1 One simple way to deliver DNA to murine liver is by high-pressure tail-vein injection, also known as hydrodynamic injection. Here, naked DNA, often plasmid DNA, is quickly injected in a large volume; typically, a 10% body weight DNA solution is injected within approximately 6 s in mice. The principle of hydrodynamic delivery relies on the mechanical force created by a transient congestion and subsequent flow back into the hepatic veins (as reviewed in Yokoo et al.2). The injected DNA solution passes through the sinusoidal structure to the portal veins and enters the hepatocytes through transient pores formed in the cell membrane.3 While hydrodynamic injection of naked DNA offers a simple and safe (non-viral) way to deliver genetic cargo to liver tissue and reaches an efficacy where nearly half of all hepatocytes are being targeted,4 this method suffers from its transient nature and gene expression drops rapidly,4, 5, 6 although inclusion of control regions may provide prolonged episomal expression.7 Gene delivery using viral vectors may offer prolonged transgene expression, and both adenovirus-, adeno-associated virus (AAV), and lentivirus-derived vectors have been adapted for potent gene transfer to liver tissue.8, 9, 10, 11, 12, 13 Whereas hydrodynamic DNA delivery relies on physical forces, virus-mediated gene transfer depends on active fusion or transport mechanisms to penetrate the outer cell membrane. Researchers have previously reported on combining these strategies and applied the hydrodynamic injection strategy for injection of lentiviral vector (LV) particles.10, 14 However, transfer efficacy of LVs to murine liver using different tail-vein injection schemes has not yet been carefully investigated in any report. Inspired by the work of Condiotti and colleagues,10 tentatively suggesting enhanced delivery by high-volume injections, we here compare reporter gene activity by in vivo bioluminescence imaging and fluorescence microscopy of liver sections after tail-vein injection of VSV-G pseudotyped LVs to mice in either a conventional, “primed,” or hydrodynamic fashion and also include an injection scheme involving surgically exposed liver tissue. Our findings demonstrate that hydrodynamic injections of lentiviral vectors through tail-vein potentiates gene delivery to mouse liver and reduce vector dissemination to most other organs or tissues.
Results
Evaluation of Promoter Activity in Liver by Hydrodynamic Plasmid Delivery
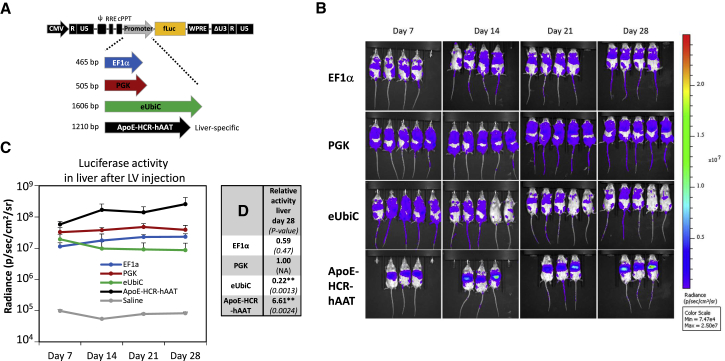
The development of potent delivery methods for transgene expression in murine liver is critical to advance gene therapy and genome-editing strategies. In order to compare efficacy and vector dissemination to non-liver tissues, we first constructed a panel of LVs each expressing the firefly luciferase reporter from a distinct ubiquitous promoter. As shown in Figure 1A, we decided to test the human elongation factor-1α (EF1α) and phosphoglycerate kinase (PGK) promoters, which are often utilized in LVs. The human EF1α promoter15 is routinely used as a promoter, expressing transgenes in multiple tissues, and is a strong promoter in mouse liver.16 Similarly, the PGK promoter17 facilitates high levels of transgene expression in many cell types,18 including liver.16 The human ubiquitin C (UbiC) promoter provides robust expression of transgenes in multiple tissues and organs.19 An optimized version of this promoter created by fusing the cytomegalovirus promoter (CMV) enhancer with the native UbiC promoter,20 here designated eUbiC (enhanced UbiC), was also included in our study. Our aim was to identify a potent and ubiquitous promoter that would allow us to identify and quantify vector spread in whole-body analysis using in vivo bioluminescence imaging. For comparison, and for later studies, we included the potent and liver-specific promoter ApoE-HCR-hAAT.21 This artificial promoter was originally constructed by Mark Kay’s group21 and is composed of the liver-specific α1-antitrypsin gene (hAAT) promoter combined with four liver-specific Apolipoprotein E (ApoE) enhancers and the ApoE hepatic control region (HCR).
Figure 1.
Quantification of Promoter Activity In Vivo after Conventional Tail-Vein Injection of Lentiviral Vector Particles
(A) Schematic diagram of the HIV-1-based third-generation LV (denoted pCCL) expressing firefly luciferase used in the present study. Different internal promoters used to drive transgene expression are shown below in distinct colors along with the length in base pairs (bp): EF1α in blue, PGK in red, eUbiC in green, and ApoE-HCR-hAAT in black. (B) Quantification of reporter activity in vivo after injection of LV vectors. Bioluminescence imaging of the mice visualize the localization and expression level of the luciferase reporter gene. The colored scale indicates radiance (photons/s/cm2/steradian). In brief, a vector dose of 3 μg p24 of the various LV vectors pCCL-EF1α-fLuc (first row; n = 4), pCCL-PGK-fLuc (second row; n = 5), pCCL-eUbiC-fLuc (third row; n = 5), and pCCL-ApoE-HCR-hAAT-fLuc (fourth row; n = 3) were delivered to the liver by conventional 0.4-mL tail-vein injections. A saline-injected animal served as control (data not shown; n = 1). The mice were analyzed in an IVIS bioluminescence scanner after subcutaneous injections of luciferin for quantification of reporter gene activity. (C) An ROI surrounding the liver was defined and radiance was quantified within each ROI. The mean and SD were calculated for each group and plotted at each sampled time point (7, 14, 21, and 28 days post-injection). (D) Table showing reporter activity in liver at day 28 normalized to the pCCL-PGK-fLuc group and p value of test statistics in brackets. Two asterisks (**) indicates significance levels below 0.01. Abbreviations: ApoE-HCR-hAAT, hybrid of Apolipoprotein E-derived enhancers and HCR fused to the human α1-antitrypsin gene promoter; CMV, cytomegalovirus promoter; cPPT, central polypurine tract; ΔU3, partial deletion of the viral unique 3′ region; EF1α, human elongation factor-1α promoter; eUbiC, chimeric CMV-enhanced human ubiquitin C promoter; fLuc, firefly luciferase gene; NA, non-applicable; PGK, human phosphoglycerate kinase promoter; Ψ, packaging signal (psi); R, repeat region; RRE, rev response element; U5, unique 5′ region; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
Transient plasmid DNA (pDNA) transfection of our vector constructs demonstrated that all expression cassettes worked well, with PGK being far superior in HEK293 kidney-derived cells and ApoE-HCR-hAAT most potent in HepG2 hepatoma cells (Figures S1A and S1B). To address the promoter strength in liver tissue, we next hydrodynamically injected plasmid DNA through the tail vein in BALB/cJ mice22 and followed luciferase activity by live bioluminescence imaging for 3 weeks (at day 3, 7, 14, and 21; Figure S1C). As expected, luciferase expression localized to the liver in all animals (Figure S1C) and showed a rapid decline consistent with previous reports using plasmid DNA without additional control elements.7, 23 Luciferase expression decreased between 40-fold (eUbiC) and 160-fold (EF1α), and the liver-specific promoter (ApoE-HCR-hAAT) displayed the highest expression at all times, whereas the EF1α, PGK, and eUbiC promoters displayed lower and overall comparable expression levels, although these differences were not statistically significant (Figure S1D).
Strong and Widespread Expression of PGK-Promoted Reporter after In Vivo Delivery of Lentiviral Vectors
Lentiviral transgene delivery has previously resulted in efficient long-term expression contrary to plasmid-based transgene expression.10 The four constructs (pCCL-EF1α-firefly luciferase (fLuc), pCCL-PGK-fLuc, pCCL-eUbiC-fLuc, and pCCL-ApoE-HCR-hAAT-fLuc, shown in Figure 1A) were utilized for production of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped lentiviral vectors for injection into the tail vein of BALB/cJ mice (Figure 1B). Luciferase activity as measured by live bioluminescence imaging was followed for 4 weeks (at day 7, 14, 21, and 28). LVs with EF1α-driven fLuc expression showed high expression, consistent with targeting of liver, spleen, and the lymph system (Figure 1B), whereas vectors with PGK-driven expression consistently resulted in an apparently stronger signal. The signal in the PGK group also localized to liver, spleen, the lymph system, tail, as well as other regions that were difficult to assign to specific organs based on bioluminescence imaging. The signals obtained with eUbiC-driven fLuc expression resembled the expression pattern obtained with the PGK promoter, albeit with a decrease over time during the 4 weeks sampling period. Vectors with the ApoE-HCR-hAAT promoter displayed a very strong signal localized to the liver and possibly the spleen. Regions of interest (ROIs) encompassing the liver were used to estimate the liver-derived signals for all groups (Figure 1C). We observed a slight increase in expression with the promoters EF1α, PGK, and ApoE-HCR-hAAT up to day 14 after injection followed by stable expression to the study endpoint. In contrast, the liver expression decreased slightly throughout the experiment for the eUbiC group. By comparing the level of expression at day 28 after injection, we determined the following hierarchy in terms of activity in the liver: ApoE-HCR-hAAT ≫ PGK ≥ EF1α > eUbiC (see values normalized to PGK in Figure 1D), consistent with previous reports on promoter activity.16 Noteworthy, reporter activity for a given type of tissue will not only depend on properties of the embedded promoter, but also on functional titers for each vector. To investigate this, we transduced HEK293 cells with an equal vector dose based on p24 and estimated vector transfer by qPCR-based titer assay. Whereas the functional titer of our reference PGK-fLuc vector was of 6.8 × 107 infectious units per μg p24, the titers of vectors harboring the large promoters, eUbiC-fLuc and ApoE-HCR-hAAT-fLuc were reduced by only 5% and 21%, respectively. The EF1α-fLuc vector showed a reduction in titer by 47%, which may at least partially explain the lower reporter activity seen in liver.
In order to pursue our aim, we needed to track vector spread, also to non-liver tissues and not least monitor the efficacy of liver delivery for different injection schemes. In this respect, the PGK promoter was clearly the best choice due to the high overall expression levels and activity in many types of tissue and organs.
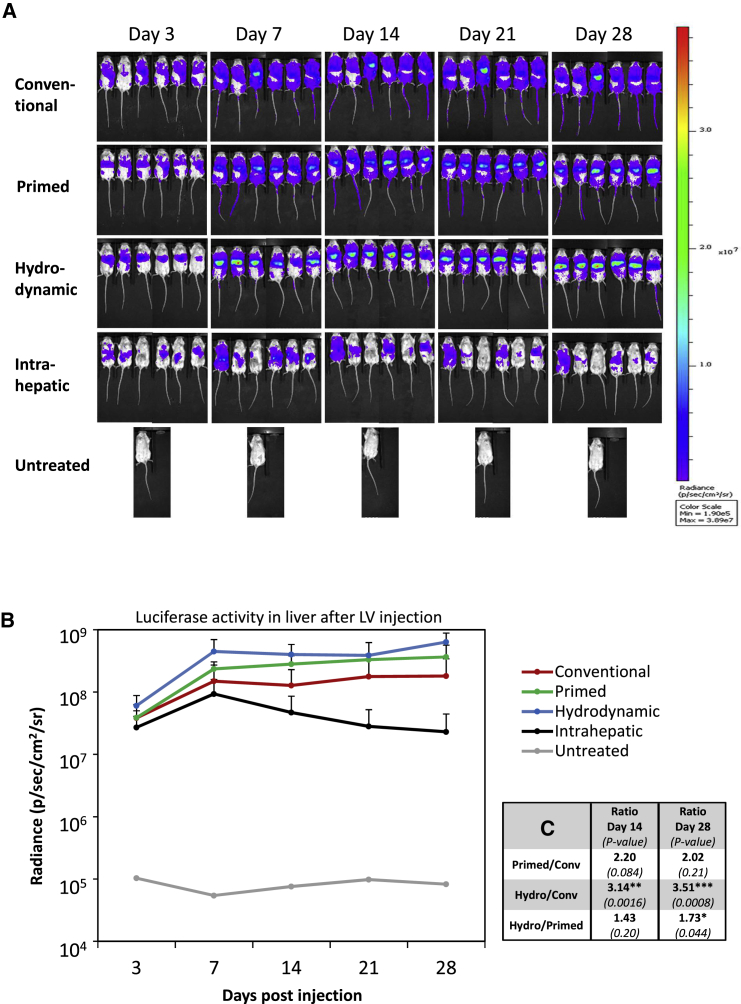
Comparison of Tail-Vein Injection Schemes for Lentiviral Vectors Shows Improved Liver Delivery by Hydrodynamic Pressure
Based on the work of Condiotti and colleagues,10 in which hydrodynamic injection of feline-immunodeficiency-virus-based vectors was utilized for delivery of GFP to the liver of mice, we decided to compare four different strategies for administering LVs to the mouse liver. For these studies and based on the above findings showing robust and widespread luciferase reporter gene expression, we used an LV with expression driven by the PGK promoter (pCCL-PGK-fLuc24). The four different strategies were as follows: (1) conventional tail-vein injection, (2) hydrodynamically “primed” injection, (3) hydrodynamic tail-vein injection, and (4) direct intrahepatic injection facilitated by an incision made through the abdominal skin and muscle. Specifically, we intravenously (i.v.) injected a dose of 4 μg p24 units of VSV-G pseudotyped LV in a total volume of 400 μL per mouse for the “conventional” group, while mice in the “hydrodynamic” group were injected within 5–10 s with a much higher volume (∼1.5 mL total) corresponding to 8% of the body weight. Inspired by the work of Brunetti-Pierri et al.25 using adenoviral-based vectors in mice, our so-called hydrodynamic “primed” group was injected in a conventional fashion half an hour after hydrodynamic “priming” each animal with Ringer solution.
Figure 2A displays the expression pattern of the four different injection schemes over a period of 28 days as investigated by live bioluminescence imaging. As shown in the top row of Figure 2A, the conventional method as expected resulted in widespread expression, consistent with targeting of the liver, spleen, the lymph system, and possibly other tissues. In the second row (Figure 2A) (“primed” injection), we observed reporter expression prominently in the liver and in the spleen. Nevertheless, significant signals from other tissues indicated a marked level of vector dissemination to, for example, the lymphatic system. In contrast, hydrodynamic delivery of LVs (Figure 2A, third row) dramatically changed the pattern, and luciferase expression localized primarily to the liver and to a smaller degree to the spleen. Finally, administration of LVs by two 50-μL injections directly into the liver (Figure 2A, fourth row), resulted in quite limited reporter activity in the liver, possibly suggesting leakage to surrounding tissue or clearance, and moreover, the expression pattern varied substantially from mouse to mouse.
Figure 2.
Increased Delivery of LVs to Murine Liver by Hydrodynamic Tail-Vein Injections
(A) Bioluminescence imaging of mice administered with a vector dose of 4 μg p24 of pCCL-PGK-fLuc vector-encoding particles visualizes the localization and expression level of the luciferase reporter gene at different time points. Administration were as follows: conventional (0.4 mL) tail-vein injections (first row), “primed” injections (second row), hydrodynamic injections (third row), intrahepatic injections (fourth row), or an untreated mouse as control (fifth row). The mice were analyzed in an IVIS bioluminescence scanner after subcutaneous injections of luciferin for quantification of reporter gene activity. The colored scale indicates radiance (photons/s/cm2/steradian). (B) An ROI surrounding the liver was defined and radiance was quantified within each ROI. The mean and SD were calculated for each group (n = 6) and plotted at each sampled time point (3, 7, 14, 21, and 28 days post-injection). (C) Table showing the ratio of reporter activities in liver between indicated groups at days 14 and 28 and the associated p value in brackets below. One (*), two (**), or three (***) asterisks indicate significance levels below 0.05, 0.01, or 0.001, respectively.
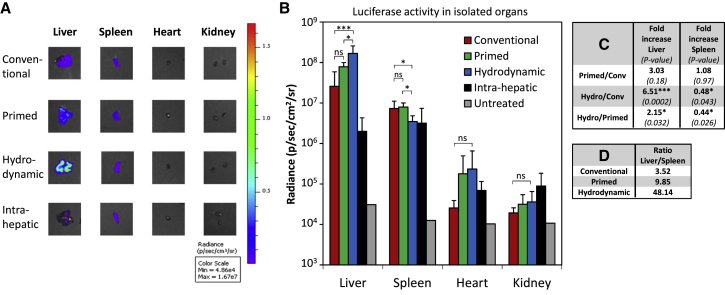
To quantify the expression levels observed with bioluminescence imaging for the different methodologies, we outlined ROIs covering the area of the liver and calculated and plotted the radiance for all time points (Figure 2B). For all the tail-vein-injected groups, we observed robust and stable expression during the 4 weeks, and only for the intrahepatic injected group did expression levels drop substantially. Notably, hydrodynamic pressure boosted reporter activity in the region of the liver by 3.5-fold at day 28 as compared to the conventionally injected group when we monitored live animals (Figure 2C). Hydrodynamic priming resulted in liver expression levels that were intermediate (see ratio of activities and test statistics in Figure 2C). To substantiate our findings and enhance the detection sensitivity, we performed bioluminescence imaging of dissected organs at day 29 (Figure 3A). When we quantified reporter activity in isolated livers (Figure 3B), our data showed the following rank in terms of efficacy for liver delivery: hydrodynamic > “priming” ≥ conventional ≫ intrahepatic. As summarized in Figure 3C, hydrodynamic “priming” resulted in an ∼3-fold increase in radiance for isolated livers as compared to conventional injections, although this is not statistically significant, while the hydrodynamic scheme augmented this by ∼6.5-fold.
Figure 3.
Vector Dissemination to Off-Target Organs Vary According to Injection Strategy
(A) Representative bioluminescence images of livers, spleens, hearts, and kidneys excised from treatment groups described in Figure 2. These end-point data were collected 29 days post-treatment, where mice were sacrificed and the organs rapidly excised and analyzed in an IVIS bioluminescence scanner. The colored scale indicates radiance (photons/s/cm2/steradian). (B) An ROI outlining the organ was defined, and radiance was quantified within each ROI. The mean and SD were calculated for each group (n = 6) and plotted. (C) Table showing the fold increase in reporter activities between indicated groups in liver or spleen and the associated p value in brackets below. (D) Table showing the ratio between reporter activity in liver and spleen for the different tail-vein-injected groups. One (*) or three asterisks (***) indicate significance levels below 0.05 or 0.001, respectively. Abbreviations: ns, non-significant.
As evident from the whole-body images in Figure 2A, applying a high-pressure strategy also seemed to reduce vector spread and reporter activity in non-liver tissues. Conventionally tail-vein-injected LV vectors clearly reached, for instance, the spleen, and to quantify this vector dissemination, we isolated and analyzed separately four internal organs, namely liver, spleen, heart, and kidney (Figure 3A). Indeed, we observed quite strong reporter activity in the spleen for the conventionally treated group and almost no activity in heart and kidney (Figure 3B). Spill-over to the spleen was unchanged if the animals were hydrodynamically primed with salt water (Figure 3C), but in contrast, the “priming” group also showed a modest targeting of the heart.
Hydrodynamic injections also resulted in some vector delivery to heart, although at a low level. For this group, however, targeting of the spleen was reduced to approximately half compared to conventional treatment (Figure 3C). Considering spleen as the most significant off-target for tail-vein-injected animals, this “on-target/off-target ratio” was improved substantially by hydrodynamic delivery as tabulated in Figure 3D. In an attempt to quantitate vector spread further, we opened up the carcasses and placed intestines, abdominal skin, and lungs separately along with exposed thorax (including ribcage and upper spinal cord), neck (including various glands and lymph nodes), and pelvic regions (including uterus, hip bones, and lower spinal cord) as illustrated in the picture insert in Figure S2. Bioluminescence scanning revealed modest to low luciferase activity in all examined tissues. We observed no major differences between all injection schemes when looking at the intestines, lungs and thorax region, or ovaries and pelvic region (Figure S2). Direct intrahepatic injections caused increased activity in the abdominal skin mainly located in the scar tissue made from the incision, while both types of high-pressure treatments reduced signals emerging from the neck and the associated lymph nodes.
All together, our data demonstrate that hydrodynamic injections boost LV delivery to liver by more than 6-fold compared to conventional injections and overall lead to less spread to non-liver tissues, in particular the spleen.
Hydrodynamic LV Injection Results in Very Efficient Delivery to Hepatocytes, with a Modest Increase in VCNs
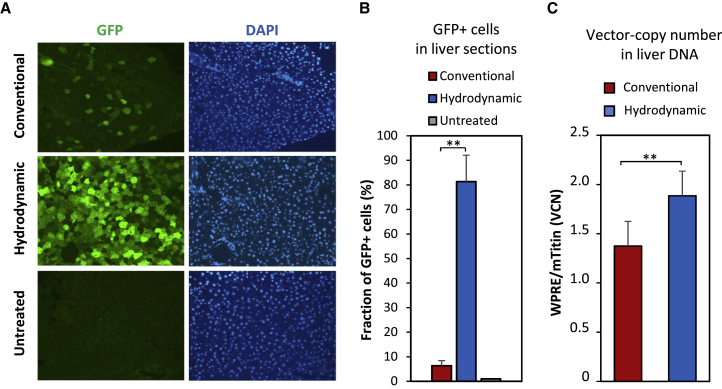
To confirm the augmented delivery to liver by high-pressure, high-volume injections and investigate the distribution and the number of transduced cells within liver tissue, we carried out a second round of in vivo experiments. Based on the above findings, we decided to only compare the hydrodynamic strategy to the conventional injection method, and moreover, we increased the dose and switched to our strongest promoter ApoE-HCR-hAAT to enhance delivery efficacy. We administered each group with a single vector dose of 15 μg p24, this time carrying an ApoE-HCR-hAAT-promoted eGFP expression cassette (pCCL-ApoE-HCR-hAAT-GFP) suitable for fluorescence-based microscopy. Thirteen weeks following treatment, we isolated liver, kidney, spleen, thymus, heart, and lung from all mice and analyzed these by epifluorescence scanning along with the remaining carcasses (Figure S3A). As apparent on the images in Figure S3A and as expected for our liver-specific promoter, only the isolated livers gave a fluorescent signal above background and quantification showed significantly enhanced delivery by hydrodynamic as compared to conventional injections (Figure S3B).
Next, we analyzed 5-μm cryosections of the liver by fluorescence microscopy. Random images were captured from sections derived from three to six different locations for each liver, and the number of GFP-positive cells was quantified from blinded images using overlay with DAPI-stained nuclei to localize individual cells. Figure 4A shows representative images from individual mice, demonstrating that the total number of GFP-positive cells was much higher for the hydrodynamically injected group. Our quantification showed a dramatic and significant increase in the fraction of GFP-positive cells from less than 10% when applying LV vectors conventionally to ∼80% for the hydrodynamic delivery (Figure 4B). In both cases, the GFP-positive cells seemed quite evenly distributed and did not vary much between for instance different lobes of the same liver (data not shown). Cell morphology and size clearly indicated that the far majority of the GFP-positive cells were hepatocytes.
Figure 4.
Increased Number of Transduced Cells in Liver Sections after Hydrodynamic Delivery
Fluorescence microscopy analysis of livers isolated from mice that were administered a single dose of 15 μg p24 of lentiviral vectors encoding GFP (pCCL-ApoE-HCR-hAAT-GFP as described in Figure S3). 93 days post-injection, mice were sacrificed, and the liver was harvested and cryosectioned. (A) Representative images of DAPI-stained liver sections from each group of mice: conventional, hydrodynamic, and untreated mice. Green color indicates GFP signal (left), and brightness above background from untreated samples hence indicates positively transduced cells. Blue color indicates DAPI stain (right) and hence localization of the cell nuclei. (B) The fraction of GFP-positive cells as quantified by inspection of several images as represented in (A). A minimum of five images, which displayed a nice even layer of cells as judged by DAPI stain, were captured at random positions on each liver sample, and the fraction of GFP-positive cells was counted manually. The mean and SD were calculated for each group (n = 3) and plotted. (C) Modest vector copy-number (VCN) increase in liver tissue after hydrodynamic delivery. Genomic DNA was isolated from the livers depicted in (A). A minimum of three samples from each liver was pooled together, and a TaqMan-based qPCR assay was used to measure VCN, as defined by the amount of vector DNA (WPRE target) relative to the amount of genomic DNA (analysis of a single copy gene, mTitin). The mean and SD were calculated for each group (n = 3) and plotted. Two asterisks (**) indicate significance levels below 0.01.
Finally, we extracted genomic DNA from tissue samples from all livers and estimated the number of vector copies per diploid genome using an established qPCR protocol based on amplification of the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) of the vector and normalization to the single-copy murine gene titin.26 Our analysis demonstrated only a modest ∼40% increase in vector copy number (VCN) from ∼1.4 to ∼1.9 in conventional- or hydrodynamic-treated animals, respectively (Figure 4C). In addition, we performed Southern blot analysis to detect a GFP-specific sequence in the genomic DNA samples. As apparent from the autoradiogram (Figure S3C), only high-molecular-weight DNA longer than 8–10 kb was detected in injected livers consistent with vector integration at random chromosomal positions and limited presence of 1-long terminal repeat (LTR) and 2-LTR circles (predicted sizes of ∼4.4 kb and ∼4.7 kb, respectively). This indicates that the bulk GFP signal originated from integrated copies of the reporter gene.
To confirm the efficacy of hydrodynamic delivery in a different mouse strain, we treated male B6:BTBR mice homozygous for the Pahenu2 mutation with a single dose (15 μg p24) of LV carrying an ApoE-HCR-hAAT-promoted GFP or mPAH (phenylalanine-4-hydroxylase) expression cassette. Four weeks after treatment, we analyzed the mice by live epifluorescent imaging, fluorescence microscopy of liver sections, and blood plasma levels of phenylalanine (Figure S4). Consistent with our earlier results, we observed massive and widespread delivery to the majority of hepatocytes in the group treated with the GFP-expressing vector, with GFP signals so intense that we were able to detect it in live animals (Figures S4A and S4B). We also induced hyperphenylalaninemia in these mice, and while we did observe a significant drop of 57% in phenylalanine (phe) plasma levels after mPAH gene transfer (Figure S4C), our experimental animal model and the limited therapeutic effect prevented direct comparison to published studies of gene therapy of phenylketonuria (PKU) mice.27, 28
Discussion
The purpose of the study was to compare the efficacy and specificity of delivering LVs to the murine liver by four different injection schemes, namely (1) conventional tail-vein injection, (2) hydrodynamically “primed” injection, (3) hydrodynamic tail-vein injection, and (4) direct intrahepatic injection. Bioluminescent imaging in the weeks following delivery of PGK-fLuc vectors clearly demonstrated that hydrodynamic injection is optimal for liver targeting. Importantly, most of the widespread radiance seen in non-liver tissues after conventional tail-vein injection of LV vectors disappears, and “on-target” delivery to the livers increases significantly. This is consistent with the findings of Brunetti-Pierri et al.,29 who found that hydrodynamic injection of adenovirus-based vectors in mice favors hepatic transduction and shows reduced dissemination to spleen and lung. One exception is vector delivery to heart, which increases from almost none to modest levels when applying high-pressure delivery. This is in accordance with the concept of hydrodynamic injection, where the injected fluid first reached the heart via the inferior vena cava before retrograde flow into the liver.30 Surprisingly, mild transduction of heart tissue is also evident from “primed” injections, which may suggest that hydrodynamic “priming” with salt water causes fenestration of the endothelial lining that lasts over 30 min and exposes heart tissue to circulating vector particles. In general, hydrodynamic “priming” did not alter the transduction profile very much as compared to conventional injections, although radiance from the liver itself tended to increase. In a key study, the method of delayed injection after hydrodynamic “priming” was compared to conventional tail-vein injection and to hydrodynamic injection of adenovirus-based vectors and was found to significantly increase the expression in the liver of mice compared to the other two methods.25 However, this particular study only investigated the liver-specific signal and did not address expression in off-target tissues. Direct injection of vector solution into surgically exposed liver tissue showed poor efficacy. In our settings, we slowly injected 50 μL at two different position of the liver, but as evident from bioluminescent imaging results, there was extensive leakage to non-liver tissues and cases with almost no liver transduction.
Quantification of isolated livers demonstrated a striking ∼6.5-fold increase in radiance for hydrodynamic delivery compared to the conventional group. This result prompted us to investigate delivery to liver and vector distribution to hepatocytes directly in a side-by-side comparison between a conventional injection strategy and hydrodynamic LV delivery. Using a high dose of a GFP vector driven by our strongest promoter in liver (ApoE-HCR-hAAT), we could confirm increased transfer to liver. Quantification of GFP-positive cells in liver sections consistently showed a marked increase in the fraction of transduced cells from less than 10% to ∼80% when using hydrodynamic delivery. In the work by Condiotti et al.,10 a transduction efficacy to hepatocytes of around 1% was reported using hydrodynamically injected feline-immunodeficiency-based LVs encoding GFP. However, substantial differences in vector properties, preparation, and biological titers prevent a direct comparison to our findings. Size, morphology, and distribution of GFP-positive cells in our liver sections clearly suggested that these were hepatocytes, and as we extended the period from vector injection to sacrificing the mice to 92 days, we are confident that detection of GFP did not stem from transient expression from un-integrated vector DNA or attracted Kupffer cells. Southern blot analysis supported the notion of vector integration following hydrodynamic delivery, consistent with the detailed analysis including partial hepatectomy by Condiotti et al.10 Surprisingly, copy-number analysis demonstrated a modest increase in the amount of vector DNA in genomic DNA samples from liver after hydrodynamic delivery compared to conventional transfer. This suggested that equal total copy numbers of integrated vector DNA were obtained following hydrodynamic and conventional LV delivery but that hydrodynamic delivery resulted in widespread transduction with a VCN close to one, whereas conventional delivery resulted in localized delivery with multiple transductions per cell. However, we cannot at this stage account for possible pressure-dependent differences in (1) the actual total vector dose reaching the liver, (2) the actual mechanism of cellular uptake or transduction process, (3) the degree of “un-productive” transfer events leading to presence of vector DNA but no GFP expression, or (4) the removal of vector DNA by the immune system.
The high efficiency of gene transfer to the liver clearly warrants the use of hydrodynamic LV delivery as a useful alternative to other efficient transfer methodologies to the liver such as AAV. Most notably, LVs offer long-term expression from the integrated provirus, high packaging capacities compared to many other viral vector systems, and have so far been limited by mediocre transduction efficacies in liver tissue. Based on mechanistic studies of hydrodynamic injections, there are several reasons why liver delivery is enhanced as compared to the conventional type also for viral particles: first, high-pressure “targets” the injected solution for passing through the liver, as retrograde entry and rapid vascular expansion of the liver clearly reduces distribution into regular circulation.30, 31 This is also in line with the marked reduction in transduction of the spleen following hydrodynamic injections, which filters circulating blood and ranks second after liver as the most transduced organ. Second, rupture of the sinusoidal endothelia causes direct exposure of hepatocytes to the injected solution.31, 32 Third, pressure-induced formation of intracellular vesicles or disruption of the plasma membrane of hepatocyte might also facilitate entry of LV particles that could reach the nucleus.31 Under normal conditions, lentivirus surface proteins mediate entry across the plasma membrane of target cells as an active process.33 The impact of pressure-induced membrane rupture and micropinocytosis-like events during uptake of vector particles is currently unknown, and we can only speculate if alternative entry routes are created and if they lead to productive transduction or vector integration. We also speculate that the kinetics of the transduction process could be faster in hydrodynamic settings, which in turn might short-circuit the restrictive type I interferon responses that limit hepatocyte delivery.34
Studies of hydrodynamic delivery have found that high-pressure and -volume injections are quite well tolerated in mice.4, 35 Consistent with delivery of naked DNA (or salt water), hydrodynamic injection of LVs do cause mild liver damage, as indicated by slightly elevated levels of AST and ALT in serum 1 day post-treatment.10 However, levels normalize within days.10 Indeed, in a more recent study using comparable doses of a hydrodynamically injected HIV-based vector similar to ours, close examinations of liver morphology, inflammatory markers, influx of immune cells, and serum levels of liver transaminases did not show any marked signs of either tissue damage, inflammation, or toxicity of the liver 1 week post-injection.14 Of note, hydrodynamic delivery can cause strong acute, but transient, hepatic toxicity in mice.25 Liver-targeted hydrodynamic delivery has been adapted for use in larger animals, including pigs and dogs, by using different closed perfusion procedures that were deemed safe.36, 37, 38, 39, 40 This has paved the way for the use of foamy virus vectors in pigs41 and adenovirus-based vectors in non-human primates,25, 42, 43 signifying, thus, the potential clinical value of pseudo-hydrodynamic LV delivery.
In summary, our data demonstrate that hydrodynamic injections boost LV delivery to liver by more than 6-fold and overall leads to less spread to non-liver tissues, in particular the spleen. Moreover, hydrodynamic injection results in extensive and widespread transduction in mouse liver with the far majority of hepatocytes being targeted with a single vector dose.
Materials and Methods
Plasmid Construction
The third generation self-inactivating (SIN) lentiviral vector named pCCL-PGK-fLuc was constructed as previously described.24 This vector backbone was used for all promoter substitutions and encodes the fLuc gene under control of the phosphoglycerate kinase (PGK) promoter. The promoters were ligated into the pCCL-PGK-fLuc vector digested by ClaI and BamHI (New England Biolabs, Ipswich, MA, USA), which precisely remove the PGK promoter. The human elongation factor-1α (EF1α) promoter was PCR-amplified with ClaI- and BamHI-tagged primers from a custom-made vector harboring a shortened version of the EF1α promoter (pUC57-mirvector, Genscript, Piscataway, USA). The novel CMV eUbiC promoter was PCR-amplified from the vector pSBT/loxP-Ei-Ubi-GIP.20 The ApoE-HCR-hAAT promoter (an artificial liver-specific promoter composed of four copies of ApoE enhancer, a HCR, and the human α1-antitrypsin promoter) was PCR amplified from vector pSB-ApoE-HCR-hAAT-FIX-bpA.21 The lentiviral vector pCCL-PGK-mPAH encoding the murine PAH (EC1.14.16.1) was generated from pCCL-PGK-fLuc by digestion with BamHI and PspXI (New England Biolabs) and insertion of murine PAH cDNA fragment (corresponding to GenBank UID X51942) amplified by RT-PCR from RNA isolated from murine fibroblasts using BamHI and PspXI-tagged primers. The HA-tag (MYPYDVPDYA) was fused N-terminally to the mPAH cDNA by QuikChangeII site-directed Mutagenesis Kit (Stratagene, Germany) to generate pCCL-PGK-HAtag-mPAH. Lentiviral vectors encoding mGTPCH (murine GTP cyclohydrolase I) and mPTPS (murine 6-pyruvoyl-tetrahydropterin synthase) were constructed using BamHI and PspXI-tagged cDNA fragments amplified by RT-PCR from RNA isolated from murine fibroblasts analogous to the pCCL-PGK-mPAH vector. All vector plasmids have been verified by sequencing and restriction enzyme digest. PCR and QuikChange primer sequences are available upon request.
Cell Culturing
The HEK293 (ATCC CRL-1573, Boras, Sweden), HEK293T (ATCC CRL-11268, Boras, Sweden), and HepG2 (hepatocellular carcinoma, ATCC HB-8065, Boras, Sweden) cells were cultured at standard conditions: 37°C in 5% CO2 in DMEM (Lonza, Verviers, Belgium) supplemented by 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (0.1 mg/mL) (Sigma-Aldrich, Broendby, Denmark).
In Vitro fLuc Reporter Assay
HEK293 and HepG2 cells were transfected in the same way. Cells were seeded at a density of 2 × 105 cells/well in 6-well dishes. The cells were transfected with 1.5-μg plasmid DNA by FuGENE6 transfection reagent (Promega, Nacka, Sweden) according to manufacturer’s protocol 24 hr after seeding. Control cells were transfected with pBlueskript-SK+ plasmid DNA devoid of luciferase activity. Forty-eight hours post-transfection, the fLuc assay (Promega) was performed according to manufacturer’s protocol, and light emission was quantified by MicroLumial Plus LB 96V (Berthold technologies, Bad Wildbad, Germany) luminometer.
Lentivirus Vector Production
HEK293T cells were seeded in 15-cm dishes (1 × 107 cells/dish) with 20 mL DMEM medium. Twenty-four hours later, the cells were transfected with 7.3 μg pRSV-Rev, 9 μg pMD.2G, 31.5 μg pMDGP-Lg/RRE, and 31.5 μg of the LV transfer plasmid using the calcium phosphate co-precipitation method as previously described.44 In brief, DNA was diluted with double distilled water (ddH2O) to a final volume of 1,089 μL. 121 μL 2.5 M CaCL2 was added to the DNA solution. 1,212 μL 2× HEPES buffer was pipetted into a 15-mL tube, and the DNA-CaCl2 solution was quickly added and vortexed thoroughly. Following incubation for 15 min at room temperature, the solution was added drop-wise to the cells and distributed by gently swirling. The medium was replaced the day after. The virion-containing media was harvested 48 hr post-transfection and filtered through a 45-μm filter (Sarstedt, Nümbrecht, Germany) to remove cell debris. The viral supernatant was purified by ultracentrifugation (2 hr, 25,000 rpm/82,705 × g, 4°C) using a 20% sucrose cushion.45 Virus pellets were re-suspended overnight in PBS−/− at 4°C in a volume of 1/300 of the original supernatant volume. A second harvest was included by adding fresh medium to the virus-producing cells and the supernatant was collected, ultracentrifuged, and re-suspended in PBS−/− the following day. In total, this results in a volume of approximately 100 μL purified virus per 15-cm dish. The titer was measured in the resulting virus preparation by HIV-1 gag p24 antigen ELISA kit (ZeptoMetrix, Buffalo, NY) according to manufacturer’s protocol. The concentrated virus was stored at −80°C in aliquots.
Injections of Mice
8- to 10-week-old BALB/cJ BomTac mice (Taconic Europe, Laven, Denmark) were used. Hydrodynamic tail-vein injections of plasmid DNA were as follows: BALB/cJ mice were anesthetized with 3.75% isofluorane (Forene, Abbott Scandinavia AB, Solna, Sweden) and injected with 30 μg plasmid suspended in Ringer solution (NaCl 147 mM, KCl 4 mM, and CaCl2 1.13 mM) and in an injection volume of 8% of the body weight (between 1.2 and 1.8 mL) in 5–10 s.4, 46 Tail-vein injections of lentivirus were as follows: BALB/cJ mice were injected with 3 μg or 4 μg p24 units of virus dissolved in 0.9% NaCl to final volume of 400 μL per mouse (Figures 1 and 2, respectively). Hydrodynamically “primed” injections were as follows: these injections were inspired by Brunetti-Pierri and coworkers.25 In brief, 30 min after “priming,” the animals with a hydrodynamic injection of Ringer solution (8% of body weight) the mice were conventionally injected with 4 μg p24 virus dissolved in 0.9% NaCl to final volume of 400 μL per mouse. Hydrodynamic tail-vein injections of lentivirus: BALB/cJ mice were anesthetized and injected with 4 μg p24 virus suspended in Ringer solution (8% of body weight). Intrahepatic injections were as follows: BALB/cJ mice were anesthetized with 3.75% isofluorane. The abdomen was shaved, and an incision was made through the abdominal skin and muscles to expose the liver. 50 μL (a total of 4 μg p24 lentivirus in 100 μL) were injected at two locations of the liver. The abdominal muscle and peritoneal layer and the skin were closed separately by interrupted sutures. Analgesia was given subcutaneously (rimadyl 5 mg/kg and temgesic 0.05–0.1 mg/kg) the first 24 hr and added in the drinking water for 3 days (temgesic approximately 0.7–1.4 mg/kg) to relieve pain. The BALB/cJ Bom Tac mice were housed in type II plastic cages (Techniplast, Italy) in temperature-controlled pathogen-free animal facility, with unrestricted access to diet (Altromin #1324, Lage, Germany) and tap water. The animal room had a 12:12-hr light-dark cycle (lights on at 06:00). Mice were given nesting material, shredded paper strips, and wooden squares as environmental enrichment. Bedding was aspen wood chips supplied by Tapvei (Kortteinen, Finland). Pahenu2 mice47 was obtained from Jackson Laboratories (stock number 002232). Experimental animals were produced by breeding B6:BTBR mice homozygous for the Pahenu2 mutation to yield homozygous PKU mice. The breeding animals were maintained on phe-free semi-synthetic diet (Teklad diet, TD.97152, Envigo, WI, USA). The diet was kept at 5°C until used. As the diet was free from phe, the drinking water was supplemented with phe (Sigma P-2126, Sigma-Aldrich Chemie) to a final concentration of 0.0625 mg/mL). Male PKU mice, 8–12 weeks of age, were tail-vein injected with 15 μg p24 units of lentivirus vector encoding either the eGFP reporter gene (pCCL-ApoE-HCR-hAAT-GFP) or mPAH (pCCL-ApoE-HCR-hAAT-mPAH or an N-terminal HA-tagged version of this construct) using a hydrodynamic strategy as described above. At the time of vector injection, the phe load was increased 4-fold to induce hyperphenylalaninemia. All animal studies were carried out with permission from the Danish experimental animal inspectorate inspectorate, and housing of the mice was carried out according to Danish legislation and the Directive 2010/63/ on the protection of animals used for scientific purposes.
In Vivo Bioluminescence and Fluorescence Imaging
Mice treated with fLuc vectors were anesthetized with 3.5% isofluorane (Forene, Abbott Scandinavia AB) and injected subcutaneously with D-luciferin potassium salt (Synchem, Felsberg-Altenburg, Germany) at a concentration of 150 μg/g body weight. Anesthesia was maintained at 2% isofluorane during the bioluminescence scan using the IVIS Spectrum imaging system (Caliper Life Sciences, Hopkinton, MA, USA). Data quantification was performed using the software Living image 4.3. Prior to scanning, one mouse was injected with luciferin and scanned for 50 min acquiring images with an interval of 2 min. ROIs were made to outline the signal and measured. The ROI intensity was plotted across time to establish the optimal interval for scanning (“peak minus 15%”) following luciferin administration in our model. The intensity of the signal (radiance) was measured in photons/s/cm2/steradian (sr). The ROIs were exactly the same size to ensure comparability. On day 29 post-injection mice were euthanized and the organs scanned following dissection. Mice treated with GFP vectors were anesthetized with 3.5% isofluorane, and anesthesia was maintained at 2% isofluorane during epifluorescence scanning using the IVIS Spectrum imaging system. Data quantification was performed using the software Living image 4.3, including the spectral unmixing tool. The intensity of the signal (total radiance efficacy) was measured in photons/s per μW/cm3.
Fluorescence Microscopy of Liver Sections
Three samples of roughly 2 × 2 × 5 mm from different locations in the liver were isolated from the mice after final examination. After overnight fixation in 4% paraformaldehyde (Hounisen, Risskov, Denmark) and overnight rehydration in 30% sucrose, samples were snap frozen in optimal cutting temperature (OCT) (Tissue-Tek, Sakura Finetek, AV Alphen aan den Rijn, the Netherlands) before cryosectioning (Microtome Cryostat, Microm HM 500 M). 5-μm sections were mounted on Superfrost-Plus object glass (Menzel-Gläser, Braunschweig, Germany) using antifade mounting medium (Vectashield H-1000), including DAPI (Sigma-Aldrich). Fluorescence microscopy and image capture was done with a Leitz microscope (DM RB, Leica Microsystems, Wetzlar, Germany) equipped with fluorescein isothiocyanate (FITC) filter (492/18X) and Leica camera (DFC 360 FX) and associated software (Leica Application Suite, version 3).
Copy Number Assay and LV Titer Assay
Genomic DNA (gDNA) from snap-frozen liver tissue samples was isolated using a standard high-salt extraction protocol and used for qPCR analysis. In brief, liver tissue was dissected into small pieces and lysed in Chorion Villis buffer (10 mM Tris, 1 mM EDTA, 150 mM NaCl, 0.5% SDS, pH 10.5) and proteinase K (18 mg/mL, Roche), before removing cell debris with 6 M NaCl and centrifugation and finally precipitating DNA with isopropanol. Genomic DNA was resuspended in Tris-EDTA buffer and stored at −20°C. Quantitative PCR was performed in triplicates with TaqMan Universal Master Mix II (Applied Biosystems) using primers and probe specific for the WPRE sequence present in the lentiviral vector, 50 ng of gDNA template, and a LightCyclerW480 real-time thermocycler (Roche Diagnostics, Mannheim, Germany). Primers and probe specific for the murine single-copy gene titin were used to normalize for the amount of gDNA. Exact PCR conditions are available upon request. Primers and probes were as follows: WPRE-forward, 5′-GGCACTGACAATTCCGTGGT-3′; WPRE-reverse, 5′-AGGGACGTAGCAGAAGGACG-3′; WPRE-probe, 5′-ACGTCCTTTCCATGGCTGCTCGC-3′ (6-carboxyfluorescein-black hole quencher [FAM-BHQ]); Titin-forward, 5′-AAAACGAGCAGTGACGTGAGC-3′; Titin-reverse, 5′-TTGCAGTCATGCTGCTAGCGC-3′; Titin-probe, 5′-TGCACGGAAGCGTCTCGTCTCAGTC-3′ (FAM-BHQ). Total proviral copy numbers were determined using the standard curve method. In brief, a standard curve for each target was made using a 10-fold serial dilution (from 108 to 101 copies) of a plasmid containing both targets, pTitin (Addgene plasmid ID 26428). The standard curves were used to calculate the copy number of WPRE sequences per cell. Infection of HEK293 cells seeded in 6-well plates in day prior (105 cells per well) with varying doses of LV (1–100 ng p24) served as gDNA template for qPCR-based titer assay performed as described above. The human Albumin gene served as a single-copy gene for quantification of the VCN per cell, and titers were calculated by counting target cells at the time of infection. Albumin forward was as follows, 5′-GCTGTCATCTCTTGTGGGCTGT-3′; albumin reverse was as follows, 5′-ACTCATGGGAGCTGCTGGTTC-3′; and albumin probe was as follows, 5′-CCTGTCATGCCCACACAAATCTCTCC-3′ (FAM-BHQ). Standard curves for detection of vector (WPRE) and cells (Albumin) were prepared using pCCL-PGK-eGFP and pAlbumin (Addgene #22037) plasmids, respectively.
Southern Blot Analysis
25 μg of genomic DNA (prepared as described above) was digested overnight at 37°C in a total volume of 200 μL with SalI restriction enzyme (New England Biolabs). The digested DNA was precipitated with sodium acetate and resuspended in 20 μL Tris EDETA (TE). The DNA fragments were resolved on a 0.8% agarose gel overnight at 1 ultraviolet (V)/cm and transferred to a nylon membrane (Amersham Hybond-N+, GE Healthcare) by vacuum suction. During transfer, the DNA was nicked, denatured, and neutralized in 0.25 M HCl, 1.5 M NaCl + 0.5 M NaOH, and 1.5 M NaCl + 1 M Tris-HCl (pH 7.6), respectively. Subsequently, the membrane was washed with 20× saline sodium citrate (SSC). The DNA was ultraviolet (UV)-cross-linked to the membrane and pre-hybridized with 0.4 mg/mL salmon sperm single-stranded DNA (ssDNA) (D1626, Sigma-Aldrich) in 5× saline sodium phosphate EDTA (SSPE) (0.75 M NaCl, 0.05 M NaH2PO4, 0.005 M EDTA), 5× Denhart’s (0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% BSA), 1% SDS, 50% formamide, and 5% dextran-sulfate for 4 hr at 42°C. A GFP probe of 740 bp was cytidine triphosphate (CTP) 32P-labeled (Prime-It II random primer labeling kit, Stratagene) and mixed with salmon sperm ssDNA (1 mg/mL) and hybridization buffer and finally incubated with the membrane at 42°C overnight. The membrane was washed in 2× SSPE for 2 × 5 min at 26°C and in 2× SSPE/0.5% SDS for 2 × 15 min at 53°C and finally in 0.2× SSPE/0.5% SDS for 1 × 5–15 min at 53°C. The membrane was air-dried and enclosed in a cassette for exposure of an X-ray film (Konica Minolta).
Quantification of Phenylalanine in Serum
Blood was withdrawn from the retro-orbital cavity of treated mice and spotted on FTA DMPK-C filter cards (Whatman) for measurements of phenylalanine and tyrosine. This was carried out by Statens Serum Institut (SSI), who is responsible for the Danish newborn screening program, by tandem mass spectrometry analysis (Lund et al.48 and references herein).
In Vitro Assay for Phenylalanine Turnover
HEK293 cells were transduced with a mixture of lentiviral vectors encoding mPTPS and mGTPCH, and the ability to reduce phenylalanine levels in the growth media was used as a measure of PAH enzyme activity from either transduced or transfected (data not shown) cultures. In brief, 105 HEK293 cells seeded 1 day earlier in 6-well dishes were transduced with different mixtures of lentiviral vectors with at total MOI of 120 transducing units per cell. pCCL-PGK-eGFP was used as stuffer vector. Two weeks later, 9 × 105 transduced cells were seeded in 6-well dishes in triplicates. The same number of HepG2 cells were seeded as a positive control. The following day, cells were transfected using FuGENE6 transfection reagent (Promega) with pCCL-PGK-mPAH or pCCL-PGK-HAtag-mPAH and pCCL-PGK-eGFP, the latter of which was used as stuffer plasmid DNA. Each well received a total of 4.25 μg DNA. The next day, medium was changed to 1 mL fresh medium. 24 hr later, 30-μL cell supernatant samples were spotted on FTA DMPK-C cards (Whatman) and sent to SSI for phenylalanine determination as described above.
Statistical Analysis
Data are presented as the mean ± SD. Statistical differences between two groups were evaluated using a two-tailed Student’s t test. p < 0.05 was considered statistically significant. Statistical differences between three or more groups were evaluated using one-way ANOVA for comparison of multiple groups (Dunnett’s). A p value of < 0.05 was considered statistically significant. Statistical differences between three groups not following Gaussian distributions were evaluated after log transformation of the data.
Author Contributions
T.D. and L.A. designed the vectors, and they were produced by T.D. and R.O.B. T.D. and P.O.A. tested the vectors in vitro. F.D.-H. injected the animals, and T.D., C.R.C., and F.D.-H. carried out IVIS scanning. The PKU mice were supplied by T.G.J., and D.H. analyzed serum phenylalanine levels. Copy-number analysis was carried out by C.R.C. and A.L.A. The study was conceived by L.A. and supervised by T.G.J., T.J.C., J.G.M., and L.A. The manuscript was drafted by L.A. and revised by T.J.C. and J.G.M. All authors read and approved the manuscript.
Conflicts of Interest
No competing financial interests exist.
Acknowledgments
We would like to thank Birgit Holm Hansen, Anne-Kerstine Thomassen, Christian Knudsen, Lisbeth Dahl Schrøder, Tina Hindkjær, and Erik Kaadt for their technical assistance at Aarhus University, and Arieh Cohen and Susan Svane Laursen at Statens Serum Institut for analyzing the serum samples. We also thank Daniel Miotto Dupont for his help with the bioluminescence scanning and for the use of the IVIS spectrum system. We would like to thank the Interdisciplinary Nanoscience Center at University of Aarhus. The project was financially supported by the Karen Elise Jensen foundation, the Riisfort foundation, the Frimodt-Heineke Foundation, and the Lundbeck foundation (R126-2012-12456).
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.07.005.
Supplemental Information
References
- 1.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoo T., Kamimura K., Abe H., Kobayashi Y., Kanefuji T., Ogawa K., Goto R., Oda M., Suda T., Terai S. Liver-targeted hydrodynamic gene therapy: Recent advances in the technique. World J. Gastroenterol. 2016;22:8862–8868. doi: 10.3748/wjg.v22.i40.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang G., Gao X., Song Y.K., Vollmer R., Stolz D.B., Gasiorowski J.Z., Dean D.A., Liu D. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 5.Zhao F., Liang S.Q., Zhou Y., Wang Y.L., Yan H., Wang X.H., Wang H.P., Du J., Zhan L.S. Evaluation of hepatitis B virus promoters for sustained transgene expression in mice by bioluminescence imaging. Virus Res. 2010;149:162–166. doi: 10.1016/j.virusres.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Umemoto Y., Kawakami S., Otani Y., Higuchi Y., Yamashita F., Hashida M. Evaluation of long-term gene expression in mouse liver using PhiC31 integrase and hydrodynamic injection. Biol. Pharm. Bull. 2012;35:1182–1186. doi: 10.1248/bpb.b12-00083. [DOI] [PubMed] [Google Scholar]
- 7.Miao C.H., Thompson A.R., Loeb K., Ye X. Long-term and therapeutic-level hepatic gene expression of human factor IX after naked plasmid transfer in vivo. Mol. Ther. 2001;3:947–957. doi: 10.1006/mthe.2001.0333. [DOI] [PubMed] [Google Scholar]
- 8.Ohashi K., Nakai H., Couto L.B., Kay M.A. Modified infusion procedures affect recombinant adeno-associated virus vector type 2 transduction in the liver. Hum. Gene Ther. 2005;16:299–306. doi: 10.1089/hum.2005.16.299. [DOI] [PubMed] [Google Scholar]
- 9.Song S., Embury J., Laipis P.J., Berns K.I., Crawford J.M., Flotte T.R. Stable therapeutic serum levels of human alpha-1 antitrypsin (AAT) after portal vein injection of recombinant adeno-associated virus (rAAV) vectors. Gene Ther. 2001;8:1299–1306. doi: 10.1038/sj.gt.3301422. [DOI] [PubMed] [Google Scholar]
- 10.Condiotti R., Curran M.A., Nolan G.P., Giladi H., Ketzinel-Gilad M., Gross E., Galun E. Prolonged liver-specific transgene expression by a non-primate lentiviral vector. Biochem. Biophys. Res. Commun. 2004;320:998–1006. doi: 10.1016/j.bbrc.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Park F., Ohashi K., Kay M.A. Therapeutic levels of human factor VIII and IX using HIV-1-based lentiviral vectors in mouse liver. Blood. 2000;96:1173–1176. [PubMed] [Google Scholar]
- 12.Cots D., Bosch A., Chillón M. Helper dependent adenovirus vectors: progress and future prospects. Curr. Gene Ther. 2013;13:370–381. doi: 10.2174/156652321305131212125338. [DOI] [PubMed] [Google Scholar]
- 13.Ding Z., Harding C.O., Rebuffat A., Elzaouk L., Wolff J.A., Thöny B. Correction of murine PKU following AAV-mediated intramuscular expression of a complete phenylalanine hydroxylating system. Mol. Ther. 2008;16:673–681. doi: 10.1038/mt.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bursill C.A., McNeill E., Wang L., Hibbitt O.C., Wade-Martins R., Paterson D.J., Greaves D.R., Channon K.M. Lentiviral gene transfer to reduce atherosclerosis progression by long-term CC-chemokine inhibition. Gene Ther. 2009;16:93–102. doi: 10.1038/gt.2008.141. [DOI] [PubMed] [Google Scholar]
- 15.Uetsuki T., Naito A., Nagata S., Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J. Biol. Chem. 1989;264:5791–5798. [PubMed] [Google Scholar]
- 16.Mikkelsen J.G., Yant S.R., Meuse L., Huang Z., Xu H., Kay M.A. Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol. Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 17.Singer-Sam J., Keith D.H., Tani K., Simmer R.L., Shively L., Lindsay S., Yoshida A., Riggs A.D. Sequence of the promoter region of the gene for human X-linked 3-phosphoglycerate kinase. Gene. 1984;32:409–417. doi: 10.1016/0378-1119(84)90016-7. [DOI] [PubMed] [Google Scholar]
- 18.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schorpp M., Jäger R., Schellander K., Schenkel J., Wagner E.F., Weiher H., Angel P. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24:1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobsen J.E., Johansen M.G., Schmidt M., Dagnæs-Hansen F., Dam K., Gunnarsson A., Liu Y., Kragh P.M., Li R., Holm I.E. Generation of minipigs with targeted transgene insertion by recombinase-mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT) Transgenic Res. 2013;22:709–723. doi: 10.1007/s11248-012-9671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao C.H., Ohashi K., Patijn G.A., Meuse L., Ye X., Thompson A.R., Kay M.A. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol. Ther. 2000;1:522–532. doi: 10.1006/mthe.2000.0075. [DOI] [PubMed] [Google Scholar]
- 22.Holm D.A., Dagnaes-Hansen F., Simonsen H., Gregersen N., Bolund L., Jensen T.G., Corydon T.J. Expression of short-chain acyl-CoA dehydrogenase (SCAD) proteins in the liver of SCAD deficient mice after hydrodynamic gene transfer. Mol. Genet. Metab. 2003;78:250–258. doi: 10.1016/s1096-7192(03)00038-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhou T., Kamimura K., Zhang G., Liu D. Intracellular gene transfer in rats by tail vein injection of plasmid DNA. AAPS J. 2010;12:692–698. doi: 10.1208/s12248-010-9231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bak R.O., Stenderup K., Rosada C., Petersen L.B., Moldt B., Dagnæs-Hansen F., Jakobsen M., Kamp S., Jensen T.G., Dam T.N., Mikkelsen J.G. Targeting of human interleukin-12B by small hairpin RNAs in xenografted psoriatic skin. BMC Dermatol. 2011;11:5. doi: 10.1186/1471-5945-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunetti-Pierri N., Stapleton G.E., Palmer D.J., Zuo Y., Mane V.P., Finegold M.J., Beaudet A.L., Leland M.M., Mullins C.E., Ng P. Pseudo-hydrodynamic delivery of helper-dependent adenoviral vectors into non-human primates for liver-directed gene therapy. Mol. Ther. 2007;15:732–740. doi: 10.1038/sj.mt.6300102. [DOI] [PubMed] [Google Scholar]
- 26.Salmon P., Trono D. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. 2006;53 doi: 10.1002/0471142301.ns0421s37. 4.21.1–4.21.23. [DOI] [PubMed] [Google Scholar]
- 27.Ding Z., Georgiev P., Thöny B. Administration-route and gender-independent long-term therapeutic correction of phenylketonuria (PKU) in a mouse model by recombinant adeno-associated virus 8 pseudotyped vector-mediated gene transfer. Gene Ther. 2006;13:587–593. doi: 10.1038/sj.gt.3302684. [DOI] [PubMed] [Google Scholar]
- 28.Yagi H., Ogura T., Mizukami H., Urabe M., Hamada H., Yoshikawa H., Ozawa K., Kume A. Complete restoration of phenylalanine oxidation in phenylketonuria mouse by a self-complementary adeno-associated virus vector. J. Gene Med. 2011;13:114–122. doi: 10.1002/jgm.1543. [DOI] [PubMed] [Google Scholar]
- 29.Brunetti-Pierri N., Palmer D.J., Mane V., Finegold M., Beaudet A.L., Ng P. Increased hepatic transduction with reduced systemic dissemination and proinflammatory cytokines following hydrodynamic injection of helper-dependent adenoviral vectors. Mol. Ther. 2005;12:99–106. doi: 10.1016/j.ymthe.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Kanefuji T., Yokoo T., Suda T., Abe H., Kamimura K., Liu D. Hemodynamics of a hydrodynamic injection. Mol. Ther. Methods Clin. Dev. 2014;1:14029. doi: 10.1038/mtm.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suda T., Gao X., Stolz D.B., Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]
- 32.Crespo A., Peydró A., Dasí F., Benet M., Calvete J.J., Revert F., Aliño S.F. Hydrodynamic liver gene transfer mechanism involves transient sinusoidal blood stasis and massive hepatocyte endocytic vesicles. Gene Ther. 2005;12:927–935. doi: 10.1038/sj.gt.3302469. [DOI] [PubMed] [Google Scholar]
- 33.Amirache F., Lévy C., Costa C., Mangeot P.E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 34.Brown B.D., Sitia G., Annoni A., Hauben E., Sergi L.S., Zingale A., Roncarolo M.G., Guidotti L.G., Naldini L. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109:2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 35.Zhang G., Song Y.K., Liu D. Long-term expression of human alpha1-antitrypsin gene in mouse liver achieved by intravenous administration of plasmid DNA using a hydrodynamics-based procedure. Gene Ther. 2000;7:1344–1349. doi: 10.1038/sj.gt.3301229. [DOI] [PubMed] [Google Scholar]
- 36.Aliño S.F., Herrero M.J., Noguera I., Dasí F., Sánchez M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- 37.Kamimura K., Suda T., Xu W., Zhang G., Liu D. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol. Ther. 2009;17:491–499. doi: 10.1038/mt.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamimura K., Kanefuji T., Yokoo T., Abe H., Suda T., Kobayashi Y., Zhang G., Aoyagi Y., Liu D. Safety assessment of liver-targeted hydrodynamic gene delivery in dogs. PLoS ONE. 2014;9:e107203. doi: 10.1371/journal.pone.0107203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sendra L., Miguel A., Pérez-Enguix D., Herrero M.J., Montalvá E., García-Gimeno M.A., Noguera I., Díaz A., Pérez J., Sanz P. Studying Closed Hydrodynamic Models of “In Vivo” DNA Perfusion in Pig Liver for Gene Therapy Translation to Humans. PLoS ONE. 2016;11:e0163898. doi: 10.1371/journal.pone.0163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabre J.W., Grehan A., Whitehorne M., Sawyer G.J., Dong X., Salehi S., Eckley L., Zhang X., Seddon M., Shah A.M. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008;15:452–462. doi: 10.1038/sj.gt.3303079. [DOI] [PubMed] [Google Scholar]
- 41.Zacharoulis D., Rountas C., Katsimpoulas M., Morianos J., Chatziandreou I., Vassilopoulos G. Efficient liver gene transfer with foamy virus vectors. Med. Sci. Monit. Basic Res. 2013;19:214–220. doi: 10.12659/MSMBR.883996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunetti-Pierri N., Stapleton G.E., Law M., Breinholt J., Palmer D.J., Zuo Y., Grove N.C., Finegold M.J., Rice K., Beaudet A.L. Efficient, long-term hepatic gene transfer using clinically relevant HDAd doses by balloon occlusion catheter delivery in nonhuman primates. Mol. Ther. 2009;17:327–333. doi: 10.1038/mt.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunetti-Pierri N., Liou A., Patel P., Palmer D., Grove N., Finegold M., Piccolo P., Donnachie E., Rice K., Beaudet A. Balloon catheter delivery of helper-dependent adenoviral vector results in sustained, therapeutic hFIX expression in rhesus macaques. Mol. Ther. 2012;20:1863–1870. doi: 10.1038/mt.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baekelandt V., Eggermont K., Michiels M., Nuttin B., Debyser Z. Optimized lentiviral vector production and purification procedure prevents immune response after transduction of mouse brain. Gene Ther. 2003;10:1933–1940. doi: 10.1038/sj.gt.3302094. [DOI] [PubMed] [Google Scholar]
- 46.Holst H.U., Dagnaes-Hansen F., Corydon T.J., Andreasen P.H., Jørgensen M.M., Kølvraa S., Bolund L., Jensen T.G. LDL receptor-GFP fusion proteins: new tools for the characterisation of disease-causing mutations in the LDL receptor gene. Eur. J. Hum. Genet. 2001;9:815–822. doi: 10.1038/sj.ejhg.5200718. [DOI] [PubMed] [Google Scholar]
- 47.Shedlovsky A., McDonald J.D., Symula D., Dove W.F. Mouse models of human phenylketonuria. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund A.M., Hougaard D.M., Simonsen H., Andresen B.S., Christensen M., Dunø M., Skogstrand K., Olsen R.K., Jensen U.G., Cohen A. Biochemical screening of 504,049 newborns in Denmark, the Faroe Islands and Greenland—experience and development of a routine program for expanded newborn screening. Mol. Genet. Metab. 2012;107:281–293. doi: 10.1016/j.ymgme.2012.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.