Abstract
Emerging evidence has proven that long noncoding RNAs (lncRNAs) play important roles in human colorectal cancer (CRC) biology, although few lncRNAs have been characterized in CRC. Therefore, the functional significance of lncRNAs in the malignant progression of CRC still needs to be further explored. In this study, through analyzing TCGA RNA sequencing data and other publicly available microarray data, we found a novel lncRNA, LINC00460, whose expression was significantly upregulated in CRC tissues compared to adjacent normal tissues. Consistently, real-time qPCR results also verified that LINC00460 was overexpressed in CRC tissues and cells. Furthermore, high LINC00460 expression levels in CRC specimens were correlated with larger tumor size, advanced tumor stage, lymph node metastasis and shorter overall survival. In vitro and in vivo assays of LINC00460 alterations revealed a complex integrated phenotype affecting cell growth and apoptosis. Mechanistically, LINC00460 repressed Krüppel-like factor 2 (KLF2) transcription by binding to enhancer of zeste homolog 2 (EZH2). LINC00460 also functioned as a molecular sponge for miR-149-5p, antagonizing its ability to repress cullin 4A (CUL4A) protein translation. Taken together, our findings support a model in which the LINC00460/EZH2/KLF2 and LINC00460/miR-149-5p/CUL4A crosstalk serve as critical effectors in CRC tumorigenesis and progression, suggesting new therapeutic directions in CRC.
Keywords: LINC00460, KLF2, CUL4A, proliferation, apoptosis, colorectal cancer
Introduction
Colorectal cancer (CRC) is ranked among the highest incidence cancers worldwide in both men and women, and there are nearly 1.6 million new cases of CRC each year.1 In 2015, China had 376,300 estimated new cases and 191,000 estimated deaths due to CRC.2 Although some studies have identified many genes involved in tumorigenesis and tumor growth, the molecular mechanisms underlying these processes remain largely unknown.3, 4 Thus, it is of urgent necessity to further investigate the mechanisms underlying CRC development and progression and identify novel promising therapeutic targets with prognostic and therapeutic value.
Recently, next-generation sequencing techniques unraveled the existence of a large portion of transcribed noncoding elements in the human whole genomes.5, 6 Among these elements, the long noncoding RNAs (lncRNAs) have recently emerged as potent regulators of gene expression.7 Generally, lncRNAs are >200 nt long with no or limited protein-coding capacity.8, 9 Increasing evidences have suggested that lncRNA expression is frequently dysregulated in cancer, and specific lncRNAs are correlated with cancer recurrence, metastasis, and poor prognosis in different kinds of cancer, including CRC.10, 11, 12 For example, lncRNA HOXA11-AS could promote gastric cancer tumorigenesis by serving as a scaffold and recruiting multiple chromosome modification enzymes to target specific genes. Alternatively, it can also act as competing endogenous RNAs (ceRNAs) targeting specific microRNA (miRNA), thereby regulating gene expression transcriptionally and post-transcriptionally.13 lncRNA MEG3 could suppress acute myeloid leukemia through both p53-dependent and p53-independent pathways.14 Our previous work also identified an lncRNA FOXP4-AS1, which is an unfavorable prognostic factor and regulates cell proliferation and apoptosis in CRC.15 However, some key issues in systematic identification and characterization of candidate lncRNAs involved in CRC, especially in prognosis for CRC patients, need to be resolved.
In the present study, we identified novel lncRNAs that were dysregulated in the development of CRC. To comprehensively characterize those aberrantly expressed lncRNAs, we analyzed The Cancer Genome Atlas (TCGA) CRC and normal tissue RNA sequencing data (49 normal and 648 cancer samples) and two independent microarray datasets from GEO: GSE33113 and GSE41328. In addition to identifying several CRC-associated lncRNAs, we also discovered a novel lncRNA termed LINC00460, which displayed significantly increasing expression levels in CRC tissues. The elucidation of its functional association and underlying molecular mechanism involved in CRC will provide better understanding of the role of lncRNA in gene regulation and tumor biology.
Results
LINC00460 Is Upregulated in Human CRC Tissues and Correlates with Poor Prognosis
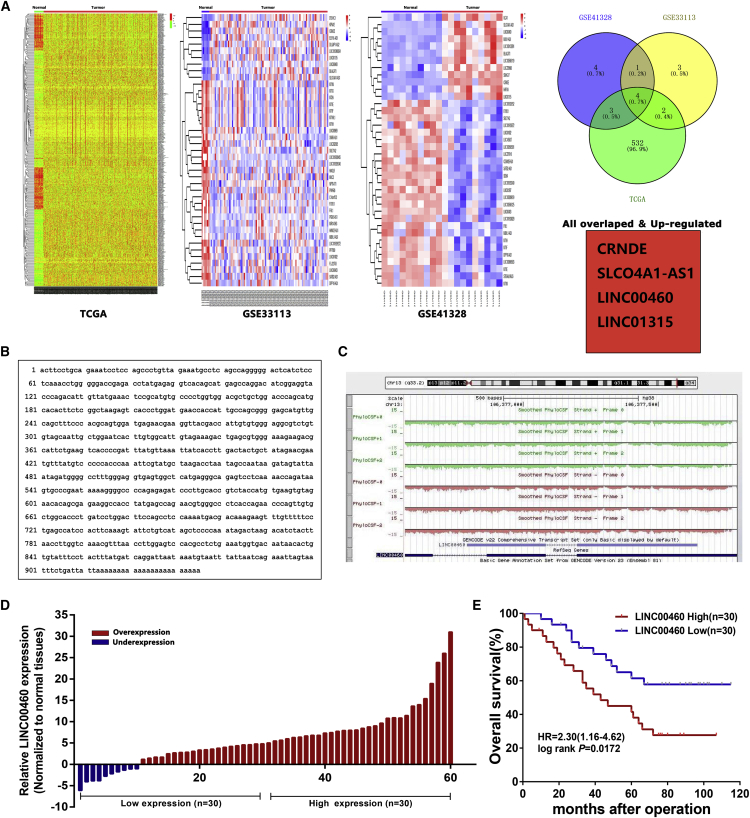
To identify the lncRNAs involved in colorectal tumorigenesis, we performed an integrative analysis of TCGA CRC and normal tissue RNA sequencing data and CRC microarray profile comprising GSE33113 and GES41328 from GEO datasets. We identified 444 lncRNAs that were upregulated in TCGA datasets, 10 in GSE33113 datasets, and 12 in GSE41328 datasets (fold change > 2.0; p < 0.05; Figure 1A). Among all, however, only four lncRNAs were consistently upregulated in all three datasets, including CRNDE, SLCO4A1-AS1, LINC00460, and LINC01315 (Figure 1A). To prioritize the most CRC biologically relevant lncRNAs, we focused on the long intergenic noncoding RNA 00460 (LINC00460), which is most highly expressed in CRC specimens. In addition, LINC00460 is a long noncoding RNA without any functional report in CRC yet. Thus, we selected LINC00460 for further investigation.
Figure 1.
Expression of LINC00460 in CRC Tissues and Its Clinical Parameters
(A) Hierarchical clustering analysis of lncRNAs that were differentially expressed (fold change > 2; p < 0.05) in CRC and normal tissues and overlap of upregulated lncRNAs in TCGA data and GEO datasets. (B) The full sequence of LINC00460 is published in the NCBI database (NR_034119.1). (C) Chromosomal location of LINC00460 and PhyloCSF predicted that LINC00460 has no protein coding potential. The tracks showed the PhyloCSF scores for each codon in each of six frames. Regions with a score greater than 0 were predicted to be coding regions, and regions with a score less than 0 were predicted to be noncoding regions. (D) LINC00460 was detected in 60 pairs of CRC tissues by real-time qPCR. The levels of LINC00460 in CRC tissues are significantly higher than those in non-tumorous tissues. (E) Patients with high levels of LINC00460 expression showed reduced survival times compared with patients with low levels of LINC00460 expression (p = 0.0172; log-rank test). *p < 0.05; **p < 0.01.
LINC00460 is located on chromosome 13q33.2 and has a transcript length of 935 nt (Figure 1B). PhyloCSF predicted that LINC00460 has no protein coding potential (Figure 1C). Next, to validate the expression results from microarray, we detected the level of LINC00460 in 60 paired CRC tissues and adjacent normal tissues by real-time qPCR. As shown in Figure 1D, LINC00460 expression was remarkably overexpressed in 83.3% (50 of 60) CRC tissues. Then, to investigate the relationship between LINC00460 expression and clinicopathological features, we divided the samples into high (above the median; N = 30) and low (below the median; N = 30) LINC00460 expression groups according to the median LINC00460 level (Figure 1D). A chi-square test was performed to compare the clinicopathological features of the two groups. As shown in Table S1, LINC00460 upregulated in CRC tissues was significantly correlated with larger tumor sizes (p = 0.029), advanced tumor node metastasis (TNM) stages (p = 0.020), and lymph node metastasis (p = 0.047) in CRC patients. However, several other clinical parameters were found to be not significantly associated with LINC00460 expression (Table S1).
To determine the relationship between LINC00460 expression and prognosis, Kaplan-Meier analysis was used to evaluate the effects of LINC00460 expression on overall survival (OS). As a result, overexpression of LINC00460 was correlated with a poor prognosis in patients with CRC (p = 0.0172). The results of multivariate analysis of the clinical variables revealed that LINC00460 expression level was considered to be potential survival predictors (Table S2).
LINC00460 Regulates CRC Cell Proliferation and Apoptosis In Vitro
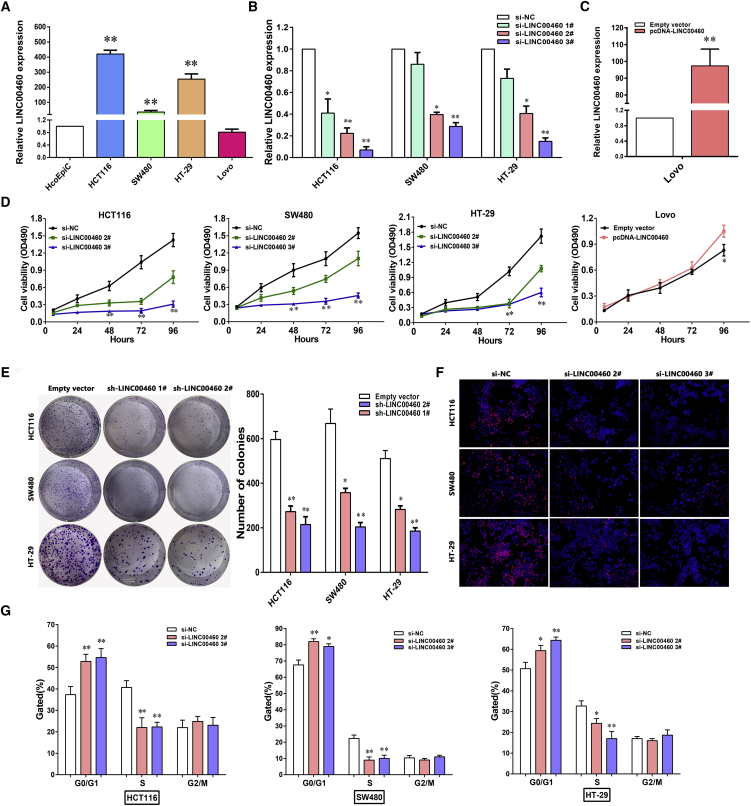
To evaluate the effects of LINC00460 on cellular processes, we first investigated the level of LINC00460 in CRC cell lines, including HCT116, SW480, HT-29, Lovo, and the human colonic epithelial cell line HcoEpiC. We found that LINC00460 expression was significantly upregulated in HCT116 (p < 0.01), SW480 (p < 0.01), and HT-29 (p < 0.01) cells compared with HcoEpiC cells (Figure 2A). Then, small interfering RNAs (siRNAs) were designed to silence LINC00460 in HCT116, SW480, and HT-29 cell lines, and plasmid-mediated overexpression was used for exogenously manipulating expression of LINC0460 in Lovo and HcoEpiC (Figures 2B, 2C, and S1A). 3-(4,5)-dimethylthiahiazo (-z-y)-3,5-di-phenytetrazoliumromide (MTT) assays revealed that knockdown of LINC00460 expression significantly inhibited cell proliferation compared with the control cells, which was also consistently confirmed by colony-formation assay and EdU assay (Figures 2E and 2F). In contrast, overexpressed LINC00460 could promote cell proliferation in Lovo, but not in HcoEpiC cell lines (Figures 2D and S1B). Consistent with the results of the MTT assay, the result of colony-formation assay and EdU assay also showed that cell growth ability was significantly inhibited following knockdown of LINC00460 (Figures 2E and 2F). Next, flow cytometric analysis was performed to further examine the effect of LINC00460 on the proliferation of CRC cells by altering cell cycle progression. The results indicated that siRNA-LINC00460 treatment induced significant G0/G1 phase arrest and decreased the percentage of cells in the S phase (Figure 2G).
Figure 2.
LINC00460 Regulates CRC Cell Proliferation In Vitro
(A) LINC00460 expression was detected by real-time qPCR in colorectal cancer cell lines (HCT116, SW480, HT-29, and Lovo) and compared with the normal human colonic epithelial cell line (HCoEpiC). (B and C) HCT116, SW480, and HT-29 cells (B) were transfected with si-LINC00460; Lovo cells (C) were transfected with pcDNA-LINC00460. (D) MTT assays were performed to determine the cell viability for si-LINC00460-transfected HCT116, SW480, and HT-29, and Lovo cells transfected with pcDNA-LINC00460. (E and F) Colony-forming assays (E) and EDU staining assays (F) were used to determine the proliferation of si-LINC00460-transfected HCT116, SW480, and HT-29 cells. (G) Flow cytometry assays were performed to analyze the cell cycle progression when CRC cells transfected with si-LINC00460. The bar chart represented the percentage of cells in G0/G1, S, or G2/M phase, as indicated. The data are presented as the mean ± SD of three independent experiments; *p < 0.05; **p < 0.01.
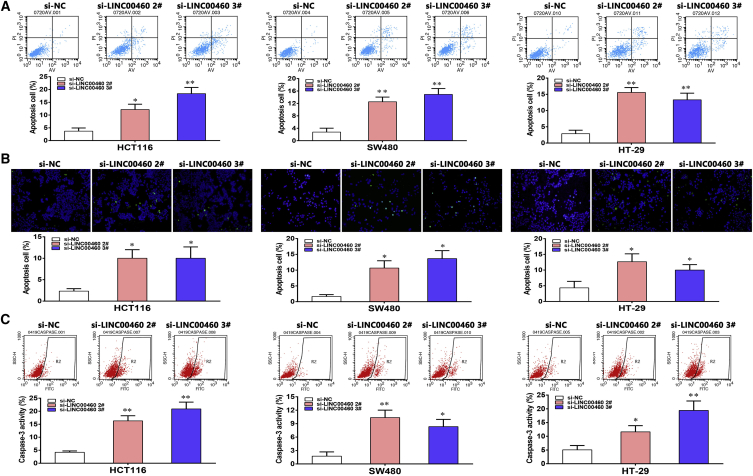
The flow cytometry results and TUNEL staining assays showed that the fraction of apoptotic cells was significantly increased among the siRNA-treated cells compared with the control cells (Figures 3A and 3B), which convinced us that CRC cell proliferation was influenced by apoptosis. Because caspase-3 plays a pivotal role in apoptosis execution phase, we next investigated whether caspase-3 was activated in CRC cells upon knockdown of LINC00460 by annexin V and propidium iodide double staining via fluorescence-activated cell sorting (FACS) analysis.16, 17 The results indicated that caspase-3 levels were increased in the siRNA-treated CRC cells, confirming that LINC00460 is involved in cell apoptosis (Figure 3C).
Figure 3.
LINC00460 Regulates CRC Cell Apoptosis In Vitro
(A) Flow cytometry assays were performed to analyze the cell apoptosis in si-LINC00460-transfected HCT116, SW480, and HT-29 cells. LL, dead cells; LR, early apoptotic cells; UL, viable cells; UR, terminal apoptotic cells. (B) Tunel staining assays were performed to analyze the cell apoptosis when LINC00460 was knocked down. Green, apoptotic cell; blue, DAPI staining for nuclear. (C) Cleaved caspase-3 was detected by annexin V+-PI staining and analyzed by flow cytometry. The data are presented as the mean ± SD of three independent experiments; *p < 0.05; **p < 0.01.
LINC00460 Promotes CRC Growth In Vivo
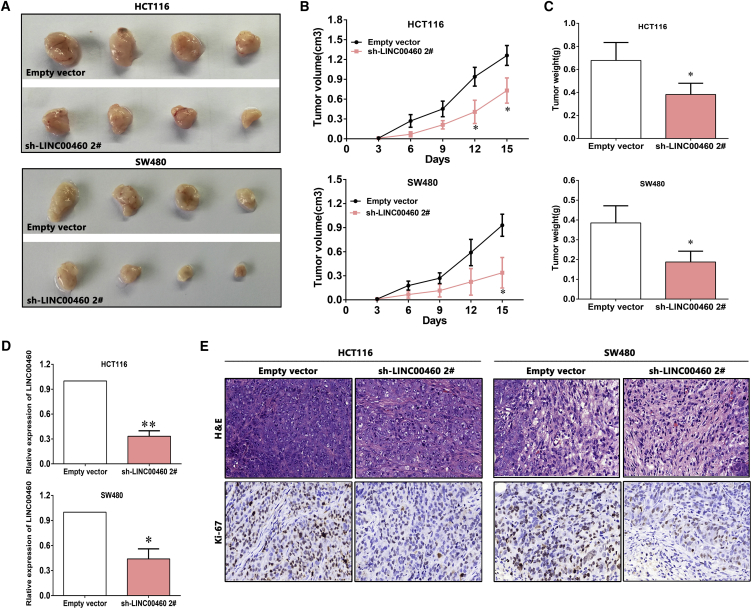
To further demonstrate the biological function of LINC00460 in CRC in vivo, we established xenograft tumor models in nude mice using both HCT116 and SW480 cells transfected with empty vector or sh-LINC00460. At 15 days post-injection, the tumor growth in the sh-LINC00460 group was significantly slower than that in the control group (Figure 4A), together with the obviously decreased tumor volumes and weights (Figures 4B and 4C), which indicated that LINC00460 regulates CRC cell proliferation in vivo. As shown in Figure 4D, real-time qPCR confirmed that the LINC00460 expression level was lower in the tumor tissues derived from the sh-LINC00460-transfected cells. Additionally, immunohistochemistry (IHC) analysis showed that the tumors formed from HCT116/sh-LINC00460 and SW480/sh-LINC00460 cells displayed weaker Ki-67 staining than those formed from the control cells (Figure 4E).
Figure 4.
LINC00460 Regulates CRC Cell Growth In Vivo
(A) The stable LINC00460 knockdown HCT116 and SW480 cells were used for the in vivo assays. The total numbers of tumors after removal from the mice is shown. (B) The tumor volume was calculated every 3 days. (C) Tumor weights from two groups are represented. (D) LINC00460 expression level in tumor tissues formed from HCT116/SW480/sh-LINC00460 and HCT116/SW480/empty vector was detected by real-time qPCR. (E) Tumors developed from sh-LINC00460-transfected HCT116/SW480 cells showed lower Ki-67 protein levels than tumors developed from control cells. Upper: H&E staining is shown; lower: immunostaining is shown. *p < 0.05; **p < 0.01.
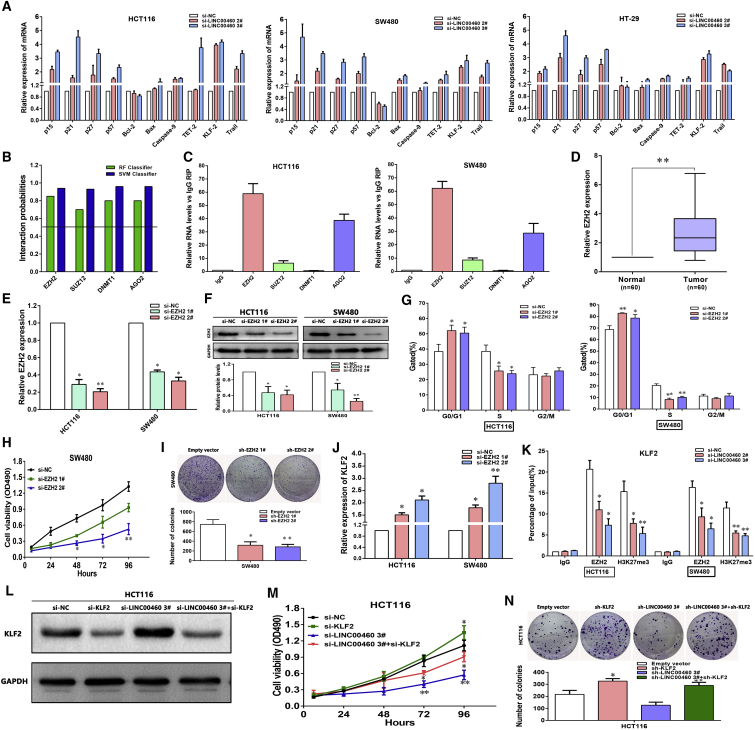
LINC00460 Promotes CRC Proliferation by Interacting with EZH2 and Repressing KLF2 Expression
To further explore the molecular mechanisms about how LINC00460 contributes to the proliferation phenotype of CRC cells, we detected the alteration of some cell-proliferation-related genes upon LINC00460 knockdown, including p15, p21, p27, p57, Bcl-2, Bax, caspase-9, TET-2, KLF2, and Trail. We found that p15, p21, p27, p57, KLF2, and Trail were consistently upregulated in all three CRC cell lines (Figure 5A). Among these altered genes, KLF2 has been identified as a well-known tumor suppressor involved in cancer cell proliferation and apoptosis.18 Therefore, we chose KLF2 for further investigation.
Figure 5.
LINC00460 Promotes CRC Cell Proliferation by Binding to EZH2 and Repressing KLF2
(A) The levels of p15, p21, p27, p57, Bcl-2, Bax, caspase-9, TET-2, KLF2, and Trail mRNA were detected by real-time qPCR when knockdown of LINC00460 in HCT116, SW480, and HT-29 cells. (B) Bioinformatics were used to predict the interaction probabilities of LINC00460 and RNA-binding proteins via RNA-protein interaction prediction (http://pridb.gdcb.iastate.edu/rpiseq/). Predictions with probabilities >0.5 were considered positive. RPISeq predictions are based on random forest (RF) or support vector machine (SVM). (C) RIP experiments were performed, and the co-precipitated RNA was subjected to real-time qPCR for LINC00460. The fold enrichment of LINC00460 in RIPs is relative to its matching IgG control RIP. (D) By real-time qPCR assays, the level of EZH2 was obviously upregulated in 60 pairs of CRC tissues. (E) The levels of EZH2 mRNA expression were determined by real-time qPCR when HCT116 and SW480 cells were transfected with si-NC and si-EZH2. (F) The EZH2 protein levels were determined by western blot in EZH2 knockdown HCT116 and SW480 cells. (G) Flow cytometry assays were performed to analyze the cell cycle progression when CRC cells transfected with si-EZH2. (H and I) MTT (H) and colony-forming assays (I) were used to determine the cell viability when SW480 cells were transfected with si-NC and si-EZH2. (J) Real-time qPCR assays were used to detect the levels of KLF2 mRNA expression in HCT116 and SW480 cells transfected with si-EZH2. (K) ChIP shows EZH2 occupancy on the KLF2 promoter regions, and knockdown of LINC00460 decreases their occupancy. (L) Western blot detection of the KLF2 protein levels in HCT116 cells after co-transfection with LINC00460, KLF2, or NC siRNAs. (M and N) Growth ability for HCT116 cells after co-transfection with LINC00460, KLF2, or NC siRNAs was determined by MTT (M) and colony-forming assays (N). The data are presented as the mean ± SD of three independent experiments; *p < 0.05; **p < 0.01.
Previous works have revealed that lncRNAs contribute to cancer cells phenotype via binding with specific RNA-binding proteins (RBPs), thus silencing certain tumor suppressors.19 Based on this finding, bioinformatics analysis was performed to predict the interaction probabilities of LINC00460 and RBPs (http://pridb.gdcb.iastate.edu/rpiseq/). The results indicated that LINC00460 could potentially bind to EZH2, SUZ12, DNMT1, and AGO2 (as the support vector machine [SVM] or random forest [RF] score >0.5; Figure 5B). We further performed RNA immunoprecipitation (RIP) assays and confirmed that LINC00460 could interact with EZH2 and AGO2, but not other RNA-binding proteins, in both HCT116 and SW480 cells (Figure 5C).
Thereafter, to investigate the roles of EZH2 in CRC, we performed real-time qPCR analysis and found that the expression level of EZH2 was significantly increased in 20 pairs of CRC tissues (Figure 5D). Consistent with the effect of silencing LINC00460, further analysis demonstrated that knockdown of EZH2 also inhibits CRC cell proliferation and promotes cell cycle arrest (Figures 5E–5I). Moreover, EZH2 siRNAs were used to transfect HCT116 and SW480 cells. The results showed that EZH2 expressions were effectively knocked down (Figures 5E and 5F), and KLF2 mRNA levels were increased (Figure 5J). To address whether LINC00460 is involved in transcriptional repression via enrichment of EZH2 to target gene KLF2 promoters, we conducted chromatin immunoprecipitation (ChIP) analysis by LINC00460-knockdown. The results showed that EZH2 could directly bind to KLF2 promoter regions and induce H3K27me3 modification in HCT116 and SW480 cells (Figure 5K).
Moreover, to investigate whether LINC00460 regulates CRC cell growth by repressing KLF2 expression, rescue assays were performed. HCT116 cells were co-transfected with si-LINC00460 and si-KLF2, which rescued the increased expression of KLF2 induced by silencing LINC00460 (Figure 5L). The results of MTT and colony-formation assays results showed that co-transfection could partially rescue si-LINC00460-suppressed growth ability in HCT116 cells (Figures 5M and 5N).
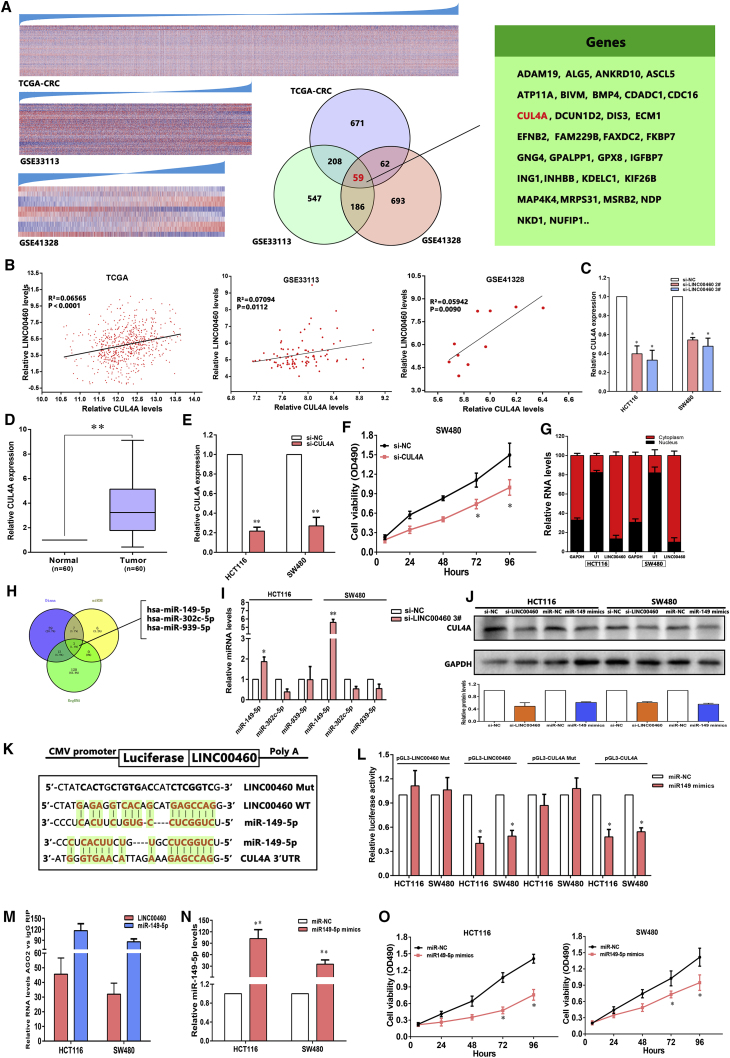
LINC00460 Promotes CUL4A Expression by Competing for miR-149-4p in Cytoplasm, Thus Enhancing CRC Cell Proliferation
Guilt-by-association (GBA) analysis has proposed that the function of a poorly characterized lncRNA gene can be inferred on the basis of known biological functions of protein-coding genes (PCGs), with which it is co-expressed.20 To verify this hypothesis, we conducted GBA analysis for LINC00460. TCGA RNA sequencing data and two independent microarray datasets from GEO (GSE33113 and GSE41328) were analyzed to identify candidate PCGs whose expression was significantly correlated with LINC00460 expression. We found that 5.9% (59 of 1,000) of LINC00460-associated PCGs were shared by all three microarray profiles (Figure 6A).
Figure 6.
LINC00460 Represses CUL4A Expression by Competing for miR-149-5p in Cytoplasm, Thus Facilitating CRC Cell Proliferation
(A) Heatmap of protein-coding genes that were significantly and positively co-expressed with LINC00460. The genes were arranged from top to bottom in ascending order of their correlation with LINC00460. Venn diagrams of LINC00460-associated genes among TCGA-CRC, GSE33113, and GSE41328 are shown. (B) The relationship between LINC00460 expression and CUL4A mRNA levels was analyzed in the profile of CRC patient tissue from TCGA and GEO. (C) The levels of CUL4A mRNA expression were determined by real-time qPCR when HCT116 and SW480 cells were transfected with si-NC and si-LINC00460. (D) By real-time qPCR assays, the level of CUL4A was remarkably upregulated in 60 pairs of CRC tissues. (E and F) Cell viability was determined upon knockdown of CUL4A in SW480 cells. (E) HCT116 and SW480 cells were transfected with si-CUL4A. (F) MTT assays were used to determine the cell viability for si-CUL4A-transfected SW480 cells. (G) Relative LINC00460 levels in HCT116 and SW480 cell cytoplasm or nucleus were detected by real-time qPCR. GAPDH was used as cytoplasm control, and U1 was used as nuclear control. (H) Potential binding sites of LINC00460-miRNA-CUL4A using the online software programs are shown. (I) Real-time qPCR assays detected the expression of miR-149-5p/miR-302c-5p/miR-939-5p after knockdown of LINC00460 in HCT116 and SW480 cells. (J) Western blot detection of the CUL4A protein levels in HCT116 and SW480 cells after knockdown of LINC00460 and overexpression of miR-149-5p is shown. (K) A photograph showing the predicted interaction between LINC00460/miR-149-5p and miR-149-5p/CUL4A mRNA through complementary base pairs is shown. (L) Both LINC00460 and CUL4A are targeted by miR-149-5p. The 3′ UTR of CUL4A mRNA and full length of LINC00460 were respectively inserted downstream of a luciferase gene. The reporter vector was co-transfected with a Renilla luciferase vector (for normalization) to SW480 and HT116, which were treated by miR-149-5p mimics or control mimics. The luciferase signals of both reporter genes were significantly decreased when cells were treated with miR-149-5p mimics. Mutant 3′ UTR of CUL4A and LINC00460 is not affected by miR-149-5p. (M) LINC00460 and miR-149-5p RNA levels in immunoprecipitates are presented as fold enrichment in AGO2 relative to IgG immunoprecipitates. (N) miR-149-5p levels were examined in HCT116 and SW480 cells after transfection with mimics. (O) MTT assays showed that cell proliferation was suppressed by miR-149-5p overexpression. The data are presented as the mean ± SD of three independent experiments; *p < 0.05; **p < 0.01.
Among these genes, the expression level of cullin 4A (CUL4A) was significantly and positively correlated with LINC00460 expression. Interestingly, it has already been confirmed that oncogenic CUL4A was involved in multiple carcinoma types by interfering with apoptosis and cell proliferation. Therefore, we further studied the interaction between CUL4A and LIN00460 in CRC.21 The real-time qPCR results showed that LINC00460 knockdown inhibited the expression of CUL4A both in HCT116 and SW480 cells (Figure 6C). To investigate the roles of CUL4A in CRC, we performed real-time qPCR analysis and found that the expression level of CUL4A was remarkably increased in 20 pairs of CRC tissues (Figure 6D). Subsequently, the results of MTT assays revealed that knockdown of CUL4A suppressed SW480 cell proliferation (Figures 6E and 6F).
Recently, growing evidence has indicated that lncRNAs may function as ceRNAs for specific microRNAs, hence modulating their downstream target genes expression in cytoplasm.22, 23 Interestingly, our above findings displayed that LINC00460 could bind to AGO2 protein both in HCT116 and SW480 cell lines (Figure 5C), suggesting that LINC00460 may also function as a ceRNA. To test this hypothesis, we first examine the distribution of LINC00460 in CRC cells. The results indicated that LINC00460 is distributed in both cytoplasm and nucleus, but the ratio of LINC00460 in cytoplasm is much higher (Figure 6G). Next, we performed a mining for potential binding sites of LINC00460-miRNA-CUL4A using the online software programs DIANA TOOLS (http://diana.imis.athena-innovation.gr), miRDB (http://www.mirdb.org/), and RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw) and found that three miRNAs (miR-149-5p, miR-302c-5p, and miR-939-5p) contain complementary sites related to LINC00460-miRNA-CUL4A (Figure 6H). Further experiments found that knockdown of LINC00460 increased miR-149-5p expression, but not others (Figure 6I). Moreover, treatment by si-LINC00460 or miR-149-5p mimics significantly reduced CUL4A protein levels (Figure 6J). To validate the effects of miR-149-5p, we cloned the LINC00460, mutant LINC0046, 3′ UTR of CUL4A, and mutant 3′ UTR of CUL4A downstream of diverse luciferase genes and co-transfected these reporters with miR-149-5p mimics both in HCT116 and SW480 cells. As expected, miR-149-5p significantly decreased the luciferase signals of reporters of LINC00460 and 3′ UTR of CUL4A. However, miR-149-5p had no effect on mutant reporters of LINC00460 and 3′ UTR of CUL4A (Figures 6K and 6L). These results directly confirmed that miR-149-5p could target LINC00460 and CUL4A.
miRNAs are known to be present in the cytoplasm in the form of miRNA ribonucleoprotein complexes containing AGO2, the core component of the RNA-induced silencing complex. To test whether miR-149-5p associates with AGO2, RIP assays were performed on HCT116 and SW480 cell extracts using antibodies against AGO2. As shown in Figure 6M, LINC00460 and miR-149-5p were both remarkably enriched in AGO2-immunoprecipitation relative to control immunoglobulin G (IgG). To investigate whether miR-149-5p functions as a tumor suppressor in CRC cells, we then performed MTT assays. As shown in Figures 6N and 6O, upregulation of miR-149-5p inhibited CRC cell proliferation.
Discussion
So far as we know, this is the first study to clarify the clinical value and biologic function of LINC00460 in CRC. In our investigation, lncRNA LINC00460 was highly expressed in CRC tissues and positively correlated with larger tumor size, advanced tumor stage, and the lymph node metastasis. Moreover, high expression of LINC00460 was an independent factor for poor prognosis in CRC patients. Furthermore, univariate and multivariate Cox regression analyses demonstrated that LINC00460 is a valuable prognostic factor independent of major clinicopathological features, which indicated that LINC00460 is a promising CRC biomarker. We also found that decreased LINC00460 expression inhibited CRC cell proliferation and induced cell apoptosis, which further suggested that LINC00460 might promote CRC progression. Although we tried overexpression of LINC00460 in HcoEpiC, the MTT assays showed that overexpression of LINC00460 expression didn’t affect HcoEpiC cell proliferation compared with the control cells. The inconsistency of LINC00460 in regulating the cell proliferation in CRC cell and normal cell line HcoEpiC may be due to its differential expression phenotype.
To clarify the molecular mechanism of LINC00460 contributing to CRC cell proliferation, we also investigated potential target genes involved in cell growth and apoptosis. Recent studies have indicated that a significant number of lncRNAs have been shown to function in cooperation with chromatin-modifying enzymes to promote epigenetic activation or silencing of gene expression.13, 24 EZH2 is a subunit of polycomb repressive complex 2 (PRC2), which has an effect of H3K27me3 and represses target gene expression.25, 26 Our results found that LINC00460 exerts oncogenic properties via recruiting EZH2 and H3K27me3 to the KLF2 promoter in nucleus, thus leading to inactivation of tumor suppressor KLF2. In addition, overexpression of EZH2 was observed in various types of tumors.26, 27, 28 It is not a surprise that knockdown of EZH2 expression significantly inhibited cell proliferation and induced G0/G1 phase arrest in CRC cell lines. The tumor suppressor roles of KLF2 have been illustrated in many kinds of cancers, including CRC.18, 29, 30, 31 To investigate whether KLF2 was involved in the LINC00460-induced CRC cell proliferation, we carried out rescue experiments, and the results indicated that co-transfection with LINC00460 and KLF2 siRNAs could partially rescue si-LINC00460-impaired proliferation in HCT116 cells. Taken together, these findings support the idea that LINC00460 may suppress KLF2 expression epigenetically and LINC00460 executes its oncogenic effects via LINC00460/EZH2/KLF2 signaling axis in nucleus.
It has been confirmed that cytoplasmic-localized lncRNAs can act as ceRNAs to regulate miRNAs.23, 32 Interestingly, we found that LINC00460 RNAs were mostly located in the cytosol rather than in the nucleus, thus suggesting LINC00460 may regulate other downstream gene by functioning as miRNA sponge. CUL4A, a member of the cullin family of proteins that composes the multifunctional ubiquitin ligase E3 complex, may act as an oncogene that is essential for cellular functions, including proliferation, differentiation, and apoptosis.21, 33, 34 Bioinformatic analysis and luciferase reporter assay proved that the post-transcriptional regulation of CUL4A is partly mediated by LINC00460 in tumor progression of CRC via competing for miR-149-5p in cytoplasm, thus facilitating cell growth of CRC. Consistent with these reports, overexpression of CUL4A was observed in 20 pairs of CRC tissues and knockdown of CUL4A could inhibit cell proliferation in SW480 cell lines. miR-149-5p plays a diverse role in different cancers. Some reports discovered that miR-149-5p, as a tumor suppressor, was downregulated in melanoma cell,35 renal cell carcinoma (RCC),36, 37 and lung adenocarcinoma.38 On the other hand, several studies supported that miR-149-5p promoted tumor growth.39 Our study demonstrated that the restoration of miR-149-5p expression using synthetic mimics suppressed CRC cell proliferation. These results indicated that miR-149-5p act as a tumor suppressor in CRC. Taken together, LINC00460 simultaneously sponges miR-149-5p and then affect the expression of CUL4A genes in the cytosol.
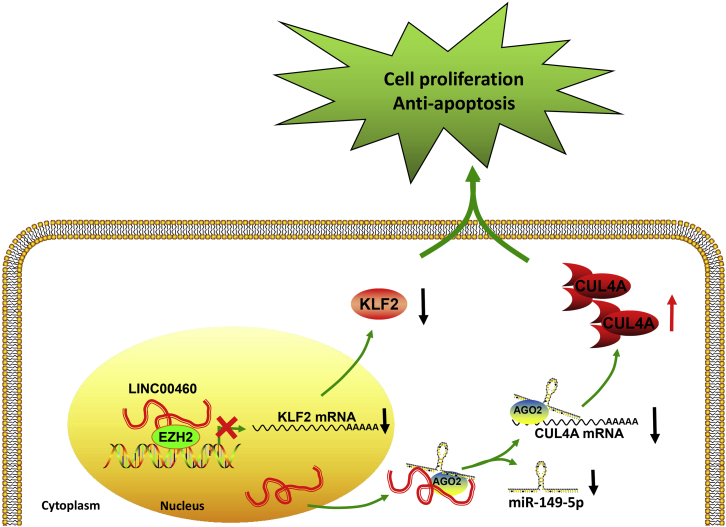
In summary, our work demonstrates that the overexpression of LINC00460 leads to CRC tumorigenesis, indicating that its knockdown can suppress CRC progression and development via both LINC00460/EZH2/KLF2 and LINC00460/miR-149-5p/CUL4A signaling axis, therefore emphasizing the potentials of this axis in CRC diagnosis and therapy (Figure 7). However, due to the complexity of lncRNA biological function, further analysis is required to achieve a more comprehensive understanding in CRC diagnosis and therapy.
Figure 7.
Proposed Model of LINC00460 Regulating Gene Expression Both in the Nucleus and Cytoplasm to Promote Tumor Growth
Materials and Methods
lncRNA Expression Profiling Data Analysis
CRC gene expression data were downloaded from the TCGA and GEO dataset. The independent datasets from GSE33113 and GSE41328 were analyzed in this study. The binary alignment/map (BAM) files and normalized probe-level intensity files were downloaded from TCGA and GEO databases, respectively. The probe sequences were downloaded from GEO or microarray manufacturers, and bowtie was used to re-annotate probes according to GENCODE Release 19 annotation for lncRNAs. For multiple probes corresponding to one gene, the mean value of the multiple probes was selected to generate expression of lncRNA.
Collection of Tissue Specimens
For this study, a total of 60 fresh CRC tissues and paired adjacent non-tumor tissues were obtained from patients who had undergone surgical resection between 2004 and 2011 at the Second Affiliated Hospital, Nanjing Medical University, China. Histopathological diagnosis was confirmed by an experienced pathologist. All of the tissue samples were snap frozen in liquid nitrogen immediately after excision and stored at −80°C until total RNA was extracted. No patients had been treated with radiotherapy or chemotherapy before surgery. All patients provided written informed consent, and the study protocol was approved by the Ethics Committee of Nanjing Medical University.
Cell Culture
Four CRC cell lines (HCT116, SW480, HT-29, and Lovo) were obtained from the American Type Culture Collection (Manassas, VA, USA). All of the cell lines were grown and maintained in RPMI 1640 Medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen, Shanghai, China) in a humidified 5% CO2 atmosphere at 37°C.
RNA Extraction and Real-Time qPCR
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Grand Island, NY, USA), following the manufacturer’s protocol. One microgram total RNA was reverse transcribed in a final volume of 20 μL using PrimerScript RT Master Mix (Takara, Dalian, China; cat. no. RR036A). Real-time qPCR was performed as previously described.40 The primers sequences for PCR amplification are described in Table S3.
Cell Transfection
Typically, CRC cells were seeded on six-well plates and then transfected the next day with specific siRNA (100 nM) or control siRNA (100 nM) using Lipofectamine 2000 (Invitrogen), following the manufacturer’s protocol (Invitrogen). After transfection, the cells were harvested for further studies. The primer sequences and siRNA or short hairpin RNA (shRNA) sequences are summarized in Table S3.
Cell Proliferation Assays
Cell viability and colony-formation assay were performed as previously reported.41 Briefly, cell viability was tested with MTT kit (Sigma) according to the manufacturer’s instruction. For colony-formation assay, a certain number of transfected cells were placed in each well of 6-well plates and maintained in proper media containing 10% FBS for two weeks, during which the medium was replaced every 3 days. Colonies were then fixed with methanol and stained with 0.1% crystal violet (Sigma) in PBS for 15 min. Colony formation was determined by counting the number of stained colonies. For the EdU incorporation assay, cells were cultured in 24-well plates. Then, 10 μM EdU was added to each well and the cells were cultured for an additional 2 hr. Then, the cells were fixed with 4% formaldehyde for 30 min. After washing, EdU can be detected with a Click-iTR EdU Kit for 30 min, and the cells were stained with DAPI for 10 min and visualized using a fluorescent microscope (Olympus). The EdU incorporation rate was expressed as the ratio of EdU-positive cells to total DAPI-positive cells (blue cells), which were counted using Image-Pro Plus (IPP) 6.0 software (Media Cybernetics).
Flow Cytometry
Cell cycle and cell apoptosis were analyzed by flow cytometry and detected as described previously.41 Briefly, Transfected cells were harvested after transfection by trypsinization. Double staining with fluorescein isothiocyanate (FITC)-annexin V and propidium iodide was performed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s recommendations. Cells were discriminated into viable cells, dead cells, early apoptotic cells, and apoptotic cells, and the relative ratio of early apoptotic cells was compared with control transfection from each experiment. For flow cytometric analysis of the cell cycle, FACS analysis was performed as described below: after 24 hr incubation, cells were washed with PBS and incubated with 0.25% trypsin for 5 min at 37°C. After centrifuging for 5 min at 1,000 g, cells were washed with PBS, resuspended in 0.9 mL PBS, and then 2.1 mL of 100% EtOH was added and cells were vortexed. Cells were incubated for 40 min on ice. After washing cells twice with PBS, 1 mL RNase A solution was added, and cells were incubated for 30 min at 37°C. Cells were then centrifuged for 5 min at 1,000 g, 1 mL protease inhibitor (PI) solution was added, and cells were incubated for 20 min at room temperature (RT). The percentages of cell cycle distribution were evaluated by PI staining, and all samples were analyzed employing FACSCalibur (BD Biosciences) with appropriate software (ModFit LT; BD Biosciences, Topsham, ME, USA).
Xenograft Mouse Model
Four-week-old athymic male mice were purchased from the Animal Center of Nanjing University (Nanjing, China) and maintained in pathogen-free conditions. HCT116 and SW480 cells were transfected with sh-LINC00460 no. 2 or empty vector and harvested from six-well plates, washed with PBS, and resuspended at a density 2 × 107 cells/mL. Each mouse was subsequently injected in the lower right flank with 100 μL of suspended cells. Tumor growth was examined every 3 days, and tumor volumes were measured as the product of length × width2 × 0.5. At 15 days post-injection, the mice were sacrificed via CO2 asphyxiation, and tumor growth was examined. This study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the NIH. Our protocol was approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University.
IHC Analysis
Tumor tissue samples were immunostained for H&E and Ki-67 as previously reported.41 Briefly, the fixed tissue specimens embedded in paraffin were cut into 4-mm-thick slices for H&E staining and IHC analysis. Following deparaffinization, dehydration, and antigen retrieval, one section for each sample was stained with H&E. For immunohistochemical studies, the specimens were incubated with anti-Ki67 antibodies (Dallas, TX, USA). against human targets at 4°C overnight. After that, the samples were washed with PBS and incubated at 37°C in a water bath for 2 hr after dropwise addition of the secondary antibody and washing with PBS. After treatment with the 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma) solution, the samples were flushed completely, counterstained with hematoxylin, washed with water, treated with dehydration and transparency media, mounted on slides, and observed under the microscope. The IHC staining results were scored by the pathologist and author separately to minimize the subjective factors, compared, and the final comprehensive results obtained. The expression was considered to be positive when ≥50% cancer cells were stained.
Western Blot Assay and Antibodies
Cells were harvested, and protein was extracted from transfected cells and quantified as previously described41 using 12% or 4%–20% polyacrylamide gradient SDS gel. Cell protein lysates were separated by 10% SDS-PAGE, transferred to 0.22 mm NC membranes (Sigma), and incubated with specific antibodies. GAPDH antibody was used as control, and anti-GAPDH and CUL4A (1:1,000) were purchased from Cell Signaling Technology; anti-KLF2 was purchased from Sigma. Protein detection was performed with Super Signal Chemiluminescence Substrate (Pierce).
Isolation of Cytoplasmic and Nuclear RNA
Separation of the nuclear and cytosolic fractions was performed using the PARIS Kit (Life Technologies) according to the manufacturer’s protocol, and RNA was used for subsequent reverse transcription reaction and real-time PCRs (SYBR Premix Ex Taq; TaKaRa). The primer sequences are described in Supplemental Experimental Procedures.
ChIP
ChIP assays were performed using the Chromatin Immunoprecipitation Kit following the manufacturer’s instructions (Millipore). Immunoprecipitations were performed using anti-H3K27me3 (Millipore) and/or normal mouse IgG used as the negative control. Primers were designed referring to the KLF2 promoter sequence, covering upstream of the transcription start site of KLF2 gene (listed in Supplemental Experimental Procedures). PCR were then performed using corresponding primers, following manufacturer’s instruction.
RIP
RIP experiments were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. HXT116 and SW480 cells were lysed in complete RIP lysis buffer, and the cell extract was incubated with magnetic beads conjugated with specific antibodies or control IgG (Millipore) for 6 hr at 4°C. Beads were washed and incubated with Proteinase K to remove proteins. Finally, purified RNA was subjected to real-time qPCR analysis. Antibody for RIP assays of EZH2 was from Abcam (ab3748).
Luciferase Reporter Assay
To construct luciferase reporter vectors, CUL4A 3′ UTR and LINC00460 cDNA fragment containing the predicted micro RNAs binding sites were amplified by PCR and then subcloned downstream of the luciferase gene in the pGL3 plasmid. Mutant of plasmids (pGL3-CUL4A-3′ UTR MUT and pGL3-LINC00460-MUT) were generated by site-directed mutagenesis PCR reaction using platinum pfx DNA polymerase following the product manual. All constructs were verified by DNA sequencing. The luciferase assays were performed using a luciferase assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Briefly, cells were first transfected with appropriate plasmids in 24-well plates. Next, the cells were collected and lysed for luciferase assay 48 hr after transfection. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity. All transfection experiments were performed in triplicates and were independently repeated three times.
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 software. The significance of differences between groups was estimated by two-tailed Student’s t test or ANOVA. Pearson’s correlation coefficient was calculated using Prism 5 software (GraphPad). A p value < 0.05 was considered statistically significant.
Author Contributions
Y.L., K.W., G.J., C.Y., and H.X. designed and performed the experiments and wrote the manuscript. J.Y. and Y.Y. performed the animal experiment and data collection. Data analysis was performed by J.Z. and Y.S. J.R. discussed and revised the manuscript. All authors approved this manuscript.
Conflicts of Interest
We declare that we have no conflicts of interest.
Acknowledgments
We thank Ming Sun, Ph.D. (Department of Bioinformatics and Computational Biology, UT MD Anderson Cancer Center, Houston, TX 77030, USA) for technical support and assay design. This work was supported by the grants from the Natural Science Foundation of Jiangsu Province of China (code: BK20151578) and the Six Talents Peak Project of Jiangsu province (code: WSN-050). This study was also supported by the National Natural Science Foundation of China (code: 81370591).
Footnotes
Supplemental Information includes four figures and three tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.06.012.
Contributor Information
Guozhong Ji, Email: nyefygz@163.com.
Keming Wang, Email: kemingwang@njmu.edu.cn.
Supplemental Information
References
- 1.Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., Brenner H., Dicker D.J., Chimed-Orchir O., Dandona R., Dandona L., Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Aziz M.A., Yousef Z., Saleh A.M., Mohammad S., Al Knawy B. Towards personalized medicine of colorectal cancer. Crit. Rev. Oncol. Hematol. 2017;118:70–78. doi: 10.1016/j.critrevonc.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Veneziano D., Di Bella S., Nigita G., Laganà A., Ferro A., Croce C.M. Noncoding RNA: current deep sequencing data analysis approaches and challenges. Hum. Mutat. 2016;37:1283–1298. doi: 10.1002/humu.23066. [DOI] [PubMed] [Google Scholar]
- 6.Reon B.J., Dutta A. Biological processes discovered by high-throughput sequencing. Am. J. Pathol. 2016;186:722–732. doi: 10.1016/j.ajpath.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunch H. Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genomics. 2018;293:1–15. doi: 10.1007/s00438-017-1370-9. [DOI] [PubMed] [Google Scholar]
- 8.Lian Y., Cai Z., Gong H., Xue S., Wu D., Wang K. HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol. Biosyst. 2016;12:3247–3253. doi: 10.1039/c6mb00475j. [DOI] [PubMed] [Google Scholar]
- 9.Deniz E., Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genomics. 2017;17:135–143. doi: 10.1007/s10142-016-0524-x. [DOI] [PubMed] [Google Scholar]
- 10.Bolha L., Ravnik-Glavač M., Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers. 2017;2017:7243968. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 12.Xie X., Tang B., Xiao Y.F., Xie R., Li B.S., Dong H., Zhou J.Y., Yang S.M. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–5239. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 14.Lyu Y., Lou J., Yang Y., Feng J., Hao Y., Huang S., Yin L., Xu J., Huang D., Ma B. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31:2543–2551. doi: 10.1038/leu.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Lian Y., Yan C., Cai Z., Ding J., Ma Z., Peng P., Wang K. Long non-coding RNA FOXP4-AS1 is an unfavourable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Cell Prolif. 2017;50:e12312. doi: 10.1111/cpr.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng X., Tian L., Zhang Z., Yu Y., Cheng J., Gong Y., Li C.Y., Huang Q. Caspase 3 in dying tumor cells mediates post-irradiation angiogenesis. Oncotarget. 2015;6:32353–32367. doi: 10.18632/oncotarget.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulda S. Targeting apoptosis for anticancer therapy. Semin. Cancer Biol. 2015;31:84–88. doi: 10.1016/j.semcancer.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Xu T.P., Liu X.X., Xia R., Yin L., Kong R., Chen W.M., Huang M.D., Shu Y.Q. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 19.Ferrè F., Colantoni A., Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief. Bioinform. 2016;17:106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P., Nag A. CUL4A ubiquitin ligase: a promising drug target for cancer and other human diseases. Open Biol. 2014;4:130217. doi: 10.1098/rsob.130217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia T., Liao Q., Jiang X., Shao Y., Xiao B., Xi Y., Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang E., Han L., Yin D., He X., Hong L., Si X., Qiu M., Xu T., De W., Xu L. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101. doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Li S., Dai X., Ma J., Wan J., Jiang H., Wang P., Liu Z., Zhang H. PRC2 regulates RNA polymerase III transcribed non-translated RNA gene transcription through EZH2 and SUZ12 interaction with TFIIIC complex. Nucleic Acids Res. 2015;43:6270–6284. doi: 10.1093/nar/gkv574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chase A., Cross N.C. Aberrations of EZH2 in cancer. Clin. Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 27.Gall Trošelj K., Novak Kujundzic R., Ugarkovic D. Polycomb repressive complex’s evolutionary conserved function: the role of EZH2 status and cellular background. Clin. Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang C.J., Hung M.C. The role of EZH2 in tumour progression. Br. J. Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie F.Q., Sun M., Yang J.S., Xie M., Xu T.P., Xia R., Liu Y.W., Liu X.H., Zhang E.B., Lu K.H., Shu Y.Q. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi H., Jacinto F.V., Villanueva A., Fernandez A.F., Yamamoto H., Carmona F.J., Puertas S., Marquez V.E., Shinomura Y., Imai K., Esteller M. Silencing of Kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene. 2012;31:1988–1994. doi: 10.1038/onc.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding J., Xie M., Lian Y., Zhu Y., Peng P., Wang J., Wang L., Wang K. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis. 2017;6:e288. doi: 10.1038/oncsis.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashid F., Shah A., Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Wen M., Kwon Y., Xu Y., Liu Y., Zhang P., He X., Wang Q., Huang Y., Jen K.Y. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520–531. doi: 10.1158/0008-5472.CAN-13-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren W., Sun Z., Zeng Q., Han S., Zhang Q., Jiang L. Aberrant expression of CUL4A is associated with IL-6/STAT3 activation in colorectal cancer progression. Arch. Med. Res. 2016;47:214–222. doi: 10.1016/j.arcmed.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Chen W., Zhang J., Xu H., Dai J., Zhang X. The negative regulation of miR-149-5p in melanoma cell survival and apoptosis by targeting LRIG2. Am. J. Transl. Res. 2017;9:4331–4340. [PMC free article] [PubMed] [Google Scholar]
- 36.Jin L., Li Y., Liu J., Yang S., Gui Y., Mao X., Nie G., Lai Y. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol. Med. Rep. 2016;13:5386–5392. doi: 10.3892/mmr.2016.5205. [DOI] [PubMed] [Google Scholar]
- 37.Okato A., Arai T., Yamada Y., Sugawara S., Koshizuka K., Fujimura L., Kurozumi A., Kato M., Kojima S., Naya Y. Dual strands of Pre-miR-149 inhibit cancer cell migration and invasion through targeting FOXM1 in renal cell carcinoma. Int. J. Mol. Sci. 2017;18:E1969. doi: 10.3390/ijms18091969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y., Qin X., Yan D., Cao H., Zhou L., Fan F., Zang J., Ni J., Xu X., Sha H. Genome-wide profiling of micro-RNA expression in gefitinib-resistant human lung adenocarcinoma using microarray for the identification of miR-149-5p modulation. Tumour Biol. 2017;39 doi: 10.1177/1010428317691659. 1010428317691659. [DOI] [PubMed] [Google Scholar]
- 39.Tian P., Yan L. Inhibition of microRNA-149-5p induces apoptosis of acute myeloid leukemia cell line THP-1 by targeting Fas ligand (FASLG) Med. Sci. Monit. 2016;22:5116–5123. doi: 10.12659/MSM.899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lian Y., Yan C., Ding J., Xia R., Ma Z., Hui B., Ji H., Zhou J., Wang K. A novel lncRNA, LL22NC03-N64E9.1, represses KLF2 transcription through binding with EZH2 in colorectal cancer. Oncotarget. 2017;8:59435–59445. doi: 10.18632/oncotarget.19738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lian Y., Xu Y., Xiao C., Xia R., Gong H., Yang P., Chen T., Wu D., Cai Z., Zhang J., Wang K. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci. Rep. 2017;7:7312. doi: 10.1038/s41598-017-07954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.