Abstract
Background & Aims
Chronic inflammation is a predisposing condition for colorectal cancer. Many studies to date have focused on proinflammatory signaling pathways in the colon. Understanding the mechanisms that suppress inflammation, particularly in epithelial cells, is critical for developing therapeutic interventions. Here, we explored the roles of transforming growth factor β (TGFβ) family signaling through SMAD4 in colonic epithelial cells.
Methods
The Smad4 gene was deleted specifically in adult murine intestinal epithelium. Colitis was induced by 3 rounds of dextran sodium sulfate in drinking water, after which mice were observed for up to 3 months. Nontransformed mouse colonocyte cell lines and colonoid cultures and human colorectal cancer cell lines were analyzed for responses to TGFβ1 and bone morphogenetic protein 2.
Results
Dextran sodium sulfate treatment was sufficient to drive carcinogenesis in mice lacking colonic Smad4 expression, with resulting tumors bearing striking resemblance to human colitis–associated carcinoma. Loss of SMAD4 protein was observed in 48% of human colitis–associated carcinoma samples as compared with 19% of sporadic colorectal carcinomas. Loss of Smad4 increased the expression of inflammatory mediators within nontransformed mouse colon epithelial cells in vivo. In vitro analysis of mouse and human colonic epithelial cell lines and organoids indicated that much of this regulation was cell autonomous. Furthermore, TGFβ signaling inhibited the epithelial inflammatory response to proinflammatory cytokines.
Conclusions
TGFβ suppresses the expression of proinflammatory genes in the colon epithelium, and loss of its downstream mediator, SMAD4, is sufficient to initiate inflammation-driven colon cancer. Transcript profiling: GSE100082.
Keywords: TGFβ, Colitis-Associated Carcinoma, Tumor Necrosis Factor
Abbreviations used in this paper: AOM, azoxymethane; APC, adenomatous polyposis coli; BMP, bone morphogenetic protein; CAC, colitis-associated carcinoma; CCL20, Chemokine (C-C motif) ligand 20; CRC, colorectal cancer; CRISPR/Cas9, Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9; DMEM, Dulbecco's modified Eagle medium; DSS, dextran sodium sulfate; FBS, fetal bovine serum; FDR, false discovery rate; GFP, green fluorescent protein; HBSS, Hank's balanced salt solution; IBD, inflammatory bowel disease; IL, interleukin; IMCS4fl/fl, immortalized mouse colonoctye cell line with loxP-flanked Smad4 alleles; IMCS4null, immortalized mouse colonocyte cell line with deletion of the Smad4 alleles; LPS, lipopolysaccharide; mRNA, messenger RNA; PBS, phosphate-buffered saline; PE, phycoerythrin; R-SMAD, Receptor-SMAD; SFG, retroviral vector; sgRNA, single-guide RNA; STAT3, signal transducer and activator of transcription 3; TGFβ, transforming growth factor β; TNF, tumor necrosis factor; UC, ulcerative colitis; WNT, wingless-type mouse mammary tumor virus integration site; YAMC, young adult mouse colon epithelial cells
Graphical abstract
See editorial on page 350.
Summary.
This study reports the novel observation that canonical TGFβ family signaling through SMAD4 inhibits inflammatory signaling within colonic epithelium. Smad4 deletion in epithelium up-regulated proinflammatory gene expression, increased submucosal immune cell numbers, and promoted colitis-associated neoplasia. This correlated with SMAD4 loss in ulcerative colitis–associated colorectal cancers, relative to sporadic colorectal cancers, in patients.
Chronic inflammation is a predisposing condition for many cancers.1 Ulcerative colitis is an inflammatory condition of the colon predisposing patients to colitis-associated carcinoma (CAC).2, 3, 4 CAC arises from a different sequence of mutation events than most sporadic colorectal cancers (CRCs). For example, sporadic CRC cases frequently have early mutation of adenomatous polyposis coli (APC) and late mutation of p53. However, in CAC, mutation of p53 is thought to be an early event and loss of APC is found late or not at all.5, 6 Although CAC clearly arises in an inflammatory microenvironment, mouse models have shown that multiple etiologies of CRC are either promoted or repressed by specific inflammatory responses.7, 8, 9, 10 Furthermore, there is compelling evidence that CRC can be triggered by a combination of microbiota-dependent and host-dependent mechanisms.7, 11 Multiple levels of regulation have evolved to precisely coordinate the extent of an inflammatory response, allowing for necessary antimicrobial and reparative responses while suppressing inappropriate and rampant responses that lead to disease. Many prior studies have focused on factors influencing the initiation and maintenance of gut inflammation. Given that minor mucosal injuries occur with regularity, it is remarkable that these rapid inflammatory responses are usually transient and extinguished promptly after the inciting cause is resolved without causing overt systemic and organism-wide inflammation with its attendant damaging effects. A better understanding of how this homeostatic balance is maintained may lead to more precise therapeutic interventions.
Transforming growth factor β (TGFβ) pathway signaling has important roles in regulating immune cell responses through its direct regulation of lymphoid and myeloid cell proliferation, differentiation, and survival,12 which in turn leads to suppression of inflammation. Homozygous germline loss of Tgfb113 resulted in a marked increase in inflammatory cell infiltration throughout alimentary tract mucosal tissues. In addition, Kim et al14 found that conditional loss of Smad4 in T cells with intact epithelial expression of Smad4 in mice caused increased T-cell expression of interleukin (IL)5, IL6, and IL13, phenocopied familial juvenile polyposis, and resulted in epithelial cancers throughout the gastrointestinal tract. In contrast, they did not observe spontaneous gastrointestinal tumors when epithelial Smad4 was disrupted using epithelial-specific promoters to drive expression of Cre recombinase (MMTV-Cre or Transthyretin-Cre). In that study, loss of epithelial Smad4 was not examined in the setting of chronic inflammation and the mice were not examined for gene expression changes in the colon epithelium.
TGFβ family members act via interaction with multimers of type I and type II receptors that then phosphorylate R-SMAD proteins in the cytoplasm.15 TGFβ1, β2, and β3 bind TGFβ receptors that, in turn, phosphorylate Receptor-SMADs (R-SMADs) SMAD2 and SMAD3 (SMAD2/3). Bone morphogenetic proteins (BMPs) are TGFβ family members that activate related receptors but lead to the phosphorylation of SMAD1/5/9. Once phosphorylated, R-SMADs bind SMAD4, translocate to the nucleus, and regulate transcription, acting as transcriptional activators of some genes and repressors of other genes. This canonical signaling activity downstream of all TGFβ family receptors is dependent on the common mediator SMAD4. These pathways have multiple levels of redundancy at the levels of ligands, receptors, and R-SMADs, but SMAD4 is uniquely required for transcriptional activity of this pathway. Thus, loss of SMAD4 abrogates all canonical signaling by TGFβ family members.
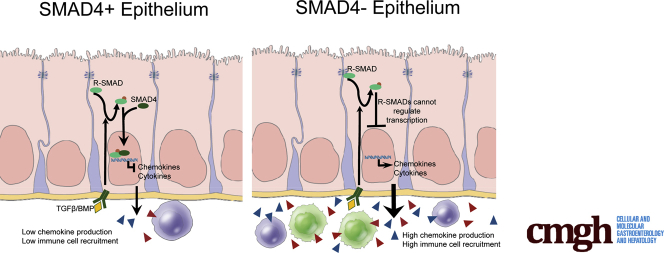
Previous studies have implicated TGFβ signaling to epithelial cells in inhibiting cell proliferation, modulating differentiation, and inducing epithelial-to-mesenchymal transition.16, 17 We previously found that tissue-specific inactivation of the Smad4 gene in adult intestinal epithelium in the context of Apc mutation led to increased Wingless-type Mouse Mammary Tumor Virus Integration Site (WNT) signaling and increased size and numbers of small intestinal and colonic adenomas as compared with Apc mutation alone.18 However, loss of Smad4 without Apc mutation did not result in increased β-catenin protein, likely owing to degradation by the β-catenin destruction complex. We now report a novel homeostatic role for TGFβ signaling in suppressing colonic epithelial cell inflammatory responses. SMAD4-mediated signaling in both human and mouse colonic epithelial cells suppresses inflammation-associated gene expression, including chemokine production, and blocks specific epithelial responses to inflammatory signals. Epithelial-specific loss of Smad4, without the introduction of any other targeted mutation, initiates inflammation-driven carcinogenesis in the colon. Furthermore, we observed a significantly increased frequency of SMAD4 loss in ulcerative colitis–associated carcinomas compared with sporadic CRCs in human beings, linking this pathway to epithelial regulation of inflammatory responses.
Materials and Methods
Mouse Models
All animal work was performed with approval from the Vanderbilt University Institutional Animal Care and Use Committee following Animal Research: Reporting of In Vivo Experiments standards. Mouse alleles CK19CreERT2 (CreERT2 inserted into the Krt19 gene), Lrig1CreERT2, and Smad4fl/fl all were genotyped as previously published19, 20, 21 and bred for at least 10 generations into the C57BL/6J background. Controls were sibling littermates. Mice were given tamoxifen (2 mg in 0.1 mL intraperitoneally 3 times on alternating days for CK19CreERT2 or a single injection or 2 injections on alternating days for Lrig1CreERT2), or corn oil vehicle control only after 8 weeks of age to ensure that gene deletion occurred in adulthood and not during development. For dextran sodium sulfate (DSS; MP Biomedicals, Santa Ana, CA) treatment, mice were given 2.0%–2.5% DSS depending on activity of individual lots in drinking water for 4 days, followed by 5 days of recovery in 3 consecutive cycles. Dosage was determined empirically for each lot, determining the DSS concentration that would cause loss of 5%–15% body weight. Azoxymethane (AOM)/DSS treatment was as described.22 After tamoxifen or vehicle treatment, bedding was mixed among all cages within each experiment once per week.
Human Tissues
De-identified tissue sections were obtained with permission from the Vanderbilt Institutional Review Board. Sporadic CRC tissues were analyzed as individual sections on slides. Tissue microarrays (TMAs) were made from de-identified inflammatory bowel disease (IBD)-associated tissues containing 27 ulcerative colitis (UC)-associated CACs, 29 UC-associated low-grade dysplasias, and 10 UC-associated high-grade dysplasias, as well as normal and inflamed colonic tissues. As a positive control, any samples that had no SMAD4 detection in stroma were excluded from the analysis.
Histology and Immunohistochemistry
Tissues were processed as described.23, 24, 25 Antibodies used were as follows: rabbit anti-SMAD4 (Abcam, Cambridge, MA, for mouse tissue), mouse anti-SMAD4 (Santa Cruz Biotechnology, Santa Cruz, CA, for human tissue), rabbit anti-Ki67 (Abcam), anti–phospho-RELA (Bethyl Laboratories, Montgomery, TX), anti–phospho–signal transducer and activator of transcription 3 (STAT3) (Cell Signaling Technology, Danvers, MA), and rat anti-F4/80 (Invitrogen). Mucin was detected with Alcian blue (Sigma Aldrich, St. Louis, MO), pH 2.5,26 and counterstained with eosin (Fisher Diagnostics, Middletown, VA). Brightfield images were captured on an Axioskop 40 microscope using Axiovision software (Carl Zeiss Microimaging, Thornwood, NY). Fluorescent images were captured with the Aperio Versa 200 Scanner (Leica Biosystems, Inc, Buffalo Grove, IL) through the Digital Histology Shared Resource at Vanderbilt University. Fluorescently labeled cells were quantified using CellProfiler cell image analysis software (Broad Institute, Cambridge, MA).27
Flow Cytometry
After euthanasia, the colon distal to the ascending colon was removed, measured, washed with phosphate-buffered saline (PBS), cut longitudinally, then cut into 1-cm lengths. Tissue then was incubated in cold 30 mL Hank’s balanced salt solution (HBSS) + 0.3 mmol/L dithiothreitol and rocked at 4°C for 20 minutes, washed with PBS, and moved to 30 mL HBSS + 1 mmol/L EDTA and again rocked at 4°C for 20 minutes. Tissue then was minced rapidly with scissors, added to prewarmed collagenase solution (1 mg/mL Collagenase I [Sigma Aldrich] + 1 mg/mL Dispase II [Sigma Aldrich] in 10 mL HBSS), and digested for 40 minutes at 37°C shaking at 350 rpm. After a brief vortex to ensure adequate dissociation of tissue, suspension was filtered through 500-μm mesh and then through 70-μm mesh. Thirty milliliters of DNase solution (30 mL HBSS + 5% fetal bovine serum [FBS; Atlanta Biologicals, Atlanta, GA] + 25 U DNase I [Sigma Aldrich]) was added and cells were incubated at room temperature for 5 minutes and then centrifuged at 1500 rpm for 10 minutes at 4°C. The pellet then was resuspended in fluorescence-activated cell sorter buffer and aliquoted for staining 2 different panels. Antibodies (all from Biolegend, San Diego, CA) for the lymphoid panel included fluorescein isothiocyanate–anti-CD45, AF700–anti-CD3, phycoerythrin (PE)–C7–anti-CD4, PE–anti-CD8, and APC–Cy7–CD19. Antibodies for the myeloid panel included fluorescein isothiocyanate–anti-CD45, AF700–anti-CD11b, PE–Cy7–anti-CD11c, PE–anti-CD103, APC–Gr1 (Ly6G/Ly6C), and PerCP5.5–anti-F4/80. 4′,6-Diamidino-2-phenylindole was used to distinguish live from dead cells. Fluorescence minus one controls were performed in parallel on separate mice to compensate for spectral overlap.28 Data were collected on a 5-laser BD Biosciences LSRII Flow Cytometer (San Jose, CA) in the Vanderbilt University Medical Center Flow Cytometry Shared Resource Core. Fluorescence-activated cell sorter gating and analysis were performed using FlowJo 10 software (FlowJo, Ashland, OR).
Cell Lines and Treatments
YAMC (Young Adult Mouse Colon epithelial Cells) cells29 and FET-1 cells30 were described previously. IMCS4fl/fl cells were derived from crossing Smad4fl/fl alleles into the Immortomouse background carrying an interferon-γ–inducible, temperature-sensitive SV40 Tag, and isolating colon epithelial cells by limiting dilution as described.29 Cells were screened by quantitative reverse transcription-polymerase chain reaction for expression of Cdh1 (E-cadherin), Vim (vimentin), and Ptprc (CD45), and even after multiple passages remained Cdh1+/Vim-/Ptprc- (data not shown), indicating that the culture contained only epithelial cells. Colonocytes were isolated under limiting dilution conditions to be free of other cell types. IMCS4null cells were derived by treating IMCS4fl/fl cells with adenoviral-Cre infection, isolating individual clones and screening for loss of Smad4 expression. All IMC and YAMC lines were maintained in RPMI1640 (Gibco, Grand Island, NY) + 10% FBS (Atlanta Biologicals) + 1× penicillin/streptomycin (Gibco) + 1 U/mL interferon-γ (Sigma Aldrich) and maintained at 33°C. For experimental analyses, cells were washed at least twice and replated without interferon-γ and were maintained at 37°C to eliminate Tag.
Cells or colonoids were treated with the indicated concentrations of TGFβ1 (R&D Systems, Minneapolis, MN), BMP2 (R&D Systems), tumor necrosis factor (TNF; R&D Systems), IL1β (Peprotech, Rocky Hill, NJ), and lipopolysaccharide (LPS; Sigma Aldrich). Vehicle controls were as follows: 4 mmol/L HCl, 0.1% bovine serum albumin (TGFβ1, BMP2), 0.1% bovine serum albumin in PBS (TNF), and water (IL1β, LPS).
Human Tumoroid Models
De-identified colorectal tumor tissue was collected from consented patients as approved by the Memorial Sloan Kettering Institutional Review Board. Mutations were determined using Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets.31, 32 Tissue was minced and digested in advanced Dulbecco's modified Eagle medium (DMEM)/F12 with 2% FBS (Atlanta Biologicals), penicillin/streptomycin (Gibco/ThermoFisher, Waltham, MA), 100 U/mL collagenase type XI (Sigma-Aldrich), and 125 μg/mL dispase type II (Invitrogen, Waltham, MA) at 37°C for 40 minutes and then digested further for 10 minutes by adding the same volume of TrypLE Express and 50 μL of DNase I (Qiagen, Germantown, MD). The tumor cells then were collected by filtering through a 70-μm cell strainer and spinning at 300×g for 5 minutes. Isolated tumor cells were embedded in Matrigel (Corning, Tewksbury, MA) and cultured as described.33 SMAD4 was deleted using Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 (CRISPR/Cas9)-mediated excision as follows. Tumoroids were harvested in Cell Recovery Solution (BD Biosciences) and suspended in tumoroid medium plus 8 μg/mL hexadimethrine bromide (Polybrene; Sigma-Aldrich) and 10 μmol/L Y27632 (Sigma-Aldrich). Colorectal cancer SFG/GFluc tumoroids were created by retrovirally introducing GFP and firefly luciferase for bioluminescence tracking.34 Retroviral vectors for SFG-GFP/firefly luciferase transduction were provided by S. P. Gao (Sloan Kettering Institute). GFP-positive cells were enriched by fluorescence-activated cell sorting and isolated. Designed SMAD4-targeting, single-guide RNA (sgRNA) oligomers were cloned into the LentiCRISPRv2 vector, and lentiviral particles were generated by transfecting HEK293T cells with the LentiCRISPRv2-sgRNA construct, psPAX2, and VSV-G.35 sgRNA sequences were designed using the online tool developed by Heigwer et al36 (www.e-crisp.org) as described by Drost et al37 (sequence used was as follows: GATCAGGCCACCTCCAGAGA). The HEK293T cells (7.25 × 106) were seeded in a 10-cm dish. LentiCRISPRv2-sgRNA construct (7.7 μg), psPAX2 (5.8 μg), and VSV-G (3.9 μg) were delivered with Lipofectamine 2000 (Invitrogen, ThermoFisher Scientific, Grand Island, NY) according to the manufacturer’s protocol. Cells were grown overnight after transfection, and medium was replaced with the standard DMEM–fetal calf serum supplemented with GlutaMax (Gibco/ThermoFisher) and Pen-Strep (Gibco/ThermoFisher). At 2 days after transfection, the virus medium was concentrated using the PEG-it Virus Precipitation Solution (System Biosciences, Palo Alto, CA) and the lentiviral particles were resuspended in 300 μL PBS. After dissociation of the SFG/GFluc tumoroids (three 50-μL Matrigel discs per viral construct) with cell recovery solution (BD Biosciences), the cell clusters were resuspended in 10 μL of infection medium (tumoroid culture medium plus 8 μg/mL hexadimethrine bromide [Polybrene; Sigma-Aldrich] and 10 μmol/L Y27632 [Sigma-Aldrich]). The cell cluster suspension and viral suspension were combined in a 48-well culture plate. The culture plate was centrifuged at 600×g at room temperature for 60 minutes and subsequently incubated for 6 hours in standard culture conditions (37°C with 5% CO2). The infection mixture was transferred to a 1.5-mL tube, and the cells were centrifuged to form a pellet, after which the infection medium was discarded. The cells were resuspended with 150 μL Matrigel and divided into 3 wells of a 24-well suspension plate. After polymerization of the Matrigel, 500 μL infection medium without Polybrene was added. Two days after infection, the medium was replaced with culture medium. The infected cells then were selected by addition of puromycin (2 μg/mL) at 6 days.
Mouse Colonoid Cultures
Intestinal crypts were isolated as follows: mouse colon was opened longitudinally, washed, cut into 2-mm segments, resuspended in Gentle Cell Dissociation Reagent (Stem Cell Technologies, Vancouver, Canada), and incubated on a rocking platform at 20 rpm for 30 minutes at room temperature. Crypts were isolated from tissue by multiple passages through a 10-mL serologic pipette, passed through a 70-μm filter, and centrifuged at 250×g for 5 minutes at 4°C. The supernatants were discarded and the colonic crypts were resuspended in DMEM + high glucose (Gibco) and evaluated by inverse microscopy. Fractions containing crypts were combined and centrifuged at 250×g for 5 minutes at 4°C. Colonic crypts then were resuspended and plated in 50-μL beads of Growth Factor Reduced Matrigel (GFR; Corning, Tewksbury, MA). Colonoid medium (advanced DMEM/F12 [Gibco] supplemented with penicillin/streptomycin [Gibco], N2 [Gibco], B27 [Gibco], Glutamax [Gibco], HEPES [Sigma Aldrich], 50 ng/mL epidermal growth factor [R&D Systems]), 40% Wnt3a-conditioned medium, 20% R-Spondin–conditioned medium, 10% Noggin-conditioned medium, and 10 μmol/L Y-27632 (Sigma Aldrich) was added to each well and colonoids were grown at 37°C in 5% CO2. Colonoids at a density of 100 per well were treated with vehicle, 100 ng/mL TNF, 3 ng/mL TGFβ1, or combinations as described.
Small Interfering RNA Transfections
YAMC cells were plated 24 hours before transfection in RPMI + 5% FBS + penicillin/streptomycin. The cells were transfected with a mixture of Dharmafect 1 (Dharmacon, Lafayette, CO) and 50 nmol/L final concentration small interfering RNA according to the manufacturer’s protocol in antibiotic-free medium. After 24 hours the medium was replaced with fresh RPMI + 5% FBS + Pen-Strep for an additional 48 hours. After a total of 72 hours of transfection, the cells were processed for RNA isolation and Western blot. The mouse-specific small interfering RNA oligonucleotides were as follows: mouse Smad4 (ON-TARGETplus SMARTpool L-040687; Dharmacon) and the nontargeting control (ON-TARGETplus SMARTpool 1 D-001810; Dharmacon).
Quantitative Reverse-Transcription Polymerase Chain Reaction
RNA was isolated and analyzed as described.18 Primer sequences are listed in Table 1.
Table 1.
Primers Used for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Primer 1 | Primer 2 | |
|---|---|---|
| Mouse genes | ||
| Ccl20 | GGTACTGCTGGCTCACCTCT | TGTACGAGAGGCAACAGTCG |
| Cxcl5 | TGCCCTACGGTGGAAGTCAT | AGCTTTCTTTTTGTCACTGCCC |
| Il18 | CAAACCTTCCAAATCACTTCCT | TCCTTGAAGTTGACGCAAGA |
| Il18bp | AGCTATTCGGGGCTTAGGAG | TGCAAGCAAGTCTGGTGTCT |
| Il34 variant 1 | CCACCCGTCCTGGAAGTAT | GGCCAATCTCCACATCCAT |
| Il34 variant 2 | CCCGTCCTGGAAGTATCTACA | GTACACCAACGGCCATGAG |
| Il1rn variant 1 | GGCAGTGGAAGACCTTGTGT | CATCTTGCAGGGTCTTTTCC |
| Il1rn variant 2 | CTCCTTCTCATCCTTCTGTTTCA | GGTCTTCTGGTTAGTATCCCAGATT |
| Il1rn variant 3 | TGTGCCAAGTCTGGAGATGA | TTCTTTGTTCTTGCTCAGATCAGT |
| Smad4 | CATTCCAGCGTGCCATTTC | TTCAAAGTAAGCAATGGAGCAC |
| Pmm1 | GGGTGGCTCTGACTACTCTAAGAT | ACACGTAGTCAAACTTCTCAATGACT |
| Human genes | Primer 1 | Primer 2 |
| CCL20 | GCTGCTTTGATGTCAGTGCT | GAAGAATACGGTCTGTGTATCCAA |
| CXCL5 | GGTCCTTCGAGCTCCTTGT | GCAGCTCTCTCAACACAGCA |
| PMM1 | TTCTCCGAACTGGACAAGAAA | CTCTGTTTTCAGGGCTTCCA |
Protein assays
Western blots
Western blot was performed as published.38 Antibodies used were as follows: SMAD4 (1:500; Santa Cruz) and β-actin (1:20,000; Sigma Aldrich).
Luminex
Colonic crypts were isolated by incubation in 2 mmol/L EDTA (Sigma Aldrich), followed by shaking and filtering through a 70-μm filter, then lysed in RIPA buffer (50 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% sodium dodecyl sulfate). Lysates from mouse colons from CK19CreERT Smad4fl/fl mice that received tamoxifen (7 mice) or vehicle control (8 mice) were isolated and applied to the Luminex as described,39 quantifying for Chemokine (C-C motif) 20 CCL20 according to the manufacturer’s instructions (Luminex Flexmap3D workstation; Millipore, Billerica, MA).
Enzyme-linked immunosorbent assay
CCL20 in cell culture media was measured per the manufacturer’s instructions (R&D Systems).
RNA Sequencing Experiments
Colonic crypts were isolated by removing and flushing the colon, opening it longitudinally, rinsing, and incubating at 4°C in 1.5 mmol/L EDTA in PBS followed by shaking for 1 minute. Crypts then were filtered through 75-μm mesh, centrifuged, and the liquid was aspirated. From this pellet, RNA was prepared as described18 and then processed using a TruSeq Stranded messenger RNA (mRNA) sample preparation kit according to the manufacturer’s instructions (Illumina, San Diego, CA). For mouse colon, 32–37 million 51–base pair, single-end reads were generated per sample. For IMCS4fl/fl colonocytes, 26–50 million 75–base pair, paired-end reads were generated per sample. For mouse colonoids, 50–72 million 75–base pair, paired-end reads were generated per sample. Reads were mapped to the mouse genome mm10 using TopHat-2.1.0,40 uniquely mapping 86%–95% single-end reads to the genome, depending on the study. The number of reads that fell into annotated genes were counted41 using samtools-1.3.142 and HTSeq-0.5.4p5.43 Count-based differential expression analysis was performed using edgeR_3.4.2.44
Data files were uploaded to the MIAME (Minimum Information About a Microarray Experiment) database, accession number GSE100082. Pathways and upstream regulators were analyzed using IPA software (Qiagen, Inc, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis).
Statistical Analyses
Results in cell lines were compared by 2-tailed t tests. Results in colonoids were compared by 2-way analysis of variance. Frequency of SMAD4 loss in human tumors was analyzed by the chi-square test with Yates correction. Flow cytometry results were analyzed by 2-tailed t tests with Holm–Sidak correction.
Ethics Statements
All animal work was performed with approval from the Vanderbilt Institutional Animal Care and Use Committee. All human tissue was acquired de-identified with approval from the Vanderbilt Institutional Review Board or the Memorial Sloan Kettering Institutional Review Board.
Results
Smad4 Loss Predisposes to Colitis-Associated Carcinoma
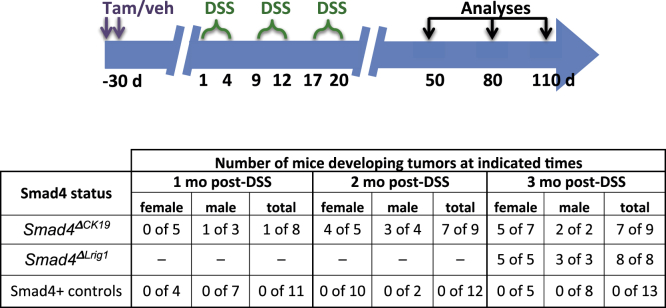
We previously published that tamoxifen-treated Ck19CreERT2 Smad4fl/fl (Smad4ΔCK19) mice had loss of the SMAD4 protein in approximately 20% of colonic crypts by 1 month after tamoxifen treatment although not affecting Smad4 expression in stromal cells.18 In the current study, we induced chronic inflammation in the colon of either Smad4ΔCK19 mice or SMAD4-positive mice (either vehicle-treated Ck19CreERT2 Smad4fl/fl or tamoxifen-treated Smad4fl/fl mice) by 3 short repeated cycles of treatment with DSS, after which the mice were followed up according to the protocol shown (Figure 1). After these 3 rounds of DSS, chronic inflammation was maintained for up to 3 months without further DSS treatment regardless of Smad4 expression (Figure 2A–D). In 78% of Smad4ΔCK19 mice, macroscopic invasive adenocarcinomas of the distal colon and rectum developed by 3 months after completion of DSS treatment, but tumors were not observed in any identically treated SMAD4+ control mice (Figure 1). For a greater efficiency of recombination, we also examined tamoxifen-treated Lrig1CreERT2 Smad4fl/fl (Smad4ΔLrig1) mice that had greater than 90% recombination in mouse colon (Figure 3). One hundred percent of these mice developed similar cancers in the distal colon and rectum (Figure 1). Alcian blue staining showed extensive mucin secretion in invading glands (Figure 2E) and the tumor cells were uniformly negative for SMAD4 protein by immunohistochemistry (Figure 2F).
Figure 1.
SMAD4 prevents tumorigenesis after DSS-induced colitis.Top: Diagram of treatment strategy. Smad4ΔCK19, Smad4ΔLrig1, or SMAD4+ control mice were injected with tamoxifen (Tam) or vehicle (veh). One month later, mice were placed on 3 rounds of DSS as indicated. One, 2, or 3 months after the last DSS, mice were analyzed histologically for tumor development. Bottom: Number of tumors observed in Smad4ΔCK19, Smad4ΔLrig1, or SMAD4+ control mice at indicated times after DSS.
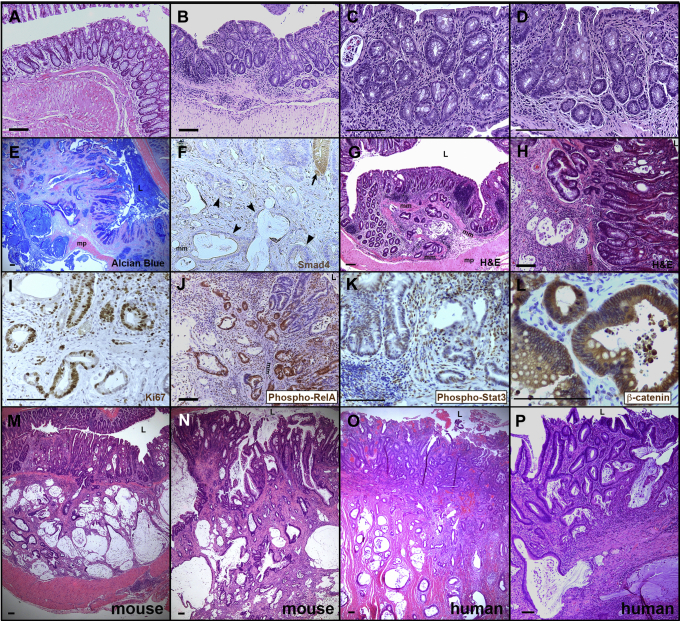
Figure 2.
Invasive mucinous carcinomas similar to human CAC arise in Smad4ΔCK19mice exposed to DSS-induced damage. (A–D) Three months after DSS, control mice show chronic inflammation but no tumorigenesis. Colons from (A) 1 age-matched mouse that received no DSS and from (B–D) 3 SMAD4+ mice that were treated with 3 rounds of DSS and then followed up for 3 months before analysis. Note increased stroma between epithelial crypts of DSS-treated mice. (E-N) Smad4ΔCK19 mice treated with three rounds DSS and analyzed three months later except as indicated. (E) At 3 months after DSS, Alcian blue shows mucinous cystic tumors invading through the muscularis propria. (F) Tumors are SMAD4-negative (arrowheads) and a nearby normal crypt (arrow) is SMAD4-positive (brown). Note that apical surfaces of mucinous cysts sometimes trap antibodies nonspecifically. (G and H) H&E shows tumors forming 2 months after DSS. Although too small to detect macroscopically, tumors already are invading through the muscularis mucosa (mm). (I) Labeling with antibodies to Ki67 (brown) indicate the proliferative nature of tumors. (J) Invading tumor cells are largely phospho-RelA positive (brown), indicating active nuclear factor-κB signaling. (K) Many tumor cells also are phospho-STAT3 positive, indicating active Janus kinase/STAT signaling. (L) β-catenin protein (brown) was not detected in nuclei. (M–P) DSS-induced tumors in Smad4ΔCK19 mice are morphologically similar to human CAC. H&E of (M and N) mouse CAC tumors or (O and P) human tumors. Luminal surfaces (L) are marked for orientation. mm, muscularis mucosa; mp, muscularis propria. Scale bars: 100 μm.
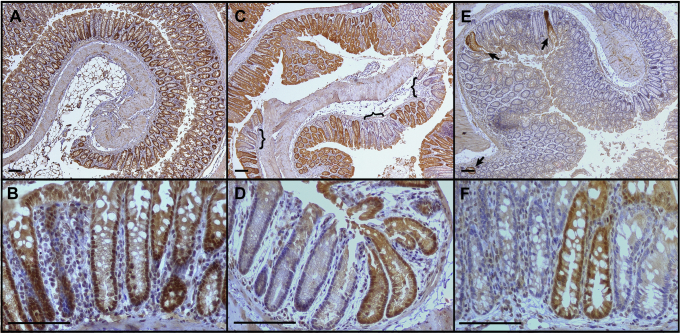
Figure 3.
Efficacy of Cre-mediated recombination of Smad4fl/flalleles. Mouse colons from mice administered tamoxifen or vehicle were stained for SMAD4 (brown). (A and B) Lrig1CreERTSmad4fl/fl mouse given vehicle. (C and D) Ck19CreERT2Smad4fl/fl mouse given tamoxifen. (E and F) Lrig1CreERT2Smad4fl/fl mouse given tamoxifen. Brackets, regions of SMAD4 loss; arrows, Smad4+ crypts surrounded by Smad4-negative crypts. Scale bars: 100 μm.
Colonic carcinomas were observed at 2 months after DSS in 78% of Smad4ΔCK19 mice (Figure 1). Although too small to detect by gross observation, they were detected histologically and already were invading into the submucosa (Figure 2G and H). At 1 month after DSS, only 1 of 8 Smad4ΔCK19 mice had an adenocarcinoma and its location adjacent to a large diverticulum suggested that it may have been exposed to inflammation for longer than 1 month. DSS-induced tumors were proliferative as shown by Ki67 immunostaining (Figure 2I). Consistent with an ongoing inflammatory response, these tumors were positive for phosphorylated RELA/p65 and phosphorylated STAT3 (Figure 2J and K). β-catenin protein was not detected in the nucleus but rather was concentrated at the cell membrane, suggesting that the WNT pathway was not up-regulated (Figure 2L), consistent with human CAC.5, 6 Thus, loss of SMAD4 in the colon epithelium in the presence of chronic inflammation is sufficient to cause neoplastic transformation and yield invasive mucinous adenocarcinomas.
These mouse tumors bore striking similarity to human colitis–associated carcinomas (Figure 2M–P). The mouse tumors were flat or slightly elevated at the mucosal surface with extensive invasion of mucinous glands into the submucosa and through the muscularis propria. Adjacent colonic epithelium showed dysplastic changes deep in crypts consistent with precursor lesions. Similar to human colitis–associated carcinomas, luminal cells in the mice maintained some crypt architecture and differentiation even as cells were invading from the base of crypts.
SMAD4 Loss Is Highly Associated With Human Colitis–Associated Carcinoma
Because of the link in our mouse model between Smad4 mutation and inflammation-induced carcinoma, we investigated whether SMAD4 loss was associated with carcinoma arising in the setting of inflammatory bowel disease. We examined SMAD4 protein expression by immunohistochemistry in CRCs from 27 UC patients and 52 sporadic CRC patients and found that UC-associated carcinomas were more likely to lack detectable SMAD4 (P < .001). Thirteen of 27 (48%) UC-associated carcinomas were negative for SMAD4 in the tumor epithelium (Table 2 and Figure 4). This is in contrast to 19% (10 of 52) of spontaneous CRC cases that were negative for SMAD4 (P < .02). To understand the temporal relationship of loss of SMAD4 expression with tumor progression, we also examined SMAD4 immunostaining in both low-grade and high-grade dysplasia associated with UC. We found that SMAD4 loss was infrequent in low-grade dysplasia (1 of 29 cases; 3%) but was more common in high-grade dysplasia (3 of 10; 30%), suggesting that SMAD4 loss occurs late in tumor progression. This number of high-grade dysplasia cases is too low to draw firm conclusions, but warrants future studies on the timing and function of SMAD4 signaling in progression to CAC.
Table 2.
Frequency of SMAD4 Loss in Colorectal Cancers
| Tumor status | Sporadic CRC | CAC | UC dysplasia low grade | UC dysplasia high grade |
|---|---|---|---|---|
| SMAD4+ | 42 | 14 (9 F, 5 M) | 28 (17 F, 11 M) | 7 (3 F, 4 M) |
| SMAD4- | 10 | 13a (6 F, 7 M) | 1 (M) | 3 (1 F, 2 M) |
F, female; M male.
P < .001.
Figure 4.
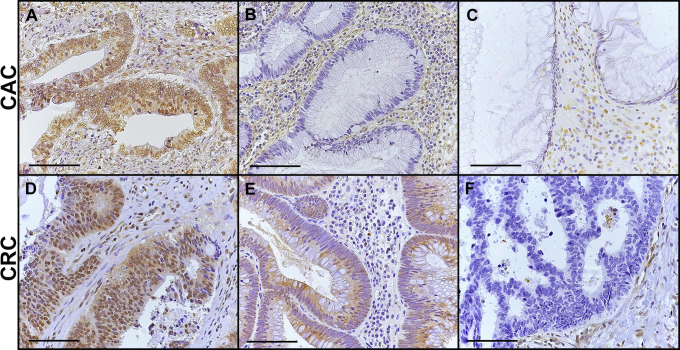
Examples of SMAD4 protein in (A–C) CAC and (D–F) sporadic CRC human tissues. In various tumors, SMAD4 protein was detected in (A and D) nuclei, (E) cytoplasm without nuclear detection, or (B, C, and F) was not detected in the tumor cells although SMAD4 routinely was detected in surrounding stromal cells. Scale bars: 100 μm.
Loss of Smad4 Expression in Mouse Colonic Epithelium Activates an Inflammatory Gene Signature
To understand the role of SMAD4 in regulating colitis-associated events, we examined alterations in gene expression that are associated with Smad4 loss. Isolated colonic epithelium from Smad4ΔLrig1 and from SMAD4-positive control mice (either Lrig1CreERT2 Smad4fl/fl mice receiving vehicle or Smad4fl/fl mice treated with tamoxifen) was analyzed by RNA sequencing. By using cut-off limits of 1.5-fold change in expression and a false discovery rate (FDR) of 0.01 or less, 911 genes were up-regulated in colonic epithelium by loss of Smad4 and 1293 genes were down-regulated. The 244 genes that were up-regulated at least 2-fold (Supplementary Table 1) or the 428 genes that were down-regulated at least 2-fold (Supplementary Table 2) were analyzed by ingenuity pathway analysis. Genes up-regulated by Smad4 loss were associated significantly with multiple pathways, approximately half of which (19 of 40) are inflammation-related (Supplementary Figure 1). Notably, genes that were down-regulated by Smad4 loss were associated with only 2 inflammation-associated pathways (of a total of 68), but were associated with many metabolic pathways (Supplementary Figure 2). Genes down-regulated by SMAD4 loss also included direct SMAD targets such as Id1, Id2, Id3, Smad7, Cdkn1a, Cdkn1d, and Thbs1, validating the functional loss of SMAD activity. Among genes regulated by Smad4 loss, there was no obvious correlation with WNT target genes. Of 33 WNT target genes detected, 6 were up-regulated significantly by Smad4 loss, 5 were down-regulated significantly, and 21 showed no significant difference. These data led to the hypothesis that SMAD4-mediated signaling is an epithelial suppressor of inflammatory responses in the colon.
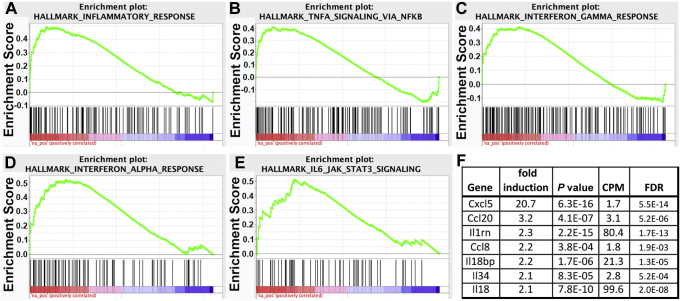
Gene pathway analysis identified 84 inflammatory response genes that were up-regulated by Smad4 loss in colonic epithelial cells (Supplementary Table 3). Upstream master regulators of genes up-regulated by Smad4 loss include a number of chemokines and cytokines and their receptors (Supplementary Table 4), whereas down-regulated genes correlated with few chemokine/cytokine regulators and more metabolic regulators (Supplementary Table 5). Gene Set Enrichment Analysis confirmed this association of Smad4 loss with up-regulation of inflammation-associated genes (Figure 5). Genes involved in inflammation, IL6/Janus kinase/STAT3 signaling, TNF/nuclear factor-κB signaling, interferon α signaling, and interferon γ signaling were over-represented significantly in up-regulated genes with a FDR less than 0.05 (Figure 5A–E). Among the genes up-regulated after Smad4 loss were a number of chemokines and cytokines (Figure 5F), suggesting that SMAD4-mediated signaling limits expression of epithelial-derived inflammatory mediators under homeostatic conditions.
Figure 5.
SMAD4 represses inflammatory gene signatures. mRNA was isolated from Smad4ΔLrig1 or SMAD4+ control mice and sequenced. Gene Set Enrichment Analysis was performed based on a gene list preranked using log(Pvalue) for genes up-regulated and -log(Pvalue) for genes down-regulated after Smad4 loss. Gene sets enriched with genes up-regulated by loss of Smad4 include (A) inflammatory response; (B) TNF signaling via nuclear factor-κB (NF-κB); (C) interferon-γ response; (D) interferon-α response; and (E) IL6/Janus kinase (JAK)/STAT signaling. (F) Chemokine and cytokine genes repressed by SMAD4. Fold induction indicates level in Smad4ΔLrig1 colon relative to control. For each group, n = 3 biological repeats (performed on 3 separate days). CPM, counts per million.
Regulation of Some Chemokines by the Smad4 Pathway Is Cell Autonomous and Counteracts Induction by Proinflammatory Cytokines
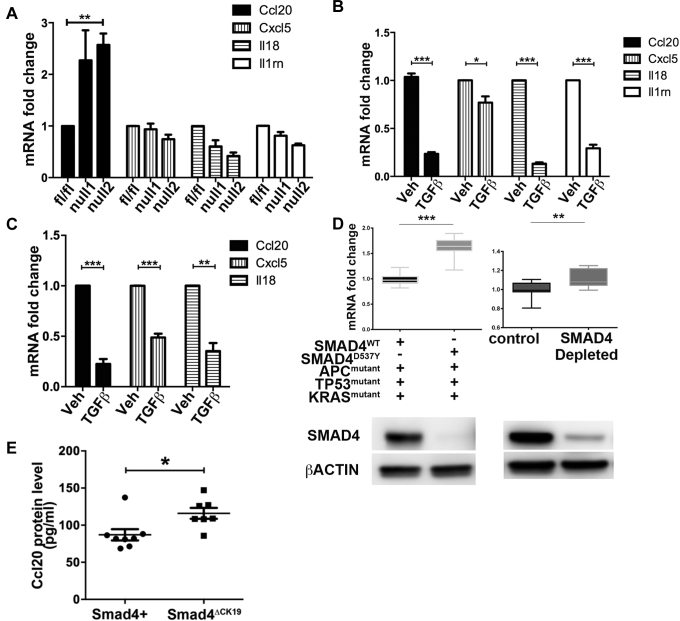
To investigate whether the chemokines up-regulated by Smad4 loss were regulated directly by this pathway or were downstream of other inflammatory responses that may have been activated in vivo, we isolated conditionally immortalized cell lines from mice bearing the Smad4fl/fl alleles in the immortomouse background (IMCS4fl/fl cells). These cells then were treated with adenoviral vectors expressing Cre recombinase to derive clones that were deleted for the Smad4 gene. As in mouse colon in vivo, mRNA expression of Ccl20 was up-regulated by loss of Smad4 in both IMCS4null clonal lines, but mRNA levels of other chemokines/cytokines up-regulated in vivo did not change (Cxcl5, Il18, and Il1rn) (Figure 6A) or were not detected (Ccl8, Il18bp, and Il34).
Figure 6.
SMAD4 signaling represses chemokine/cytokine expression. (A) Chemokine and cytokines regulated by SMAD4 in vivo were compared in IMCS4fl/fl colonocytes that expressed Smad4 or IMCS4null1 and IMCS4null2 colonocytes that had a Smad4 deletion. (B) Chemokine expression was compared in colonocytes treated with vehicle or 3 ng/mL Tgfβ1 for 24 hours. (C) Chemokine expression was compared in YAMC colonocytes treated with vehicle or 3 ng/mL TGFβ1 for 24 hours. (A–C) Data shown represent 3 biological replicates (performed on 3 separate days). (D) Two colorectal cancer tumoroid lines were cultured and RNA was isolated (left panel). The mutational pattern for each tumoroid is shown below. The SMAD4mutant colorectal cancer tumoroids have significantly higher levels of CCL20 than SMAD4wildtype colorectal cancer tumoroids. Colorectal cancer tumoroids were cultured, SMAD4 was depleted via CRISPR/Cas9 methodology, and then RNA was isolated (right panel). CCL20 levels were normalized to ACTB and compared between SMAD4-retained and SMAD4-depleted tumoroids. SMAD4 depletion was associated with up-regulation of CCL20. SMAD4 protein loss in the respective tumoroids was validated by Western blot and is shown. Data shown represent the average of 3 biologic replicates (experiments performed on 3 separate days) and at least 3 technical replicates for each tumoroid. P values by 2-tailed t tests. (E) CCL20 protein levels were quantified by enzyme-linked immunosorbent assay from the colonic mucosa of 8 control (Smad4+) and 8 Smad4ΔCK19 mice. Veh, vehicle. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Lack of response to Smad4 loss could be owing to little or no endogenous activation of SMAD4-mediated pathways. Therefore, we treated IMCS4fl/fl cells (Smad4-expressing) with TGFβ1 or vehicle for 24 hours and determined the effect on the chemokines/cytokines that were identified as up-regulated in vivo after Smad4 loss. We found that TGFβ1 treatment inhibited expression of Ccl20, Cxcl5, Il18, and Il1rn (Figure 6B), whereas Ccl8, Il18bp, and Il34 remained undetectable in the cultured colonocytes. Similar results were seen in the YAMC conditionally immortalized colonocyte line except that Il1rn also was not detected (Figure 6C).
We further confirmed that loss of SMAD4 expression correlated with increased CCL20 expression in human colorectal cancer–derived tumoroids growing in 3-dimensional culture conditions. Two tumoroid lines were examined that had similar mutations in APC, TP53, and KRAS, but 1 retained SMAD4 expression while the other had a D536Y point mutation, eliminating SMAD4 production. The SMAD4 null line had increased CCL20 mRNA as compared with the SMAD4-positive line (Figure 6D). To ensure that this difference was not caused by unknown mutations that may have varied between the lines, a separate SMAD4-positive tumoroid line was depleted for SMAD4 using CRISPR/Cas-mediated excision of the SMAD4 gene. An 80% reduction of SMAD4 protein resulted in a significant increase in CCL20 mRNA (Figure 6D).
In mouse colon, we validated that CCL20 protein was increased significantly in colonic mucosa when Smad4 expression was lost in Smad4ΔCK19 mouse colon, consistent with the earlier-described increase in Ccl20 mRNA (Figure 6E). Although the increase in protein was relatively modest, it should be noted that these mice only lose expression of Smad4 in approximately 20% of colonic crypts.
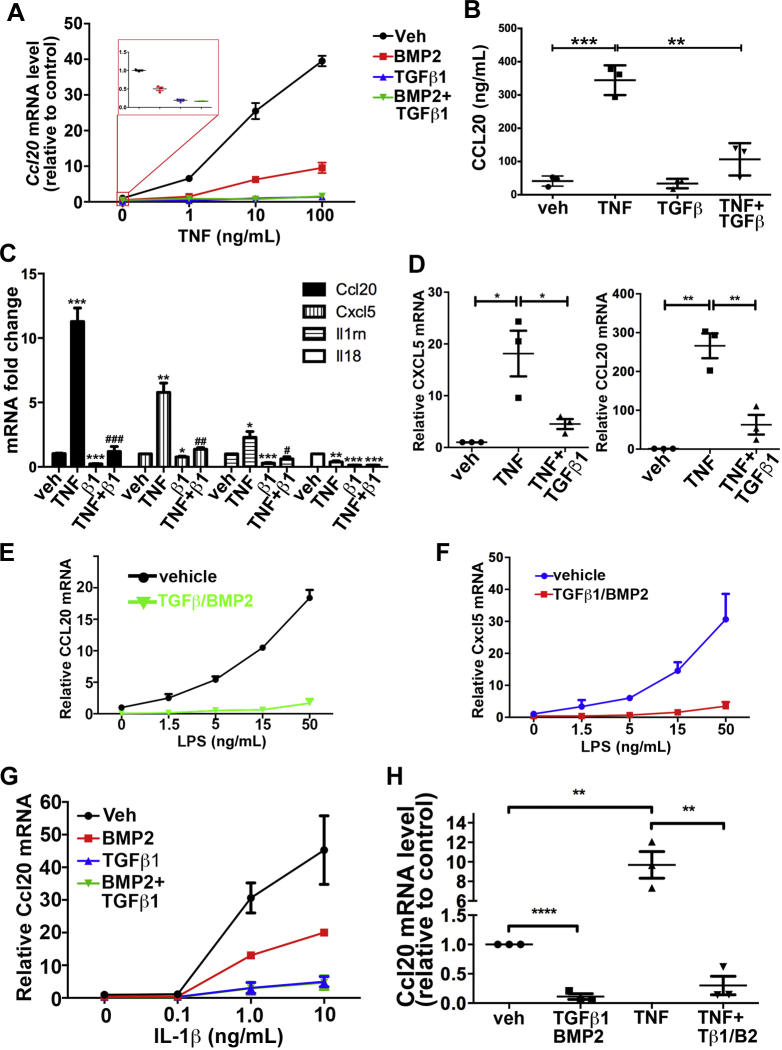
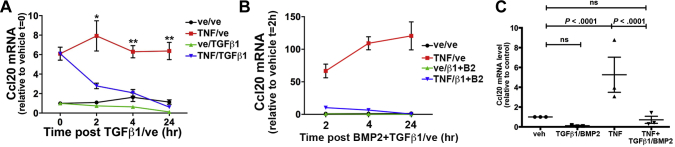
Some chemokines such as CCL20 are up-regulated by inflammatory signaling pathways,45, 46 raising the question of whether or not TGFβ1 or BMP2 could block induction by cytokines TNF and IL1β, or endotoxins such as LPS. IMCS4fl/fl colonocytes were treated with TGFβ1 and/or BMP2 for 6 hours, followed by addition of TNF (Figure 7A). TNF alone induced Ccl20 up to 40-fold in a concentration-dependent manner, but BMP2 partially blocked and TGFβ1 almost completely blocked this induction. The induction of Ccl20 by TNF and its repression by TGFβ1 were reflected in increased and decreased levels of CCL20 protein, respectively (Figure 7B). TNF also induced expression of Cxcl5 and Il1rn, and this induction also was blocked by TGFβ1 pretreatment (Figure 7C). In human FET-1 CRC cells, TNF induced expression of both CCL20 and CXCL5 and co-treatment with TGFβ1 blocked that induction (Figure 7D). LPS induced Ccl20 and Cxcl5 by 20- to 30-fold, and pretreatment with combined TGFβ1 and BMP2 blocked that induction (Figure 7E and F). Similarly, IL1β induced Ccl20 expression in IMCS4fl/fl cells as much as 50-fold in a concentration-dependent manner, and pretreatment with TGFβ1 and to some extent BMP2 inhibited that induction (Figure 7G). We also examined Ccl20 expression in mouse colonoids derived from crypts of wild-type mice and cultured in a 3-dimensional matrix. As in immortalized cell lines, TNF induced expression of Ccl20 9.68 ± 1.36-fold in colonoids and addition of TGFβ1 and BMP2 blocked that induction (Figure 7H). These combined data suggest that, in colonic epithelium, the SMAD4 pathway inhibits expression of some chemokines and cytokines in a cell-autonomous manner independently of effects from surrounding immune cells or from microbiota.
Figure 7.
SMAD4-mediated signaling blocks the effect of proinflammatory cytokines and endotoxin. (A) IMCS4fl/fl colonocytes were treated for 6 hours with 3 ng/mL TGFβ1 and 10 ng/mL BMP2 or vehicle, followed by the addition of indicated concentrations of TNF or vehicle for 24 hours followed by quantitative reverse-transcription polymerase chain reaction analysis for Ccl20 mRNA. (B) IMCS4fl/fl cells were treated as indicated with vehicle, 10 ng/mL TNF, and/or 3 ng/mL TGFβ1 for 6 hours. Conditioned media was collected and analyzed by enzyme-linked immunosorbent assay for CCL20 protein levels. Three biological replicates (collected on 3 different days) were analyzed. (C) IMCS4fl/fl colonocytes were co-treated with vehicle, 20 ng/mL TNF, and/or 3 ng/mL TGFβ1 for 24 hours and indicated genes were measured for mRNA levels. (D) Human CRC FET-1 cells were co-treated with 10 ng/mL TNF and/or 3 ng/mL TGFβ1 or vehicle controls. mRNA was isolated and analyzed 24 hours later for Cxcl5 and Ccl20. Values are relative to vehicle-only controls. (E and F) IMCS4fl/fl cells were pretreated similarly with 3 ng/mL TGFβ1 and 10 ng/mL BMP2 for 6 hours followed by the addition of LPS at indicated concentrations. Cells were harvested 24 hours later and analyzed as described earlier for (E) Ccl20 or (F) Cxcl5. (G) IMCS4fl/fl cells were treated with 3 ng/mL TGFβ1 and/or 10 ng/mL BMP2 or vehicle for 6 hours followed by the addition of IL1β in the indicated doses. Cells were harvested 24 hours later and analyzed for Ccl20 as described earlier. (H) Mouse colonoids were treated with 3 ng/mL TGFβ1 and 10 ng/mL BMP2 or vehicle for 4 hours, followed by the addition of 100 ng/mL TNF or vehicle for 24 hours followed, by analysis of Ccl20 levels. (A and C–H) Three biological replicates were assayed for each arm and mRNA levels were normalized to invariant Pmm1 and presented as fold change compared with (A) IMCS4fl/fl or (C–H) vehicle-treated. For each group, n = 3 (3 independent experiments on 3 separate days); *P < .05; **P < .01; ***P < .001 relative to vehicle only (2-tailed t tests). (E) #P < .05; ##P < .01; ###P < .001 relative to TNF-treated. Veh, vehicle.
We next asked whether TGFβ1 and BMP2 treatment could extinguish Ccl20 induction after TNF treatment because this effect would have important clinical implications. YAMC cells were treated with TNF or vehicle for 24 hours followed by TGFβ1 or vehicle in addition to TNF for the indicated times. By 4 hours of TGFβ1 treatment, the Ccl20 level in the presence of TNF was reduced to the level seen with no TNF treatment (Figure 8A). In IMCS4fl/fl cells, which have a higher induction of Ccl20 by TNF, TGFβ1 and BMP2 similarly were able to extinguish the TNF-mediated induction of Ccl20 (Figure 8B). Similar results were obtained in mouse colonoids (Figure 8C). Taken together, these data support that TGFβ and BMP inhibit specific chemokine and cytokine responses induced by proinflammatory cytokines and LPS in colon epithelial cells.
Figure 8.
TGFβ1 reverses TNF-induced induction of Ccl20 mRNA. (A) YAMC colonocytes were treated with 10 ng/mL TNF or vehicle for 24 hours followed by the addition of 3 ng/mL TGFβ1 or vehicle for the indicated times followed by mRNA analysis. (B) IMCS4fl/fl cells were treated with TNF or vehicle for 24 hours followed by the addition of 3 ng/mL TGFβ1 and 100 ng/mL BMP2 for the indicated times. Colonoids were treated with vehicle or 100 ng/mL TNF for 24 hours followed by the addition of 3 ng/mL TGFβ1 for 8 hours. mRNA was quantified as described in Figure 7. For each group, n = 3 biological repeats (performed on 3 separate days). ∗P < .05, ∗∗P < .01. ve or veh, vehicle.
Smad4 Loss Increases Macrophage Infiltration in Mouse Colon
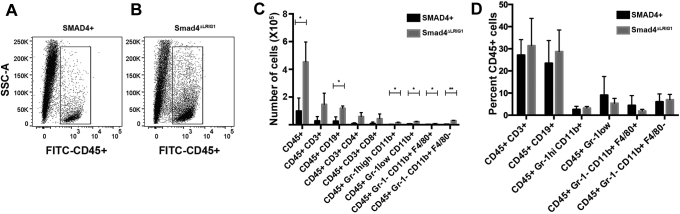
Multiple chemokines were up-regulated by Smad4 loss in mouse colon epithelium (Figure 5). Therefore, we used flow cytometry of the lamina propria to determine if there was a concomitant alteration in immune cell infiltrate when Smad4 was lost. We found that loss of Smad4 specifically in the epithelium led to an overall increase in CD45+ cells in the surrounding tissue (Figure 9A and B). This overall 4-fold increase in leukocytes was reflected in significant increases in CD19+ B cells, Gr-1high/CD11b+ neutrophils, Gr-1low/CD11b+ monocytes, GR-1-/CD11b+/F4/80- dendritic cells, and GR-1-/CD11b+/F4/80+ macrophages (Figure 9C). When these cell populations were normalized to the number of CD45+ leukocytes, no specific alteration in any 1 cell type was observed (Figure 9D), suggesting that the array of chemokines regulated by SMAD4 act broadly on many components of the immune microenvironment.
Figure 9.
Loss of epithelial Smad4 results in increased leukocyte infiltration in the surrounding lamina propria. Lamina propria was isolated from SMAD4+ control mice or from Smad4ΔLrig1 mice and analyzed by flow cytometry for the indicated markers. (A and B) Scattergram showing the relative number of live cells that were CD45+. (C) The number of specific cell populations per colon are shown for the indicated markers. (D) The percentage of each population normalized to the number of CD45+ cells. CD45+Gr-1- CD11b+ F4/80- cells included cells that were CD11c+ and/or CD103+, but not cells that were CD11c- CD103-. For each group, n = 3 (performed 1 control and 1 Smad4ΔLrig1 mouse on each of 3 separate days). *P < .05; **P < .01. FITC, fluorescein isothiocyanate; SSC-A, side scatter-area.
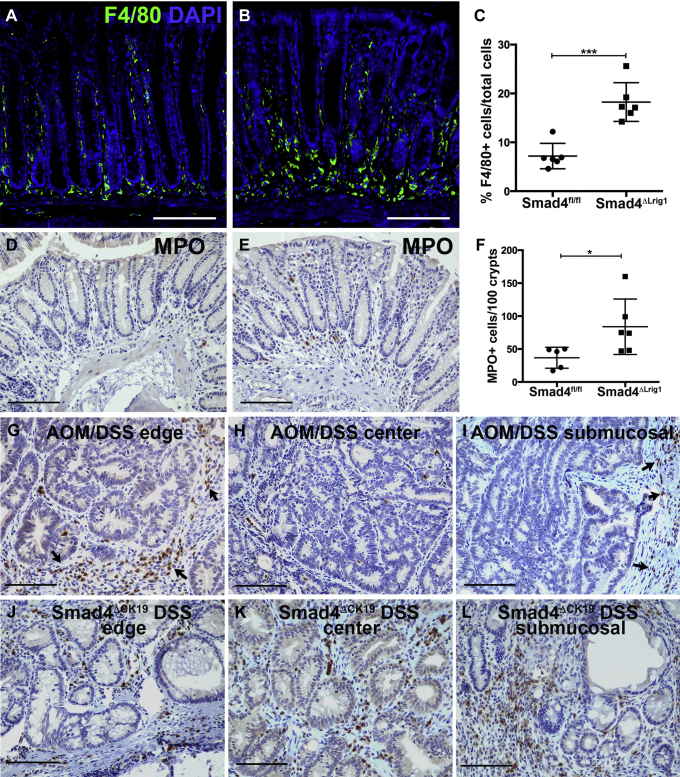
Although flow cytometry indicated that loss of Smad4 resulted in an overall increase in CD45+ leukocytes, we also determined if loss of Smad4 altered localization of specific leukocyte populations. The top 2 chemokine genes up-regulated by Smad4 loss were Cxcl5 and Ccl20. Ccl20 encodes a chemokine for lymphoid cells and dendritic cells, although a recent publication indicated that loss of CCR6, the receptor for CCL20, decreased macrophage infiltration in association with decreased tumor burden in Apc mutant mice.47 Therefore, we examined macrophage infiltration in the colons of 6 Smad4ΔLrig1 mice with increased Ccl20 expression and 6 control mice. We found that Smad4ΔLrig1 mice had significantly increased macrophage infiltration compared with SMAD4+ control mice (Figure 10A–C). CXCL5 is a chemokine for neutrophils. Therefore, we examined neutrophil infiltration in mice with or without epithelial SMAD4. Neutrophils were not abundant in control colons with an average of less than 1 myeloperoxidase+ neutrophil per crypt. We quantified the total number of neutrophils surrounding the distal-most 100 crypts of each colon. Smad4ΔLrig1 mice had a 2.3-fold increase in the number of neutrophils compared with control mice (Figure 10D–F). We also examined neutrophil infiltration in colon tumors with and without Smad4 loss. For Smad4+ tumors, we examined wild-type mice treated with AOM and DSS. We found that neutrophils were abundant in both AOM/DSS tumors and in Smad4ΔCK19/DSS tumors. However, neutrophil localization varied between these different tumor types. AOM/DSS tumors had large infiltrations of neutrophils around the tumor periphery but few neutrophils within the tumor mass, both within the mucosal layer and in regions of submucosal invasion. Smad4ΔCK19/DSS tumors contained numerous myeloperoxidase+ cells throughout the tumor mass rather than being restricted to the tumor periphery (Figure 10G–L). In summary, loss of SMAD4 in colon epithelium significantly increases inflammatory cell infiltration of neutrophils, macrophages, lymphocytes, and dendritic cells into the colonic submucosa. Our data are consistent with a role for SMAD4-mediated signaling as a homeostatic inhibitory regulator of inflammatory signaling in colonic epithelium and associated inflammatory cell infiltration into the submucosa of the colon.
Figure 10.
Loss of epithelial Smad4 alters the immune microenvironment. (A–C) Sections from colons of Smad4+ controls (Smad4fl/fl) or Smad4ΔLrig1 mice were immunolabeled for F4/80 as a marker of macrophages. (A and B) Representative images of F4/80 staining (green) and nuclear counterstain (blue) in (A) control or (B) Smad4ΔLrig1 colons. (C) Quantification of F4/80+ cells in each condition. (D–F) Sections from colons of Smad4+ controls (Smad4fl/fl) or Smad4ΔLrig1 mice were immunolabeled for myeloperoxidase (MPO) as a marker for neutrophils. (D and E) Representative images of MPO staining (brown) and nuclear counterstain (blue) in comparable regions of distal colon from (D) control or (E) Smad4ΔLrig1 mice. (F) Quantification of MPO+ cells surrounding distal-most 100 crypts in each condition. (G–L) MPO immunolabeling in Smad4+ tumors from AOM/DSS-treated mice and in Smad4- tumors from Smad4ΔLrig1 DSS-treated mice. Regions shown represent outer edges, central tumor regions, and regions of submucosal invasion, as indicated. Arrows indicate MPO+ cells at the tumor periphery. For each group, n = 5 or 6. *P < .05; ***P < .001 (2-tailed t tests). Scale bars: 100 μm. DAPI, 4′,6-diamidino-2-phenylindole.
SMAD-Mediated Signaling Represses Only a Subset of TNF-Regulated Genes
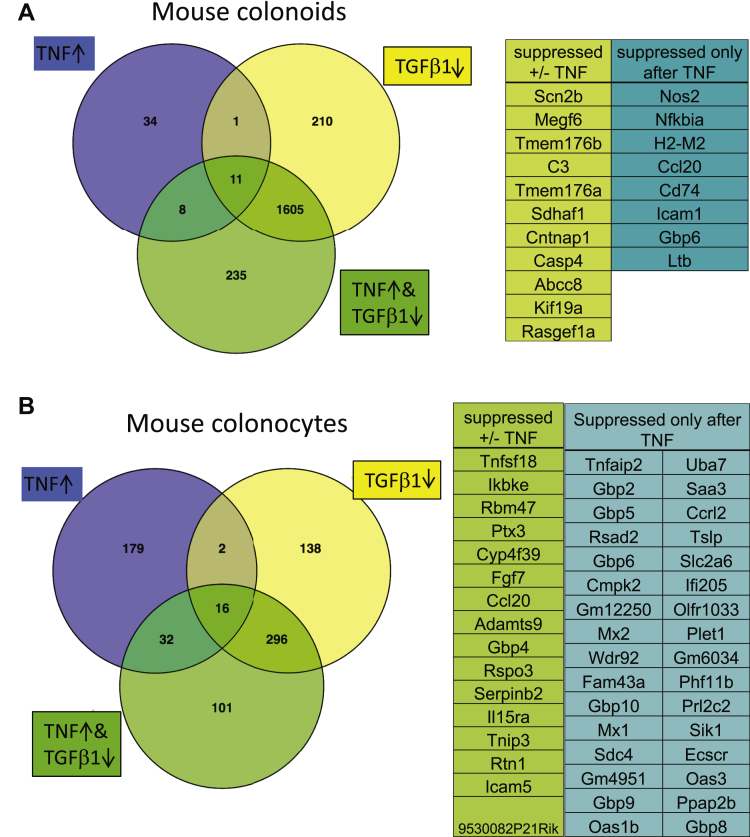
The ability of the SMAD4 pathway to block TNF-mediated induction of Ccl20, Cxcl5, and Il1rn suggests that this pathway may act as a gatekeeper for induction of other inflammation-associated genes. To determine if SMAD-mediated signaling blocks all or specific subsets of TNF-induced genes, we analyzed mouse colonoids treated with or without TNF and TGFβ1 by RNA sequencing using parameters of a FDR of 0.01 or less and fold-change of 1.5 or greater. Treatment of colonoids with TGFβ1 decreased the expression of 1827 genes compared with vehicle control (Figure 11A). TNF treatment alone increased the expression of 54 genes. Co-treatment with TGFβ1 reduced the expression of 19 (35%) of the genes that had been induced by TNF alone. Similar results were obtained with IMCS4fl/fl colonocytes in which TGFβ1 and BMP2 together decreased expression of 452 genes (FDR, ≤0.01; fold-change, ≥1.5) compared with vehicle control, whereas treatment with TGFβ1 and BMP2 before and during TNF treatment prevented the induction of 48 of the 229 (21%) genes induced by TNF alone (Figure 11B). Thus, SMAD4-mediated signaling inhibits expression of a subset of the genes induced by TNF in colonocytes rather than blocking all TNF signaling.
Figure 11.
TGFβ1 signaling inhibits expression of a subset of TNF-induced genes. (A) Mouse colonoids were treated with vehicle, 100 ng/mL TNF, and/or 3 ng/mL TGFβ1, as indicated for 24 hours followed by RNA isolation and sequencing. The Venn diagram shows the overlap between genes up-regulated by TNF treatment alone compared with vehicle control (blue), down-regulated by TGFβ1 treatment alone compared with vehicle control (yellow), and down-regulated by co-treatment with TGFβ1 and TNF treatment compared with TNF treatment alone (green). (B) IMCS4fl/fl SMAD4+ colonocytes were treated with vehicle, 100 ng/mL TNF, 3 ng/mL TGFβ1, or both TNF and TGFβ1 for 6 hours followed by RNA isolation and sequencing. The Venn diagram shows the overlap between genes up-regulated by TNF treatment compared with vehicle control (blue), down-regulated by TGFβ1/BMP treatment compared with vehicle control (yellow), and down-regulated by both TNF and TGFβ1/BMP treatment compared with TNF treatment alone. Numbers indicate the number of genes regulated with FDR of 0.01 or less, fold-change of 1.5 or greater. For each group, n = 3 biological repeats (isolated on 3 separate days).
Discussion
The colonic epithelium provides an important barrier to prevent the influx of microorganisms into underlying tissues and provides the first line of defense when that barrier is breached through damage or infection. Part of that function is the ability to up-regulate production of chemokines to induce a rapid immune response in the event of damage or infection. The epithelium also increases chemokine production in response to cytokines such as TNF and IL1β produced by immune cells to further amplify the inflammatory response. This epithelial mechanism of activation/amplification could result in inappropriate and uncontrolled inflammation if not tightly regulated. Therefore, it is critical that regulatory pathways exist to prevent inappropriate activation of innate signaling pathways and to suppress inflammatory reactions when infection is cleared and/or barrier function restored. TGFβ is a known mediator of immune suppression in T cells. Tumor-suppressive macrophages or myeloid-derived suppressor cells produce TGFβ, which then activates T-regulatory cells, inducing FoxP3 expression via SMAD binding sites on the FoxP3 promoter.48 FoxP3, another transcription factor, then can up-regulate expression of CTLA4 and other immune-suppressive proteins that inhibit T-effector cells. We have uncovered a novel target of TGFβ-mediating immune suppression. In addition to acting directly on T regulatory cells, TGFβ can signal directly to epithelial cells to suppress expression of chemokines and cytokines that otherwise could exacerbate inflammation.
We found that multiple chemokine/cytokine and other inflammation-associated genes are regulated by TGFβ/BMP signaling in colonic epithelial cells. In mouse colon, loss of Smad4 increased the expression of chemokines for T cells (Ccl20, Il18), neutrophils (Cxcl5), and monocytes (Ccl8), suggesting a broad range of inflammatory mediation. Within this group, Ccl20, is particularly intriguing with known functions in colon inflammation and association with both IBD49, 50 and colon cancer.51 Future studies will examine the importance of SMAD4-mediated regulation of CCL20 and these other chemokines in progression to CAC. Similar to our previous work,18 Principe et al52 found that broad epithelial expression via a metalloproteinase promoter of a dominant-negative Tgfbr2 transgene resulted in increased adenomas in Apc mutant mice. These mice also had increased numbers of mast cells and macrophages in the tumor microenvironment as compared with Apc mutation alone, supporting a role for TGFβ/SMAD4 signaling in immune suppression.
We have shown a direct role of TGFβ canonical pathway signaling in the repression of CCL20 gene expression in colonic epithelial cells. Interestingly, our results are in contrast to those of Brand et al,45 who recently reported that TGFβ signaling increases the expression of CCL20 in lung fibroblasts. Our differing observations may be explained by the tissue context and differential responses to specific activating and inhibitory factors that may be found in these 2 distinct cell types. Further studies are ongoing to determine the mechanism by which TGFβ canonical signaling and SMAD4 binding to regulatory elements differ in fibroblasts compared with colonic epithelial cells.
There are many sources of TGFβ and BMP production in the colon that could support homeostasis by inhibiting inflammation in a microenvironment of abundant commensal bacteria and immune cells. All 3 isoforms of TGFβ are expressed in the epithelium of the small intestine and colon,53 as well as by immune cells as discussed earlier.54 Multiple BMPs are expressed in either the crypt epithelium or the adjacent stroma, whereas BMP inhibitors are produced largely by the colonic stroma.55, 56, 57 We found that nuclear SMAD4 (indicative of active TGFβ pathway signaling) is normally abundant throughout the murine colonic gland (Figure 3), indicating sufficient ligand and receptor interaction to activate signal transduction in the normal colon. Loss of Smad4 specifically in the colonic epithelial cells disrupts this homeostatic signaling and results in up-regulation of multiple proinflammatory genes. Thus, interactions among multiple cell and tissue types, including epithelial and immune cells, impinge on this pathway to fine tune the inflammatory response.
The regimen of DSS exposure that we used for C57BL/6J mice does not lead to tumorigenesis without loss of the Smad4 gene in the time course of our studies. However, there are reports that different regimens of DSS can lead to tumorigenesis in other strains of mice. In CBA/J mice, up to 9 rounds of DSS induced invasive carcinoma in 8% of mice after 6 months without any genetically engineered mutations.58 In Swiss Webster mice, 10% to 25% of mice developed cancer after 4 rounds of DSS and followed up for varying lengths of time.6 The different observations in those studies as compared with our study may be owing to differences in mouse strain and/or in DSS regimen.
The invasive mucinous tumors that arose in the colons of Smad4ΔCK19 mice after DSS treatment were reminiscent of the tumors observed in ulcerative colitis–associated colon carcinomas. This observation led us to examine SMAD4 expression in human UC-associated colon cancers in comparison with sporadic colon cancers. We found that 48% of the human UC-associated colon cancers were negative for SMAD4 immunostaining in the cancer epithelial cells, whereas only 19% of sporadic colon cancers were negative for SMAD4 immunostaining. Our finding of an increasing rate of SMAD4 loss in high-grade as compared with low-grade dysplasia suggests that this loss may be an important factor in progression from premalignant lesions to invasive malignancy. Our findings of the high frequency of SMAD4 protein loss are somewhat surprising in light of a recent report by Robles et al59 showing that only 13% of IBD-associated CRCs have SMAD4 gene mutations, compared with the 10% of SMAD4 mutations in the TCGA collection of mostly sporadic CRCs.60 Therefore, the rate of identified mutations in SMAD4 in either sporadic or IBD-associated cancers does not fully explain the loss of SMAD4 immunostaining that we have observed in 19% of sporadic CRCs and 48% of UC-associated cancers. The higher frequency of SMAD4 protein loss compared with gene mutation in both sporadic and UC-associated carcinomas suggests that there is some other mechanism for silencing SMAD4 protein levels in CRC. Future studies will identify the mechanisms underlying the observed SMAD4 protein loss in UC-associated CRC.
Loss of a single tumor-suppressor gene rarely leads to carcinogenesis, however, we have found that loss of Smad4 alone is sufficient to initiate inflammation-driven carcinogenesis in the colon. Our observation that chronic inflammation combined with loss of SMAD4 in the adult colonic epithelium is sufficient to drive carcinogenesis seems, on the surface, to be inconsistent with the five-hit hypothesis of colorectal carcinogenesis proposed by Fearon and Vogelstein.61 However, we suspect that chronic inflammation in the presence of SMAD4 loss results in further genotoxic stress and mutations that drive the carcinogenic process. Future studies will determine whether there are consistent additional genetic lesions that account for the invasive mucinous adenocarcinomas that we have observed and their similarity to those identified in human CAC.
Footnotes
Current affiliations of T.J.F.: University of Pittsburgh Medical Center, Pittsburgh, PA; R.C.: Jawaharlal Nehru University, New Delhi, India; and T.D.B.: ICON PLC, Nashville, TN.
Author contributions Anna L. Means and R. Daniel Beauchamp supervised, designed, and analyzed experiments, and wrote the manuscript; Anna L. Means, Tanner J. Freeman, Connie J. Weaver, Luke G. Woodbury, Hanbing An, Jinghuan Zi, Bronson C. Wessinger, and Tasia D. Brown performed mouse, cell line, and colonoid experiments; Jing Zhu and Chandrasekhar Padmanabhan analyzed RNA sequencing data; Paula Marincola-Smith performed flow cytometry; Sergey V. Novitskiy designed and interpreted flow cytometry experiments; Chao Wu performed the tumoroid experiments; J. Joshua Smith and Charles L. Sawyers supervised tumoroid experiments; Anne R. Meyer performed macrophage analysis; James R. Goldenring supervised macrophage analysis; Rupesh Chaturvedi performed the Luminex experiment; Keith T. Wilson supervised the Luminex experiment; Natasha G. Deane established the IMCS4fl/fl cell line and supervised the development of the IMCS4null lines; Robert J. Coffey provided the Lrig1CreERT2 mice and unpublished data; and M. Kay Washington and Chanjuan Shi interpreted all pathology and provided human tissue slides and tissue microarrays.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Institutes of Health (NIH) grants CA069457 (R.D.B.), DK058404 (K.T.W., C.S., M.K.W.), AT004821 (K.T.W.), CA095103 (C.S., M.K.W., R.J.C.), F30 CA165726 (T.J.F.), CAR35197570 (R.J.C.), DK071590 (J.R.G.), T32 GM008554 (A.R.M.), CA200681 (S.V.N.); VA Merit Awards I01BX000930 (J.R.G.) and I01BX001453 (K.T.W.); NIH P30CA068485 (Vanderbilt-Ingram Cancer Center core services) and NIH P30DK058404 (Vanderbilt Digestive Diseases Research Center core services); Joel J. Roslyn Research Award/Association for Academic Surgery (J.J.S.); Limited Project and Career Development grants/American Society of Colon and Rectal Surgeons (J.J.S.); Franklin Martin, MD, FACS, Faculty Research Fellowship/American College of Surgeons (J.J.S.); Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant AACR-DR22-17 (J.J.S; administered by the American Association of Cancer Research); Memorial Sloan Kettering Cancer Center grant (J.J.S.); Memorial Sloan Kettering Cancer Center Surgery Faculty Research award (J.J.S.); and the Howard Hughes Medical Institute funding (C.L.S.).
Contributor Information
Anna L. Means, Email: anna.means@vanderbilt.edu.
R. Daniel Beauchamp, Email: daniel.beauchamp@vanderbilt.edu.
Supplementary Material
References
- 1.Elinav E., Nowarski R., Thaiss C.A., Hu B., Jin C., Flavell R.A. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein C.N., Blanchard J.F., Kliewer E., Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91:854–862. doi: 10.1002/1097-0142(20010215)91:4<854::aid-cncr1073>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Eaden J.A., Abrams K.R., Mayberry J.F. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutter M.D., Saunders B.P., Wilkinson K.H., Rumbles S., Schofield G., Kamm M.A., Williams C.B., Price A.B., Talbot I.C., Forbes A. Thirty-year analysis of a colonoscopic surveillance program for neoplasia in ulcerative colitis. Gastroenterology. 2006;130:1030–1038. doi: 10.1053/j.gastro.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Aust D.E., Terdiman J.P., Willenbucher R.F., Chang C.G., Molinaro-Clark A., Baretton G.B., Loehrs U., Waldman F.M. The APC/beta-catenin pathway in ulcerative colitis-related colorectal carcinomas: a mutational analysis. Cancer. 2002;94:1421–1427. doi: 10.1002/cncr.10334. [DOI] [PubMed] [Google Scholar]
- 6.Cooper H.S., Murthy S., Kido K., Yoshitake H., Flanigan A. Dysplasia and cancer in the dextran sulfate sodium mouse colitis model. Relevance to colitis-associated neoplasia in the human: a study of histopathology, B-catenin and p53 expression and the role of inflammation. Carcinogenesis. 2000;21:757–768. doi: 10.1093/carcin/21.4.757. [DOI] [PubMed] [Google Scholar]
- 7.Arthur J.C., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maggio-Price L., Treuting P., Zeng W., Tsang M., Bielefeldt-Ohmann H., Iritani B.M. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006;66:828–838. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiechl G., Bauer B., Fuss I., Lang S.A., Moser C., Ruemmele P., Rose-John S., Neurath M.F., Geissler E.K., Schlitt H.J., Strober W., Fichtner-Feigl S. Tumor development in murine ulcerative colitis depends on MyD88 signaling of colonic F4/80+CD11b(high)Gr1(low) macrophages. J Clin Invest. 2011;121:1692–1708. doi: 10.1172/JCI42540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan Y., Chen P.J., Sadler W.D., Wang F., Poe S., Nunez G., Eaton K.A., Chen G.Y. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013;73:7199–7210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song X., Gao H., Lin Y., Yao Y., Zhu S., Wang J., Liu Y., Yao X., Meng G., Shen N., Shi Y., Iwakura Y., Qian Y. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40:140–152. doi: 10.1016/j.immuni.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Pang Y., Moses H.L. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni A.B., Huh C.G., Becker D., Geiser A., Lyght M., Flanders K.C., Roberts A.B., Sporn M.B., Ward J.M., Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci U S A. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim B.G., Li C., Qiao W., Mamura M., Kasprzak B., Anver M., Wolfraim L., Hong S., Mushinski E., Potter M., Kim S.J., Fu X.Y., Deng C., Letterio J.J. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7 doi: 10.1126/scisignal.2005189. re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moustakas A., Heldin C.H. Mechanisms of TGFbeta-induced epithelial-mesenchymal transition. J Clin Med. 2016;5:7. doi: 10.3390/jcm5070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman T.J., Smith J.J., Chen X., Washington M.K., Roland J.T., Means A.L., Eschrich S.A., Yeatman T.J., Deane N.G., Beauchamp R.D. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012;142:562–571 e2. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardeesy N., Cheng K.H., Berger J.H., Chu G.C., Pahler J., Olson P., Hezel A.F., Horner J., Lauwers G.Y., Hanahan D., DePinho R.A. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Means A., Xu Y., Zhao A., Ray K., Gu G. A CK19-CreERT knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46:318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Greene N.T., Means A.L., Lu P., Jiang A., Schmidt C.R., Chakravarthy A.B., Merchant N.B., Washington M.K., Zhang B., Shyr Y., Deane N.G., Beauchamp R.D. Four jointed box 1 promotes angiogenesis and is associated with poor patient survival in colorectal carcinoma. PLoS One. 2013;8:e69660. doi: 10.1371/journal.pone.0069660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaine S.A., Ray K.C., Anunobi R., Gannon M.A., Washington M.K., Means A.L. Adult pancreatic acinar cells give rise to ducts but not endocrine cells in response to growth factor signaling. Development. 2010;137:2289–2296. doi: 10.1242/dev.048421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Means A.L., Ray K.C., Singh A.B., Washington M.K., Whitehead R.H., Harris R.C., Jr., Wright C.V., Coffey R.J., Jr., Leach S.D. Overexpression of heparin-binding EGF-like growth factor in mouse pancreas results in fibrosis and epithelial metaplasia. Gastroenterology. 2003;124:1020–1036. doi: 10.1053/gast.2003.50150. [DOI] [PubMed] [Google Scholar]
- 25.Ray K.C., Moss M.E., Franklin J.L., Weaver C.J., Higginbotham J., Song Y., Revetta F.L., Blaine S.A., Bridges L.R., Guess K.E., Coffey R.J., Crawford H.C., Washington M.K., Means A.L. Heparin-binding epidermal growth factor-like growth factor eliminates constraints on activated Kras to promote rapid onset of pancreatic neoplasia. Oncogene. 2014;33:823–831. doi: 10.1038/onc.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luna L.G. 3rd ed. McGraw Hill Publications; New York: 1968. Manual of histologic staining methods of the Armed Forces Institute of Pathology. [Google Scholar]
- 27.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumgarth N., Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 29.Whitehead R.H., VanEeden P.E., Noble M.D., Ataliotis P., Jat P.S. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc Natl Acad Sci U S A. 1993;90:587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grau A.M., Datta P.K., Zi J., Halder S.K., Beauchamp R.D. Role of Smad proteins in the regulation of NF-kappaB by TGF-beta in colon cancer cells. Cell Signal. 2006;18:1041–1050. doi: 10.1016/j.cellsig.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M., Hellmann M.D., Barron D.A., Schram A.M., Hameed M., Dogan S., Ross D.S., Hechtman J.F., DeLair D.F., Yao J., Mandelker D.L., Cheng D.T., Chandramohan R., Mohanty A.S., Ptashkin R.N., Jayakumaran G., Prasad M., Syed M.H., Rema A.B., Liu Z.Y., Nafa K., Borsu L., Sadowska J., Casanova J., Bacares R., Kiecka I.J., Razumova A., Son J.B., Stewart L., Baldi T., Mullaney K.A., Al-Ahmadie H., Vakiani E., Abeshouse A.A., Penson A.V., Jonsson P., Camacho N., Chang M.T., Won H.H., Gross B.E., Kundra R., Heins Z.J., Chen H.W., Phillips S., Zhang H., Wang J., Ochoa A., Wills J., Eubank M., Thomas S.B., Gardos S.M., Reales D.N., Galle J., Durany R., Cambria R., Abida W., Cercek A., Feldman D.R., Gounder M.M., Hakimi A.A., Harding J.J., Iyer G., Janjigian Y.Y., Jordan E.J., Kelly C.M., Lowery M.A., Morris L.G.T., Omuro A.M., Raj N., Razavi P., Shoushtari A.N., Shukla N., Soumerai T.E., Varghese A.M., Yaeger R., Coleman J., Bochner B., Riely G.J., Saltz L.B., Scher H.I., Sabbatini P.J., Robson M.E., Klimstra D.S., Taylor B.S., Baselga J., Schultz N., Hyman D.M., Arcila M.E., Solit D.B., Ladanyi M., Berger M.F. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M., Hellmann M.D., Barron D.A., Schram A.M., Hameed M., Dogan S., Ross D.S., Hechtman J.F., DeLair D.F., Yao J., Mandelker D.L., Cheng D.T., Chandramohan R., Mohanty A.S., Ptashkin R.N., Jayakumaran G., Prasad M., Syed M.H., Rema A.B., Liu Z.Y., Nafa K., Borsu L., Sadowska J., Casanova J., Bacares R., Kiecka I.J., Razumova A., Son J.B., Stewart L., Baldi T., Mullaney K.A., Al-Ahmadie H., Vakiani E., Abeshouse A.A., Penson A.V., Jonsson P., Camacho N., Chang M.T., Won H.H., Gross B.E., Kundra R., Heins Z.J., Chen H.W., Phillips S., Zhang H., Wang J., Ochoa A., Wills J., Eubank M., Thomas S.B., Gardos S.M., Reales D.N., Galle J., Durany R., Cambria R., Abida W., Cercek A., Feldman D.R., Gounder M.M., Hakimi A.A., Harding J.J., Iyer G., Janjigian Y.Y., Jordan E.J., Kelly C.M., Lowery M.A., Morris L.G.T., Omuro A.M., Raj N., Razavi P., Shoushtari A.N., Shukla N., Soumerai T.E., Varghese A.M., Yaeger R., Coleman J., Bochner B., Riely G.J., Saltz L.B., Scher H.I., Sabbatini P.J., Robson M.E., Klimstra D.S., Taylor B.S., Baselga J., Schultz N., Hyman D.M., Arcila M.E., Solit D.B., Ladanyi M., Berger M.F. Erratum: mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:1004. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 34.Chang Q., Bournazou E., Sansone P., Berishaj M., Gao S.P., Daly L., Wels J., Theilen T., Granitto S., Zhang X., Cotari J., Alpaugh M.L., de Stanchina E., Manova K., Li M., Bonafe M., Ceccarelli C., Taffurelli M., Santini D., Altan-Bonnet G., Kaplan R., Norton L., Nishimoto N., Huszar D., Lyden D., Bromberg J. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia. 2013;15:848–862. doi: 10.1593/neo.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 37.Drost J., van Jaarsveld R.H., Ponsioen B., Zimberlin C., van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H., Korving J., van de Wetering M., Schwank G., Logtenberg M., Cuppen E., Snippert H.J., Medema J.P., Kops G.J., Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 38.Shiou S.R., Datta P.K., Dhawan P., Law B.K., Yingling J.M., Dixon D.A., Beauchamp R.D. Smad4-dependent regulation of urokinase plasminogen activator secretion and RNA stability associated with invasiveness by autocrine and paracrine transforming growth factor-beta. J Biol Chem. 2006;281:33971–33981. doi: 10.1074/jbc.M607010200. [DOI] [PubMed] [Google Scholar]
- 39.Carbo A., Olivares-Villagomez D., Hontecillas R., Bassaganya-Riera J., Chaturvedi R., Piazuelo M.B., Delgado A., Washington M.K., Wilson K.T., Algood H.M. Systems modeling of the role of interleukin-21 in the maintenance of effector CD4+ T cell responses during chronic Helicobacter pylori infection. mBio. 2014;5 doi: 10.1128/mBio.01243-14. e01243–e01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 42.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand O.J., Somanath S., Moermans C., Yanagisawa H., Hashimoto M., Cambier S., Markovics J., Bondesson A.J., Hill A., Jablons D., Wolters P., Lou J., Marks J.D., Baron J.L., Nishimura S.L. Transforming growth factor-beta and interleukin-1beta signaling pathways converge on the chemokine CCL20 promoter. J Biol Chem. 2015;290:14717–14728. doi: 10.1074/jbc.M114.630368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiie S., Hieshima K., Izawa D., Nakayama T., Fujisawa R., Ohyanagi H., Yoshie O. Proinflammatory cytokines induce liver and activation-regulated chemokine/macrophage inflammatory protein-3alpha/CCL20 in mucosal epithelial cells through NF-kappaB [correction of NK-kappaB] Int Immunol. 2001;13:1255–1263. doi: 10.1093/intimm/13.10.1255. [DOI] [PubMed] [Google Scholar]
- 47.Nandi B., Shapiro M., Samur M.K., Pai C., Frank N.Y., Yoon C., Prabhala R.H., Munshi N.C., Gold J.S. Stromal CCR6 drives tumor growth in a murine transplantable colon cancer through recruitment of tumor-promoting macrophages. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1189052. e1189052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmetterer K.G., Neunkirchner A., Pickl W.F. Naturally occurring regulatory T cells: markers, mechanisms, and manipulation. FASEB J. 2012;26:2253–2276. doi: 10.1096/fj.11-193672. [DOI] [PubMed] [Google Scholar]
- 49.Kaser A., Ludwiczek O., Holzmann S., Moschen A.R., Weiss G., Enrich B., Graziadei I., Dunzendorfer S., Wiedermann C.J., Murzl E., Grasl E., Jasarevic Z., Romani N., Offner F.A., Tilg H. Increased expression of CCL20 in human inflammatory bowel disease. J Clin Immunol. 2004;24:74–85. doi: 10.1023/B:JOCI.0000018066.46279.6b. [DOI] [PubMed] [Google Scholar]
- 50.Lee A.Y., Eri R., Lyons A.B., Grimm M.C., Korner H. CC Chemokine ligand 20 and its cognate receptor CCR6 in mucosal T cell immunology and inflammatory bowel disease: odd couple or axis of evil? Front Immunol. 2013;4:194. doi: 10.3389/fimmu.2013.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frick V.O., Rubie C., Kolsch K., Wagner M., Ghadjar P., Graeber S., Glanemann M. CCR6/CCL20 chemokine expression profile in distinct colorectal malignancies. Scand J Immunol. 2013;78:298–305. doi: 10.1111/sji.12087. [DOI] [PubMed] [Google Scholar]
- 52.Principe D.R., DeCant B., Staudacher J., Vitello D., Mangan R.J., Wayne E.A., Mascarinas E., Diaz A.M., Bauer J., McKinney R.D., Khazaie K., Pasche B., Dawson D.W., Munshi H.G., Grippo P.J., Jung B. Loss of TGFbeta signaling promotes colon cancer progression and tumor-associated inflammation. Oncotarget. 2017;8:3826–3839. doi: 10.18632/oncotarget.9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnard J.A., Warwick G.J., Gold L.I. Localization of transforming growth factor beta isoforms in the normal murine small intestine and colon. Gastroenterology. 1993;105:67–73. doi: 10.1016/0016-5085(93)90011-z. [DOI] [PubMed] [Google Scholar]
- 54.Wahl S.M. Transforming growth factor-beta: innately bipolar. Curr Opin Immunol. 2007;19:55–62. doi: 10.1016/j.coi.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Haramis A.P., Begthel H., van den Born M., van Es J., Jonkheer S., Offerhaus G.J., Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 56.Kosinski C., Li V.S., Chan A.S., Zhang J., Ho C., Tsui W.Y., Chan T.L., Mifflin R.C., Powell D.W., Yuen S.T., Leung S.Y., Chen X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., Madison B.B., Zacharias W., Kolterud A., States D., Gumucio D.L. Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol Genomics. 2007;29:290–301. doi: 10.1152/physiolgenomics.00269.2006. [DOI] [PubMed] [Google Scholar]
- 58.Okayasu I., Yamada M., Mikami T., Yoshida T., Kanno J., Ohkusa T. Dysplasia and carcinoma development in a repeated dextran sulfate sodium-induced colitis model. J Gastroenterol Hepatol. 2002;17:1078–1083. doi: 10.1046/j.1440-1746.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 59.Robles A.I., Traverso G., Zhang M., Roberts N.J., Khan M.A., Joseph C., Lauwers G.Y., Selaru F.M., Popoli M., Pittman M.E., Ke X., Hruban R.H., Meltzer S.J., Kinzler K.W., Vogelstein B., Harris C.C., Papadopoulos N. Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology. 2016;150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.