Abstract
Background & Aims
Uncontrolled liver proliferation is a key characteristic of liver cancer; however, the mechanisms by which this occurs are not well understood. Elucidation of these mechanisms is necessary for the development of better therapy. The oncogene Gankyrin (Gank) is overexpressed in both hepatocellular carcinoma and hepatoblastoma. The aim of this work was to determine the role of Gank in liver proliferation and elucidate the mechanism by which Gank promotes liver proliferation.
Methods
We generated Gank liver-specific knock-out (GLKO) mice and examined liver biology and proliferation after surgical resection and liver injury.
Results
Global profiling of gene expression in GLKO mice showed significant changes in pathways involved in liver cancer and proliferation. Investigations of liver proliferation after partial hepatectomy and CCl4 treatment showed that GLKO mice have dramatically inhibited proliferation of hepatocytes at early stages after surgery and injury. In control LoxP mice, liver proliferation was characterized by Gank-mediated reduction of tumor-suppressor proteins (TSPs). The failure of GLKO hepatocytes to proliferate is associated with a lack of down-regulation of these proteins. Surprisingly, we found that hepatic progenitor cells of GLKO mice start proliferation at later stages and restore the original size of the liver at 14 days after partial hepatectomy. To examine the proliferative activities of Gank in cancer cells, we used a small molecule, cjoc42, to inhibit interactions of Gank with the 26S proteasome. These studies showed that Gank triggers degradation of TSPs and that cjoc42-mediated inhibition of Gank increases levels of TSPs and inhibits proliferation of cancer cells.
Conclusions
These studies show that Gank promotes hepatocyte proliferation by elimination of TSPs. This work provides background for the development of Gank-mediated therapy for the treatment of liver cancer. RNA sequencing data can be accessed in the NCBI Gene Expression Omnibus: GSE104395.
Keywords: Liver, Proliferation, Cancer, Tumor-Suppressor Proteins, Progenitor Cells
Abbreviations used in this paper: BrdU, bromodeoxyuridine; cDNA, complementary DNA; C/EBP, CCAAT/enhancer binding protein; Co-IP, co-immunoprecipitation; CUGBP1, CUG triplet repeat binding protein 1; DEN, diethylnitrosamine; FXR, farnesoid X receptor; Gank, Gankyrin; HCC, hepatocellular carcinoma; GLKO, Gankyrin liver-specific knock-out; HNF4α, hepatocyte nuclear factor 4α; LKO, liver-specific knock-out; mRNA, messenger RNA; Opn, osteopontin; PCNA, proliferating cell nuclear antigen; PH, partial hepatectomy; Rb, retinoblastoma; RT-PCR, reverse-transcriptase polymerase chain reaction; TSP, tumor-suppressor protein; 2D, 2-dimensional; UPS, ubiquitin proteasome system; WT, wild-type
Graphical abstract
See editorial on page 345.
Summary.
The mechanisms by which gankyrin promotes hepatic proliferation are not known. This study shows that gankyrin promotes proteosomal degradation of tumor-suppressor proteins. Gankyrin deletion restored tumor-suppressor protein expression and delayed regenerative hepatocyte proliferation in vivo. Furthermore, proteasome inhibition limited growth of human- and mouse-derived liver cancer cell lines in vitro.
The development of liver cancer is associated with multiple alterations in cellular function and gene expression.1 One of the main hallmarks of liver cancer is uncontrolled proliferation, which is owing in part to damage of pathways essential to cell-cycle control. In addition, regulation in the coordinated expression of oncogenes and tumor-suppressor proteins (TSPs) is vital to tumor proliferation. One of the key oncogenes and promoters of liver proliferation is a small subunit of 26S proteasome, Gankyrin (Gank). This non–adenosine triphosphatase subunit of the ubiquitin proteasome system (UPS) is a notorious oncogene expressed in several cancer types, including hepatocellular carcinoma (HCC), in which it was first discovered.2, 3, 4 In agreement with these observations, Gank has been identified as the driver oncogene in the early development of liver cancer through chemical models as well as age-dependent hepatic tumorigenesis.2, 3, 4, 5, 6, 7
Gank promotes the development of HCC through several mechanisms, including the neutralization of TSPs. TSPs are the main proteins that support the quiescent status of the liver, and it has been shown that the activities of more than 20 different TSPs are lost in HCC because of mutations or hypermethylation of their promoters.8 In addition, the elimination of TSPs by Gank is essential to carcinogenesis.4 Specifically, Gank leads to the neutralization of essential TSPs such as tumor-suppressor protein p53, through stabilization of murine double minute 2 ligase and subsequent enhanced ubiquitination, and retinoblastoma, by direct interaction, both of which trigger UPS-mediated degradation.2, 3, 9 Studies of liver cancer have identified 2 additional targets of Gank: CCAAT/enhancer binding protein α (C/EBPα) and hepatocyte nuclear factor 4α (HNF4α).6, 7, 9 C/EBPα belongs to the C/EBP family of proteins, basic leucine zipper proteins, which contain basic region and leucine zipper regions.10 C/EBPα has been shown to be a strong inhibitor of proliferation and a strong TSP.4, 6, 10, 11 In fact, several recent reports with activation of the C/EBPα gene in animal models of liver carcinogenesis showed that its activation leads to inhibition of liver proliferation and carcinogenesis as well as normalization of liver function.12, 13, 14 HNF4α is also a strong TSP and expression of this protein correlates with the epithelial–mesenchymal transition involved in metastatic tumor formation.15 It also has been shown that deletion of HNF4α promotes hepatocyte proliferation and diethylnitrosamine (DEN)-induced liver cancer.16 In addition to these known TSPs, recent studies have identified RNA CUG triplet repeat binding protein 1 (CUGBP1) as a tumor suppressor, whose activity depends on phosphorylation/dephosphorylation at serine 302.17 Generation of CUGBP1-S302A KI mice showed that this TSP protects the liver from the development of cancer and that during liver carcinogenesis, Gank eliminates this isoform of CUGBP1.17 In agreement with this, livers of CUGBP1 knock-out mice show a molecular signature of hepatoblastoma and express increased levels of stem cell markers and reduced levels of markers of hepatocytes.9
Increasing evidence has shown how Gank is responsible for the activation of additional pathways critical to liver cancer. For example, in addition to its effect on TSPs, Gank also stabilizes the stem cell marker octamer binding transcription factor 4 through elimination of WW domain-containing E3 ubiquitin protein ligase 2, the ubiquitin ligase that normally marks octamer binding transcription factor 4 for degradation.18 To promote uncontrolled proliferation, Gank also binds to D-type kinase, cyclin dependent kinase 4, and replaces p16INK4a from cyclin dependent kinase 4, leading to the activation of cyclin dependent kinase 4 and cell-cycle progression.2 In addition, Gank increases levels of oncogene Nrf2 by the elimination of Keap1 ligase, which triggers degradation of Nrf2.19
Regulation of activities of Gank in the liver is quite complicated. In the quiescent liver, farnesoid X receptor (FXR) partially represses Gank, however, with DEN-mediated carcinogenesis, there is a reduction of FXR, activation of Gank, and subsequent activation of the cascade of Gank-dependent pathways including loss of TSPs.7 Our recent article showed that activation of FXR by GW4064 inhibits the development of liver cancer and that the FXR–Gank axis is involved in the development of pediatric liver cancer.9 In agreement with these findings, a recent report showed that DEN-mediated liver cancer is reduced significantly in mice with liver-specific deletion of Gank.20
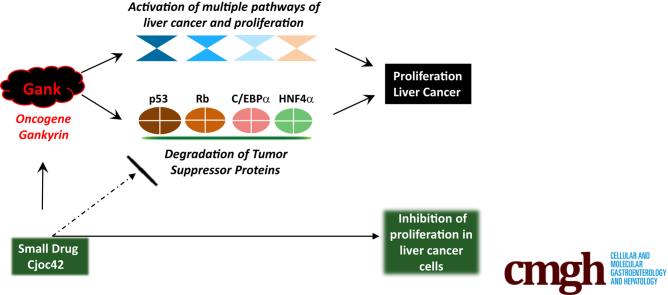
In this study, we examined the proliferative activities of Gank in recently generated liver-specific Gank liver-specific knock-out (LKO) mice. By using 2 models of liver proliferation/regeneration, partial hepatectomy (PH), and CCl4 treatments, we obtained evidence showing that Gank promotes liver proliferation via direct interaction and elimination of at least 5 TSPs. We also found that inhibition of Gank by a small drug, cjoc42, inhibits proliferation of liver cancer by blocking the Gank–TSPs axis, suggesting that cjoc42 might be considered a novel therapy approach.
Methods
Animals
Experiments with animals were approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital (protocol IACUC2014-0042). A Gank LKO (GLKO) mouse model was created using the Cre–Lox system. Mice expressing the Cre recombinase protein driven by the albumin promoter were crossed with mice that had LoxP sequences flanking exons 2–4 of the Gank gene. The resulting offspring had the Gank gene excised only in cells expressing albumin.
Histology
Liver tissue was taken from the left lobe and fixed in 4% formaldehyde. Mice were injected intraperitoneally with 66.5 mg/kg bromodeoxyuridine (BrdU) 4 hours before harvest. BrdU incorporation was measured using a commercially available kit (93-3943; Invitrogen, Carlsbad, CA).
Partial Hepatectomy and CCl4-Induced Liver Injury
LoxP and GLKO mice were maintained as previously described.17 Experiments were performed on young mice (age, 2–3 mo).
Partial hepatectomy
Surgery was performed as described in our previous publications and review articles.6, 21, 22 Four to 5 animals per each time point were analyzed. BrdU was injected 4 hours before death, livers were collected, and proteins and messenger RNA (mRNA) were isolated and analyzed as described later.
Acute CCl4 Treatments
Treatment of mice with CCl4 was performed as described in our article.23 Male mice were injected intraperitoneally with a single dose of 10% CCl4 in olive oil (5.0 mL/kg). Data represent summaries of work with 4–5 mice per time point. Experiments with animals were approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital (protocol IACUC2014-0042).
Cjoc42 Studies in Cancer Cell Lines
Cjoc42 was synthesized as described in the article first describing this drug.24 Successful synthesis was confirmed by mass spectrometry. Cjoc42 was dissolved in 100% dimethyl sulfoxide to make a 10 mmol/L stock solution. Huh6 cells (kindly gifted from Dimiter-Karl Bissig) were grown in Dulbecco’s modified Eagle medium (Fisher, Waltham, MA) and Hepa1c1c7 (European Collection of Authenticated Cell Cultures 95090613) were grown in Alpha-Minimum Essential Medium + GlutaMAX without nucleosides (Fisher). All media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were treated with 1, 5, and 10 μmol/L of cjoc42 at a final dilution of 0.1% dimethyl sulfoxide for 48–72 hours. Cells were incubated at 37°C in a CO2 incubator.
RNA Sequencing: Total RNA Was Isolated From 3 LoxP and 3 Gank LKO Mice
RNA sequencing libraries were prepared using an Illumina TruSeq RNA preparation kit and sequenced on the Illumina HiSeq 2500, using paired-end, 100-bp reads (Illumina, San Diego, CA). After removal of primers and barcodes, reads were aligned using mm10 annotations produced by University of California Santa Cruz, and quantified using Kallisto (Pasadena, CA), which accurately quantifies read abundances (in transcripts per million) through pseudoalignment. All statistical analysis was performed in GeneSpring (Santa Clara, CA) 13.0. Raw counts were thresholded at 1, normalized using a 75th percentile shift, and baselined to the median of all samples (n = 31,253 transcripts). A filtration was applied to ensure analysis of reasonably expressed transcripts, requiring at least 2 reads in >50% of samples in at least 1 experimental condition (n = 10,427 transcripts). Differential expression was assessed between treatment and genotype conditions using a 2-way analaysis of variance, with a significance cut-off value of FC > 1.5 (n = 672 transcripts across 2 comparisons). RNA sequencing data may be accessed in NCBI’s Gene Expression Omnibus: GSE104395.
Real-Time Quantitative Reverse-Transcriptase Polymerase Chain Reaction
Total RNA was isolated from mouse and human livers using the RNEasy Plus mini kit (Qiagen, Hilden, Germany). Complementary DNA (cDNA) was synthesized with 2 ug of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). cDNA was diluted 5 times with diethyl pyrocarbonate-treated water and subsequently used for reverse-transcription polymerase chain reaction (RT-PCR) assays with the TaqMan Gene Expression system (Applied Biosystems). Gene expression analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) in a total volume of 10 μL containing 5 μL Master Mix, 1.5 μL water, 3 μL cDNA template, and 0.5 μL of the gene-specific TaqMan Assay probe mixture. The cycling profile was 50°C for 2 minutes, 95° for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute as recommended by the manufacturer. TaqMan probe mixtures were purchased from ThermoFisher (Waltham, MA). The following probes were used: cytochrome (CYP)7A1, Mm00484150_m1; CYP2E1, Mm00491127_m1; Monoacylglycerol O-Acyltransferase 1, Mm00503358_m1; Peroxisome proliferator-activated receptor gamma, Mm00440940_m1; CUGBP1 (CELF1), Mm04279608_m1; C/EBPα, Mm01265914_s1; HNF4α, Mm01247712_m1; C/EBPB, Mm00843434_s1; JUN, Mm00495062_s1; and nuclear factor-κB1, Mm00476361_m1. Levels of all mRNAs were normalized to β-actin.
Protein Isolation, Western Blot, Co-Immunoprecipitation, and 2-Dimensional Gel Electrophoresis
Nuclear and cytoplasmic extracts were prepared as previously described.6, 7 Lysates (50–100 μg) were loaded onto 4%–20% gradient gels (Bio-Rad, Hercules, CA) and transferred to nitrocellulose membranes (Bio-Rad). Membranes were probed with corresponding antibodies. Co-immunoprecipitations and 2-dimensional (2D) examinations of the proteins were performed as previously described.6, 7 The following antibodies were used: CUGBP1 (sc-20003; Santa Cruz, Dallas, TX), C/EBPα (sc-61; Santa Cruz), Gank (12985S; Cell Signaling Technologies, Danvers, MA), HNF4α (PP-K9218-00; Perseus Proteomics), Rb (sc-50; Santa Cruz), E2F1 (sc-193; Santa Cruz), pRb (3590S; Cell Signaling Technologies), p53 serine 6 (sc-135630; Santa Cruz), β-actin (A5316; Sigma, St. Louis, MO), cyc D1 (RM-9104-S1; Thermo), proliferating cell nuclear antigen (PCNA) (sc-7907; Santa Cruz), Cyclin-dependent kinase 1 (cdc2) (sc-954; Santa Cruz), cyclin E (sc-481; Santa Cruz), cyclin A (sc-751; Santa Cruz), p21 (sc-6246; Santa Cruz), and p27 (sc-1641; Santa Cruz). Membranes then were incubated with the corresponding secondary horseradish-peroxidase–coupled antibodies: sc-2031 and sc-2313 (Santa Cruz) or 18-8817-31 and 18-8816-33 (Rockland, Limerick, PA).
Proliferation Assay
Huh6 and Hepa1c1c7 cells were seeded in 96-well plates at 5.0 × 104. Images were taken at 24 hours after seeding before treatment. Cells were treated with 10 μmol/L cjoc42, and at 72 hours after treatment media was removed and cells were washed with 1× phosphate-buffered saline. As per the CyQUANT Cell Proliferation Assay Kit (MP07026; Invitrogen), 200 μL CyQUANT GR dye/cell lysis buffer was added to each well and incubated for 2–5 minutes at room temperature, protected from light. Fluorescence was measured using a fluorescence plate reader with filters for 480 nm excitation and 520 nm emission maxima.
Immunohistochemistry
Liver sections were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and sectioned (6-μm sections). Four hours before tissue harvest mice were injected intraperitoneally with 66.5 mg/kg BrdU for histologic examination. BrdU staining was performed using a BrdU up-take assay kit (Invitrogen). Glutamine synthetase (610517) antibody was purchased from BD Biosciences (San Jose, CA). The Apoptag kit was from Millipore (St. Louis, MO) for terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling staining. For detection of proliferating progenitor cells in livers after PH, we performed co-staining of markers of progenitor cell epithelial cell adhesion molecule and osteopontin (Opn), and BrdU/4′,6-diamidino-2-phenylindole staining as described previously.25 Epithelial cell adhesion molecule antibody (ab71916) and BrdU (ab2284) antibody were purchased from Abcam (Cambridge, MA). Opn antibody (AF808) was purchased from R&D Systems (Minneapolis, MN).
Statistical Analysis
All values are presented as means ± SEM. An unpaired Student t test was applied for comparison of normally distributed data. Two-way analysis of variance was used with a Bonferroni test for multiple comparisons between different time points if the P value was < .05. Statistical significance was defined as follows: *P < .05, **P < .01, and ***P < .001, and ****P < .0001. All statistical analyses were performed using GraphPad Prism 6.0 (La Jolla, CA).
All authors had access to the study data and reviewed and approved the final manuscript.
Results
Generation and Characterization of Gank LKO Mice
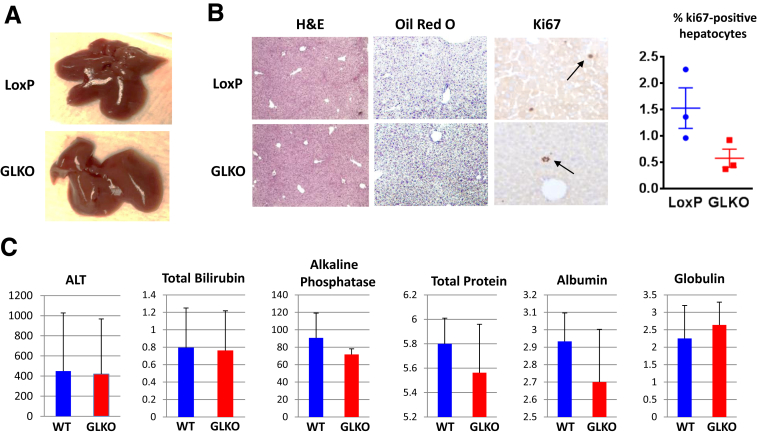
Previous studies of the role of Gank in liver biology have been performed in wild-type (WT) animals and presented only correlative observations. To understand the causal role of Gank in the regulation of liver biology and liver proliferation, we generated GLKO mice using the Cre–Lox system. Albumin-Cre mice were crossed with mice that had LoxP sequences flanking exons 2–4 of the Gank gene, resulting in deletion of the Gank gene only in cells expressing albumin. We partially described these mice in our previous article in which we found that Gank interacts and eliminates the tumor-suppressor isoform of CUGBP1.17 Figure 1 presents the main characteristics of the Gank LKO mice. The Gank LKO mice did not show any notable phenotype within 1 year of life. Gross liver examination of 2-month-old Gank LKO mice showed no morphologic differences compared with control LoxP mice (Figure 1A). In addition, there were no histologic differences in H&E, Oil Red O, and marker of proliferation ki67 (ki67) staining, illustrating that the deletion of Gank in hepatocytes does not change hepatocyte morphology and fat deposition (Figure 1B). Although we detected a slight reduction of ki67-positive hepatocytes in Gank LKO mice (Figure 1B, bar graphs), this reduction was not statistically significant. Note that the number of ki67-positive hepatocytes in control LoxP mice was approximately 1%, indicating very low or no proliferation. Examination of blood parameters also showed no differences in hepatic function (Figure 1C). It is important to note that at the time of writing this article, Sakurai et al20 published an article with the generation of Gank LKO mice and also did not detect any abnormalities in these mice under normal conditions.
Figure 1.
Characterization of Gank LKO mice. (A) Typical pictures of LoxP and Gank LKO livers at the age of 2 months. (B) H&E, Oil Red O, and Ki67 staining show no differences between LoxP and Gank LKO mice. Histologic analysis was performed in 3 LoxP and 3 GLKO mice, examining 4–5 fields/animal. Ki67 quantitation was performed in 3 mice per group, counting the number of Ki67-positive hepatocytes in 3 fields/mice (P > .09) (C) Blood parameters of LoxP and Gank LKO mice using 6 LoxP mice and 8 GLKO mice. P values were as follows: alanine aminotransferase (ALT) (P > .46), total bilirubin (P > .92), alkaline phosphatase (P > .09), total protein (P > .2), albumin (P > .1), and globulin (P > .9).
Deletion of Gank in the Liver Leads to Changes in Expression of Genes Within Several Critical Pathways of Liver Biology
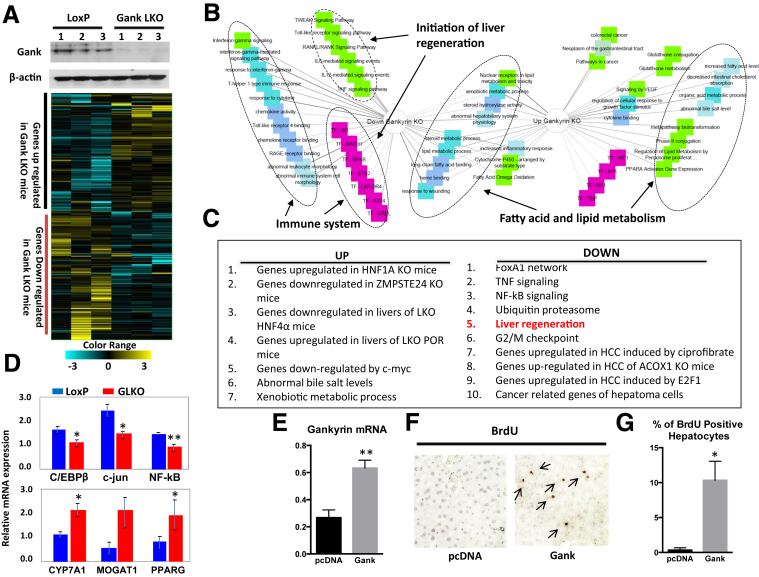
Although no significant abnormalities of liver biology were found in the initial characterization of Gank LKO mice, it is likely that the deletion of such an important molecule of the UPS in adult mice should lead to alterations in gene expression. To test this possibility, we performed RNA sequencing and compared transcriptome profiling in livers of Gank LKO and LoxP mice. A heat map diagram clearly showed that a large body of mRNAs are up-regulated and down-regulated in Gank LKO mice compared with LoxP mice (Figure 2A). Ontologic examination showed that altered genes belong to the pathways involved in the immune system, fat metabolism, liver proliferation, and carcinogenesis (Figure 2B). The list of major up-regulated and down-regulated pathways is presented in Figure 2C. The alterations of key genes were confirmed by the quantitative RT-PCR approach, for which an example is shown in Figure 2D. Overall, some of the most significantly down-regulated genes by RNA sequencing involved pathways in liver cancer and regeneration, strongly suggesting that Gank promotes liver proliferation. mRNA levels of liver regeneration genes C/EBPβ, c-jun, and nuclear factor-κB were reduced in the majority of examined GLKO mice (Figure 2D). However, we have found variability in some animals of both genotypes. Based on RNA sequencing results, we further investigated in-depth the proliferative capacities of the Gank LKO livers. Before the examination of liver proliferation in Gank LKO mice, we asked if the ectopic expression of Gank could initiate proliferation in WT livers. To study this, Gank-expressing plasmid was administered by tail vein injection to WT mice and DNA replication was examined by BrdU staining. BrdU was injected 24 hours after injection of Gank plasmid and 4 hours before killing mice. Figure 2E shows that the injection of Gank leads to a 3- to 4-fold increase of Gank mRNA in the liver. Measurements of BrdU uptake showed that ectopic expression of Gank initiated hepatocyte proliferation and that up to 15% of hepatocytes proliferate 24 hours after injection of Gank plasmid. No proliferation was detected in mice injected with empty plasmid complementary (DNA) vector (Figure 2F and G). These studies showed that Gank alone is able to initiate DNA replication in normal hepatocytes, indicating that Gank is a very strong initiator of liver proliferation and that its overexpression in cancer might be a critical event in malignant transformation.
Figure 2.
Examination of signaling pathways that are altered in livers of Gank LKO mice. (A) RNA sequencing–based heat map of genes with altered expression in livers of Gank LKO mice. Three animals of each genotype were used. Upper image shows deletion of Gank by Western blot in the livers of mice used for RNA sequencing. (B) Schematic presentation of networks that are changed in livers of Gank LKO mice. (C) List of main pathways altered in Gank LKO mice. (D) An example of confirmation of genes overexpressed in livers of Gank LKO mice using quantitative RT-PCR with 3 mice per genotype. Two technical replicates were performed for each biological sample. Upper: mRNA levels for genes involved in liver regeneration. Bottom: Levels of mRNAs that are increased in GLKO mice. P values were as follows: C/EBPβ (P < .04); c-jun (P < .02); nuclear factor-κB (NF-κB) (P < .002), CYP7A1 (P < .04), MOGAT1 (P > .08), and PPARG (P < .04). (E) Levels of Gank mRNA in the livers transfected with empty vector plasmid DNA vector and with vector expressing Gank (P < .0015). (F) Immunostaining of the livers with antibodies to BrdU. Arrows show BrdU-positive hepatocytes. (G) Bar graphs show the percentage of BrdU-positive hepatocyte in livers transfected with empty vector and with plasmid expressing Gank (P < .02). Three mice were used for the empty vector and 3 mice were used for the plasmid expressing Gank experiment, with 3 fields analyzed per mouse. An unpaired Student t test. ∗P < .05, ∗∗P < .01 LoxP vs GKO.
Livers of Gank LKO Mice Have Reduced Liver Proliferation After Partial Surgical Resection
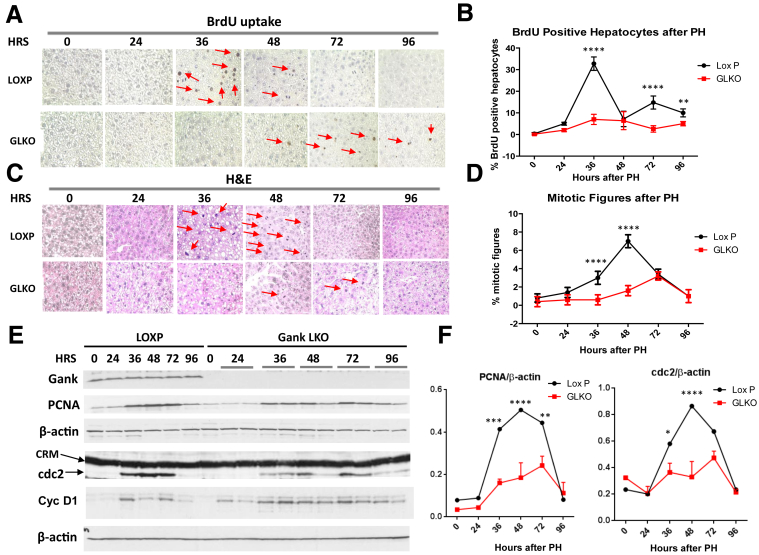
To study the role of Gank in liver proliferation, we first used a PH model. We performed two-thirds PH with Gank LKO mice and LoxP control mice. Animals were killed at 24, 36, 48, 72, and 96 hours after surgery. Four mice were used for each time point. As shown in Figure 3A and B, DNA replication (BrdU uptake) was inhibited dramatically in livers of Gank LKO mice at 36 hours compared with LoxP mice and shifted to 72 hours after PH. Consistent with this result, examination of mitotic figures showed a dramatic reduction of mitosis in livers of Gank LKO mice compared with LoxP control mice (Figure 3C and D). To obtain additional evidence for the reduction of liver proliferation, we examined levels of cell-cycle proteins PCNA, cyclin D1, cdc2, cyclin E, and cyclin A. These studies supported our histologic findings, with reduced levels of these proteins in livers of Gank LKO mice compared with livers of LoxP mice (Figure 3E and F for PCNA, cdc2, and cyclin D1; and Figure 4A for cyclins A and E). Note that although the difference in cyclin D1 expression was minor, other cell-cycle proteins showed much less increase in Gank LKO mice. Overall, the PH model of liver regeneration confirmed that Gank is required for liver proliferation.
Figure 3.
Livers of Gank LKO mice do not proliferate after PH. (A) Examination of liver proliferation by BrdU uptake. Livers of LoxP and Gank LKO mice harvested at different time points after PH (hours are shown on the top) were stained with antibodies to BrdU. Four to 5 mice of each genotype per each time point were analyzed with 4–5 fields examined per mouse. (B) Percentage of BrdU-positive hepatocytes. Quantitation was performed using 3 mice per genotype and time point and 3 fields per mouse. P values were as follows: 0 hours (P > .99), 24 hours (P > .26), 36 hours (P < .0001), 48 hours (P > .99), 72 hours (P < .0001), and 96 hours (P < .0069). (C) H&E staining of the livers at different time points after PH. Arrows show mitotic figures. Four to 5 mice of each genotype per each time point were analyzed with 4–5 fields examined per mouse. (D) Percentage of mitotic figures in livers of LoxP and Gank LKO mice. Quantitation was performed using 3 mice per genotype and time point and 3 fields per mouse. P values were as follows: 0 hours (P > .99), 24 hours (P > .23), 36 hours (P < .0001), 48 hours (P < .0001), 72 hours (P > .9999), and 96 hours (P > .9999). (E) Western blot analysis of Gank and cell-cycle proteins PCNA, cyclin D1, and cdc2. Membranes were reprobed with β-actin. Analysis included 3 biological replicates for each genotype and time point along with 2–3 technical replicates. (F) Levels of PCNA and cdc2 were calculated as ratios to β-actin. P values for PCNA were as follows: 0 hours (P > .99), 24 hours (P > .99), 36 hours (P < .0002), 48 hours (P < .0001), 72 hours (P < .0019), and 96 hours (P > .99). P values for cdc2 were as follows: 0 hours (P > .99), 24 hours (P > .99), 36 hours (P < .03), 48 hours (P < .0001), 72 hours (P > .05), and 96 hours (P > .99). CRM; cross-reactive molecule. Two way analysis of variance ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 LoxP vs GKO.
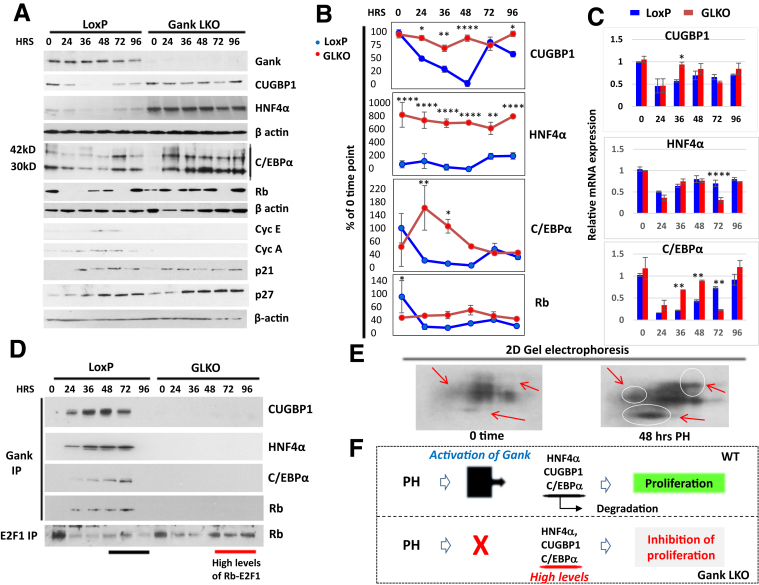
Figure 4.
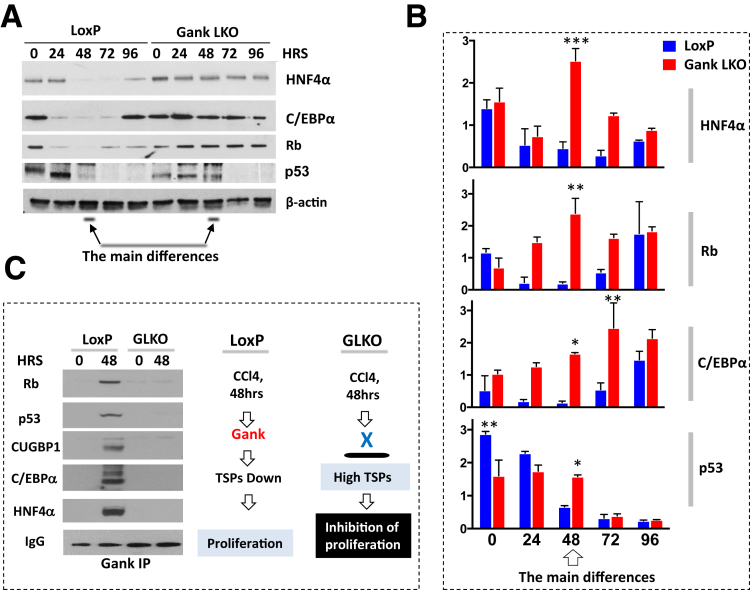
The inhibition of liver proliferation in Gank LKO mice is mediated by high levels of tumor-suppressor proteins. (A) Western blot analysis of nuclear extracts for expression of proteins (right) in livers of LoxP and Gank LKO mice after PH. C/EBPα is expressed as 2 isoforms with a molecular weight of 42 and 30 kilodaltons. All Western blots of TSPs had 2 biological replicates and 1–2 technical replicates. (B) Calculations of levels of TSPs as ratios to β-actin. Statistical analysis was performed with biological replicates. P values for CUGBP1 were as follows: 0 hours (P > .99), 24 hours (P < .03), 36 hours (P < .01), 48 hours (P < .0001), 72 hours (P > .99), and 96 hours (P < .03). P values for HNF4α were as follows: 0 hours (P < .0001), 24 hours (P < .0001), 36 hours (P < .0001), 48 hours (P < .0001), 72 hours (P < .0019), and 96 hours (P < .0001). P values for C/EBPα were as follows: 0 hours (P > .13), 24 hours (P < .001), 36 hours (P < .02), 48 hours (P > .49), 72 hours (P > .99), and 96 hours (P > .99). P values for Rb were as follows: 0 hours (P < .01), 24 hours (P > .35), 36 hours (P > .19), 48 hours (P > .16), 72 hours (P > .99), and 96 hours (P > .99). (C) Levels of C/EBPα, CUGBP1, and HNF4α mRNAs after PH as ratios to β-actin mRNA. Experiments were performed with 2 biological replicates per genotype and time point and 2 technical replicates. Statistical analysis was performed with biological replicates. P values for CUGBP1 were as follows: 0 hours (P > .99), 24 hours (P > .99), 36 hours (P < .01), 48 hours (P > .85), 72 hours (P > .99), and 96 hours (P > .99). P values for HNF4α were as follows: 0 hours (P > .99), 24 hours (P > .09), 36 hours (P > .34), 48 hours (P > .99), 72 hours (P < .0001), and 96 hours (P > .99). P values for C/EBPα were as follows: 0 hours (P > .88), 24 hours (P > .59), 36 hours (P < .003), 48 hours (P < .004), 72 hours (P < .001), and 96 hours (P > .07). (D) Gank interacts and triggers degradation of TSPs in livers of LoxP mice, but not in the livers of Gank LKO mice. Gank was immunoprecipitated from nuclear extracts isolated from LoxP and Gank LKO livers and the IPs were probed with antibodies to CUGBP1, HNF4α, C/EBPα, and Rb. The bottom panel shows IP of E2F1 and Western blotting of IPs with antibodies to Rb. The most notable differences between LoxP and Gank LKO in the E2F1 IP were at 48–96 hours (illustrated with a black line). The Co-IP with C/EBPα had 1 technical replicate and all others were performed once. (E) Examination of post-translational modifications of Gank by 2D gel electrophoresis–Western blot assay. Nuclear extracts from livers of LoxP mice (0-hour time point and 48 hours after PH) were separated by 2D technique and probed with antibodies to Gank. Red arrows and circles show isoforms that are increased significantly in livers at 48 hours after PH. (F) A hypothesis for the inhibition of liver proliferation after PH in Gank LKO mice (see text). Two way analysis of variance ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 LoxP vs Gank LKO. HRS, hours.
Reduction of Proliferative Capacities of Gank LKO Mice Is Owing to the Lack of Reduction of Tumor-Suppressor Proteins After PH
Because Gank triggers degradation of TSPs in proliferating cancer cells,7, 9 we examined if the deletion of Gank might change the expression of 4 TSPs: CUGBP1, C/EBPα, HNF4α, and Rb. CUGBP1 has been included in these studies because we recently have found that it is a TSP that is degraded by Gank in DEN-mediated cancer as well as pediatric liver cancer.17 Western blot analyses of nuclear extracts with antibodies to these TSPs is shown in Figure 4A. We found that all 4 examined TSPs are reduced in livers of LoxP mice after PH; however, no reduction of these proteins was observed in Gank LKO mice. Quantitation of protein levels of TSPs as ratios to β-actin showed that, although the degree of reduction for each protein is different, Gank LKO livers have much higher levels of TSPs after PH during the entire time period examined (Figure 4B). It is important to note that protein levels of HNF4α are increased in quiescent livers of GLKO mice before PH. Examination of levels of mRNAs for TSPs showed that the patterns of expression of these mRNAs are mostly similar in LoxP and Gank LKO mice (Figure 4C), strongly suggesting that the differences in protein levels are owing to degradation of proteins. Because cell-cycle inhibitors p21 and p27 regulate liver proliferation, we also examined their expression in GLKO mice and found that overall levels of these proteins were slightly higher in GLKO mice within 96 hours after PH (Figure 4A). However, these differences are not as high as differences for targets of Gank: CUGBP1, C/EBPα, HNF4α, and Rb. Therefore, we further examined if Gank interacts with and reduces these TSPs in LoxP mice after PH. For this goal, we used co-immunoprecipitation (Co-IP). These investigations showed that Gank interacts with all examined TSPs within 24–72 hours after PH, whereas no interaction was observed in Gank LKO mice (Figure 4D). To examine consequences of the Gank-mediated degradation of Rb, we determined levels of Rb-E2F1 complexes that repress cell-cycle progression. We found that the amount of these complexes was higher in livers of Gank LKO mice, supporting the reduced cell-cycle progression and proliferation (Figure 4D, bottom). Interestingly, although Gank levels were not increased and remained stable at all time points after PH, the interaction of Gank with TSPs was not detected in LoxP mice before PH (time 0) and at 96 hours after PH, suggesting that some additional mechanisms regulate these interactions. To examine this possibility, we examined post-translational modifications of Gank at 0 and 48 hours after PH using 2D gel electrophoresis. These studies showed that Gank undergoes post-translational modifications after PH and there are at least 3 additional isoforms that differ by charge (Figure 4E, circles). We suggest that these modifications change the activity of Gank and increase its interactions with TSPs. In summary, studies of liver proliferation after PH showed that Gank triggers degradation of 4 strong TSPs that allow for liver proliferation. This Gank-mediated reduction does not take place in Gank LKO mice within 96 hours after surgery and the high levels of these TSPs inhibit liver proliferation within this time frame (Figure 4F).
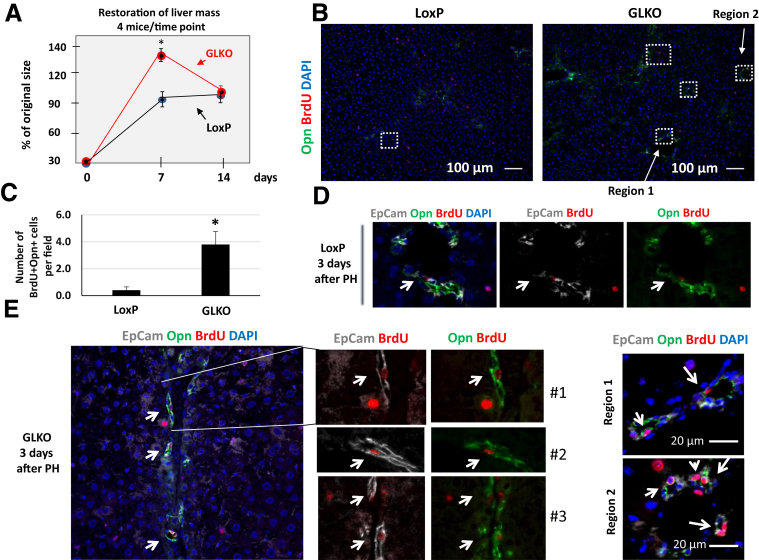
Failure of GLKO Hepatocytes to Proliferate After PH Causes Proliferation of Hepatic Progenitor Cells, Which Leads to Restoration of Original Size 14 Days After PH
Because hepatocytes of the liver of GLKO mice have very weak proliferation (or do not proliferate) within 96 hours after PH, we expected that GLKO mice would not be able to restore their original liver size or would have a delay in restoration. To test this suggestion, we examined liver size (liver/body ratio) at 7 and 14 days after PH in LoxP and GLKO mice. We examined 4 mice for each time point and for each genotype and surprisingly found that, opposite to our expectations, all GLKO mice had an increased liver size at 7 days, which then went back to normal at 14 days after PH (Figure 5A). Given this unexpected result, we further examined possible mechanisms that are used by GLKO mice to increase liver mass above normal size. It has been shown previously that if hepatocyte proliferation is inhibited, there may be an appearance of progenitor cells that start proliferation and subsequently differentiate into hepatocytes.26 Therefore, we next examined if progenitor cells proliferate in GLKO mice after PH. For these studies, we selected the 72-hour time point after PH, with the belief that initiation of proliferation at that time likely would contribute to the pattern of liver mass restoration seen at 7 days. A typical picture of big fields of LoxP and Gank LKO livers with Opn-BrdU–4′,6-diamidino-2-phenylindole staining is shown in Figure 5B (regions with proliferating progenitor cells are marked by white squares). Calculations of BrdU-positive progenitor cells showed that up to 5–6 proliferating progenitor cells were detected in a 0.28-mm2 field of livers of GLKO mice; although many fields of the same size in LoxP mice did not have proliferating progenitor cells. Recalculation per several fields showed that LoxP livers contained 0.2–0.3 progenitor cells per field (Figure 5C). For further examinations, livers were co-stained with markers of progenitor cells EpCam and Opn, and then with BrdU. The results for LoxP mice are shown in Figure 5D. Control livers of LoxP mice contained just a few small cells positive for these markers of progenitor cells and BrdU; however, GLKO livers contained a much higher number of BrdU-positive progenitor cells. Figure 5E shows examples of these studies with 3 GLKO mice. The right panel shows high magnifications of regions 1 and 2 of Figure 5B. These results suggest that oversized livers of GLKO mice at 7 days may be the result of small cell proliferation and perhaps further differentiation into hepatocytes. Further studies are required for detailed investigations of the role of progenitor cells in the restoration of liver mass in GLKO mice at 7–14 days after PH.
Figure 5.
Livers of GLKO mice restore the original size by increasing proliferation of progenitor cells. (A) Restoration of liver mass in LoxP and GLKO mice at 7 and 14 days after PH. Liver/body ratios were calculated and presented as a percentage of original ratios that were measured before surgery. Four mice of each genotype were used for each time point. P values were as follows: 0 days (P > .99), 7 days (P < .02), and 14 days (P > .99). (B) Typical pictures of the Opn/BrdU/4′,6-diamidino-2-phenylindole (DAPI) staining (proliferating progenitor cells) of big fields of the liver. Locations of proliferating progenitor cells are shown by white squares. (C) Calculations of BrdU-positive progenitor cells in 0.28-mm2 fields of livers. Calculation was performed using 3 LoxP mice and 3 GLKO mice, examining 7 fields per animal (P < .04). (D) Example of co-immunostaining of livers of 3 LoxP mice with EpCam, Opn, BrdU, and DAPI at 3 days after PH. Seven fields per mouse were examined. (E) Immunostaining of 3 GLKO mice 3 days after PH with the earlier-mentioned antibodies. Seven fields per mouse were examined. White arrows show progenitor cells. Right panel represents high magnifications of regions 1 and 2 of panel B stained with EpCam, Opn, BrdU, and DAPI. Two way analysis of variance ∗P < .05 LoxP vs Gank LKO.
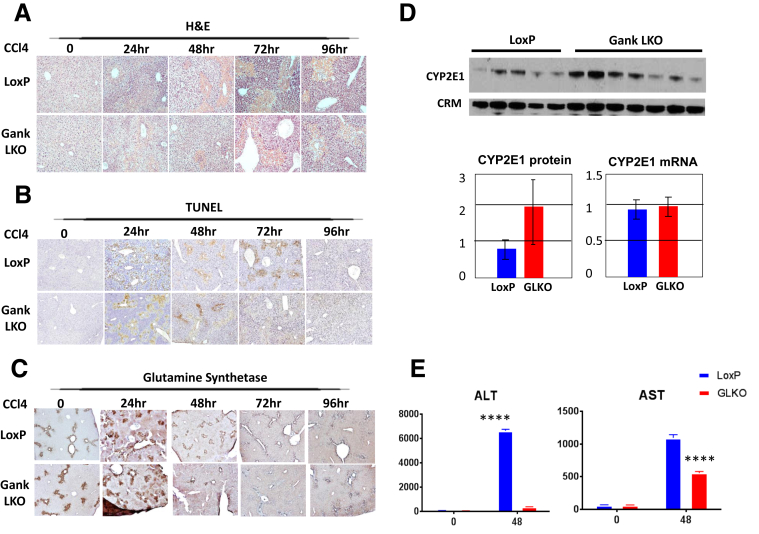
Gank LKO Mice Have Identical Levels of Liver Injury After CCl4 Injections, but Have Inhibited Liver Proliferation
To obtain additional proof that livers of Gank LKO mice have reduced proliferative capacities, we applied a second approach to induce proliferation: acute treatment with CCl4. LoxP and Gank LKO mice were injected with CCl4 as described in our reports21, 23 and animals were killed at 24, 48, 72, and 96 hours after injection. Because the proliferative response of the liver to CCl4 injections strongly depends on the CCl4–mediated liver injury and on the number of dead hepatocytes, we first performed careful examination of liver injury in LoxP and Gank LKO mice. H&E staining, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling staining, and glutamine synthase staining showed no significant differences (Figure 6A–C), indicating that deletion of Gank does not affect the response to liver injury with CCl4. Note that a small difference in apoptotic hepatocytes was observed at 72 hours, at which time Gank LKO mice showed less staining. To further compare liver injury in LoxP and Gank LKO mice, we measured levels of CYP2E1, an enzyme that converts CCl4 into a toxic CCl3+ molecule. No significant differences in expression of CYP2E1 mRNA and proteins were detected (Figure 6D). We also examined liver injury through measurement of serum transaminase levels. Despite no changes by histology or CYP2E1 expression, there was a significant difference in liver injury determined by aspartate aminotransferase and alanine aminotransferase, with LoxP mice having higher transaminase levels 48 hours after PH compared with Gank LKO mice (Figure 6E).
Figure 6.
Deletion of Gank does not affect the histologic response of mice to CCl4-mediated liver injury. (A) H&E staining, (B) terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining, and (C) glutamine synthetase staining of livers of LoxP and Gank LKO mice at different time points after CCl4 injection. The majority of glutamine synthetase staining is observed around central veins. Three animals per genotype per time point were analyzed with 5 fields examined per mouse. (D) Examination of baseline levels of CYP2E1 in livers of LoxP and Gank LKO mice by Western blot and by quantitative RT-PCR. P values were as follows: protein level (P > .2) and mRNA level (P > .4). Each sample on Western blot represented a biological replicate (5 LoxP mice and 7 Gank LKO mice), which also was used for quantitative RT-PCR. Both experiments had 2 technical replicates. (E) Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) from LoxP and Gank LKO mice at the 0-hour time point and at 48 hours after CCl4 injection. Serum analysis was performed on 3 mice per genotype and per time point. P values for ALT were as follows: 0 hours (P > .99) and 48 hours (P < .0001). P values for AST were as follows: 0 hours (P > .99) and 48 hours (P < .0001). Two way analysis of variance. ∗∗∗∗P < .0001 LoxP vs Gank LKO.
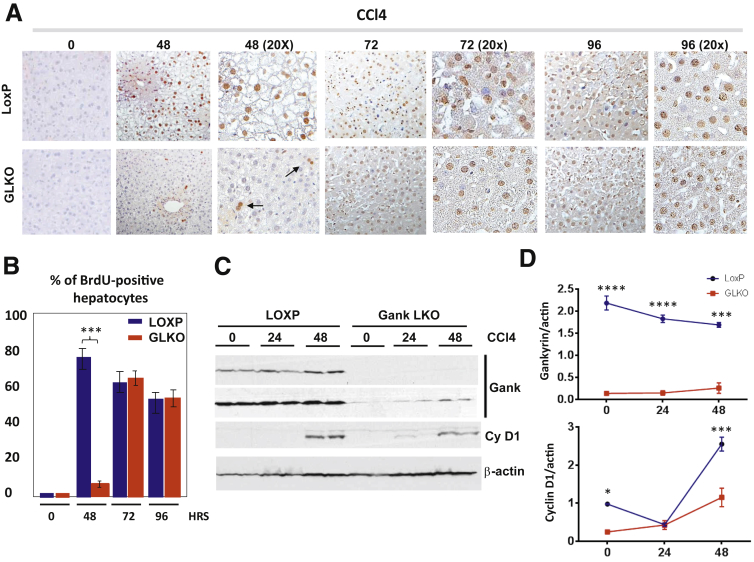
To examine proliferation of the liver, we injected CCl4-treated mice with BrdU and examined the number of BrdU-positive hepatocytes. BrdU uptake showed quite complicated patterns of liver proliferation. Consistent with data for PH, proliferation of hepatocytes was inhibited dramatically in Gank LKO mice at 48 hours; whereas LoxP mice had a very high level of proliferation of hepatocytes that reached up to 80%–90% (Figure 7A and B). However, in contrast to PH, hepatocytes of GLKO mice start a robust proliferation at 72 and 96 hours after CCl4 injections. Given the dramatic difference in proliferation of hepatocytes at 48 hours, we further investigated molecular events at 48 hours. Examination of expression of Gank showed that protein levels of Gank were not increased after CCl4 treatment in LoxP mice (Figure 7C and D). Interestingly, a very long exposure of Gank film showed an increase of Gank in Gank LKO mice, suggesting that, similar to late time points after PH, nonparenchymal or progenitor cells (Alb Cre eliminates Gank in hepatocytes and cholangiocytes) might proliferate and express Gank, or progenitor cell proliferation is not regulated by Gank. Cyclin D1 was increased in both groups at 48 hours; however, the level of increase was significantly higher in livers of LoxP mice than in livers of Gank LKO mice (Figure 7C and D). Taken together, we found that CCl4 treatment caused similar liver injury in LoxP and Gank LKO mice; however, the proliferation of hepatocytes in livers of Gank LKO mice was reduced significantly at 48 hours, but was initiated at later time points.
Figure 7.
Liver proliferation is inhibited in livers of Gank LKO mice after CCl4-mediated injury. (A) Examination of liver proliferation by measuring DNA replication (BrdU uptake). Livers were stained with antibodies to BrdU. Arrows show rare BrdU-positive hepatocytes in Gank LKO mice at 48 hours after CCl4 injections. (B) Calculations of BrdU-positive hepatocytes in livers of LoxP and Gank LKO mice after CCl4 injections using 2 mice per genotype and per time point and analyzing 3 fields per mouse. P values were as follows: 0 hours (P > .99), 48 hours (P < .001), 72 hours (P > .27), and 96 hours (P > .28). (C) Expression of Gank and cyclin D1 after CCl4 injury in LoxP and Gank LKO mice using 2 biological replicates. Western blot was performed with nuclear extracts from livers of LoxP and Gank LKO mice. The membrane was reprobed with antibodies to β-actin. Dark and light exposures are shown for Gank. (D) Calculation of levels of Gank and cyclin D1 as ratios to β-actin. Note that low levels of Gank are detected in Gank LKO mice at 24 and 48 hours after CCl4 injections, perhaps because of proliferation of nonparenchymal cells in which Cre is not expressed. P values for Gankyrin were as follows: 0 hours (P < .0001), 24 hours (P < .0001), and 48 hours (P < .001). P values for cyclin D1 were as follows: 0 hours (P < .25), 24 hours (P > .99), and 48 hours (P < .0009). Two way analysis of variance. ∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001 LoxP vs Gank LKO.
Inhibition of Proliferation in CCl4-Treated Gank LKO Mice at 48 Hours Is Associated With High Levels of Tumor-Suppressor Proteins
We next examined mechanisms by which deletion of Gank inhibits liver proliferation at 48 hours after CCl4. Because the failure to reduce TSPs causes inhibition of proliferation after PH (Figure 4), we tested if a similar mechanism was involved in the inhibition of proliferation at 48 hours after CCl4 treatment. Western blot with TSPs HNF4α, C/EBPα, Rb, and p53 showed complex patterns of expression; however, the consistent result for these proteins is that all of them were reduced significantly in LoxP mice, but they were increased in Gank LKO mice at 48 hours (Figure 8A). Densitometric calculations of their levels as ratios to β-actin showed up to a 10-fold difference for HNF4α, C/EBPα, and Rb, and a 2-fold difference for p53 at 48 hours after CCl4 treatment (Figure 8B). Note that the 48-hour time point is the time when liver proliferation was most notable in LoxP mice; however, it was reduced dramatically in Gank LKO livers compared with LoxP mice (Figure 7). Given that the dramatic difference in TSPs occurs 48 hours after CCl4 injection, we examined if Gank triggers degradation of the TSPs at that time. The Co-IP approach showed that Gank interacts with all examined TSPs at 48 hours in LoxP livers, but no interaction was found in livers of Gank LKO mice (Figure 8C). Note that for these Co-IP studies we used 5 times more proteins from LoxP mice. It also is interesting to note that levels of TSPs at 72 and 96 hours after CCl4 did not differ significantly in LoxP and GLKO mice, which is in agreement with observations showing comparative hepatocyte proliferation at these time points (Figure 7A and B). Based on these results, we suggest that the inhibition of liver proliferation in Gank LKO mice at 48 hours after CCl4 treatments is attributed to high levels of TSPs in the absence of Gank (Figure 8C, diagrams on the right).
Figure 8.
The failure of livers of Gank LKO mice to proliferate after CCl4injections is caused by high levels of tumor-suppressor proteins, which are eliminated by Gank in control LoxP mice. (A) Expression of TSPs in LoxP and Gank LKO mice after CCl4 injury. The experiment was performed with 3 biological replicates. (B) Calculations of levels of TSPs as ratios to β-actin using biological replicates. P values for HNF4α were as follows: 0 hours (P > .99), 24 hours (P > .99), 48 hours (P < .0002), 72 hours (P > .05), and 96 hours (P > .99). P values for Rb were as follows: 0 hours (P > .99), 24 hours (P > .18), 48 hours (P < .009), 72 hours (P > .33), and 96 hours (P > .99). P values for C/EBPα were as follows: 0 hours (P > .99), 24 hours (P > .19), 48 hours (P < .03), 72 hours (P < .009), and 96 hours (P > .86). P values for p53 were as follows: 0 hours (P < .002), 24 hours (P > .23), 48 hours (P < .02), 72 hours (P > .99), and 96 hours (P > .99). (C) Gank interacts with TSPs in livers of LoxP mice at 48 hours after CCl4 injury. Co-IP assay was applied for these studies. Nuclear extracts of LoxP and Gank LKO mice (200 μg for LoxP 0-hour time point and Gank LKO at 0 and 48 hours and 1 mg for LoxP at 48 hours) were incubated with antibodies to Gank and rabbit IgG beads (TrueBlot). After washing, IPs were loaded on the gel and probed with antibodies to Rb, p53, CUGBP1, C/EBPα, and HNF4α. The IPs were performed 3 times. Every IP was examined by Western blot with the earlier-mentioned antibodies. Right: Hypothetical mechanism for the failure of livers of Gank LKO mice to proliferate after CCl4-mediated injury. Two way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 LoxP vs Gank LKO.
Block of Interactions of TSPs With Gank by Small Drug cjoc42 Rescues TSPs and Inhibits Proliferation of Cancer Cells
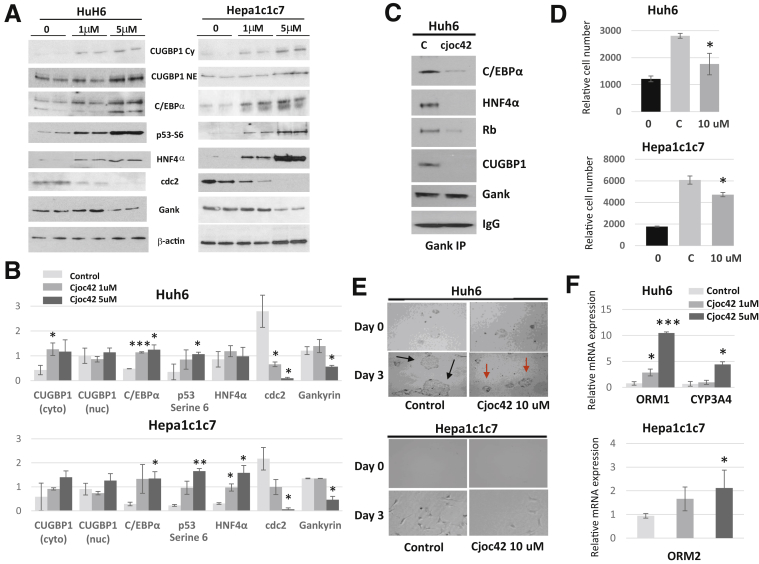
Although experiments with Gank LKO mice provided strong support for the hypothesis that Gank inhibits liver proliferation through elimination of TSPs, the causal role of Gank in this elimination needed additional confirmation in a different setting. To obtain such evidence, we performed studies in cultured liver cancer cells Huh6 (human) and Hepa1c1c7 (mouse). A recent report described the identification of the small drug cjoc42, which binds to Gank and blocks its interactions with TSPs such as p53.24 We synthesized cjoc42 and treated cancer cells with 1 and 5 μmol/L cjoc42. Western blot analysis showed that the cjoc42-mediated block of interactions of Gank with proteasome significantly increased the levels of CUGBP1 in the cytoplasm (Huh6 cells) and nuclear levels of C/EBPα, p53-Ser6-ph, and HNF4α (Figure 9A). We chose to look at p53 Ser6 because of our recent finding showing that this modification is a strong characteristic of liver cancer. Calculations of these proteins as ratios to β-actin showed a significant increase of TSPs in cells treated with cjoc42 (Figure 9B). Interestingly, there was a statistically significant reduction in Gank after treatment with 5 μmol/L of cjoc42. In addition, Co-IP studies showed that Gank interacts with all of these TSPs in untreated Huh6 cells, whereas weak or no interactions were detected in cjoc42-treated Huh6 cells (Figure 9C). These results were unexpected because the original report showed that cjoc42 inhibited interactions of Gank with the rest of the 26S proteasome only. Therefore, our data show that, in addition to inhibition of interactions with the proteasome, cjoc42 also blocks the interaction between Gank and TSPs and may directly reduce Gankyrin expression as well. We suggest that cjoc42 causes conformational changes in the 3-dimensional structure of Gank that prevents its interactions with both TSPs and the 26S proteasome. Taken together, the inhibition of the Gank–TSP interaction by cjoc42 leads to the increase of TSPs.
Figure 9.
Small-molecule cjoc42 inhibits interactions of Gank with TSPs, increases levels of TSPs, and inhibits proliferation of cancer cells. (A) Examination of protein expression in cancer cells after inhibition of Gank by cjoc42. Human Huh6 and mouse Hepa1c1c7 cancer cells were treated with 1 and 5 μmol/L cjoc42, and nuclear extracts were isolated and examined by Western blot with antibodies shown in the middle. NE CUGBP1 and CE CUGBP1 show examination of CUGBP1 in nuclear extracts and cytoplasmic extracts correspondingly. There were 3 biological replicates and 1–2 technical replicates for each Western blot. (B) Calculation of protein levels as ratios to β-actin. P values for Huh6 cells treated with 1 umol/L cjoc42 were as follows: CUGBP1 (cyto) (P < .04), CUGBP1 (nuc) (P > .61), C/EBPα (P < .0008), p53 serine 6 (P > .09), HNF4α (P > .18), cdc2 (P < .04), and Gankyrin (P > .49). P values for Huh6 cells treated with 5 umol/L cjoc42 were as follows: CUGBP1 (cyto) (P > .12), CUGBP1 (nuc) (P > .64), C/EBPα (P < .03), p53 serine 6 (P < .04), HNF4α (P > .38), cdc2 (P < .03), and Gankyrin (P < .03). P values for Hepa1c1c7 cells treated with 1 umol/L cjoc42 were as follows: CUGBP1 (cyto) (P > .5), CUGBP1 (nuc) (P < .45), C/EBPα (P > .13), p53 serine 6 (P > .06), HNF4α (P < .02), cdc2 (P > .09), and Gankyrin (P > .65). P values for Hepa1c1c7 cells treated with 5 umol/L cjoc42 were as follows: CUGBP1 (cyto) (P > .21), CUGBP1 (nuc) (P < .32), C/EBPα (P < .03), p53 serine 6 (P < .003), HNF4α (P < .02), cdc2 (P < .02), and Gankyrin (P < .02). (C) Examination of interactions of TSPs with Gankyrin using a Co-IP approach in untreated cells and in cells treated with 5 μmol/L cjoc42. This experiment was repeated once. (D) Proliferation of cells treated with 10 μmol/L cjoc42 was examined by cell proliferation assay. The bar graph illustrates the number of cells on day 0 (0), as well as control cells (C) and treated cells (10 μmol/L) at 72 hours: Huh6 (P < .02) and Hepa1c1c7 (P < .01). There were 4–6 biological replicates used for this experiment and 2 technical replicates. (E) Microscopic images of cells at 20× magnification treated with 10 μmol/L cjoc42 at day 0 and day 3. Black arrows show big multicell colonies of cells in the control experiment without treatment. Red arrows show significantly smaller Huh6 colonies on cjoc42-treated plates. (F) Quantitative RT-PCR–based examination of mRNA levels of mature hepatocyte markers in Huh6 cells (top panel) and Hepa1c1c7 cells (bottom panel) treated with cjoc42. This experiment was performed with 2 biological replicates and 2 technical replicates. Statistical analysis was performed with biological replicates. P value for Orosomucoid 1 expression in Huh6 cells: 1 umol/L cjoc42 (P < .05) and 5 umol/L cjoc42 (P < .0008); CYP3A4 expression in Huh6 cells: 1 umol/L cjoc42 (P > .57) and 5 umol/L cjoc42 (P < .01). P value for Orosomucoid 2 expression in Hepa1c1c7 cells: 1 umol/L cjoc42 (P < .07) and 5 umol/L cjoc42 (P < .05). Two way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 LoxP vs Gank LKO.
To determine the biological consequences of Gank inhibition, we evaluated the proliferation of cancer cells treated with cjoc42. A 72-hour proliferation assay using 10 μmol/L of cjoc42 showed a significant reduction in the proliferation of Huh6 and Hepa1c1c7 cells (Figure 9D). Examination of cell images showed that untreated Huh6 and Hepa1c1c7 cells formed big colonies at 3 days after plating, whereas treated cells stayed as small colonies (Figure 9E, black arrows show big colonies, red arrows show small colonies). In addition, cell-cycle protein cdc2 was reduced by Western blot in cjoc42-treated cells at both 1- and 5-umol/L doses (Figure 9A and B). Given that Gank previously was shown to promote the loss of hepatocyte markers,18 we examined such markers using quantitative RT-PCR. We found that after treatment of Huh6 cells with cjoc42, there was an increase in mature hepatocyte markers Orosomucoid 1 and CYP3A4 (Figure 9F, top) and an increase in Orosomucoid 2 after treatment in Hepa1c1c7 cells (Figure 9F, bottom). This finding shows how Gank inhibition through cjoc42 not only reduces proliferation, but has other biological implications that weaken carcinogenesis. Figure 10 summarizes studies in cultured cancer cells. Based on our data, we suggest that Gank-mediated degradation of TSPs by UPS promotes proliferation of cancer cells. We found that cjoc42 inhibits both interactions of Gank with the 26S proteasome and with TSPs, which lead to the rescue of TSPs and inhibition of proliferation of cancer cells (Figure 10).
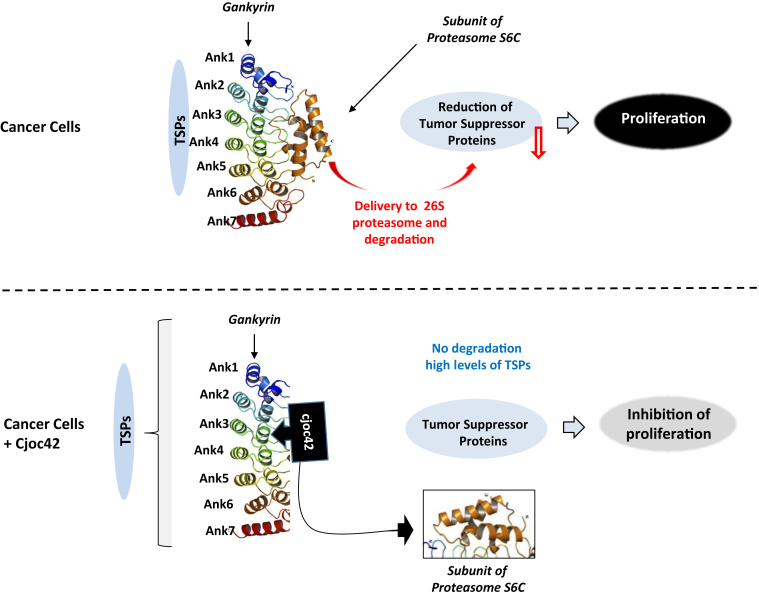
Figure 10.
Mechanistic hypothesis for cjoc42-mediated inhibition of proliferation of cancer cells. In cancer cells, the oncogene Gank binds to TSPs and delivers them to the UPS for degradation (top panel). The loss of TSPs eliminates their growth-inhibitory effect and allows proliferation of cancer cells. Cjoc42 blocks interactions of Gank with the 26S proteasome and with TSPs. This leads to the rescue of TSPs, which, in turn, inhibits proliferation (bottom panel).
Discussion
Hallmarks of cancer include sustaining proliferative signaling and evading growth suppressors, which commonly is achieved through activation of oncogenes and elimination of TSPs.27 The quiescent liver expresses up to 20 TSPs, which are severely reduced in liver cancer9, 28; however, the causal role of the oncogene–TSP pathways in the initiation of liver proliferation is not well understood. Previous studies have shown that increase of the oncogene Gank correlates with the proliferation seen in liver cancer, but the exact role of Gank in liver proliferation had not been examined because of a lack of appropriate animal models. Therefore, we generated a mouse line (GLKO) to further understand the mechanistic role of this oncogene. Initial examination of liver morphology and phenotype showed no significant changes in Gank LKO mice compared with LoxP mice, suggesting that Gank does not seem to play a role in the maintenance of liver biology and liver function in adult mice. However, examination of the global transcriptome profiling showed significant changes in multiple pathways including inflammation, fatty liver biology, carcinogenesis, and liver proliferation. Consistent with these findings, overexpression of Gank in quiescent livers stimulated proliferation of hepatocytes, supporting that Gank is an oncogene that promotes liver proliferation.
Regulation of liver proliferation is quite complex and includes cooperation of multiple signaling networks, which include neutralization of TSPs and other proteins that inhibit liver proliferation. The most powerful systems to investigate liver proliferation are PH and acute treatment with CCl4. We found that in both of these models, Gank is a key promoter of liver proliferation and the deletion of Gank dramatically inhibits hepatocyte proliferation. In the course of our studies with PH, we surprisingly found that Gank LKO livers have a significant increase in liver mass at 7 days after surgery and return to their original size by day 14. Our studies showed that this phenomenon is associated with the appearance of progenitor cells and increased proliferation of these cells in Gank LKO livers. However, it is likely that additional mechanisms of growth are involved in restoration of liver mass after PH in GLKO mice. Regarding molecular mechanisms by which Gank promotes liver proliferation after PH and CCl4 treatments, we obtained convincing evidence that Gank interacts with and reduces levels of several inhibitors including C/EBPα, HNF4α, Rb, and CUGBP1. It is interesting that in control LoxP mice, expression of Gank was not increased after PH and acute CCl4 treatment, but its ability to interact with TSPs was increased dramatically. Although we do not know the mechanisms that initiate interaction of Gank with TSPs, one possibility is post-translational modifications of Gank after PH and CCl4 that enhance its activity and interaction with TSPs. In this regard, 2D examination of Gank after PH showed the appearance of additional immune-reactive forms of Gank. In addition, there may be post-translational modifications of TSPs. It has been shown that after PH, C/EBPα and Rb undergo phosphorylation at Ser193 and Ser780, respectively, and that these modifications initiate interactions of Rb and C/EBPα with Gank.6, 9 Independent of the underlying mechanisms, TSPs are eliminated by Gank in LoxP mice, allowing for proliferation of the liver; however, this elimination does not happen in Gank LKO mice, leading to the inhibition of liver proliferation (Figures 3E and 7C). In support of our mechanistic work with the Gank LKO mouse model, our studies with the small molecule binder of Gank, cjoc42, clearly show that the elimination of TSPs by Gank is a causal key event in the inhibition of hepatocyte proliferation.
Oncogenic activities of Gank in liver cancer have been well described starting in the early 2000s,29 initially in HCC and more recently in hepatoblastoma.9 The development of Gank-based approaches for the treatment of liver cancer, especially HCC and chemoresistant hepatoblastoma, is a promising direction.30, 31 In this regard, it is important to note our recent work with inhibition of Gank by activating FXR. In the quiescent liver, Gank is partially repressed by FXR. However, FXR is reduced dramatically in both human HCC and mouse models of liver cancer, leading to an increase of Gank.6, 32 We have found that the activation of FXR by agonist GW4064 and subsequent inhibition of Gank is sufficient to inhibit the development of liver cancer under conditions of DEN-mediated liver cancer.9 In the current article, we initiated investigations of antitumor activities of the small molecule cjoc42 as a potential drug for Gank-mediated inhibition of liver cancer. Experiments in 2 cancer cell lines showed that the cjoc42-mediated inhibition of Gank leads to the inhibition of cell proliferation, which correlates with the rescue of TSPs. Further examination of this drug in animals is underway and will show if cjoc42 might be considered for additional preclinical studies and eventually clinical trials. In summary, our work showed a critical role of Gank in liver proliferation and provides evidence that the inhibition of Gank should be considered as a potential approach for the treatment of liver cancer.
Acknowledgments
The authors thank Katie Glaser and Alex Bondoc for discussion of the results.
Footnotes
Author contributions Amber M. D’Souza generated the hypotheses and design, performed experiments, and wrote the manuscript; Leila Valanejad performed real-time quantitative reverse transcription polymerase chain reaction studies and RNA sequencing analysis; Ashley Cast performed immunostaining analyses; Mary Wright and Meenasri Kumbaji performed real-time quantitative reverse-transcriptase polymerase chain reaction studies; Yanjun Jiang generated Gankyrin liver-specific knock-out mice and performed partial hepatectomy studies; Kyle Lewis was involved in partial hepatectomy studies and analyses of gene expression; Sheeniza Shah and David Smithrud synthesized cjoc42 and were involved in the design development and interpretation of data with cjoc42; Rebekah Karns performed biostatistical analyses of RNA sequencing and was involved in the design and interpretation of results; Soona Shin examined progenitor cells in Gankyrin liver-specific knock-out mice; and Nikolai Timchenko generated overall hypotheses and the design for the entire project and manuscript, along with providing funds.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R01DK102597 and R01CA159942 (N.T.), and by internal development funds from Cincinnati Children’s Hospital Medical Center (N.T. and S.S.). This project also was supported in part by National Institutes of Health grant P30 DK078392 (R.K.).
References
- 1.Castelli G., Pelosi E., Testa U. Liver cancer: molecular characterization, clonal evolution and cancer stem cells. Cancers (Basel) 2017;9:9. doi: 10.3390/cancers9090127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson S.P. Hepatocellular carcinoma and the ubiquitin-proteasome system. Biochim Biophys Acta. 2008;1782:775–784. doi: 10.1016/j.bbadis.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Iakova P., Timchenko L., Timchenko N.A. Intracellular signaling and hepatocellular carcinoma. Semin Cancer Biol. 2011;21:28–34. doi: 10.1016/j.semcancer.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timchenko N.A., Lewis K. Elimination of tumor suppressor proteins during liver carcinogenesis. Cancer Studies Mol Med Open J. 2015;1:27–38. [Google Scholar]
- 5.Lim I.K. Spectrum of molecular changes during hepatocarcinogenesis induced by DEN and other chemicals in Fisher 344 male rats [Mechanisms of Ageing and Development 123 (2002) 1665-1680] Mech Ageing Dev. 2003;124:697–708. doi: 10.1016/s0047-6374(02)00087-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang G.-L., Shi X., Haefliger S., Jin J., Major A., Iakova P., Finegold M., Timchenko N.A. Elimination of C/EBPalpha through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J Clin Invest. 2010;120:2549–2562. doi: 10.1172/JCI41933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y., Iakova P., Jin J., Sullivan E., Sharin V., Hong I.-H., Anakk S., Mayor A., Darlington G., Finegold M., Moore D., Timchenko N.A. Farnesoid X receptor inhibits gankyrin in mouse livers and prevents development of liver cancer. Hepatology. 2013;57:1098–1106. doi: 10.1002/hep.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revill K., Wang T., Lachenmayer A., Kojima K., Harrington A., Li J., Hoshida Y., Llovet J.M., Power S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424–1435. doi: 10.1053/j.gastro.2013.08.055. e1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valanejad L., Lewis K., Wright M., Jiang Y., D’Souza A., Karns R., Sheridan R., Gupta A., Bove K., Witte D., Geller J., Tiao G., Nelson D.L., Timchenko L., Timchenko N.A. FXR-Gankyrin axis is involved in development of pediatric liver cancer. Carcinogenesis. 2017;38:738–747. doi: 10.1093/carcin/bgx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson P.F. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 11.Yoon S., Huang K.-W., Reebye V., Mintz P., Tien Y.-W., Lai H.-S., Saetrom P., Reccia I., Swiderski P., Armstrong B., Jozwiak A., Spalding D., Jiao L., Habib N., Ross J.J. Targeted delivery of C/EBPα-saRNA by pancreatic ductal adenocarcinoma-specific RNA aptamers inhibits tumor growth in vivo. Mol Ther. 2016;24:1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voutila J., Reebye V., Roberts T.C., Protopapa P., Andrikakou P., Blakey D.C., Habib R., Huber H., Saetrom P., Rossi J.J., Habib N.A. Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol Ther. 2017;25:2705–2714. doi: 10.1016/j.ymthe.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reebye V., Sætrom P., Mintz P.J., Huang K.-W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D., Kostomitsopoulos N., Kasahara N., Nicholls J.P., Jiao L.R., Pai M., Spalding D.R., Mizandari M., Chikovani T., Emara M.M., Haoudi A., Tomalia D.A., Rossi J.J., Habib N.A. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huan H., Wen X., Chen X., Wu L., Liu W., Habib N.A., Bie P., Xia F. C/EBPα short-activating rna suppresses metastasis of hepatocellular carcinoma through inhibiting EGFR/β-catenin signaling mediated EMT. PLoS One. 2016;11:e0153117. doi: 10.1371/journal.pone.0153117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M., Li S.-N., Anjum K.M., Gui L.-X., Zhu S.-S., Liu J., Chen J.-K., Liu Q.-F., Ye G.-D., Wang W.-J., Wu J.-F., Cai W.-Y., Sun G.-B., Liu Y.-J., Liu R.-F., Zhang Z.-M., Li B.-A. A double-negative feedback loop between Wnt-β-catenin signaling and HNF4α regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J Cell Sci. 2013;126:5692–5703. doi: 10.1242/jcs.135053. [DOI] [PubMed] [Google Scholar]
- 16.Walesky C., Edwards G., Borude P., Gunewardena S., O’Neil M., Yoo B., Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis K., Valanejad L., Cast A., Wright M., Wei C., Iakova P., Stock L., Karns R., Timchenko L., Timchenko N.A. RNA binding protein CUGBP1 inhibits liver cancer in a phosphorylation-dependent manner. Mol Cell Biol. 2017;37:16. doi: 10.1128/MCB.00128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian Y.-W., Chen Y., Yang W., Fu J., Cao J., Ren Y.-B., Zhu J.-J., Su B., Luo T., Zhao X.-F., Dai R.-Y., Li J.-J., Sun W., Wu M.-C., Feng G.-S., Wang H.-Y. p28(GANK) prevents degradation of Oct4 and promotes expansion of tumor-initiating cells in hepatocarcinogenesis. Gastroenterology. 2012;142:1547–1558.e14. doi: 10.1053/j.gastro.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 19.Yang C., Tan Y.-X., Yang G.-Z., Zhang J., Pan Y.-F., Liu C., Fu J., Chen Y., Ding Z.-W., Dong L.-W., Wang H.-Y. Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma. J Exp Med. 2016;213:859–875. doi: 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakurai T., Yada N., Hagiwara S., Arizumi T., Minaga K., Karnata K., Takenaka M., Minami Y., Watanabe T., Nishida N., Kudo M. Gankyrin induces STAT3 activation in tumor microenvironment and sorafenib resistance in hepatocellular carcinoma. Cancer Sci. 2017;108:1996–2003. doi: 10.1111/cas.13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breaux M., Lewis K., Valanejad L., Iakova P., Chen F., Mo Q., Medrano E., Timchenko L., Timchenko N.A. p300 Regulates liver functions by controlling p53 and C/EBP family proteins through multiple signaling pathways. Mol Cell Biol. 2015;35:3005–3016. doi: 10.1128/MCB.00421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevzorova Y.A., Tolba R., Trautwein C., Liedtke C. Partial hepatectomy in mice. Lab Anim. 2015;49(Suppl):81–88. doi: 10.1177/0023677215572000. [DOI] [PubMed] [Google Scholar]
- 23.Hong I.-H., Lewis K., Iakova P., Jin J., Sullivan E., Jawanmardi N., Timchenko L., Timchenko N.A. Age-associated change of C/EBP family proteins causes severe liver injury and acceleration of liver proliferation after CCl4 treatments. J Biol Chem. 2014;289:1106–1118. doi: 10.1074/jbc.M113.526780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay A., O’Connor C.J., Zhang F., Galvagnion C., Galloway W.R.J.D., Tan Y.-S., Stokes J.E., Rahman T., Verma C., Spring D.R., Itzhaki L.S. Discovery of a small-molecule binder of the oncoprotein gankyrin that modulates gankyrin activity in the cell. Sci Rep. 2016;6:23732. doi: 10.1038/srep23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin S., Wangensteen K.J., Teta-Bissett M., Wang Y.J., Mosleh-Shirazi E., Buza E.L., Greenbaum L.E., Kaestner K.H. Genetic lineage tracing analysis of the cell of origin of hepatotoxin-induced liver tumors in mice. Hepatology. 2016;64:1163–1177. doi: 10.1002/hep.28602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michalopoulos G.K. Liver regeneration: alternative epithelial pathways. Int J Biochem Cell Biol. 2011;43:173–179. doi: 10.1016/j.biocel.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Martin J., Dufour J.-F. Tumor suppressor and hepatocellular carcinoma. World J Gastroenterol. 2008;14:1720–1733. doi: 10.3748/wjg.14.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higashitsuji H., Itoh K., Nagao T., Dawson S., Nonoguchi K., Kido T., Mayer R.J., Arii S., Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 30.Zamani P., Matbou Riahi M., Momtazi-Borojeni A.A., Jamialahmadi K. Gankyrin: a novel promising therapeutic target for hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2017 doi: 10.1080/21691401.2017.1388250. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 31.Wang C., Cheng L. Gankyrin as a potential therapeutic target for cancer. Invest New Drugs. 2017;35:655–661. doi: 10.1007/s10637-017-0474-8. [DOI] [PubMed] [Google Scholar]
- 32.Su H., Ma C., Liu J., Li N., Gao M., Huang A., Wang X., Huang W., Huang X. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1245–G1253. doi: 10.1152/ajpgi.00439.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]