Abstract
The use of transcriptional factors as cell fate regulators are often the primary focus in the direct reprogramming of somatic cells into neurons. However, in human adult fibroblasts, deriving functionally mature neurons with high efficiency requires additional neurogenic factors such as microRNAs (miRNAs) to evoke a neuronal state permissive to transcription factors to exert their reprogramming activities. As such, increasing evidence suggests brain-enriched miRNAs, miR-9/9∗ and miR-124, as potent neurogenic molecules through simultaneously targeting of anti-neurogenic effectors while allowing additional transcription factors to generate specific subtypes of human neurons. In this review, we will focus on methods that utilize neuronal miRNAs and provide mechanistic insights by which neuronal miRNAs, in synergism with brain-region specific transcription factors, drive the conversion of human fibroblasts into clinically relevant subtypes of neurons. Furthermore, we will provide insights into the age signature of directly converted neurons and how the converted human neurons can be utilized to model late-onset neurodegenerative disorders.
Keywords: microRNA, chromatin, neuronal conversion, reprogramming, neurogenesis, disease modeling, human neurons
Introduction
Overcoming epigenetic barriers through direct cellular reprogramming has allowed scientists to rapidly acquire cell types of interest for regenerative therapies and disease modeling. Direct conversion of mouse fibroblasts into functional neurons have been demonstrated through the use of transcription factors (Vierbuchen et al., 2010; Son et al., 2011; Chanda et al., 2014; Blanchard et al., 2015). Empirically, however, obtaining mature human neurons from adult human fibroblasts with transcription factors have been challenging (Caiazzo et al., 2011). To enhance reprogramming efficiency and to promote neuronal maturation, small chemical molecules (Ladewig et al., 2012; Liu et al., 2013; Pfisterer et al., 2016; Smith et al., 2016) and RNA molecules, miRNAs (Ambasudhan et al., 2011; Yoo et al., 2011; Victor et al., 2014; Abernathy et al., 2017), have been used in conjunction with transcription factors to robustly generate functional neurons from human fibroblasts. The mechanism(s) by which miR-9/9∗ and miR-124 collectively drive robust neuronal fate conversion remains an ongoing investigation, but through examining transcriptome and epigenetic changes at the genome-wide level, we attempt to elucidate how miRNAs promote the neuronal identity during direct conversion of human fibroblasts to neurons.
miRNAs as Potent Cell Fate Regulators
Traditionally, transcription factors, in particular, pioneer transcription factors, have been viewed as regulators and determinants of cell fate. With domains that can interact directly with chromatin and/or other modifier proteins, transcription factors have been widely used for cellular reprogramming (Iwafuchi-Doi and Zaret, 2014, 2016), including the generation of induced pluripotent stem cells (iPSC) from somatic cells (Takahashi and Yamanaka, 2006). Increasing studies across different cellular contexts have revealed that miRNAs are also potent cell fate regulators as miRNAs not only target large repertoire of genes in genetic networks but also epigenetic regulators necessary for the remodeling of the chromatin (Yoo et al., 2009; Ivey and Srivastava, 2010; Gruber and Zavolan, 2013; Rajman and Schratt, 2017). Subsequently, miRNAs have been used to generate iPSCs (Anokye-Danso et al., 2011), cardiomyocytes (Jayawardena et al., 2012), and neurons (Yoo et al., 2011) from fibroblasts.
miR-9/9∗ and miR-124 are Neurogenic Molecules
The acquisition of neuronal fate requires the downregulation of the neuron-restrictive silencer factor (NRSF) or repressor element-1 silencing transcription factor (REST) that represses neuronal genes in non-neuronal cells, including the neuron-specific miRNAs, miR-9/9∗, and miR-124 (miR-9/9∗-124) (Lagos-Quintana et al., 2002; Lim et al., 2005; Conaco et al., 2006; Deo et al., 2006). Both miR-9/9∗ and miR-124 are highly abundant in neuronal tissues (Lagos-Quintana et al., 2002; Lim et al., 2005; He et al., 2012), and are essential for neuronal differentiation (Cheng et al., 2009; Dajas-Bailador et al., 2012; Xue Q. et al., 2016) and the maintenance of neuronal identity through the repression of anti-neural genes including cofactors of the REST complex, RCOR1 and SCP1 (Visvanathan et al., 2007; Packer et al., 2008). As overexpression of miR-9/9∗ (Leucht et al., 2008; Zhao et al., 2009) and/or miR-124 (Krichevsky et al., 2006; Cheng et al., 2009; Akerblom et al., 2012) in stem cells or neural progenitors resulted in the precocious acquisition of neuronal fate, demonstrating the function of miRNAs in the activation of neuronal program (Lim et al., 2005). Ectopic expression of miR-9/9∗-124 was also shown to drive the direct conversion of primary human dermal fibroblasts into functional neurons (Yoo et al., 2011). Therefore, knockdown of REST is sufficient to promote neuronal identity in part due to miR-9/9∗-124-dependent mechanisms (Drouin-Ouellet et al., 2017). Furthermore, miR-9/9∗-124 orchestrates the reduction of REST protein stability during neuronal reprogramming to promote chromatin accessibility of neuronal loci (Lee et al., 2018) and induction of neuronal genes (Abernathy et al., 2017; Drouin-Ouellet et al., 2017; Lee et al., 2018). Interestingly, miR-124 alone has also been used in neuronal conversion with the help of transcription factors (Ambasudhan et al., 2011; Jiang et al., 2015). Here, we review current understanding of the properties of miR-9/9∗ and miR-124 in both developmental and cellular reprogramming contexts highlighting their synergistic roles in coordinating the molecular switching of several critical non-neuronal to neuronal components during mammalian neurogenesis. We will mainly focus on the molecular switches critical in epigenetic regulation such as chromatin remodeling and DNA methylation, and transcriptome dynamics such as alternative splicing underlying the adoption of the neuronal identity. Our discussion will also include molecular pathways that occur during in vivo neurogenesis and are also recapitulated in the miRNA-directed reprogramming of human fibroblasts into neurons for the successful overcoming of cell fate barriers.
miRNAs Orchestrate the Composition of BAF Chromatin Remodeling Complexes
Spatial and temporal reciprocity of homologous gene or isoform expression during neurogenesis is a recurring theme. Previous studies have indicated that the neurogenic and reprogramming activity of miR-9/9∗ and miR-124 may be in part through the direct targeting of subunits of the ATP-dependent BRG/BRM associated factor (BAF) chromatin remodeling complexes (Yoo et al., 2009; Staahl and Crabtree, 2013; Staahl et al., 2013). Mammalian BAF complexes are large multi-subunit complexes combinatorically assembled in a cell type-dependent manner. The combinatorial assembly of different homologs and splice variants of BAF subunit families confers functional specificity as each subunit contains functional domains that recognized DNA and/or modified histones (Wu et al., 2009; Zheng et al., 2012). For example, embryonic stem cell (ESC) BAF (esBAF) is characterized by BAF53a and a homodimer of BAF155, as opposed to a heterodimer of BAF155 and BAF170 in differentiated cells (Wang et al., 1996a,b; Ho et al., 2009b). The esBAF complex is involved in maintaining pluripotency by establishing an ESC-specific chromatin state permissive for transcription factors and signaling molecules to access ESC-associated genes (Ho et al., 2009a; Kidder et al., 2009). Although BAF complexes are traditionally known to antagonize the function of polycomb repressive complexes (PRC) to promote chromatin accessibility for gene activation (Kennison, 1995; Ho et al., 2011), studies have also suggested that BAF complexes can synergize with PRC for gene regulation (Ho et al., 2011).
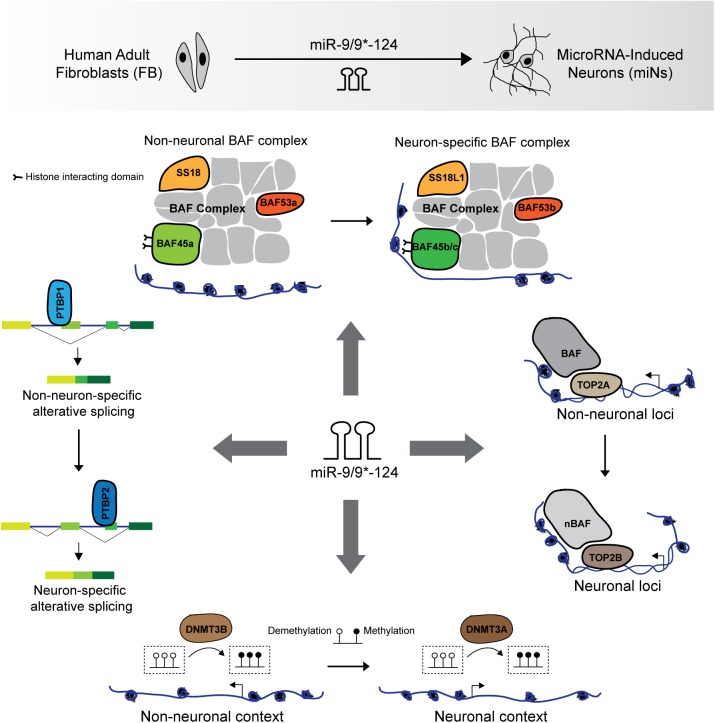
The BAF complex is crucial for mammalian nervous system as mutations in BAF subunits have been implicated in neurological disorders such as Coffin-Siris syndrome due to mutations in BRG1 and BRM (Tsurusaki et al., 2012; Ronan et al., 2013), and SS18L1/CREST in amyotrophic lateral sclerosis (ALS) (Chesi et al., 2013). During neural development, several BAF complex subunit switches to form the neuron-specific BAF (nBAF) complex (Staahl and Crabtree, 2013). The assembly of the nBAF complex requires the switching of progenitor subunits (BAF53a, BAF45a, and SS18) to neuronal subunits (BAF53b, BAF45b or BAF45c, and SS18L1/CREST) between the proliferating ventricular zone and the post-mitotic zone (Olave, 2002; Lessard et al., 2007; Wu et al., 2007; Yoo et al., 2009; Staahl et al., 2013). These molecular switches also occur during miRNA-mediated direct conversion of human fibroblasts into neurons (Staahl et al., 2013), in which the reciprocal switching of BAF53a to BAF53b is directly orchestrated by miR-9/9∗ and miR-124 (Yoo et al., 2009; Staahl et al., 2013; Figure 1). The assembly of the nBAF complex is essential for proper neuronal function in learning and memory as loss of function of either BAF53b or SS18L1/CREST dramatically reduced dendritic outgrowth and morphology (Aizawa et al., 2004; Wu et al., 2007; Staahl et al., 2013; Vogel-Ciernia et al., 2013). The function of chromatin remodeling by BAF complex was also found to be critical for neuronal reprogramming as loss of BRG1 during miRNA-mediated reprogramming abolished the chromatin landscape permissive to the activation of the neuronal program (Abernathy et al., 2017).
FIGURE 1.
miR-9/9∗-124-mediated neuronal reprogramming of human adult fibroblasts. Ectopic expression of miR-9/9∗-124 in human adult fibroblasts leads to the switching of several non-neuronal to neuronal components for neural fate acquisition. miRNAs orchestrate the genetic switching of epigenetic regulators including the BAF complex subunits (BAF53a, BAF45a, and SS18 to BAF53b, BAF45b/c, and SS18L1/CREST), TOP2A to TOP2B, and DNMT3B to DNMT3A. These coordinated molecular switches underlie the establishment of a neuronal epigenetic landscape during neuronal reprogramming. Additionally, the switching of PTB proteins, from PTBP1 to PTBP2, mediated by miR-124 activates neuron-specific alternative splicing program.
Switching of Chromatin Modifiers are Crucial for Cell Fate Conversion
The switching of other homologous epigenetic regulators also occur during neurogenesis and neuronal reprogramming, though may not be direct targets of miR-9/9∗-124. These include the switching of DNA topoisomerase II (TOP2) that functions to decatenate and catenate chromatin, from non-neuronal TOP2A to neuronal TOP2B (Watanabe et al., 1994; Tsutsui et al., 2001a; Tiwari et al., 2012; Thakurela et al., 2013). TOP2A is expressed in mitotic cells and interacts with BAF complexes to modulate chromatin accessibility (Tsutsui et al., 2001b; Dykhuizen et al., 2013; Wijdeven et al., 2015; Miller et al., 2017). On the other hand, TOP2B is required for neuronal differentiation in vitro and in vivo (Yang, 2000; Tsutsui et al., 2001b; Tiwari et al., 2012). Consistent with neuronal differentiation, miR-9/9∗-124 instruct the similar switch of TOP2 homologs during neuronal reprogramming of human adult fibroblasts by inducing a rapid reduction of TOP2A in fibroblasts for the selective expression TOP2B in converted neurons (Abernathy et al., 2017; Figure 1).
Similarly, the reconfiguration of the epigenetic landscape during neuronal reprogramming also involves changes in DNA methylation patterns (Abernathy et al., 2017). Regarding the reprogramming activities of the miRNAs, it has been shown that ectopic expression of miR-9/9∗-124 in fibroblasts recapitulated the molecular switching of de novo methyltransferases, DNMT3B to DNMT3A, similarly, to neural differentiation in vivo (Feng et al., 2005; Watanabe et al., 2006; Abernathy et al., 2017; Figure 1). DNMT3A has been implicated in various aspects of neuronal development, including synaptic plasticity (Feng et al., 2010; Colquitt et al., 2014), but it remains unclear how DNMT3A modulates gene expression in the nervous system. Although DNA methylation is viewed as a repressive mark, methylation marks deposited by DNMT3A have also been associated with enhanced gene expression through antagonizing PRC2 activity (Wu et al., 2010). The dramatic change in DNA methylation profile in miRNA-induced neurons also involves the induction of a family of demethylase, ten–eleven translocation (TET) family proteins (TET1/2/3) (Abernathy et al., 2017), implicated in neuronal development (Hahn et al., 2013; Zhang et al., 2013). It should be noted, however, what additional molecules interact with DNMT3A and TET proteins during neuronal reprogramming to influence DNA methylation at specific loci remains largely unknown.
Although the mechanisms underlying the switching of epigenetic effectors remain to be precisely defined, it is clear that miR-9/9∗-124 promote neuronal identity during the direct reprogramming of human fibroblasts through establishing an epigenetic state permissive for the downstream acquisition of neuronal fate. The reciprocal temporal and spatial switching of chromatin modifiers observed both during neurogenesis and neuronal reprogramming highlight the complex and dynamic epigenetic regulations required to overcome cell fate barriers.
miR-124-Mediated Ptb Switching Regulates Neuronal Splicing Profile
In addition to epigenetic regulators, post-transcriptional regulation of gene expression appears to be integral for neuronal reprogramming. The expression of PTB (polypyrimidine tract-binding) proteins, PTBP1 and PTBP2, are mutually exclusive and exhibit reciprocal switching during neural fate acquisition (Boutz et al., 2007). PTB proteins are RNA-binding proteins that bind to U-rich tracts primarily in introns for the post-transcriptional regulation of mRNAs, including alternative splicing (Wagner and Garcia-Blanco, 2001; Keppetipola et al., 2012). PTBP1 is expressed in non-neuronal cells and neural progenitors whereas the expression of its neuronal homolog, PTBP2 (nPTB), a splicing target of PTBP1, is primarily restricted to post-mitotic neurons in the nervous system (Boutz et al., 2007; Makeyev et al., 2007). PTBP1 represses PTBP2 expression by introducing a premature stop through the skipping of PTBP2 exon (Boutz et al., 2007). During development, the expression of miR-124 at the onset of neurogenesis mediates the switching of PTB proteins by targeting the 3′UTR of PTBP1, thereby alleviating PTBP1-mediated repression of PTBP2 in neurons (Makeyev et al., 2007). Although PTB proteins exhibit functional redundancy (Spellman et al., 2007), PTBP2 in neurons are essential for the proper splicing of various transcripts involved in neuronal function (Boutz et al., 2007; Licatalosi et al., 2012; Zheng et al., 2012; Li et al., 2014). Interestingly, ablating PTBP1 function in several cell types, including mouse embryonic fibroblasts, though insufficient in human fibroblasts, led to the direct conversion into neurons (Xue et al., 2013; Xue Y. et al., 2016), suggesting the significance of PTBP2 for the induction of neuronal fate. In addition to the activation of PTBP2 upon neural fate acquisition, PTBP2 level attenuates later in development for neuronal maturation (Li et al., 2014; Xue Y. et al., 2016). The attenuation of PTBP2 can be recapitulated with sequential knockdown of both PTB proteins resulting in the reprogramming of human fibroblasts into neurons (Xue Y. et al., 2016). The proposed mechanism is that PTBP2 reduction initiates a regulatory loop that activates downstream BRN2 for miR-9 expression, which dampens PTBP2 activity through 3′UTR targeting (Xue Y. et al., 2016). Altogether, PTBP2 level is dynamically regulated throughout neuronal differentiation and is essential as PTBP2 knockout results in neuronal death (Li et al., 2014).
The Use of miRNA-Induced Neuronal Ground State for Subtype-Specific Neuronal Reprogramming
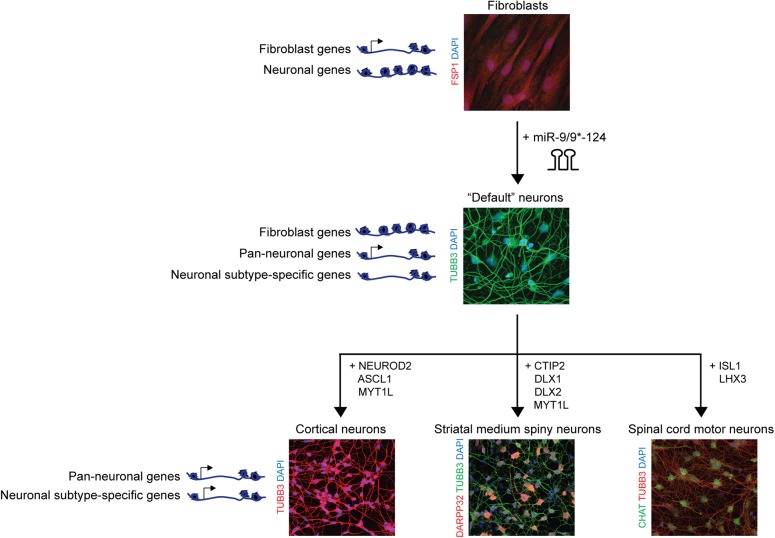
As neurological disorders affect distinct neuronal subtypes, the generation of neuronal subtypes has been of interest not only for dissecting the underlying mechanisms behind subtype-specific neuronal conversion, but also for the implication of the reprogrammed neurons in disease modeling. miR-9/9∗-124 have been shown to induce a “default” neuronal state characterized by enhanced accessibility of chromatin regions encompassing neuronal genes (Figure 2). These regions include genes specifically expressed in distinct neuronal subtypes, yet remain inactivated, thereby providing the chromatin environment that is open and permissive for subtype-defining inputs of transcription factors (Abernathy et al., 2017). Furthermore, unlike iPSC-based reprogramming methods, direct neuronal conversion bypasses an embryonic intermediate (Lapasset et al., 2011; Miller et al., 2013), thereby retaining the age signatures of starting fibroblasts including the epigenetic clock (Horvath, 2013), age-associated changes in transcriptome and microRNAs, reactive oxygen species (ROS) levels, DNA damage and telomere lengths (Mertens et al., 2015; Huh et al., 2016; Tang et al., 2017). As direct neuronal conversion can faithfully recapitulate age-associated phenotypes, directly reprogrammed neurons hold promise in the modeling of adult-onset neurodegenerative diseases and necessitates the control of subtype-specificity during neuronal reprogramming.
FIGURE 2.
Synergism between microRNAs and transcription factors for subtype-specific neuronal reprogramming of human adult fibroblasts. miR-9/9∗-124 mediate the erasure of fibroblast fate through remodeling of a fibroblast-specific chromatin landscape and transcriptome profile and promote a neuronal chromatin landscape and transcriptome. miRNA alone generates a default neuronal state characterized by the reconfiguration of the chromatin state permissive to the subtype-defining inputs of transcription factors. With distinct combinations of transcription factors, the miRNA-based conversion have been successfully employed to generate cortical neurons (Yoo et al., 2011), striatal medium spiny neurons (Victor et al., 2014), and spinal cord motor neurons (Abernathy et al., 2017).
The use of synergism between miRNAs and subtype-defining transcription factors to obtain subtype-specific neurons has been successful in generating cortical neurons (Yoo et al., 2011), striatal medium spiny neurons (MSN) (Victor et al., 2014), and spinal cord motor neurons (Abernathy et al., 2017; Figure 2). For instance, heterogeneous population of excitatory and inhibitory neurons belonging to the cortex can be obtained with the use of miR-9/9∗-124 in combination with NEUROD2, ASCL1, and MYT1L (DAM) cocktail from adult fibroblasts (Yoo et al., 2011). However, it remains to be tested whether layer-enriched transcription factors would be able to further guide the cortical lineage to neurons with layer-specific identities. Since previous studies demonstrated the plasticity of cortical neurons being able to transition between cortical layer fates (Rouaux and Arlotta, 2010, 2013; De la Rossa et al., 2013), it raises the potential that a similar approach may be taken in a cellular reprogramming context.
An enriched population of striatal MSN, the neuronal subtype primarily degenerated in Huntington’s disease (HD), can be derived using miR-9/9∗-124 in conjunction with CTIP2, DLX1/2, and MYT1L (CDM) factors (Victor et al., 2014). More than 70% of cells express DARPP32, a marker of MSNs, and when injected into the stratum of mouse pups, the converted MSNs incorporate and project to the substantia nigra and globus pallidus in vivo with electrophysiological properties similar to neighboring endogenous mouse MSNs (Victor et al., 2014). Interestingly, applying the MSN-specific neuronal conversion approach in fibroblast samples from symptomatic patients has proven to be successful in generating patient-specific MSNs manifesting hallmark HD pathology, including HTT aggregation, spontaneous neuronal death, and increased DNA damage (Victor et al., 2018). Importantly, the manifestation of HD-associated phenotypes was dependent on the specificity of the type of neurons generated and the age status in converted neurons, further highlighting the importance of age and subtype-specificity in modeling adult-onset diseases (Victor et al., 2018).
Spinal cord motor neurons are most susceptible to degeneration in amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA) diseases, and devising a conversion protocol could be instrumental to the study and modeling of motor neuron (MN) diseases. Using a combination of NEUROG1, SOX11, ISL1, and LHX3 (NSIL), MNs can be generated from fibroblasts of ALS patients in which the patient-derived MNs manifest various ALS pathologies, including FUS protein mislocalization and neuronal degeneration (Liu et al., 2016). Alternatively, miRNAs in conjunction with two transcription factors, ISL1 and LHX3, have been shown to generate functional mature MNs that display transcriptional signatures similar to in vivo mouse spinal MNs (Abernathy et al., 2017). Despite the robustness in generating a highly enriched population of spinal cord MNs, it remains to be demonstrated whether the MNs derived through the miRNA-induced neuronal state can be used to model ALS or SMA.
The use of transcription factors only, ASCL1, NURR1, and LMX1A, to directly convert fibroblasts of healthy and Parkinson’s disease (PD) patients into dopaminergic cells is possible but with limited efficiency in human cells (Caiazzo et al., 2011). Interestingly, reprogramming efficiency improved with the addition of neuronal miRNA, miR-124, and shRNA against p53, to the transcription factor cocktail using adult fibroblasts (Jiang et al., 2015). One of the proposed mechanisms behind this enhancement is due to the activation of TET proteins, in particular TET1, during reprogramming, as knockdown of TET1 results in increased cell death while overexpression enhances the overall number of TUBB3 and TH positive cells (Jiang et al., 2015). The induction of TET family members has also been observed in miR-9/9∗-124-mediated reprogramming (Abernathy et al., 2017).
The generation of additional neuronal subtypes, including serotonergic neurons for the study of neuropsychiatric disorders such as schizophrenia (Vadodaria et al., 2016; Xu et al., 2016) and sensory neurons for the study of pain sensation (Blanchard et al., 2015; Wainger et al., 2015) have been demonstrated using the transcription factor approach. It remains to be tested whether miR-9/9∗-124 could be combined with similar transcription factors to enhance overall conversion efficiency in human cells.
Summary
miR-9/9∗-124-mediated direct conversion of human adult fibroblasts into functional neurons reconfigures and establishes a pan-neuronal epigenetic landscape permissive on which brain region-enriched transcription factors can act and generate specific neuronal subtype. miR-9/9∗-124 are potent neurogenic molecules as they mediate numerous genetic switches that occur during neurogenesis, in which many include epigenetic players and pro-neurogenic effectors that are important to overcome cell fate barriers and activate neuronal fate programs. As miRNAs regulate expression of multiple genes, the pro-neural environment established by the miRNAs allow for the use of this paradigm for the study of neural fate acquisition. To better understand and address the role of brain-enriched miRNAs, examining miRNA-mRNA network would provide invaluable insights to the acquisition of neuronal fate. Though much remains to be uncovered, with the maintenance of age of starting fibroblasts preserved after cellular conversion, modeling age-dependent neurodegenerative diseases through direct reprogramming allows for the faithful recapitulation of age-associated pathogenesis for mechanistic studies of the disease.
Author Contributions
Y-LL planned, researched, and wrote the manuscript. AY planned and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Matheus B. Victor for providing images of reprogrammed cortical and medium spiny neurons, and Daniel G. Abernathy for providing the image of reprogrammed motor neurons.
Footnotes
Funding. AY is supported by the Andrew B and Virginia C. Craig Faculty Fellowship Endowment, NIH Director’s Innovator Award (DP2NS083372-01), Missouri Spinal Cord Injury/Disease Research Program (SCIDRP), Cure Alzheimer’s Fund (CAF), Presidential Early Career Award for Scientists and Engineers (PECASE), and NIA (RF1AG056296).
References
- Abernathy D. G., Kim W. K., McCoy M. J., Lake A. M., Ouwenga R., Lee S. W., et al. (2017). MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21 332.e9–348.e9. 10.1016/j.stem.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H., Hu S.-C., Bobb K., Balakrishnan K., Ince G., Gurevich I., et al. (2004). Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303 197–202. 10.1126/science.1089845 [DOI] [PubMed] [Google Scholar]
- Akerblom M., Sachdeva R., Barde I., Verp S., Gentner B., Trono D., et al. (2012). MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 32 8879–8889. 10.1523/JNEUROSCI.0558-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., et al. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9 113–118. 10.1016/j.stem.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F., Trivedi C. M., Juhr D., Gupta M., Cui Z., Tian Y., et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8 376–388. 10.1016/j.stem.2011.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J. W., Eade K. T., Szűcs A., Lo Sardo V., Tsunemoto R. K., Williams D., et al. (2015). Selective conversion of fibroblasts into peripheral sensory neurons. Nat. Neurosci. 18 25–35. 10.1038/nn.3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P. L., Stoilov P., Li Q., Lin C.-H., Chawla G., Ostrow K., et al. (2007). A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21 1636–1652. 10.1101/gad.1558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476 224–227. 10.1038/nature10284 [DOI] [PubMed] [Google Scholar]
- Chanda S., Ang C. E., Davila J., Pak C., Mall M., Lee Q. Y., et al. (2014). Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3 282–296. 10.1016/j.stemcr.2014.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L.-C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12 399–408. 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi A., Staahl B. T., Jovičić A., Couthouis J., Fasolino M., Raphael A. R., et al. (2013). Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 16 851–855. 10.1038/nn.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquitt B. M., Markenscoff-Papadimitriou E., Duffié R., Lomvardas S. (2014). Dnmt3a regulates global gene expression in olfactory sensory neurons and enables odorant-induced transcription. Neuron 83 823–838. 10.1016/j.neuron.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.-J., Mandel G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103 2422–2427. 10.1073/pnas.0511041103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajas-Bailador F., Bonev B., Garcez P., Stanley P., Guillemot F., Papalopulu N. (2012). microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15 697–699. 10.1038/nn.3082 [DOI] [PubMed] [Google Scholar]
- De la Rossa A., Bellone C., Golding B., Vitali I., Moss J., Toni N., et al. (2013). In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat. Neurosci. 16 193–200. 10.1038/nn.3299 [DOI] [PubMed] [Google Scholar]
- Deo M., Yu J.-Y., Chung K.-H., Tippens M., Turner D. L. (2006). Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235 2538–2548. 10.1002/dvdy.20847 [DOI] [PubMed] [Google Scholar]
- Drouin-Ouellet J., Lau S., Brattås P. L., Rylander Ottosson D., Pircs K., Grassi D. A., et al. (2017). REST suppression mediates neural conversion of adult human fibroblasts via microRNA-dependent and -independent pathways. EMBO Mol. Med. 9 1117–1131. 10.15252/emmm.201607471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen E. C., Hargreaves D. C., Miller E. L., Cui K., Korshunov A., Kool M., et al. (2013). BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 497 624–627. 10.1038/nature12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Chang H., Li E., Fan G. (2005). Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79 734–746. 10.1002/jnr.20404 [DOI] [PubMed] [Google Scholar]
- Feng J., Zhou Y., Campbell S. L., Le T., Li E., Sweatt J. D., et al. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13 423–430. 10.1038/nn.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. J., Zavolan M. (2013). Modulation of epigenetic regulators and cell fate decisions by miRNAs. Epigenomics 5 671–683. 10.2217/epi.13.65 [DOI] [PubMed] [Google Scholar]
- Hahn M. A., Qiu R., Wu X., Li A. X., Zhang H., Wang J., et al. (2013). Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 3 291–300. 10.1016/j.celrep.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Liu Y., Wang X., Zhang M. Q., Hannon G. J., Huang Z. J. (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73 35–48. 10.1016/j.neuron.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. (2009a). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106 5187–5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R., Crabtree G. R. (2011). esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13 903–913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., et al. (2009b). An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106 5181–5186. 10.1073/pnas.0812889106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14:R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh C. J., Zhang B., Victor M. B., Dahiya S., Batista L. F., Horvath S., et al. (2016). Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. eLife 5:e18648. 10.7554/eLife.18648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey K. N., Srivastava D. (2010). MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7 36–41. 10.1016/j.stem.2010.06.012 [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28 2679–2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K. S. (2016). Cell fate control by pioneer transcription factors. Development 143 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena T. M., Egemnazarov B., Finch E. A., Zhang L., Payne J. A., Pandya K., et al. (2012). MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110 1465–1473. 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Xu Z., Zhong P., Ren Y., Liang G., Schilling H. A., et al. (2015). Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat. Commun. 6:10100. 10.1038/ncomms10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A. (1995). The polycomb and trithorax group proteins of drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29 289–303. 10.1146/annurev.ge.29.120195.001445 [DOI] [PubMed] [Google Scholar]
- Keppetipola N., Sharma S., Li Q., Black D. L. (2012). Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 47 360–378. 10.3109/10409238.2012.691456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S., Knott J. G. (2009). SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27 317–328. 10.1634/stemcells.2008-0710 [DOI] [PubMed] [Google Scholar]
- Krichevsky A. M., Sonntag K.-C., Isacson O., Kosik K. S. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24 857–864. 10.1634/stemcells.2005-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J., Mertens J., Kesavan J., Doerr J., Poppe D., Glaue F., et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9 575–578. 10.1038/nmeth.1972 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12 735–739. 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- Lapasset L., Milhavet O., Prieur A., Besnard E., Babled A., Ait-Hamou N., et al. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 25 2248–2253. 10.1101/gad.173922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Oh Y. M., Lu Y.-L., Kim W. K., Yoo A. S. (2018). MicroRNAs overcome cell fate barrier by reducing EZH2-controlled rest stability during neuronal conversion of human adult fibroblasts. Dev. Cell 46 73.e7–84.e7. 10.1016/j.devcel.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., et al. (2007). An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55 201–215. 10.1016/j.neuron.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C., Stigloher C., Wizenmann A., Klafke R., Folchert A., Bally-Cuif L. (2008). MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11 641–648. 10.1038/nn.2115 [DOI] [PubMed] [Google Scholar]
- Li Q., Zheng S., Han A., Lin C.-H., Stoilov P., Fu X.-D., et al. (2014). The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife 3:e01201. 10.7554/eLife.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D. D., Yano M., Fak J. J., Mele A., Grabinski S. E., Zhang C., et al. (2012). Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 26 1626–1642. 10.1101/gad.191338.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Liu M.-L., Zang T., Zhang C.-L. (2016). Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep. 14 115–128. 10.1016/j.celrep.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-L., Zang T., Zou Y., Chang J. C., Gibson J. R., Huber K. M., et al. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4:2183. 10.1038/ncomms3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27 435–448. 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Paquola A. C. M., Ku M., Hatch E., Böhnke L., Ladjevardi S., et al. (2015). Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17 705–718. 10.1016/j.stem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. L., Hargreaves D. C., Kadoch C., Chang C.-Y., Calarco J. P., Hodges C., et al. (2017). TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 24 344–352. 10.1038/nsmb.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. D., Ganat Y. M., Kishinevsky S., Bowman R. L., Liu B., Tu E. Y., et al. (2013). Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13 691–705. 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I. (2002). Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16 2509–2517. 10.1101/gad.992102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer A. N., Xing Y., Harper S. Q., Jones L., Davidson B. L. (2008). The bifunctional microRNA miR-9/miR-9∗ regulates rest and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 28 14341–14346. 10.1523/JNEUROSCI.2390-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Ek F., Lang S., Soneji S., Olsson R., Parmar M. (2016). Small molecules increase direct neural conversion of human fibroblasts. Sci. Rep. 6:38290. 10.1038/srep38290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajman M., Schratt G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144 2310–2322. 10.1242/dev.144337 [DOI] [PubMed] [Google Scholar]
- Ronan J. L., Wu W., Crabtree G. R. (2013). From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 14 347–359. 10.1038/nrg3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C., Arlotta P. (2010). Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat. Neurosci. 13 1345–1347. 10.1038/nn.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C., Arlotta P. (2013). Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 15 214–221. 10.1038/ncb2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. K., Yang J., Liu M.-L., Zhang C.-L. (2016). Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Rep. 7 955–969. 10.1016/j.stemcr.2016.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., et al. (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9 205–218. 10.1016/j.stem.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Llorian M., Smith C. W. J. (2007). Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 27 420–434. 10.1016/j.molcel.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl B. T., Crabtree G. R. (2013). Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr. Opin. Neurobiol. 23 903–913. 10.1016/j.conb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl B. T., Tang J., Wu W., Sun A., Gitler A. D., Yoo A. S., et al. (2013). Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33 10348–10361. 10.1523/JNEUROSCI.1258-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu M.-L., Zang T., Zhang C.-L. (2017). Direct reprogramming rather than iPSC-based reprogramming maintains aging hallmarks in human motor neurons. Front. Mol. Neurosci. 10:359. 10.3389/fnmol.2017.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S., Garding A., Jung J., Schübeler D., Burger L., Tiwari V. K. (2013). Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nat. Commun. 4:2478. 10.1038/ncomms3478 [DOI] [PubMed] [Google Scholar]
- Tiwari V. K., Burger L., Nikoletopoulou V., Deogracias R., Thakurela S., Wirbelauer C., et al. (2012). Target genes of topoisomerase II regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. U.S.A. 109 E934–E943. 10.1073/pnas.1119798109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurusaki Y., Okamoto N., Ohashi H., Kosho T., Imai Y., Hibi-Ko Y., et al. (2012). Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44 376–378. 10.1038/ng.2219 [DOI] [PubMed] [Google Scholar]
- Tsutsui K., Tsutsui K., Hosoya O., Sano K., Tokunaga A. (2001a). Immunohistochemical analyses of DNA topoisomerase II isoforms in developing rat cerebellum. J. Comp. Neurol. 431 228–239. [DOI] [PubMed] [Google Scholar]
- Tsutsui K., Tsutsui K., Sano K., Kikuchi A., Tokunaga A. (2001b). Involvement of DNA topoisomerase IIβ in neuronal differentiation. J. Biol. Chem. 276 5769–5778. 10.1074/jbc.M008517200 [DOI] [PubMed] [Google Scholar]
- Vadodaria K. C., Mertens J., Paquola A., Bardy C., Li X., Jappelli R., et al. (2016). Generation of functional human serotonergic neurons from fibroblasts. Mol. Psychiatry 21 49–61. 10.1038/mp.2015.161 [DOI] [PubMed] [Google Scholar]
- Victor M. B., Richner M., Hermanstyne T. O., Ransdell J. L., Sobieski C., Deng P.-Y., et al. (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84 311–323. 10.1016/j.neuron.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor M. B., Richner M., Olsen H. E., Lee S. W., Monteys A. M., Ma C., et al. (2018). Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 21 341–352. 10.1038/s41593-018-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463 1035–1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J. W., Lee S.-K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21 744–749. 10.1101/gad.1519107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A., Matheos D. P., Barrett R. M., Kramár E. A., Azzawi S., Chen Y., et al. (2013). The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16 552–561. 10.1038/nn.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. J., Garcia-Blanco M. A. (2001). Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21 3281–3288. 10.1128/MCB.21.10.3281-3288.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B. J., Buttermore E. D., Oliveira J. T., Mellin C., Lee S., Saber W. A., et al. (2015). Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat. Neurosci. 18 17–24. 10.1038/nn.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cote J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., et al. (1996a). Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., Kuo A., Cairns B. R., Crabtree G. R. (1996b). Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10 2117–2130. 10.1101/gad.10.17.2117 [DOI] [PubMed] [Google Scholar]
- Watanabe D., Uchiyama K., Hanaoka K. (2006). Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience 142 727–737. 10.1016/j.neuroscience.2006.07.053 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Tsutsui K., Tsutsui K., Inoue Y. (1994). Differential expressions of the topoisomerase IIa and IIb mRNAs in developing rat brain. Neurosci. Res. 19 51–57. 10.1016/0168-0102(94)90007-8 [DOI] [PubMed] [Google Scholar]
- Wijdeven R. H., Pang B., van der Zanden S. Y., Qiao X., Blomen V., Hoogstraat M., et al. (2015). Genome-wide identification and characterization of novel factors conferring resistance to topoisomerase II poisons in cancer. Cancer Res. 75 4176–4187. 10.1158/0008-5472.CAN-15-0380 [DOI] [PubMed] [Google Scholar]
- Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., et al. (2010). Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329 444–448. 10.1126/science.1190485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. I., Lessard J., Crabtree G. R. (2009). Understanding the words of chromatin regulation. Cell 136 200–206. 10.1016/j.cell.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. I., Lessard J., Olave I. A., Qiu Z., Ghosh A., Graef I. A., et al. (2007). Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56 94–108. 10.1016/j.neuron.2007.08.021 [DOI] [PubMed] [Google Scholar]
- Xu Z., Jiang H., Zhong P., Yan Z., Chen S., Feng J. (2016). Direct conversion of human fibroblasts to induced serotonergic neurons. Mol. Psychiatry 21 62–70. 10.1038/mp.2015.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q., Yu C., Wang Y., Liu L., Zhang K., Fang C., et al. (2016). miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a. Sci. Rep. 6:26781. 10.1038/srep26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152 82–96. 10.1016/j.cell.2012.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Qian H., Hu J., Zhou B., Zhou Y., Hu X., et al. (2016). Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 19 807–815. 10.1038/nn.4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. (2000). DNA topoisomerase II and neural development. Science 287 131–134. 10.1126/science.287.5450.131 [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Staahl B. T., Chen L., Crabtree G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460 642–646. 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476 228–231. 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.-R., Cui Q.-Y., Murai K., Lim Y. C., Smith Z. D., Jin S., et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13 237–245. 10.1016/j.stem.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16 365–371. 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Gray E. E., Chawla G., Porse B. T., O’Dell T. J., Black D. L. (2012). PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 15 381–388. 10.1038/nn.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]