Abstract
Vertebrate gut microbiota mediate critical physiological processes known to affect host fitness, but the mechanisms that expose wildlife to pioneer members of this important microbial community are not well understood. For example, oviparous vertebrates are thought to acquire gut microbiota through post-natal exposure to the external environment, but recent evidence from placental mammals suggests that the vertebrate reproductive tract harbours microbiota that may inoculate offspring in utero. These findings suggest that oviparous vertebrates may be capable of acquiring pioneer microbiota in ovo, but this phenomenon remains unexplored. To fill this knowledge gap, we used culture-independent inventories to determine if the eggs of wild birds and lizards harboured in ovo microbial communities. Our approach revealed distinct in ovo bacterial communities, but fungal communities were indistinguishable from controls. Further, lizard eggs from the same clutch had bacterial community structures that were more similar to each other than to unrelated individuals. These results suggest that oviparous vertebrates may acquire maternal microbiota in ovo, possibly through the inoculation of egg yolk prior to shelling. Therefore, this study may provide a first glimpse of a phenomenon with substantial implications for our understanding of the ecological and evolutionary factors shaping gut microbial communities.
Keywords: microbiome, eggs, birds, lizards, bacteria, maternal transmission
1. Introduction
A recent surge in research has demonstrated that the archaeal, bacterial and fungal communities of the vertebrate gastrointestinal tract, collectively known as gut microbiota, affect critical host physiological processes [1,2]. Microbial exposure in early-life is thought to influence the trajectory of microbial community assembly in the vertebrate gut, thereby affecting long-term host fitness [3]. While evidence for these phenomena are primarily derived from experiments in model organisms [4], recent ecological studies on wild vertebrates suggest that disruptions to incipient gut microbial communities affect host performance later in life [5]. Nevertheless, the initial acquisition of gut microbiota remains an understudied aspect of animal ecology.
For more than a century, vertebrate embryos were thought to develop in a sterile environment and to acquire pioneer gut microbiota through post-natal contact with their mother and the external environment [3]. However, recent culture-independent molecular investigations suggest that placental mammals harbour in utero microbiota [6], though these microbial communities may be in low abundance, and thus difficult to distinguish from environmental controls [7]. While these findings suggest that microbial transmission during development may be universal among vertebrates [3], the possibility of in ovo microbial exposure among oviparous vertebrates has not been sufficiently explored. Currently, evidence for in ovo microbial communities is limited to research on chickens [8], with most studies focused on pathogenic bacterial infections [3], rather than maternal microbial transmission in an ecological context.
To fill this knowledge gap, we used culture-independent bacterial and fungal inventories to determine if the eggs of wild birds and lizards harboured in ovo microbial communities. We predicted that (i) eggs would harbour distinct microbial communities compared with environmental controls and (ii) eggs from the same clutch would have similar microbial communities, suggesting that wild oviparous vertebrates may acquire pioneer gut microbiota of maternal origin in ovo.
2. Material and methods
(a). Sample collection and molecular processing
We collected 11 wild bird eggs (representing four species) from nest-boxes approximately 1–5 days after oviposition, and 14 Eastern Fence Lizard (Sceloporus undulatus) eggs approximately 4–12 h after oviposition (two eggs each from seven clutches) from wild-caught individuals housed in captivity for a maximum of 38 days prior to egg collection (electronic supplementary material, table S1). Wild bird eggs were placed on ice after collection, then processed immediately upon returning to the field station. All eggs were processed by (i) washing the external surface with 70% ethanol, (ii) puncturing the eggshell using a 1000 µl barrier pipette tip and (iii) transferring egg contents into a sterile 1.5 ml tube. Environmental contamination controls were generated by transferring sterile water (alongside bird eggs; four total) and sterile saline (alongside lizard eggs; 14 total). As an additional environmental control, we collected sand from four lizard enclosures. All egg contents and controls were immediately frozen at −80°C.
(b). Molecular analyses and bioinformatics
We extracted DNA from controls and eggs using the Qiagen PowerFecal Kit. Because commercial extraction kits contain microbial DNA [9], we included four additional ‘blank' extraction controls. We used polymerase chain reaction to amplify the 16S and internal transcribed spacer (ITS) rRNA genes for Illumina sequencing (see electronic supplementary material for detailed molecular protocols). Sequence reads were filtered and processed using the DADA2 pipeline [10]. We identified bacterial and fungal sequence variants (hereafter, operational taxonomic units or OTUs) using the Greengenes [11] and UNITE [12] reference databases, respectively. To ensure that our analyses were restricted to bacterial and fungal taxa, we removed OTUs identified as archaea, chloroplasts and mitochondria. We rarefied OTU tables to 557 sequences for bacteria and 512 for fungi before comparisons of alpha- and beta-diversity. Last, we removed OTUs detected in extraction, saline and water controls from all egg inventories to describe egg-specific microbiota and identify core members (OTUs occurring in 50% or more of a given egg type) of in ovo microbial communities. All bioinformatic analyses were conducted in QIIME2 [13].
(c). Statistical analyses
To determine if bird and lizard eggs harbour in ovo microbial communities, we used Kruskal–Wallis tests to investigate differences in OTU richness and phylogenetic diversity between eggs and controls. Differences in community membership (OTU presence/absence via unweighted UniFrac distances) and structure (OTU relative abundance via weighted UniFrac distances) were investigated using principal coordinate analysis and permutational multivariate analysis of variance (PERMANOVA), which was conducted using one egg from each clutch to avoid pseudo-replication. Similarities between the bacterial communities of lizard eggs from the same clutch were investigated using analysis of similarity (ANOSIM). All statistical analyses were conducted in QIIME2 [13].
3. Results
(a). Bacterial community analysis
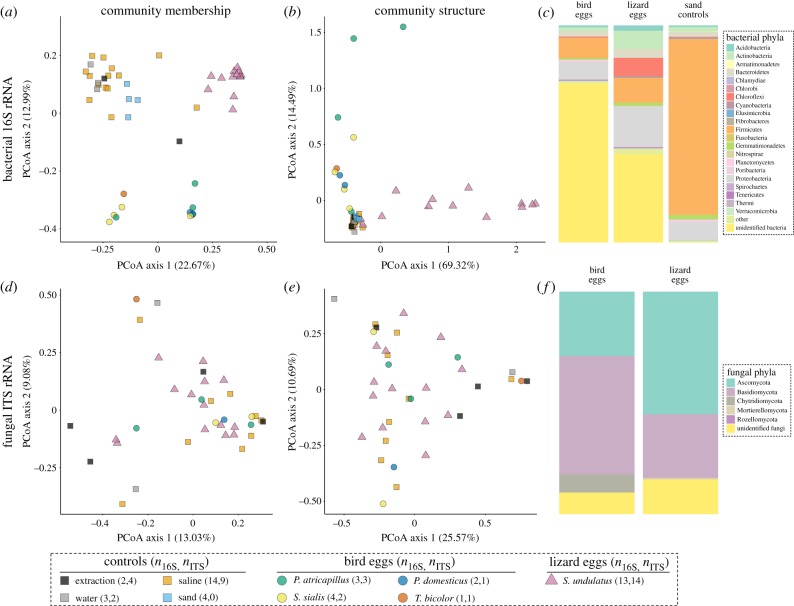
Illumina sequencing generated 885 903 16S rRNA sequences (mean of 17 371 ± 30 404 s.d.) and 1733 OTUs after DADA2 processing and removal of non-bacterial OTUs. The in ovo bacterial communities of bird and lizard eggs were significantly more phylogenetically diverse than controls (extraction, saline and water), but exhibited no significant differences in OTU richness (electronic supplementary material, figure S1). Further, in ovo bacterial community membership (figure 1a) and structure (figure 1b) differed significantly from these same controls (electronic supplementary material, table S2). In a separate analysis, sand bacterial communities differed significantly from those of lizard eggs in both membership (PERMANOVA: pseudo-F = 5.42, p = 0.003; figure 1a) and structure (PERMANOVA: pseudo-F = 6.2, p = 0.02; figure 1b), sharing only 13 of 1390 total OTUs (electronic supplementary material, table S3). Bird and lizard eggs differed significantly in bacterial community membership (PERMANOVA: pseudo-F = 8.2, p = 0.001; figure 1a; electronic supplementary material, table S2) and structure (PERMANOVA: pseudo-F = 12.9, p = 0.001; figure 1b; electronic supplementary material, table S2). Lizard eggs from the same clutch were more similar in bacterial community structure than unrelated individuals (ANOSIM: R = 0.69, p = 0.004; electronic supplementary material, figure S2), but not in community membership (ANOSIM: R = 0.14, p = 0.12; electronic supplementary material, figure S2).
Figure 1.
Summary of bird and lizard in ovo microbial communities using bacterial 16S rRNA (a–c) and fungal ITS rRNA (d–f) amplicon sequences. (a) Principal coordinate analysis (PCoA) of bacterial community membership using unweighted UniFrac distances. (b) PCoA of bacterial community structure using weighted UniFrac distances. (c) Relative abundances of egg-specific bacterial phyla (after removal of OTUs detected in extraction, saline and water controls) compared with sand controls. (d) PCoA of fungal community membership using unweighted UniFrac distances. (e) PCoA of fungal community structure using weighted UniFrac distances. (f) Relative abundances of egg-specific fungal phyla (after removal of OTUs detected in extraction, saline and water controls). Replicates for each sample type differed between 16S (n16S) and ITS (nITS) libraries due to independent rarefaction of OTU tables.
From a total of 1079 total egg-specific OTUs, we detected the presence of 13 bacterial phyla in bird eggs and 19 in lizard eggs (figure 1c). Despite representing a relatively small fraction of in ovo 16S sequences, Firmicutes was the most dominant identified bacterial phylum in bird eggs (9%), while Proteobacteria was most dominant in lizard eggs (19%; figure 1c). OTUs classified as unidentified bacteria (129 in total) composed the majority of both bird (74%) and lizard eggs (41%; figure 1c). By contrast, the bacterial communities of lizard enclosure sand were almost entirely composed of identifiable bacterial taxa in the phyla Firmicutes (81%) and Proteobacteria (9%; figure 1c). As for core members of in ovo bacterial communities, 55% of bird eggs harboured an unidentified bacterial OTU, whereas 50% of lizard eggs harboured Morganella morganii (phylum Proteobacteria).
(b). Fungal community analysis
Illumina sequencing generated 137 061 ITS rRNA sequences (mean of 2916 ± 5984 s.d.) and 338 OTUs after DADA2 processing and removal of non-fungal OTUs. Fungal OTU richness and phylogenetic diversity did not differ significantly between eggs and controls (electronic supplementary material, figure S1). Further, there were no differences between fungal community membership (figure 1d) or structure (figure 1e) in any pairwise comparisons (electronic supplementary material, table S2).
We detected the presence of three fungal phyla for birds, and four for lizards (288 total egg-specific OTUs), with Ascomycota and Basidiomycota being the most dominant (figure 1f). Unidentified fungal OTUs represented a substantial proportion of sequence reads for both bird (9.8%) and lizard eggs (15.6%; figure 1f). No egg-specific fungal OTUs occurred in 50% or more of either egg type.
4. Discussion
Here, we demonstrated that bird and lizard eggs harbour bacterial communities that are distinct from each other and controls. Further, the in ovo bacterial community structures of lizard eggs from the same clutch were more similar to each other than to unrelated individuals. These results suggest that the transmission of maternal microbiota in ovo may be a potential mechanism shaping incipient gut microbial communities of oviparous vertebrates.
Our detection of in ovo bacterial communities is consistent with recent studies suggesting that vertebrate reproductive tracts harbour microbiota [6]. We found that Firmicutes and Proteobacteria were the most dominant bacterial phyla in the eggs of wild birds and lizards (figure 1c), which is consistent with a recent culture-independent inventory of chicken embryos [8]. The detection of the known commensal gut bacterium M. morganii [14] in 50% of lizard eggs suggests that oviparous vertebrates may be capable of transmitting gut microbiota to their offspring in ovo. While we were unable to determine whether eggs harboured fungi (figure 1d and e), the detection of 288 egg-specific fungal OTUs suggests that we should not rule out the possibility of a low-abundance fungal community.
We speculate that the presence of in ovo microbiota may be the result of bacterial colonization during egg formation. While bacteria are capable of trans-shell colonization or ascension from the cloaca [15], the limited number of OTUs shared between lizard eggs and their immediate sand environment suggests that these mechanisms were not influential in our study. Rather, our results suggest that, like placental mammals, the reproductive tract of oviparous vertebrates may harbour microbiota typically found in the digestive system [6]. Studies on pathogenic bacterial infections in chickens have suggested that immune cells may transport intestinal microbiota into the infundibulum, thereby inoculating the egg yolk prior to shell deposition [15]. This mechanism suggests the possibility of in ovo microbial inheritance, but further experimental evidence is necessary to determine if eggs harbour viable microbiota of maternal origin.
5. Conclusion
We provide evidence that in ovo bacterial communities are more similar among siblings compared to unrelated individuals, suggesting that wild oviparous vertebrates may transmit gut microbiota to their offspring. Therefore, this study may provide a first glimpse of a phenomenon with substantial implications for our understanding of the ecological and evolutionary factors shaping gut microbial communities.
Supplementary Material
Supplementary Material
Acknowledgements
We thank the University of Minnesota Itasca Biological Station, Solon Dixon Forestry Education Center and Land Between the Lakes National Recreation Area for the use of their facilities. David Ensminger and Cameron Venable assisted with wild lizard collection. Dustin Owen, Heather Engler, Thomas Adams and Jennifer Heppner assisted with lizard egg and environmental sample collection. We thank Dr Stefan Green and the DNA Services Facility of University of Illinois at Chicago for conducting Illumina sequencing.
Ethics
Animal protocols were approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Minnesota (protocol no. 1502-32331A) and the Pennsylvania State University (protocol no. 44595). Permits for lizard collections were approved by the Alabama Department of Conservation and Natural Resources, and the Kentucky Department of Fish and Wildlife.
Data accessibility
Sequences are accessible from GenBank at PRJNA445840 [16].
Authors' contributions
S.A.K. and K.J.M. collected the samples. B.K.T. extracted DNA, analysed the data and wrote the manuscript. K.D.K. and T.L. assisted in the analysis and interpretation of data. All authors contributed to the editing of the manuscript, agree to be held accountable for the content herein and approved the final version.
Competing interests
The authors declare no competing interests.
Funding
This project was funded in part by NSF grant 1456655.
References
- 1.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg E, Zilber-Rosenberg I. 2018. The hologenome concept of evolution after 10 years. Microbiome 6, 78 ( 10.1186/s40168-018-0457-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 11, e1001631 ( 10.1371/journal.pbio.1001631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascoe EL, Hauffe HC, Marchesi JR, Perkins SE. 2017. Network analysis of gut microbiota literature: an overview of the research landscape in non-human animal studies. ISME J. 11, 2644–2651. ( 10.1038/ismej.2017.133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knutie SA, Wilkinson CL, Kohl KD, Rohr JR. 2017. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 8, 86 ( 10.1038/s41467-017-00119-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. 2017. A critical assessment of the ‘sterile womb’ and ‘in utero colonization’ hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5, 48 ( 10.1186/s40168-017-0268-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauder AP, et al. 2016. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29 ( 10.1186/s40168-016-0172-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding J, et al. 2017. Inheritance and establishment of gut microbiota in chickens. Front. Microbiol. 8, 1967 ( 10.3389/fmicb.2017.01967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salter SJ, et al. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 ( 10.1186/s12915-014-0087-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. ( 10.1038/nmeth.3869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. ( 10.1128/AEM.03006-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kõljalg U, et al. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. ( 10.1111/mec.12481) [DOI] [PubMed] [Google Scholar]
- 13.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shroff KE, Meslin K, Cebra JJ. 1995. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immunity 63, 3904–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, Humphrey TJ, Van Immerseel F. 2009. Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol. Rev. 33, 718–738. ( 10.1111/j.1574-6976.2008.00161.x) [DOI] [PubMed] [Google Scholar]
- 16.Trevelline BK, MacLeod KJ, Knutie SA, Langkilde T, Kohl KD. 2018. Data from: In ovo microbial communities: a potential mechanism for the initial acquisition of gut microbiota among oviparous vertebrates GenBank (Accession: PRJNA445840). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Trevelline BK, MacLeod KJ, Knutie SA, Langkilde T, Kohl KD. 2018. Data from: In ovo microbial communities: a potential mechanism for the initial acquisition of gut microbiota among oviparous vertebrates GenBank (Accession: PRJNA445840). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Sequences are accessible from GenBank at PRJNA445840 [16].