Abstract
Wolbachia is an endosymbiotic bacterium that can block viral infections in arthropods, generating interest in its potential to control the spread of mosquito-borne disease. Drosophila melanogaster is model organism for Wolbachia infection, and the wMel strain of Wolbachia can improve host survival following viral infection. However, it is unclear whether wMel induces anti-viral blocking against the broader native virome of D. melanogaster, or whether the major effect of Wolbachia is a reduction in viral abundance rather than viral clearance. We examined the effect of Wolbachia on viral abundance by comparing the total transcriptome of wMel-positive and wMel-negative D. melanogaster populations sampled from six locations in Australia. In addition, we examined the impact of wMel on individual flies by obtaining transcriptome data from 20 wMel-positive and 20 wMel-negative D. melanogaster from the location (Melbourne) with highest density of wMel. These data revealed high viral abundance in both Wolbachia-positive and -negative populations and individuals. Notably, none of the viral species identified, representing RNA viruses from at least nine families/floating genera, showed evidence of protection by wMel. Although the viral loads of picorna-like viruses are reduced by wMel under experimental conditions, we observed no such effect here. These data show that D. melanogaster can harbour abundant RNA viruses regardless of its Wolbachia status and imply that the interaction between Wolbachia and viruses in nature is more complex than simple blocking.
Keywords: Wolbachia, virus, Drosophila, evolution, transcriptomics
1. Introduction
Wolbachia is an endosymbiotic bacterium that can have a range of fitness effects on its insect hosts including the blocking of viral infections. The propensity for virus blocking underpins programmes aimed at using Wolbachia to reduce disease transmission. Most notably, a Wolbachia strain initially identified in natural populations of Drosophila melanogaster flies [1], designated wMel, has been manually transferred to Aedes aegypti mosquitoes where it blocks the replication of dengue, Zika and other human viruses and greatly reduces their transmission [2,3]. Other Wolbachia strains from Drosophila and mosquitoes that reduce viral densities have also been identified [4,5]. Two Wolbachia strains, including wMel, are currently being released in a variety of locations with the aim of replacing existing A. aegypti populations with Wolbachia-infected mosquitoes and hence reducing the burden of human disease [6].
wMel has been shown to block viral transmission in mosquitoes [7]. A similar effect has been observed for wMel in its natural host, D. melanogaster, under experimental conditions, where it reduced both viral load and mortality rates associated with Drosophila C virus (DCV) infection when introduced into flies [8]. This appears to be a common phenomenon [9–11] across several experimental Wolbachia infections in Drosophila with an efficacy likely linked to Wolbachia density [11,12]. In addition to DCV, wMel shows anti-viral protection against Nora virus and Flock House virus (FHV) [8,11], although the major effect of the latter is most likely a reduction in host mortality rather than a reduction in viral load or viral clearance [8,11]. As there is no evidence for the blocking of DNA viruses [8,13], these effects appear to be specific to RNA viruses.
While the virus blocking attributed to Wolbachia has only considered a limited number of viral species (DCV, FHV and Nora virus), D. melanogaster harbours a much larger viral diversity. Many of these viruses remain partially characterized, with little information on key aspects of phenotype including virulence. Two of the first D. melanogaster viruses to be isolated were Sigma virus and Drosophila A virus [14], although their interaction with Wolbachia is yet to be determined. Recently, a major study using an RNA metagenomics approach [15] identified more than 20 species of previously undescribed RNA viruses in D. melanogaster, representing more than 12 families or floating genera, most of which were found in wild fly populations. Importantly, strong host structure and a global distribution suggest that many of these viral species have been associated with flies for extended periods [15,16].
Webster et al. [15] also examined the viral infection status within the context of presence or absence of Wolbachia, targeting the natural viromes of wild fly populations, including DCV and Nora virus against which wMel has repeatedly demonstrated its protective effect under experimental conditions [8,10,11]. However, no association was found between the prevalence of Wolbachia and that of any virus, suggesting a lack of effect in natural D. melanogaster populations [15]. Importantly, while the prevalence data reflect the presence of viruses in a population, they do not indicate the level of viral abundance within individual flies. Indeed, experiments suggest that the blocking effect of Wolbachia occurs through a reduced viral load rather than complete clearance [11]. Whether this is also true of the natural virome of D. melanogaster is unknown.
In Australia, wMel is prevalent in D. melanogaster populations. Partial cytoplasmic incompatibility generated by Wolbachia in D. melanogaster was first identified in crosses between tropical and temperate populations from Australia [1]. The wMel infection responsible increases in frequency clinally from temperate to subtropical areas, and northern populations all carry a high frequency of this infection [17], while wMel-CS is absent or has a low frequency [18]. This geographical pattern has been stable over at least 20 years [19]. Although it is not clear why Wolbachia frequencies are lower in colder areas, under field conditions the Wolbachia infection is not transmitted with complete fidelity [17,19]. Wolbachia-infected females from eastern Australia can also have a smaller adult body size on occasion [17], as well as reduced fecundity and egg viability after overwintering [19].
Our aim was to examine the effect of Wolbachia on both viral load and diversity of the natural virome of D. melanogaster in Australia. To achieve this, we used a meta-transcriptomics approach to analyse Wolbachia-virus interactions in passaged (mostly F1) flies derived from wild populations in eastern Australia. We focused on passaged flies because the vertically transmitted virome in these animals should maintain a longer host relationship and contain fewer environmentally acquired viromes. Accordingly, we first examined the total transcriptomes of wMel-positive and wMel-negative D. melanogaster populations from six sampling locations in Australia. To increase the power of the analyses, we also compared the transcriptomes of individual wMel-positive and wMel-negative D. melanogaster from the sampled population with the highest Wolbachia density.
2. Material and methods
(a). Study locations
Drosophila melanogaster flies were collected from the field in 2008 along a latitudinal climate gradient on the east coast of Australia using nets from banana baits. This gradient extends from temperate Tasmania to tropical Queensland (electronic supplementary material, figure S1). Populations of D. melanogaster are genetically differentiated along the gradient for various phenotypic traits including thermal tolerance and life-history traits [20,21]. Frequencies of the Wolbachia infection wMel (Wolbachia pipientis) that causes weak cytoplasmic incompatibility vary stably along this gradient from a low frequency of infection in Tasmania to a high frequency in tropical Queensland [17,19], although infected and uninfected flies are found along the entire gradient. Offspring from individual field females were reared to the F1 or F3 generation (depending on the sample) before testing for virus. Field females were tested for Wolbachia and one offspring per female was used to establish the Wolbachia-positive and -negative pools. Samples were stored at −80°C.
(b). F1 fly rearing
Each F1 or F3 used to form the pooled sample was raised separately under the same conditions without a priori knowledge of Wolbachia status. All flies were cultured at a constant 19°C under 12 L : 12 D cycle in glass vials (50 mm height × 12 mm diameter) containing laboratory medium composed of dextrose (7.5% w/v), cornmeal (7.3% w/v), inactive yeast (3.5% w/v), soy flour (2% w/v), agar (0.6% w/v), methyl 4-hydroxybenzoate (1.6%) and acid mix (1.4%, 10 : 1 propionic acid : orthophosphoric acid).
(c). Wolbachia screening
Heads were removed from individual flies for the purpose of Wolbachia screening and to ensure sufficient tissue was available for subsequent RNA viral work. DNA was extracted from fly heads using the Chelex 100 Resin (Bio-Rad Laboratories, Hercules, CA, USA) method of Endersby et al. [22] but with the following alterations: tissue was ground in a Mixer Mill (Retsch MM300, Hahn, Germany) (25 Hz; 2 min) with four 2 mm glass beads (Lomb Scientifix, Taren Point, NSW, Australia; Cat. no. SIF1295/2G) and incubated at an elevated temperature and time (60°C for 2 h). DNA was diluted 1 : 3 or 1 : 10 with water and screened for Wolbachia infection using the RT/HRM assay (real-time PCR/high-resolution melt assay) first developed by Lee et al. [23] to detect wMel infection in A. aegypti. This assay was run on a Roche LightCycler® 480 system (using a 384-well format). The wMel target and wMel primers and the PCR reagent and cycling conditions used here to detect wMel in D. melanogaster are the same as those reported in Lee et al. [23]. However, we also developed primers for a Drosophila control gene for confirmation of both the host genus and quality of the DNA extraction prior to final typing of Wolbachia: Dros_RpL40_F (5′-CAA CTG CCG CAA GAA GAA GTG-3′) and Dros_RpL40_R (5′-CTA CTT CAA CTT CTT CTT GGG-3′) target the conserved RpL40 gene region in Drosophila species/detect the presence of Drosophila DNA and the amplified product is 64 bp. The Dros_RpL40 target was amplified with the same PCR reagent and cycling conditions as wMel.
Screened individuals were typed ‘wMel infected’ when robust amplification occurred for both primer pairs. A robust amplification was defined as a Cp or ‘crossing-point’ value less than 30 generated by the ‘Abs Quant/2nd Derivative Max’ quantification analysis mode and a Tm or ‘melting temperature’ within the acceptable range as determined by positive controls and previous light cycler results for D. melanogaster infected with wMel. Individuals were typed as ‘wMel uninfected’ when robust amplification in multiple replicates was observed for the Dros_RpL40 primers, no amplification or amplification attributed to ‘background’ only (blank Cp or Cp value of 35 and Tm outside the acceptable range) was observed for the wMel primers, and there was no amplification in controls lacking DNA. Melting temperatures for the two targeted gene regions (Dros_RpL40 and wMel) showed distinct light cycler profiles with a separation of at least 6°C. Wolbachia density was estimated for each individual by subtracting the Cp value of the Wolbachia gene from the Cp value of the control gene (Dros_RpL40) to allow a comparison of density in infected individuals from the different source populations. Wolbachia densities were compared using a non-parametric Kruskal–Wallis test.
(d). RNA extraction and sequencing
RNA sequencing was first performed on pooled flies. A total of 122 fly samples (entire individuals less head) were organized into 12 pools based on the wMel infection status and geographical locations. Each pool was washed three times with 1 ml sterile, RNA and DNA-free PBS solution (GIBCO). The samples were then homogenized in 600 µl lysis buffer for 1 min using TissueRuptor (Qiagen). Total RNA was then extracted using an RNeasy Plus Mini Kit (Qiagen) following the manufacturer's instructions. The quality of extracted RNA was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). The libraries were constructed using a TruSeq total RNA Library Preparation Kit (Illumina) with rRNA removed using a Ribo-Zero-Gold (Human-Mouse-Rat) Kit (Illumina). Paired-end (100 bp) sequencing of each RNA library was performed on the Hiseq2500 platform (Illumina). Detailed information of each library/pool is presented in electronic supplementary material, table S1.
To further examine the role of wMel in protecting the hosts against viruses, RNA sequencing was performed on individual flies. This analysis considered an additional 20 wMel-positive and 20 wMel-negative flies sampled from the Melbourne population, using the same procedures for RNA extraction, library preparation and sequencing as described above.
(e). Total transcriptome annotation and virus discovery
To reduce the possibility of in silico cross-library contamination, all sequence reads were de-multiplexed with 0 mismatches to the index sequences and quality screened using Trimmomatic [24]. De novo assembly of the remaining reads was performed using Trinity [25] with default parameters. The resulting contigs were then compared against the non-redundant nucleotide (nt) and non-redundant protein (nr) databases using the blastn and diamond blastx program, respectively, with e-value cut-offs of 1 × 10−10 and 1 × 10−5. For both blast searches, all hits to ‘Viruses’ (Taxonomy database) were retained. All virus contigs with unassembled overlaps were merged using the SeqMan program implemented in Lasergene v. 7.1 (DNAstar). The assembled virus genome sequences were then compared with reference genomes from the same virus species to check for assembly errors. To discover viruses at relatively low abundance, raw reads from each library were mapped to a genome dataset that contained all the Drosophila virus sequences documented to date [15,16].
(f). Sequence quantification
The abundance of viral genome or host/bacteria genes was estimated as the percentage of non-rRNA reads mapped to the corresponding sequences. The genome/gene sequences for mapping comprised: (i) all virus genomes detected in this study, (ii) the gapdh, rpl32 and cox1 genes of D. melanogaster, (iii) selected wMel protein coding genes, namely, cox1, recA and gyrB, and (iv) the small unit ribosomal rRNA (16S) genes for all bacteria (including wMel) identified in this study. For each library, we first removed any reads associated with host ribosomal RNA. The remaining reads were then mapped to these sequence sets using Bowtie2 [26] and inspected using the Integrated Genomics Viewer [27].
(g). Phylogenetic analysis
We used nucleotide sequences to determine the phylogenetic relationship among the viruses discovered in different Australian locations as well as those obtained from GenBank. For each viral species, complete or partial genome sequences were aligned using the E-INS-i algorithm implemented in MAFFT v. 7 [28]. Phylogenetic trees were inferred from these alignments using the maximum-likelihood method implemented in PhyML v. 3.0 [29], using the GTR+Γ substitution model and the Subtree Pruning and Regrafting branch-swapping algorithm. Support for individual nodes in the tree was assessed using an approximate likelihood ratio test in PhyML.
(h). Statistical analysis
As the data were not normally distributed, the protective effect of wMel on individual viral abundance was assessed using a Mann–Whitney U-test. The effect on individual viral presence/absence was tested using a Fisher exact test. A logistic regression was used to test for an association between virus presence/absence and wMel presence/absence after adjusting for virus type. Analyses were carried out in R: a language and environment for statistical computing, v. 3.4.2.
3. Results
(a). Diversity and distribution of RNA viruses in Australian Drosophila melanogaster
We first examined a total of 122 F1 and F3 D. melanogaster individuals derived from a 2008 sample collection along the east coast of Australia. The Wolbachia (wMel) infection status of each individual fly was determined using a real-time PCR assay prior to RNA extraction and sequencing. Based on the assay results, the flies were subsequently divided into 12 pools, representing ‘wMel-positive’ (wMel+) and ‘wMel-negative’ (wMel−) groups from six sampling locations: Innisfail, Maryborough, Coffs Harbour, Hunter Valley, Melbourne and Tasmania (electronic supplementary material, figure S1 and table S1). Each pool contained 10–12 individuals, from which high-quality total RNA (RIN > 7.7) was extracted and used to prepare ribosomal-rRNA depleted cDNA libraries. Sequencing of these libraries was then performed on an Illumina Hiseq2500 platform, with wMel+ and wMel− libraries assigned to separate lanes.
To confirm the genetic background of wMel strain used in this study, we mapped the RNA sequencing reads of each library to an MLST gene set (i.e. 16S, aspC, atpD, ftsZ, sucB, groEL and coxA) used in [11], all of which show 100% nucleotide identity to the reference wMel strain (AE017196) at regions with greater than 3× coverage. However, these genes are too conserved to distinguish between different wMel variants, such as the wMel prototype and wMel-CS strains that confer differing degrees of viral protection [10,11], although wMel-CS was not identified in recent sampling of D. melanogaster in eastern and southeastern Australia [19,30]. To distinguish between these different variants, we compared the genome sequences of existing variants and identified 31 genes that contained one or more positions that differ between the wMel prototype and wMel-CS. Among these, five genes (i.e. rplF, rplB, rpoBC, fusA and rpsC) with an average of greater than 10X coverage were compared. The results suggested that the wMel population examined here exhibited greater similarity to the wMel prototype than to wMel-CS (electronic supplementary material, figure S2), with the exception of the Tasmanian population that shared the same mutation with wMel-CS in the fusA and rpsC genes (electronic supplementary material, figure S2).
All sequencing reads were assembled de novo and compared against the non-redundant nucleotide (nt) and protein (nr) databases to determine the presence of RNA virus genomes as well as a number of stably expressed marker genes of D. melanogaster and wMel. In total, we discovered 11 RNA virus species across all libraries and their genomes were all sequenced to complete length. These viruses comprised nine families/floating genera from the positive-sense (n = 5), negative-sense (n = 1) and double-stranded RNA (n = 5) groups of RNA viruses (figure 1 and table 1). All these viruses have been previously described in D. melanogaster or related hosts [15,16]. Among them, we confirmed that Galbut virus (two segments) is in fact likely to comprise two additional segments of Drosophila-associated Partitiviridae-like virus 3 (one segment) [15], based on their synchronized appearance and matching abundance levels. We, therefore, used the name ‘Galbut virus’ to refer to all three segments. Similarly, we updated the genome of Torrey Pines virus from six to nine segments.
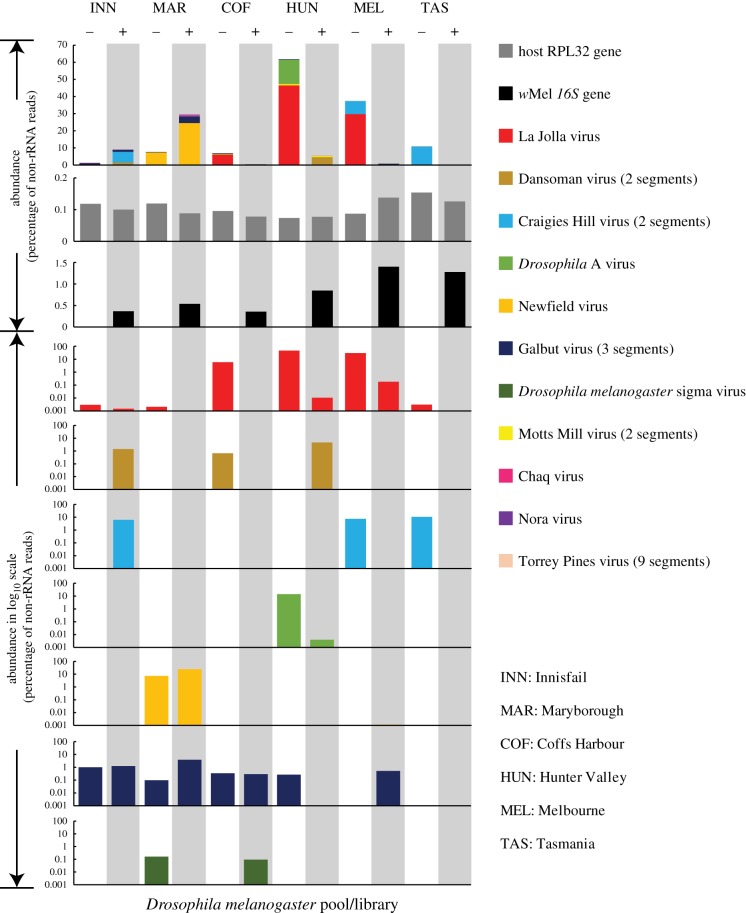
Figure 1.
Comparison of the natural RNA viromes in wMel+ and wMel− D. melanogaster populations from six Australian locations. The first three rows show the abundance of all viruses, the wMel 16S gene and the Drosophila rpl32 gene in Wolbachia-positive and -negative libraries, with viral abundance (i.e. percentage of total reads in the library) shown on the y-axis on a normal numeric scale. The 4th–10th rows show the abundance of individual virus species, with abundance on a logarithmic scale on the y-axis. For clarity, only seven higher abundance are presented. In all cases, we used one set of colour panels to represent different viral species (11 colours), the host gene (grey) and the Wolbachia gene (black). All Wolbachia-positive columns have a grey-shaded background.
Table 1.
The presence and abundance of viruses, bacteria (Wolbachia) and host genes (% total reads) in Australian D. melanogaster.
| organism/gene name | classification | genome architecture (RNA virus) | Innisfail |
Maryborough |
Coffs Harbour |
Hunter Valley |
Melbourne |
Tasmania |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wol− | wol+ | wol− | wol+ | wol− | wol+ | wol− | wol+ | wol− | wol+ | wol− | wol+ | |||
| La Jolla virus | Iflaviridae (Picornavirales) | positive-sense | 0.003 | 0.002 | 0.002 | 0.000 | 5.899 | 0.000 | 46.510 | 0.011 | 29.748 | 0.182 | 0.003 | 0.000 |
| Nora virus | Noravirus (Picornavirales) | positive-sense | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Dansoman virus (2 segments) | Tombusviridae-like | positive-sense | 0.000 | 1.444 | 0.000 | 0.000 | 0.668 | 0.000 | 0.000 | 4.602 | 0.000 | 0.000 | 0.000 | 0.000 |
| Craigies Hill virus (2 segments) | Nodaviridae-like | positive-sense | 0.000 | 6.249 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 7.555 | 0.000 | 10.733 | 0.000 |
| Motts Mill virus (2 segments) | Luteoviridae-like | positive-sense | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.790 | 0.775 | 0.000 | 0.000 | 0.000 | 0.000 |
| Drosophila A virus | Permutotetra-like | double-stranded | 0.001 | 0.000 | 0.001 | 0.000 | 0.001 | 0.000 | 14.164 | 0.004 | 0.000 | 0.000 | 0.001 | 0.000 |
| Newfield virus | Permutotetra-like | double-stranded | 0.000 | 0.001 | 7.320 | 24.454 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.001 | 0.000 | 0.001 |
| Galbut virus (or Drosophila-associated Partitiviridae-like 3, 3 segments) | Patitiviridae-like | double-stranded | 1.014 | 1.244 | 0.097 | 3.869 | 0.339 | 0.292 | 0.265 | 0.000 | 0.000 | 0.524 | 0.000 | 0.001 |
| Chaq virus | Patitiviridae-like | double-stranded | 0.255 | 0.000 | 0.000 | 1.219 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Torrey Pines virus (9 segments) | Reoviridae | double-stranded | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.150 | 0.000 | 0.000 | 0.000 |
| Drosophila melanogaster sigma virus | Rhabdoviridae | negative-sense | 0.000 | 0.000 | 0.160 | 0.000 | 0.000 | 0.094 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| all viruses | 1.272 | 8.940 | 7.580 | 29.543 | 6.907 | 0.388 | 61.751 | 5.392 | 37.454 | 0.708 | 10.737 | 0.002 | ||
| Drosophila melanogaster gapdh | Insecta (host) | 0.185 | 0.171 | 0.178 | 0.127 | 0.156 | 0.169 | 0.080 | 0.133 | 0.107 | 0.179 | 0.117 | 0.138 | |
| Drosophila melanogaster rpl32 | Insecta (host) | 0.118 | 0.100 | 0.119 | 0.088 | 0.095 | 0.078 | 0.074 | 0.077 | 0.087 | 0.138 | 0.154 | 0.126 | |
| Drosophila melanogaster cox1 | Insecta (host) | 1.164 | 1.532 | 1.719 | 1.172 | 1.541 | 2.048 | 0.698 | 2.195 | 0.844 | 1.676 | 0.874 | 1.123 | |
| Wolbachia pipientis wMel 16S | bacteria | 0.000 | 0.367 | 0.000 | 0.536 | 0.000 | 0.357 | 0.000 | 0.844 | 0.000 | 1.400 | 0.000 | 1.277 | |
| Wolbachia pipientis wMel cox1 | bacteria | 0.0000 | 0.0006 | 0.0000 | 0.0013 | 0.0000 | 0.0004 | 0.0000 | 0.0008 | 0.0000 | 0.0008 | 0.0000 | 0.0012 | |
| Wolbachia pipientis wMel recA | bacteria | 0.0000 | 0.0004 | 0.0000 | 0.0005 | 0.0000 | 0.0002 | 0.0000 | 0.0006 | 0.0000 | 0.0007 | 0.0000 | 0.0008 | |
| Wolbachia pipientis wMel gyrB | bacteria | 0.0000 | 0.0005 | 0.0000 | 0.0009 | 0.0000 | 0.0005 | 0.0000 | 0.0011 | 0.0000 | 0.0009 | 0.0000 | 0.0011 | |
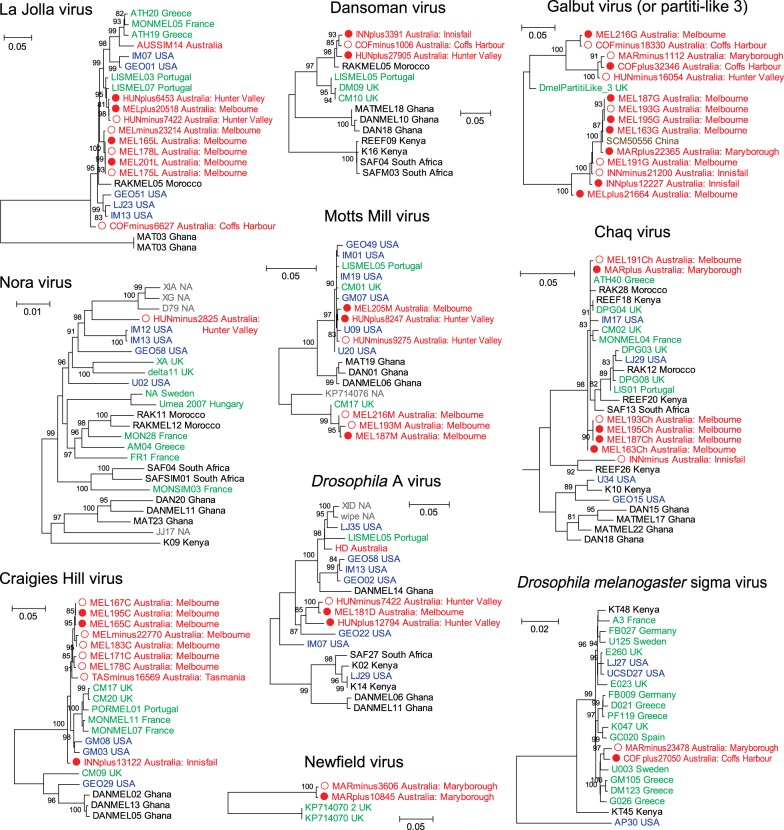
A further examination of the intra-species diversity of some of these viruses revealed a close relationship between viruses circulating in Australia and other geographical localities [15], although in some cases, the Australian sequences comprised separate lineages (figure 2). Importantly, these viruses are unlikely to represent endogenous viral elements because (i) none appear in the D. melanogaster genome, (ii) none appear in all libraries, and (iii) all are present as complete genomes without interruption by nonsense mutations, frame-shifts. The distribution of these viruses differed substantially among D. melanogaster populations. For example, La Jolla virus and Galbut virus had a wide distribution across different libraries, whereas Torrey Pines virus and Nora virus were only found in a single library (table 1). Generally, a single library contained two to five virus species, although in the case of the wMel-positive library from Tasmania, no viruses were detected with the exception of Newfield virus at a very low frequency (table 1).
Figure 2.
Evolutionary relationships and intra-species genetic diversity of the RNA viruses identified in this study and those from previous studies. Virus sequences from wMel-positive samples are indicated by red solid circles, while those from wMel-negative samples are shown by red hollow circles. The virus sequence names contain information on the strain name and sampling location, while the colours of sequence names indicate their geographical locations, including Australia (red), Africa (black), Europe (green), North America (blue), Asia (brown) or unknown (grey). All horizontal branch lengths are scaled to the number of nucleotide substitutions per site and trees are mid-point rooted.
(b). Virus abundance in relation to wMel status
For each library, we estimated virus abundance as the percentage of total RNA after the removal of fly rRNA (i.e. non-ribosomal total RNA). Interestingly, these results revealed that total viral abundance varied from 0% (wMel+, Tasmania) to 61.8% (wMel−, Hunter Valley) (figure 1), with nine of the 12 libraries showing relatively high abundance (greater than 1%, table 1). Hence, RNA viruses commonly comprise a substantial part of the host non-rRNA transcriptome, as seems to be true of invertebrates in general [31,32]. The great variation in viral RNA concentration is unlikely to be an artefact of sample processing or nucleic acid extraction because the examination of stably expressed host genes (rpl32 and gapdh genes) and the abundantly expressed host gene (cox1) revealed consistent RNA levels across different libraries (figure 1). In addition, all extracted RNA was at high concentration, with clear 18S and 28S rRNA peaks and RIN values greater than 7.7 (electronic supplementary material, table S1), suggesting that the extraction is of high quality and should include both viral and cellular RNA. Furthermore, we examined the expression of the 16S, cox1, recA and gyrB genes of wMel: the presence/absence of these genes was consistent with our experimental settings and confirmed the Wolbachia status within each library (figure 1 and table 1). In addition, the abundance levels of non-rRNA Wolbachia genes, which were comparable across different libraries, were much lower than those of either host genes or RNA viruses (table 1).
We next examined the influence of the presence/absence of wMel on the total abundance of viruses, particularly whether the presence of wMel is associated with a reduction in viral load. Generally, high viral titres (greater than 1% of non-ribosomal total RNA) were observed in both wMel-positive and wMel-negative groups, suggesting an overall lack of effect on viral abundance. A similar pattern was observed for individual viral species. With the exception of Nora virus and Torrey Pines virus, which only appeared once in the dataset, all other viruses appeared in both wMel-positive and wMel-negative libraries (table 1). In the case of Dansoman virus, Craigies Hill virus, Motts Mill virus and Galbut virus, there was no evidence for protection against high viral loads as relatively high abundance levels (greater than 1% or greater than 0.1% of total non-ribosomal RNA) were observed in both wMel-negative and wMel-positive libraries (figure 1). However, there was a trend suggesting that wMel may have protected against a high titre of La Jolla virus, a member of Iflaviridae family from the order Picornavirales. Specifically, among the 12 libraries, only wMel-negative libraries had ‘high’ titres of La Jolla virus: 5.9%, 46.5% and 29.7% for Coffs Harbour, Hunter Valley and Melbourne negative subpopulations, respectively. In comparison, their wMel-positive counter-parts from the matching geographical regions all possessed lower titres of La Jolla virus, with those from Innisfail at least two orders of magnitude lower (figure 1 and table 1).
(c). Transcriptome analysis of individual Drosophila melanogaster
As samples from each wMel group were pooled at each geographical location, there was insufficient statistical power to understand the effect of wMel on viral titre for each virus with sufficient precision. To increase analytical power, we performed a second round of RNA sequencing on un-pooled samples of individual D. melanogaster from the Melbourne population, which has the highest average wMel density and in theory the strongest virus blocking. The samples analysed comprised 20 wMel+ and 20 wMel– flies, which were subsequently sequenced with wMel+ and wMel– groups assigned to separate runs.
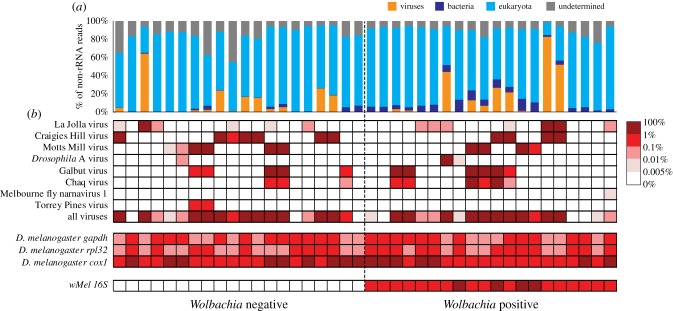
In general, these results demonstrated that individual D. melanogaster can experience multiple infections of up to five virus species simultaneously, and that they can contain extremely high viral loads (up to 82.65% of total non-ribosomal RNA) under natural conditions and regardless of wMel infection status (figure 3; electronic supplementary material, table S2). Indeed, high overall viral loads (greater than 1% of total non-ribosomal RNA) were observed in 11 wMel− individuals and 10 wMel+ individuals, respectively (figure 3). Similarly, at the level of viral species, both wMel+ and wMel– individuals experienced high viral titres of La Jolla virus (one wMel− versus two wMel+ individuals), Craigies Hill virus (six versus four), Motts Mill virus (three versus three) and Galbut virus (two versus six) (figure 3). Notably, for several other virus species, only wMel+ individuals experienced high viral titre, including Drosophila A virus (n = 1) and Chaq virus (n = 2), although the numbers were small (figure 3). It is also noteworthy that Chaq virus always co-occurs with Galbut virus, although in some of the libraries, only Galbut virus was found (figure 3). This suggests that Chaq virus might be a satellite virus of Galbut virus.
Figure 3.
Comparison of the RNA viromes of 20 wMel+ and 20 wMel– individual D. melanogaster. (a) A comparison of the relative abundance of major taxonomic groups, including viruses (orange), bacteria (dark blue), eukaryotes (light blue) and undetermined (grey). (b) A heat map showing the abundance level for all virus genomes, the Drosophila marker genes (gapdh, rpl32 and cox1) and the wMel 16S gene in wMel+ and wMel− individuals.
We next examined the ability of wMel to reduce the abundance of RNA viruses in individual flies. As very low virus abundance may be a product of index swapping in the sequencing runs, we tested the association between wMel and virus abundance and virus presence/absence at thresholds of 1, 0.1 and 0.01% of the total non-ribosomal RNA. This revealed no evidence of wMel protection against viral load for all viruses combined (p = 0.88, odds ratio (OR) = 1.05, 95% CI = 0.57–1.95) or for individual viruses including La Jolla virus (table 2). Collectively, these results suggest wMel is unlikely to release D. melanogaster from the intensive replication and transcription of viral nucleic acid, although it remains unclear if high viral RNA loads impose large fitness costs on their hosts [33,34].
Table 2.
The effect of the presence/absence of wMel on the presence/abundance of viruses at different levels of virus abundance. n.a. indicates cases in which there were insufficient samples to perform the calculation.
| virus | abundance p-value |
presence/absence p-value (OR, 95% CI) |
||||
|---|---|---|---|---|---|---|
| threshold | 0.01 | 0.1 | 1 | 0.01 | 0.1 | 1 |
| La Jolla virus | 0.19 | 0.57 | 0.57 | 0.27 (0.34, 0.05–1.84) | 1 (0.48, 0.01–10.02) | 1 (0.48, 0.01–10.02) |
| Craigies Hill virus | 0.34 | 0.34 | 0.49 | 0.48 (2.11, 0.42–12.15) | 0.48 (2.11, 0.42–12.15) | 0.72 (1.69, 0.32–9.94) |
| Motts Mill virus | 0.81 | 0.98 | 0.79 | 1 (1.32, 0.23–8.04) | 1 (1, 0.16–6.4) | 1 (1.4, 0.2–11.13) |
| Drosophila A virus | 0.57 | 0.34 | 0.34 | 1 (0.48, 0.01–10.02) | 1 (0, 0–39) | 1 (0, 0–39) |
| Galbut virus | 0.50 | 0.50 | 0.17 | 1 (0.78, 0.15–3.89) | 1 (0.78, 0.15–3.89) | 0.41 (0.34, 0.03–2.47) |
| Chaq virus | 0.18 | 0.09 | 0.16 | 0.45 (0.42, 0.06–2.41) | 0.24 (0.27, 0.02–1.8) | 0.49 (0, 0–5.28) |
| Melbourne fly narnavirus 1 | 0.34 | n.a. | n.a. | 1 (0, 0–39) | n.a. | n.a. |
| Torrey Pines virus | 0.16 | 0.16 | n.a. | 0.49 | 0.49 | n.a. |
| total | 0.70 | 0.80 | 0.92 | 0.69 (1.86, 0.3–14.05) | 0.75 (1.5, 0.36–6.57) | 1 (1.22, 0.3–5.04) |
(d). Impact of Wolbachia density
It is possible that the effects of wMel on virus abundance differ because of variation in Wolbachia density, with the expectation that higher Wolbachia density increases viral blocking [10,35]. We, therefore, estimated average Wolbachia density in pooled wMel+ flies from the different locations as well as from individual flies. Density differences among sites in the pooled data were non-significant for host (Kruskal–Wallis test, χ2 = 9.35, d.f. = 4, p = 0.053) but significant for wMel (χ2 = 20.580, d.f. = 5, p = 0.001) as well as for the Cp differences between host and wMel (χ2 = 14.494, d.f. = 5, p = 0.013), which suggests there is a variation of wMel density between different sites. However, it is evident from our results that higher average Wolbachia density was not necessarily associated with lower viral abundance for the six locations studied here (Pearson's r = −0.528, CI: −0.938–0.496, p = 0.2815, figure 2; electronic supplementary material, tables S3 and S4). In addition, there was no significant association based on the individual fly samples from Melbourne (Pearson's r = 0.087, CI: −0.370–0.510, p = 0.7157, figure 3; electronic supplementary material, table S5), where flies with high viral abundance exhibited relatively high Wolbachia densities. Hence, these results suggest that Wolbachia density does not impact the abundance of the natural D. melanogaster virome in the populations studied here.
(e). Impact of phylogenetic diversity
Finally, we examined whether the presence of the wMel shapes patterns and levels of intra-specific virus genetic variation, comparing the virus genome sequences from wMel+ flies with those from wMel− flies. Based on the sequence alignment, we found no consistent nucleotide substitutions corresponding to either the presence or absence of wMel. This was also apparent from the phylogenetic analyses, where there was no evidence of clustering according to wMel status, with the virus sequences obtained from both positive and negative populations showing mixed distributions on the trees (figure 2).
4. Discussion
The anti-viral properties of Wolbachia have been tested against a wide range of viruses under experimental conditions, and it is clear that there is substantial variation according to both Wolbachia strain and host background [6,10,11]. Although the wMel strain does not have the strongest blocking phenotype, it has been repeatedly demonstrated that in the case of D. melanogaster it is associated with an increased survival rate, although not necessarily a significant reduction in viral load [8,11]. Interestingly, those viruses in which the reduction in virus load associated with wMel is greatest—DCV and Nora virus—are both picorna-like viruses, although Nora virus is currently classified as a floating genus [36]. Two picorna-like viruses were also present in our dataset—Nora virus and La Jolla virus. While the Nora virus dataset is uninformative because it only appeared in one wMel– library and with low virus titre (figure 1), La Jolla virus appeared in the majority of the libraries, and it is important to note that Wolbachia presence was associated with neither a reduction in prevalence nor load of this virus. Hence, not all picorna-like viruses may be affected by Wolbachia in the same way. This again suggests that there is no universal protective effect against a broad range of viruses and further narrows the diversity of those viruses whose abundance is significantly reduced by the presence of wMel.
Alternatively, it is possible the lack of virus blocking by wMel observed here is due to differences in the response to the natural Drosophila virome, or with virus strains established in the field compared to those introduced only recently or under experimental settings. Under natural conditions, the lack of reduction in viral load may reflect a complex intra-host interaction shaped by co-adaptation of host, virus and bacterial symbionts over long time periods [37]. Indeed, the antiviral effect of Wolbachia often appears to be strongest following their transinfection into new host species [38–40], whereas weaker effects have been observed in the case of longer evolutionary associations between arthropods and Wolbachia [12,41,42]. Over time, viruses may evolve resistance to Wolbachia blocking, although we cannot exclude the possibility that those viruses that are most susceptible to Wolbachia infection have already been cleared from the population and hence were not detected in this study. As such, our study may have implications for the long-term effect of Wolbachia release programmes on viral eradication, even though the presence of wMel following introduction into A. aegypti [2] mosquitoes so far appears stable following field release [7].
Although Wolbachia density is reported to be negatively correlated with virus density [11,12], we did not observe such an effect in our data. However, even under experimental conditions, Wolbachia density alone may not explain the blocking against all virus strains [11]. Indeed, when comparing different Wolbachia strains, wAu results in a more significant reduction in viral titre compared to wMel, although the two strains are close in density values [11]. Furthermore, wAu induces a reduction in viral titre in the case of FHV, whereas wMel failed to deliver the same effect. It will, therefore, be interesting to determine how a more potent strain, such as wAu or wMel-CS, impacts virome prevalence and abundance under natural conditions.
Our study also shows that individual D. melanogaster are characterized by an exceptionally high prevalence rate and virus titre, although they share a similar diversity of viruses as revealed in a previous survey of this species [15]. High viral abundance is not uncommon in arthropods, and it has been repeatedly demonstrated that levels of viral RNA can exceed those of the most abundantly expressed non-rRNA genes such as cox1 gene to the extent that they represent a major part of the transcriptome [31,32,43]. It is still unclear how such high levels of viral abundance impact flies and their well-being, although it has been suggested that high viral abundances may reduce host fitness [33,34]. However, it has also been shown that Wolbachia might increase host tolerance to high viral loads. Specifically, the host survival rate with viruses increases substantially in the presence of wMel, although the viral load of FHV remained similar to that of the control (i.e. no wMel) group [11]. Such tolerance, however, is expected to result in an increase in viral abundance and prevalence in Wolbachia-infected Drosophila populations, which is not observed in the data generated here.
Finally, our study examined the anti-viral effect of Wolbachia using a meta-transcriptomics approach, simultaneously characterizing the virome, microbiome and host RNA transcriptome [32]. As this method provides a comprehensive overview of all the essential transcriptomic information within each individual D. melanogaster, be it from viruses, bacteria or the host, it may be a useful way to reveal the mechanisms that underpin Wolbachia-mediated virus blocking, either experimentally or in nature.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Rayners Orchard (Woori Yallock) for access to samples and Kelly Richardson for assistance in sample collection and rearing. We thank the reviewers for insightful comments.
Data accessibility
All sequence reads generated are available on the NCBI Sequence Read Archive (SRA) database under the BioProject accession PRJNA454554. All virus genome sequences have been assigned GenBank accession nos. MH384268–MH384387. Sequence alignments and phylogenetic trees are deposited in figshare under the link: https://doi.org/10.6084/m9.figshare.c.4159325.
Authors' contributions
A.A.H. and E.C.H. contributed to conceptualization; M.S., V.L.W., T.S., J.-S.E., A.A.H. and E.C.H. contributed to methodology; M.S., V.L.W., T.S., J.-S.E., A.A.H. and E.C.H. contributed to investigation; M.S. and E.C.H. contributed to writing—original draft; M.S., V.L.W., T.S., J.-S.E., A.A.H. and E.C.H. contributed to writing—review and editing. All authors gave their final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by NHMRC Program grant 1037003 and NIH grant no. 1R01GM104325-01. E.C.H. is funded by an ARC Australian Laureate Fellowship (FL170100022) and A.A.H. by an NHMRC Research Fellowship (1118640).
References
- 1.Hoffmann AA. 1988. Partial cytoplasmic incompatibility between two Australian populations of Drosophila melanogaster. Entomol. Exp. Appl. 48, 61–67. ( 10.1111/j.1570-7458.1988.tb02299.x) [DOI] [Google Scholar]
- 2.Walker T, et al. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453. ( 10.1038/nature10355) [DOI] [PubMed] [Google Scholar]
- 3.Dutra HL, Rocha MN, Dias FB, Mansur SB, Caragata EP, Moreira LA. 2016. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe 19, 771–774. ( 10.1016/j.chom.2016.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser JE, De Bruyne JT, Iturbe-Ormaetxe I, Stepnell J, Burns RL, Flores HA, O'Neill SL.. 2017. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog. 13, e1006751 ( 10.1371/journal.ppat.1006751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP.. 2018. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathog. 14, e1006815 ( 10.1371/journal.ppat.1006815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann AA, Ross PA, Rasic G. 2015. Wolbachia strains for disease control: ecological and evolutionary considerations. Evol. Appl. 8, 751–768. ( 10.1111/eva.12286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frentiu FD, Zakir T, Walker T, Popovici J, Pyke AT, van den Hurk A, McGraw EA, O'Neill SL. 2014. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl. Trop. Dis. 8, e2688 ( 10.1371/journal.pntd.0002688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teixeira L, Ferreira A, Ashburner M.. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6, e1000002 ( 10.1371/journal.pbio.1000002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne SE, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL, Johnson KN. 2012. Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl. Environ. Microbiol. 78, 6922–6929. ( 10.1128/AEM.01727-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chrostek E, Marialva MS, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L.. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9, e1003896 ( 10.1371/journal.pgen.1003896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez J, Longdon B, Bauer S, Chan YS, Miller WJ, Bourtzis K, Teixeira L, Jiggins FM. 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog. 10, e1004369 ( 10.1371/journal.ppat.1004369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu P, Bian G, Pan X, Xi Z.. 2012. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl. Trop. Dis. 6, e1754 ( 10.1371/journal.pntd.0001754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer WH, Medd N, Beard PM, Obbard DJ. 2018. Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. PLoS Pathog. 14, e1007050 ( 10.1371/journal.ppat.1007050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brun G, Plus N. 1978. The viruses of Drosophila. In The genetics and biology of Drosophila (eds Ashburner M, Wright TRF), pp. 625–702. New York, NY: Academic Press. [Google Scholar]
- 15.Webster CL, et al. 2015. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol . 13, e1002210 ( 10.1371/journal.pbio.1002210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster CL, Longdon B, Lewis SH, Obbard DJ. 2016. Twenty-five new viruses associated with the Drosophilidae (Diptera). Evol. Bioinform. Online 12(Suppl. 2), 13–25. ( 10.4137/EBO.S39454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann AA, Clancy DJ, Merton E. 1994. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 136, 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riegler M, Sidhu M, Miller WJ, O'Neill SL. 2005. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 15, 1428–1433. ( 10.1016/j.cub.2005.06.069) [DOI] [PubMed] [Google Scholar]
- 19.Kriesner P, Conner WR, Weeks AR, Turelli M, Hoffmann AA. 2016. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70, 979–997. ( 10.1111/evo.12923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitrovski P, Hoffmann AA. 2001. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. R. Soc. B 268, 2163–2168. ( 10.1098/rspb.2001.1787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann AA, Weeks AR. 2007. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica 129, 133–147. ( 10.1007/s10709-006-9010-z) [DOI] [PubMed] [Google Scholar]
- 22.Endersby NM, McKechnie SW, Vogel H, Gahan LJ, Baxter SW, Ridland PM, Weeks AR. 2005. Microsatellites isolated from diamondback moth, Plutella xylostella (L.), for studies of dispersal in Australian populations. Mol. Ecol. Notes 5, 51–53. ( 10.1111/j.1471-8286.2004.00827.x) [DOI] [Google Scholar]
- 23.Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. 2012. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Appl. Environ. Microbiol. 78, 4740–4743. ( 10.1128/AEM.00069-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. ( 10.1093/bib/bbs017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 30.Riegler M, O'Neill SL, Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E.. 2006. The genus Wolbachia. In The prokaryotes: volume 5: proteobacteria: alpha and beta subclasses (eds S Falkow, E Rosenberg, K-H Schleifer, E Stackebrandt), pp. 547–561. New York, NY: Springer. [Google Scholar]
- 31.Shi M, et al. 2016. Redefining the invertebrate RNA virosphere. Nature 540, 539–543. ( 10.1038/nature20167) [DOI] [PubMed] [Google Scholar]
- 32.Shi M, Neville P, Nicholson J, Eden JS, Imrie A, Holmes EC. 2017. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in western Australia. J. Virol. 91, e00680-17 ( 10.1128/JVI.00680-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yampolsky LY, Webb CT, Shabalina SA, Kondrashov AS. 1999. Rapid accumulation of a vertically transmitted parasite triggered by relaxation of natural selection among hosts. Evol. Ecol. Res. 1, 581–589. [Google Scholar]
- 34.Wilfert L, Jiggins FM. 2013. The dynamics of reciprocal selective sweeps of host resistance and a parasite counter-adaptation in Drosophila. Evolution 67, 761–773. ( 10.1111/j.1558-5646.2012.01832.x) [DOI] [PubMed] [Google Scholar]
- 35.Stevanovic AL, Arnold PA, Johnson KN. 2015. Wolbachia-mediated antiviral protection in Drosophila larvae and adults following oral infection. Appl. Environ. Microbiol. 81, 8215–8223. ( 10.1128/AEM.02841-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy: 9th report of the international committee on taxonomy of viruses. San Diego, CA: Elsevier Academic Press. [Google Scholar]
- 37.Wang A. 2015. Dissecting the molecular network of virus–plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. ( 10.1146/annurev-phyto-080614-120001) [DOI] [PubMed] [Google Scholar]
- 38.Bian G, Xu Y, Lu P, Xie Y, Xi Z.. 2010. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6, e1000833 ( 10.1371/journal.ppat.1000833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blagrove MS, Arias-Goeta C, Failloux AB, Sinkins SP. 2012. Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc. Natl Acad. Sci. USA 109, 255–260. ( 10.1073/pnas.1112021108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagrove MS, Arias-Goeta C, Di Genua C, Failloux AB, Sinkins SP.. 2013. A Wolbachia wMel transinfection in Aedes albopictus is not detrimental to host fitness and inhibits Chikungunya virus. PLoS Negl. Trop. Dis. 7, e2152 ( 10.1371/journal.pntd.0002152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mousson L, Martin E, Zouache K, Madec Y, Mavingui P, Failloux AB. 2010. Wolbachia modulates Chikungunya replication in Aedes albopictus. Mol. Ecol. 19, 1953–1964. ( 10.1111/j.1365-294X.2010.04606.x) [DOI] [PubMed] [Google Scholar]
- 42.Mousson L, Zouache K, Arias-Goeta C, Raquin V, Mavingui P, Failloux AB.. 2012. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl. Trop. Dis. 6, e1989 ( 10.1371/journal.pntd.0001989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Remnant EJ, Shi M, Buchmann G, Blacquiere T, Holmes EC, Beekman M, Ashe A. 2017. A diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 91, e00158-17 ( 10.1128/JVI.00158-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence reads generated are available on the NCBI Sequence Read Archive (SRA) database under the BioProject accession PRJNA454554. All virus genome sequences have been assigned GenBank accession nos. MH384268–MH384387. Sequence alignments and phylogenetic trees are deposited in figshare under the link: https://doi.org/10.6084/m9.figshare.c.4159325.