Abstract
Introductions of non-native lineages increase opportunities for hybridization. Non-native lineages of the common wall lizard, Podarcis muralis, are frequently introduced in cities where they hybridize with native populations. We aimed at unravelling the invasion history and admixture of native and non-native wall lizards in four German cities using citywide, comprehensive sampling. We barcoded and genotyped 826 lizards and tested if gene flow in populations composed of admixed native and introduced lineages is facilitated by similar environmental factors to those in native populations by comparing fine-scale landscape genetic patterns. In cities with non-native lineages, lizards commonly occurred in numerous clusters of hybrid swarms, which showed variable lineage composition, consisting of up to four distinct evolutionary lineages. Hybrid swarms held vast genetic diversity and showed recent admixture with other hybrid swarms. Landscape genetic analyses showed differential effects of cityscape structures across cities, but identified water bodies as strong barriers to gene flow in both native and admixed populations. By contrast, railway tracks facilitated gene flow of admixed populations only. Our study shows that cities represent unique settings for hybridization, caused by multiple introductions of non-native taxa. Cityscape structure and invasion histories of cities will determine future evolutionary pathways at these novel hybrid zones.
Keywords: urban biodiversity, species distribution models, landscape resistance, dispersal, corridor, reptile
1. Introduction
In cities, individuals experience unique selective forces that can make long-term persistence of populations particularly difficult [1–3]. Anthropogenic factors such as roads, traffic, human disturbances or buildings may act as barriers to gene flow within cities [4,5]. Consequently, genetic drift may increase differentiation of urban populations [6–9] through the combined effects of isolation and reduction in population size. Nonetheless, many species thrive in cities [5,10,11], such as urban resident and human commensal species [12], and may disperse well throughout the cityscape [13–16].
The effect of barriers on gene flow can be quantified using landscape genetics. Studies on urban gene flow have shown disparate results depending on the species' ecology. For example, green spaces facilitate gene flow in European hedgehogs, Erinaceus europaeus [17], white footed mice, Peromyscus leucopus [14] and striped field mice, Apodemus agrarius [18]. Canopy cover has been identified as a barrier to gene flow in common wall lizards, Podarcis muralis [16] and the garden snail, Cornu aspersum [19]. Finally, rivers represent corridors for fire salamanders, Salamandra salamandra [20], but barriers to European hedgehogs and common wall lizards [16,17]. Interestingly, major anthropogenic structures (e.g. buildings) appear to be less important for gene flow in cities than expected, although buildings hampered gene flow in common wall lizards and the garden snail [16,19]. On the contrary, multi-scale analyses in the garden snail revealed that transportation infrastructure, such as roads, can enhance urban gene flow [19]. These few examples show that the way in which environmental factors act as barriers or corridors is expected to vary between species because of their different habitat requirements and dispersal abilities. However, few landscape genetic studies used replicated settings for a single species [5,21], and the single one on gene flow within cities demonstrated differential effects of environmental factors across urban landscape replicates [19]. This suggests that the idiosyncratic structures of cityscapes may play a major role affecting urban gene flow.
Introductions of non-native lineages (species, subspecies or other evolutionary divergent units) are frequent in cities [22,23]. Concerns have been put forward warning of a global biotic homogenization driven by the spread of invasive species and urbanization [24], while native species decrease [25,26]. In addition to the changes in communities, introductions have also led to an unprecedented number of encounters between lineages previously separated by natural barriers [27–29]. These encounters facilitate hybridization and initiate gene flow between previously isolated lineages, leading to genetic homogenization [30–32].
Hybridization is a key process shaping the genetic structure of populations [23,33,34]. It is defined as ‘reproduction between members of genetically distinct populations, producing offspring of mixed ancestry’, which applies to both inter- and intraspecific interactions [35]. While founder effects usually increase genetic drift and differentiation caused by a loss of genetic diversity [36], hybridization increases genetic diversity [37]. Depending on hybrid fitness, fitness of parental taxa and relative hybrid abundances, hybridization can result in heterosis and adaptive introgression [38], formation of stable hybrid zones [39], admixture, formation of hybrid swarms [40] or hybrid speciation [35]. A recent review on outcomes of hybridization caused by anthropogenic habitat disturbance most commonly detected admixture and the emergence of hybrid swarms [41]. Hybrid swarms are characterized by extensive admixture of parental taxa, whose genotypes are no longer present but have merged into a unique population of hybrids [42]. They are an intermediate evolutionary phase, which may lead to complete genomic extinction of parental taxa or formation of hybrid species [40]. Therefore, they are interesting systems for the study of evolutionary trajectories. However, despite the importance of urban areas for novel species interactions, hybridization has so far rarely been studied in cities (but see a study on chipmunks [43]).
To examine environmental drivers of hybridization in cities, the common wall lizard is a well-suited study species. These lizards are common in cities [44] and introductions of non-native evolutionary lineages within cities have been reported frequently [45]. Furthermore, hybridization readily occurs upon secondary contact of lineages, potentially leading to the formation of hybrid swarms [34,46,47]. The processes that involve the formation of a hybrid swarm will not only depend on the invasion history, but also on environmental characteristics of the respective cityscape. As environmental characteristics vary among cityscapes, they can influence dispersal of lineages and determine opportunities for admixture. Environmental drivers have been shown to facilitate the spread of a non-native wall lizard population along railway tracks in a non-urban area in southern Germany [48], while a river was identified as a major barrier to gene flow in the native population of the city of Trier, western Germany [16]. However, it remains unknown whether landscape genetic results can be generalized across cities [19]. Furthermore, the extent of introductions and the degree of admixture have not been fully quantified within cities.
Making use of the unique and dynamic situation of common wall lizards within and among German cities, we barcoded and genotyped 826 lizards from four German cities, all within the range of the native ‘Eastern France’ evolutionary lineage [34]. We chose two cities to represent only native populations and two cities with a known presence of non-native evolutionary lineages [34]. We aimed to (1) unravel the invasion history within cities and quantify past and present admixture by determining the evolutionary origin of individuals using mitochondrial haplotypes, identifying genotypic population clusters and by quantifying contemporary admixture between these clusters. Furthermore, (2) we employed landscape genetic analyses to test which environmental factors determine gene flow within cities, to compare the extent to which the landscape and environmental predictors of gene flow are consistent across cities and how these drivers relate to patterns of admixture and invasion history. With regard to the invasion history (1), we expected to find genetic clusters within cities that either represent only the native lineage, only non-native lineages or admixed hybrid swarms of native and non-native lineages. We predicted finding four different types of ongoing admixture: (i) between pure clusters of individuals belonging to the same evolutionary lineage (admixture type 0), (ii) between two pure clusters of different evolutionary origin (admixture-type 1), (iii) between a pure single-lineage cluster and a hybrid swarm of multiple lineage origin (admixture-type 2), and (iv) between two hybrid swarms (admixture-type 3). For the landscape genetics part (2), we hypothesized that for both native and non-native populations, (v) water cover would represent the most important barrier to gene flow, and (vi) buildings and canopy cover additionally increase landscape resistance, albeit to a lesser degree. For non-native populations, we predicted that (vii) railway tracks and adjacent habitats would enhance gene flow.
2. Material and methods
(a). Species account
The common wall lizard, P. muralis, is a small lacertid lizard with a snout–vent length of up to 8 cm and a body weight of 4–10 g. It is well adapted to rocky habitats, including artificial stone walls and is distributed throughout southern Europe, from Spain to Turkey and southern Italy to southwestern Germany. The species comprises a large number of distinct evolutionary lineages stemming from multiple Mediterranean refugia [49,50]. At its northern-most distribution, the common wall lizard frequents urban habitats and disturbed sites, such as road margins, stone quarries or railway tracks [44,48]. It has successfully established populations outside its natural range in Switzerland, Austria, the UK, Canada and the USA [44], and 106 non-native populations have been confirmed in Germany [45]. Multiple evolutionary lineages of the common wall lizard co-occur in several German cities [34], and hybridization is common upon secondary contact [34,46,47].
(b). Field methods
All four studied cities are located at the northern range margin of the species (with a mean of 140 km ± s.d. 52 km between cities; electronic supplementary material, figure S1), where the mitochondrial Eastern France lineage is native [49]. Trier (TR) and Saarbrücken (SB) presumably harboured only native populations, while Freiburg (FR) and Mannheim (MA) were known to contain mixed introduced populations [34].
We followed the same method as previously applied to the native Trier population [16], the results of which are included for comparison. We produced fine-scale distribution maps covering the respective citywide distribution of lizards, from which 200 random sampling points were produced across each city, weighted by the relative local abundance of lizards (details are given in electronic supplementary material, section 1). Individual-based sampling schemes are regarded superior to population-based sampling in continuously distributed species and in areas of uncertain dispersal success, where a priori assignment of populations is difficult or even impossible [51–53]. Sampling took place from April to July 2012 in Mannheim (N = 203), from July to September 2012 and April to May 2013 in Trier (N = 223), and from April to September 2013 in Saarbrücken (N = 197) and Freiburg (N = 203). Multiple samplings of single individuals were avoided by sampling each locality within cities only once or within a few days and using colour marks that persisted for approximately two to three weeks. Furthermore, if a locality had to be resampled at a later stage, previously taken individual photographs were referred to in order to avoid duplicate sampling.
(c). Barcoding evolutionary lineages
We obtained DNA by buccal swabbing individuals using sterile dry swabs (Copan Diagnostics Inc., ‘Sterile R’) [54]. Samples were stored within 12 h of collection at −20°C until DNA extraction. DNA extraction was carried out according to the manufacturer's protocol of the Qiagen DNEasy blood and tissue kit. Evolutionary lineages were identified using a 500 bp sequence of the mitochondrial cytochrome b gene for a total of 819 individuals (221 out of 223 from Trier; all 203 from MA; 195 out of 197 from Saarbrücken; 200 out of 203 from Freiburg). Sequences were aligned with sequences of known native geographical origin of all lineages known to have established in Germany, and a phylogenetic tree was constructed using Podarcis siculus and Podarcis melisellensis as outgroups. We applied a maximum composite likelihood substitution model and the neighbour-joining method with 2000 bootstrap replicates to assign lineages using MEGA6 [55]. Details for PCR conditions and GenBank accession numbers of used sequences are given in the electronic supplementary material (section 2).
(d). Microsatellite analysis of population structure
All individuals were genotyped at 17 microsatellite loci. Details on loci, PCR conditions and determination of fragment lengths are given in electronic supplementary material, section 2. Population structure was inferred using STRUCTURE (v2.3.4; [56]). We used the correlated allele frequency model and the admixture model in STRUCTURE and ran Markov chain Monte Carlo simulations with a burn-in of 100 000 followed by 1 000 000 simulations. First, we ran simulations across cities for K = 1–25 with 10 iterations per K. Following a hierarchical approach, we continued exploration within cities with the same settings but with K = 1–15. We used STRUCTURE HARVESTER [57] to determine the second-order rate of change (ΔK) [58]. However, the ΔK method has recently been shown to underestimate population structure, identifying only the highest level of genetic differentiation even within hierarchically structured populations [59]. Therefore, we adopted the approach by Schulte et al. [34] to detect further population structure beyond max ΔK, accepting the highest value of K that produces clusters that all contain at least one individual with a Q-value of at least 0.9. This method has been shown to produce similar results to independent structure runs within structure clusters [34]. Q is the assignment probability for each individual to a cluster (scaled from 0 to 1). Clusters to which no individuals were assigned with a high probability are therefore ignored [34]. Replicate STRUCTURE runs were combined in CLUMPP to identify optimal clustering solutions across runs (v.1.1.2; [60]) using standard parameter settings and FullSearch algorithm for K = 2–3, Greedy for K = 4–7 and FullKGreedy for K ≥ 8. We plotted results for spatial representation using ArcGIS (v.10.2.1 ©Esri Inc.). For each genetic cluster detected by STRUCTURE, we calculated the number of mtDNA lineages and estimated π-diversity using the maximum composite likelihood model [61]. Using GenAlEx (v.6.501; [62]), we calculated a principal coordinate analysis based on genotypic distances, unbiased expected heterozygosity, allelic richness, FST-values and deviations from Hardy–Weinberg equilibrium (HWE). Deviations from HWE were detected in between one and 11 clusters per locus (or between zero and six loci per cluster), but no locus showed a general pattern of deviation from HWE. Deviations from HWE were expected, as populations still in the process of expansion and admixture are unlikely to be in equilibrium. Using FSTAT (v.2.9.3.2; [63]), no genotypic disequilibrium was detected within cities at the 5% significance level based on 2400 permutations.
(e). Analyses of parental clusters and admixture-types
Owing to the unknown invasion histories of urban wall lizard populations, we inferred clusters, determined their lineage composition and distinguished four different admixture-types. The Q-values generated for each individual by CLUMPP formed the basis for this. In a first step, we characterized ‘parental cluster’ by referring only to individuals with Q ≥ 0.9 for any given cluster. If all individuals of a parental cluster carried haplotypes of the same mtDNA lineage, it was considered of single lineage origin in further analyses. If two or more mtDNA lineages were found among individuals of a parental cluster, it was considered a hybrid swarm. Admixture-types were calculated for all individuals with maximum Q-values of less than 0.8. We applied this conservative threshold of Q-values, omitting individuals with Q-values between 0.8 and 0.9, to prevent overestimation of admixed individuals [34,64]. The following four admixture-types were distinguished: admixture-type 0 indicates admixture between two clusters consisting of individuals belonging to the same evolutionary lineage. Admixture-type 1 stems from two single-lineage clusters of two different evolutionary lineages. Admixture-type 2 is admixture of a single-lineage cluster with a hybrid swarm, i.e. a cluster of multiple lineage origin. Finally, admixture-type 3 is admixture of two hybrid swarms. Of these four admixture-types, we expected to find admixture-type 0 in Trier and Saarbrücken because these cities are thought to be comprised of only native populations. In other cities, admixture-type 0 may also be found, either between native or non-native populations. Admixture-type 1 should be most prevalent if multiple introductions of different lineages had occurred. Admixture-types 2 and 3 should prevail when hybridization has been common between lineages, and the resulting hybrid swarms constitute either one or both parental clusters, respectively. This should be dependent on the degree of previous hybridization, and the latter type is presumably unique to areas with frequent introductions and exceptionally high levels of hybridization among lineages.
(f). Landscape and species distribution modelling
We digitized the sampling extent for Trier, Saarbrücken, Mannheim and Freiburg using the world imagery embedded in ArcGIS (basemap; taken on 11 August 2012; ArcGIS v10.2.1; Esri Inc.) at a scale of 1:2000 for all environmental factors. For further analyses, we converted the digitized layers of environmental factors into a grid layer using the majority rule in ArcGIS. The grid size was set to 25 m × 25 m, with each grid cell covering 625 m2. This scaling results in a reasonable computation time. It also represents the scale at which wall lizards can be expected to assess habitat quality according to available information on home-range sizes of P. muralis of up to 50 m2, which regularly change between years [44]. We created 10 of the 12 layers specifically for this study, to encompass all habitat requirements essential for P. muralis. Environmental factors included: (i) aspect; (ii) substrate type; (iii) canopy cover; (iv) vegetation type; (v) structural diversity; (vi) south-facing walls; (vii) buildings; (viii) roads; (ix) traffic volume; (x) water cover; (xi) railway tracks. Furthermore, we created (xii) habitat suitability maps (SDMs). We used a presence-only (PO) method (Maxent v.3.3.3 [65]) to build SDMs. We applied a procedure to prevent overestimation of resistances due to effects of artificial boundaries [66]. More details on environmental factors and grid creation are given in the electronic supplementary material, section 3, and in section 4 for SDMs.
(g). Landscape genetic analysis: optimization of grids
Effects of environmental factors on gene flow were assessed using pairwise genetic distances of individuals and the R-package ResistanceGA v.3.1-1 [67]. We calculated Nei's genetic distance [68] between individuals using Alleles in Space (v.1.0) as a proxy for functional connectivity [69]. Employing a genetic algorithm, ResistanceGA transforms resistance surfaces to maximize fit to the specified genetic distances, based on AICc values of linear mixed effect models [67]. This circumvents typical issues of subjectivity in assigning resistance values and makes a wider parameter space disposable for the process of optimization [70,71]. It uses the commuteDistance function of the R-package gdistance v.1.2-1 [72] to calculate pairwise resistance distances between individuals similar to CIRCUITSCAPE [73]. As only one sample location can be specified per grid cell, and due to small inter-sample distances, we thinned samples randomly (Trier = 199; Saarbrücken = 128; Mannheim = 193; Freiburg = 161). As recommended, ResistanceGA was run twice for each environmental factor and included null and distance models for [67]. Runs were checked for convergence and AICc values were compared between runs for each environmental factor. There were mostly marginal differences in AICc values between runs and no changes in the ranks of the best-performing factors. We report the highest AICc scores for all models within cities, marginal R2 values of models (R2m) and average rank of models across cities. Additional analyses became necessary when optimized SDMs, termed PO-optim models, ranked highest within cities. To identify environmental factors most relevant within PO-optim models [16], all environmental factors were correlated to the PO-optim model and, additionally, resistance values of the PO-model were extracted per subcategory of environmental factors to identify those subcategories with high or low values.
3. Results
(a). Invasion history and admixture of native and non-native wall lizards
Native haplotypes were found in all cities (Trier: 100% of individuals, Saarbrücken: 54.9%, Mannheim: 9.9% and Freiburg: 34%). We detected two non-native lineages in Saarbrücken, three in Mannheim and four in Freiburg; this was unexpected for Saarbrücken, which was supposed to harbour only a native population (see above). Among the introduced lineages, the Western France lineage was encountered at low frequencies in all cities (up to 5% per city), the Southern Alps lineage occurred in all cities at higher frequencies (Saarbrücken: 41.5%, Mannheim: 68%, Freiburg: 28.5%), the Venetian lineage was found in Mannheim and Freiburg (19.7% and 1%, respectively) and the Tuscany lineage was detected only in Freiburg, but at a relatively high frequency (31.5%).
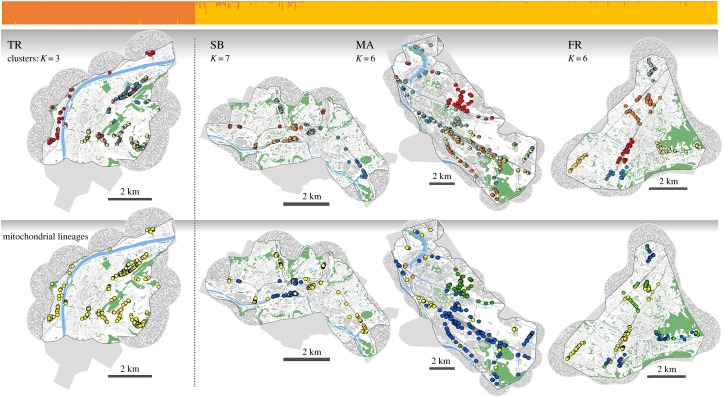
Combined Bayesian clustering of all cities showed the strongest differentiation at K = 2, based upon maximum ΔK (figures 1 and 2), and the largest likelihood estimate at K = 17 (electronic supplementary material, figure S2). Within cities, ΔK was maximal at K = 2 in Trier, Saarbrücken, Mannheim and K = 4 in Freiburg (electronic supplementary material, figure S3). Using the Q-value approach, we identified K = 3 in Trier, K = 7 in Saarbrücken and K = 6 in Freiburg and Mannheim (electronic supplementary material, figure S3 shows plots of ΔK and L(K) per city). The spatial distribution of these clusters is shown in figure 2. Cities with non-native lineages had a stronger population genetic structure than the native population in Trier (K = 3, versus K = 6–7). Saarbrücken and Mannheim also had higher intra-city FST values between clusters (Kruskal–Wallis χ2 = 23.12, d.f. = 3, p < 0.001; electronic supplementary material, figure S4).
Figure 1.
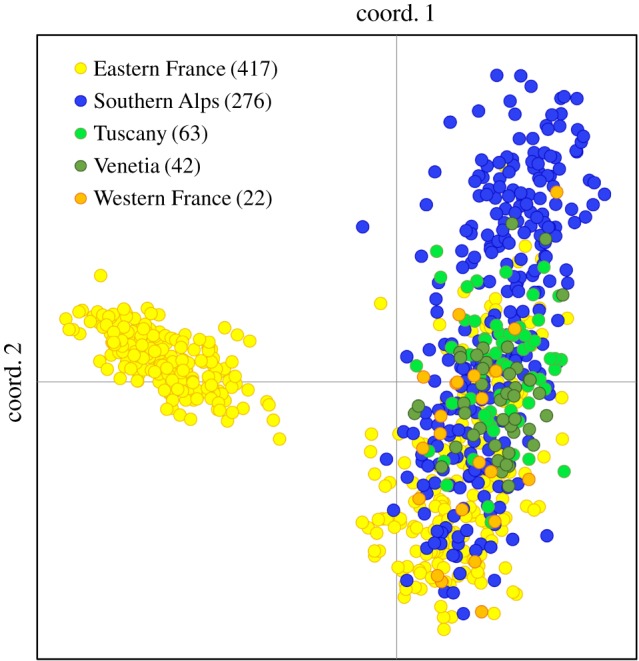
All lizards of the four cities are shown in a PCoA, calculated from genotypic genetic distances. The evolutionary lineage of lizard haplotypes is depicted by different colours, the native lineage being the Eastern France lineage (yellow). Lizards from Trier (native individuals only), group separately at the left. The other three cities also contain introduced lizards belonging to non-native evolutionary lineages and are grouped on the right side of the plot. In these cities, high levels of hybridization between lizards of all lineages have led to the emergence of multiple hybrid swarms.
Figure 2.
Molecular results for all lizards of the four cities. A barplot at the top, above the maps, depicts genotypic results for K = 2 (ΔK = max; generated using CLUMPP) showing strongest differentiation between the city of Trier (TR: native haplotypes only) and all other cities where non-native lineages have also been found (SB: Saarbrücken; MA: Mannheim; FR: Freiburg; see electronic supplementary material, figure S1 for geographical location in Germany). The upper row of maps shows genotypic results of analyses conducted within cities, at values of K above ΔK = max; pie-charts represent cluster membership of individuals at the location of capture (based on Q-values). Lower maps show evolutionary lineages of lizards (cyt-b haplotype). The native Eastern France lineage is shown in yellow, while all other colours depict non-native lineages: Southern Alps (blue); Venetian (dark green); Tuscany (light green); Western France (orange). Within all maps, water is shown in blue, canopy cover in green. Linear, dark grey structures depict railway tracks and buildings are shown in a lighter grey. Outside of coloured maps, uniform grey areas show the sampling extent where no lizards were found. Areas without lizards were omitted from landscape genetic analyses if they fell outside of a 1 km buffer around sampled individuals. Buffers became necessary due to specifics of landscape genetic analyses and if they extended beyond the sampling extent, we generated random environmental data (represented by ‘noisy’ grey pixels inside buffers; details are given in the electronic supplementary material, section 3).
Across cities, 13 of 22 clusters were classified as hybrid swarms, i.e. being composed of two or more evolutionary lineages (electronic supplementary material, table S1). At least one single lineage cluster was found in all cities, but of different origin. In Trier, only single lineage clusters were found, all of native origin. In Saarbrücken, the only single lineage cluster was of native origin. All four single-lineage clusters in Mannheim were of non-native origin (Southern Alps and Venetian lineages). In Freiburg, one single lineage cluster of native origin was detected. With the exception of the one cluster in Saarbrücken and all clusters in Trier, we also found admixture between each single-lineage cluster and a cluster of another lineage or a hybrid swarm. Genetic diversity within clusters reflected lineage composition and levels of π-diversity, and unbiased expected heterozygosity and allelic richness all increased significantly with the number of lineages present in hybrid swarms (Pearson's correlation for π-diversity: r = 0.88, d.f. = 20, p < 0.001; for HE: r = 0.638, d.f. = 20, p = 0.001; for allelic richness: r = 0.543; d.f. = 20; p = 0.009).
Admixture-types calculated for clusters based on maximum ΔK and higher values of K showed similarly high levels of admixture-types 2 + 3 in cities with introduced lizards, while distribution of admixture-types at maximum ΔK values was less nuanced (electronic supplementary material, table S2). As justified above, we will focus on results for higher values of K per city. As presumed, all individuals in Trier were of admixture-type 0. In Saarbrücken, all admixed individuals were of admixture-types 2 and 3, i.e. with hybrid swarms as either one or both parental clusters, respectively. In Mannheim, all admixture-types were found, with type 2 being most frequent (type 0 = 10.3%; type 1 = 22.1%; type 2 = 64.7%; type 3 = 2.9%). In Freiburg, all admixed individuals were of admixture-types 2 and 3, as in Saarbrücken. With 69.2% in Saarbrücken and 91.3% in Freiburg, admixture-type 3 dominated in both cities.
(b). Landscape predictors of gene flow and population structure of native and introduced wall lizards
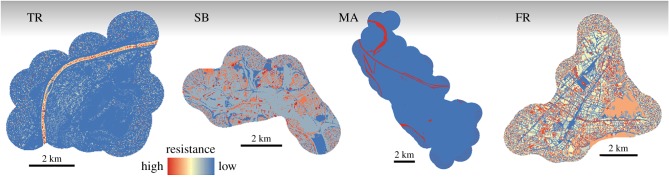
In all cities, the null model ranked lowest. The distance model ranked lower than a large proportion of environmental factors, varying in number between cities (11 environmental factors ranked above the distance model in Trier, 4 in Saarbrücken, 10 in Mannheim and Freiburg, see electronic supplementary material, table S3; all optimized models are shown in electronic supplementary material, figures S5–S8). The best model for gene flow varied among cities (figure 3). The PO-optim model performed best in Trier (marginal R2 = 0.1, figure 3TR), vegetation type in Saarbrücken (R2m = 0.04, figure 3SB), water cover in Mannheim (R2m = 0.66, figure 3MA) and the PO-optim model in Freiburg (R2m = 0.41, figure 3FR). The underlying environmental factors of the PO-optim model were easily determined in Trier. The three environmental factors correlating most strongly to the PO-optim model also contained subcategories, which received resistance values most deviant from the overall median resistance value of the PO-optim model (1.0): water cover was a major barrier (resistance value of 171.3; r = 0.83) and to a lesser degree also buildings (34.1; r = 0.383) and canopy cover (28.9; r = 0.329) reduced gene flow (more details are given in [16]). In Saarbrücken, semi-natural, ruderal vegetation facilitated gene flow and received 27 times lower resistance values than other types of vegetation. In Mannheim, water cover, as a barrier to gene flow, received resistance values 305 times that of non-water surfaces. In Freiburg, correlation coefficients and resistance values of the underlying environmental factors of the PO-optim model were less congruent. In ascending order, walls (r = −0.238), buildings (r = −0.25), structural diversity (r = −0.279), substrate (r = −0.291) and railway tracks (r = 0.295) correlated most strongly with the PO-optim model, but coefficients were generally low (electronic supplementary material, table S4). Of these environmental factors, resistance values of subcategories were lower in comparison to the overall median resistance of 77.8 of the PO-optim model (electronic supplementary material, table S5). Resistance values below 2.5 suggest railway tracks, rocky + gravel substrate type as well as low and medium structural diversity facilitate gene flow. The highest median resistance value of all subcategories was registered for high traffic volume (182.5), indicating a barrier effect of traffic on gene flow, while the correlation coefficient to the PO-optim model was low (r = 0.024).
Figure 3.
Best models of gene flow for each city, following landscape genetic optimization with ResistanceGA. Colours of plots are scaled to highest and lowest resistance values (from red to blue) within each city. The PO-optim model performed best in Trier (TR), which is derived from a transformed species distribution model. It depicts foremost water cover and to a lesser degree buildings and canopy cover to impede gene flow. In Saarbrücken (SB), vegetation type was the best model, with semi-natural, ruderal vegetation facilitating gene flow. In Mannheim (MA), water cover was the single best model, representing a barrier to gene flow. In Freiburg (FR), the PO-optim model performed best. Lowest resistance values were assigned to railway tracks, rocky and gravel substrate type and areas of low and intermediate structural diversity, while high traffic volume received highest resistance values.
Averaging ranks of models across cities, water cover ranked first, followed by PO-optim models, traffic and railway tracks (electronic supplementary material, table S6). Water cover was among the top three models in Trier, Saarbrücken and Mannheim. Water cover had high resistance values and acted as a barrier to gene flow, but resistance values decreased with average river width (Pearson's correlation: r = 0.976; d.f. = 2; p = 0.023). Railway tracks were among the top three models in Mannheim and Freiburg, but did not perform better than the distance model in Trier and Saarbrücken. Railway tracks received higher resistance values than the surrounding matrix in the native population (Trier: 1.8 times), while they enhanced gene flow in all other cities: this effect was lower in Saarbrücken (2.2 times lower resistance values) than in Mannheim and Freiburg (80 and 40 times lower, respectively). Electronic supplementary material, figures S5–S8 show all optimized models per city.
4. Discussion
(a). Introductions and admixture
Our results show that introductions of non-native wall-lizards are more frequent than expected in German cities, and proportions of non-native haplotypes are extremely high in Saarbrücken, Freiburg and Mannheim (45%, 65% and 90%, respectively; from here on referred to as ‘admixed populations’). This was unexpected for Saarbrücken, which we had included for comparison as a second ‘native population’. In admixed populations, the high number of hybrid swarm clusters (13 out of 19) indicates extensive hybridization following introduction events. The spatial distribution of mitochondrial haplotypes within cities suggests that the number of introduction events greatly exceeds the number of mitochondrial lineages. Intraspecific hybridization upon introductions from multiple non-native sources has been documented previously, e.g. in seven out of eight Anolis species in Florida [74] and in nine out of 23 populations of common wall lizards in England [46]. In contrast to these two studies, cities sampled here are situated within the native range of common wall lizards. When we combined all populations in a single STRUCTURE analysis, the highest level of differentiation (max. ΔK) clustered all individuals of admixed populations together but separate from the native population. As postulated previously [34], our data emphasize a fast genetic displacement of the native gene pool in admixed populations (figures 1 and 2).
Following patterns of recurrent admixture in cities consisting of multiple evolutionary lineages, the most common admixture-types were type 3 (43.6%), namely admixture between two hybrid swarms, and type 2 (42.3%), i.e. admixture between a single-lineage cluster and a hybrid swarm. However, cities varied in the proportion of admixture-types, and all types were found in Mannheim, as predicted by a scenario where a combination of admixture, founder effects and multiple introductions shape population structure. In Saarbrücken and Freiburg, admixture-type 3 dominates, and genetically differentiated clusters of hybrid swarms prevail. In both cities, admixture dominates population structure, and, as expected, intra-city FST values are lower compared to Mannheim, while only significantly so for Freiburg (electronic supplementary material, figure S4). This suggests that in Saarbrücken and Freiburg, rapid admixture of hybrid swarms leads to a completely new genetic compositions of the population, whereas the river as a major barrier in Mannheim temporarily maintains genetic population structure with different evolutionary legacies in different parts of the city.
Contrary to natural hybrid zones, the hybrid swarms encountered here frequently consist of multiple lineages (six out of 13 clusters contain haplotypes of three to four lineages; electronic supplementary material, table S1), some of them not occurring parapatrically in their native ranges [49]. Although populations that contain up to three evolutionary lineages have previously been found in the common wall lizard [46], the hybrid swarms encountered here occur at very fine spatial scales, e.g. within only hundreds of metres. Furthermore, lineage composition varies between hybrid swarms. For example, all five hybrid swarms in Freiburg are composed of a different set of evolutionary lineages. Potentially, frequent human-mediated introductions in cities are prerequisite to these novel hybrid zones. If the elevated genetic diversity of hybrid swarms relates to adaptive traits, they have high potential to adapt to ecological conditions. Common wall lizards have been demonstrated to adapt quickly to changing environments [75] and testing persistence and local adaptation, e.g. to urban environmental stressors [76], of hybrid swarms could be a promising direction of future research. Little is known about the long-term persistence of hybrid swarms as typically, only single points in time have documented their existence [41].
(b). Comparative cityscape genetics
Similar to a study on garden snails, the only landscape genetic study that also analysed replicate urban landscapes for a single species [19], our results highlight locally important environmental drivers for intraspecific gene flow. The PO-optim models explained gene flow best in Trier and Freiburg, vegetation type in Saarbrücken and water cover in Mannheim (figure 3). Whether PO-optim models only perform well in settings with high genetic–environmental associations remains unknown, but Trier and Freiburg had significantly lower intra-city FST-values than the other cities. The environmental factors we identified also shape gene flow in other species: water cover is a strong barrier to gene flow in Trier (figure 3TR) and Mannheim (figure 3MA) and also in European hedgehogs [17]. Gene flow of several small mammal species is facilitated by green spaces [14,17,18], and semi-natural vegetation was least resistant to gene flow in Saarbrücken (figure 3SB). While most studies that have demonstrated a negative effect of roads on gene flow were conducted outside urban environments [4], high traffic volume was most resistant to gene flow in Freiburg (figure 3FR).
However, despite the different drivers of gene flow between cities, we also detected common patterns. Water cover received between three and 305 times higher resistance values than non-water cover in cities (electronic supplementary material, figures S5–S8). Therefore, the hypothesized barrier effect of water surfaces is confirmed across cities and we assume that variation in the strength of this barrier is caused by differences in river width. While a strong correlation between resistance values of water cover and river width supports this, this association requires further corroboration due to the low sample size of four cities. In contrast to the native population, railway tracks in admixed populations had 2–80 times lower resistance values than surrounding areas. Thus, our data reveal differential utility of railway tracks and highlight the importance of these cityscape elements for gene flow in non-native populations only. Because of their suitability as habitats for the wall lizard, railway embankments probably play a major role for dispersal of non-native wall lizards in cities [48]. Intriguingly, gene flow in the native population does not benefit from railway tracks. Speculatively, either colonization took place before railway tracks were built in Trier or differential dispersal by hybrids, e.g. spatial sorting, is responsible for this contrasting patterns [77].
5. Conclusion
In the Anthropocene, introductions of non-native taxa are common in cities. Therefore, cities are likely to become major playgrounds for hybridization. Using a comparative cityscape genetic approach, we demonstrated recurrent and extensive admixture in all cities after introduction of non-native evolutionary lineages. Consistently, the native lineage was genetically displaced in all three cities with introduced populations. Furthermore, admixture between hybrid swarms of differential lineage composition frequently occurred at small spatial scales. Based upon our models, we expect introduced lizards to spread along railway tracks, and hybridize with other wall lizard populations, regardless of their evolutionary origin. While water cover is a strong barrier to gene flow within cities, even wide rivers do not prevent hybridization. Subsequently, human-mediated introductions have repeatedly led to the emergence of novel hybrid zones and these novel admixture processes will determine future evolutionary pathways of urban lizards.
Supplementary Material
Acknowledgements
We would like to thank Marc Johnson and James Santangelo as well as two anonymous reviewers for their recommendations, which greatly improved this manuscript. We also owe thanks to William Peterman for valuable suggestions on his R package ResistanceGA and the NABU Mannheim for making available data on local lizard distribution.
Ethics
Permits for catching and sampling of lizards were obtained from the local authorities (Trier: SGD-Nord 425-104.211.1202 and 425-104.211.1303; Saarbrücken: ZfB/DD 15/02/2012 and 30/01/2013; Mannheim: 56b-8851.15/Reptilien; Freiburg: 55-8851.44/030).
Data accessibility
Input files for ResistanceGA and microsatellite genotypes are available on Dryad (http://dx.doi.org/10.5061/dryad.p564p61) [78].
Authors' contributions
A.H., J.B. and M.V. designed the research. J.B. conducted field and laboratory work. J.B. and S.F. conducted analyses. J.B. wrote a first draft of the paper and all authors contributed substantially to revisions.
Competing interests
We declare we have no competing interests.
Funding
J.B. was funded by the DFG interdisciplinary research training group 1319.
References
- 1.Björklund M, Ruiz I, Senar JC. 2010. Genetic differentiation in the urban habitat: the great tits (Parus major) of the parks of Barcelona city. Biol. J. Linn. Soc. 99, 9–19. ( 10.1111/j.1095-8312.2009.01335.x) [DOI] [Google Scholar]
- 2.Rézouki C, Dozières A, Le Cœur C, Thibault S, Pisanu B, Chapuis J-L, Baudry E. 2014. A viable population of the European red squirrel in an urban park. PLoS ONE 9, e105111 ( 10.1371/journal.pone.0105111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sumasgutner P, Nemeth E, Tebb G, Krenn HW, Gamauf A. 2014. Hard times in the city—attractive nest sites but insufficient food supply lead to low reproduction rates in a bird of prey. Front. Zool. 11, 48 ( 10.1186/1742-9994-11-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holderegger R, Di Giulio M. 2010. The genetic effects of roads. A review of empirical evidence. Basic Appl. Ecol. 11, 522–531. ( 10.1016/j.baae.2010.06.006) [DOI] [Google Scholar]
- 5.Johnson MTJ, Munshi-South J. 2017. Evolution of life in urban environments. Science 358, eaam8327 ( 10.1126/science.aam8327) [DOI] [PubMed] [Google Scholar]
- 6.Noël S, Ouellet M, Galois P, Lapointe F-J. 2007. Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv. Genet. 8, 599–606. ( 10.1007/s10592-006-9202-1) [DOI] [Google Scholar]
- 7.Munshi-South J, Zolnik CP, Harris SE. 2016. Population genomics of the Anthropocene. Urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evol. Appl. 9, 546–564. ( 10.1111/eva.12357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lourenço A, Álvarez D, Wang IJ, Velo-Antón G. 2017. Trapped within the city: integrating demography, time since isolation and population-specific traits to assess the genetic effects of urbanization. Mol. Ecol. 26, 1498–1514. ( 10.1111/mec.14019) [DOI] [PubMed] [Google Scholar]
- 9.Evans KL, et al. 2009. Independent colonization of multiple urban centres by a formerly forest specialist bird species. Proc. R. Soc. B 276, 2403–2410. ( 10.1098/rspb.2008.1712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sol D, Lapiedra O, González-Lagos C. 2013. Behavioural adjustments for a life in the city. Anim. Behav. 85, 1101–1112. ( 10.1016/j.anbehav.2013.01.023) [DOI] [Google Scholar]
- 11.Harris SE, Munshi-South J. 2017. Signatures of positive selection and local adaptation to urbanization in white-footed mice (Peromyscus leucopus). Mol. Ecol. 26, 6336–6350. ( 10.1111/mec.14369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonnell MJ, Hahs AK. 2015. Adaptation and adaptedness of organisms to urban environments. Annu. Rev. Ecol. Evol. Syst. 46, 261–280. ( 10.1146/annurev-ecolsys-112414-054258) [DOI] [Google Scholar]
- 13.Angold PG, et al. 2006. Biodiversity in urban habitat patches. Sci. Total Environ. 360, 196–204. ( 10.1016/j.scitotenv.2005.08.035) [DOI] [PubMed] [Google Scholar]
- 14.Munshi-South J. 2012. Urban landscape genetics: canopy cover predicts gene flow between white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 21, 1360–1378. ( 10.1111/j.1365-294X.2012.05476.x) [DOI] [PubMed] [Google Scholar]
- 15.Brashear WA, Ammerman LK, Dowler RC. 2015. Short-distance dispersal and lack of genetic structure in an urban striped skunk population. J. Mammal. 96, 72–80. ( 10.1093/jmammal/gyu004) [DOI] [Google Scholar]
- 16.Beninde J, Feldmeier S, Werner M, Peroverde D, Schulte U, Hochkirch A, Veith M. 2016. Cityscape genetics: structural vs. functional connectivity of an urban lizard population. Mol. Ecol. 25, 4984–5000. ( 10.1111/mec.13810) [DOI] [PubMed] [Google Scholar]
- 17.Braaker S, Kormann U, Bontadina F, Obrist MK. 2017. Prediction of genetic connectivity in urban ecosystems by combining detailed movement data, genetic data and multi-path modelling. Landsc. Urban Plan. 160, 107–114. ( 10.1016/j.landurbplan.2016.12.011) [DOI] [Google Scholar]
- 18.Gortat T, Rutkowski R, Gryczyńska A, Pieniążek A, Kozakiewicz A, Kozakiewicz M. 2015. Anthropopressure gradients and the population genetic structure of Apodemus agrarius. Conserv. Genet. 16, 649–659. ( 10.1007/s10592-014-0690-0) [DOI] [Google Scholar]
- 19.Balbi M, Ernoult A, Poli P, Madec L, Guiller A, Martin M-C, Nabucet J, Beaujouan V, Petit EJ. 2018. Functional connectivity in replicated urban landscapes in the land snail (Cornu aspersum). Mol. Ecol. 27, 1357–1370. ( 10.1111/mec.14521) [DOI] [PubMed] [Google Scholar]
- 20.Straub C, Pichlmüller F, Helfer V. 2015. Population genetics of fire salamanders in a pre-Alpine urbanized area (Salzburg, Austria). Salamandra 51, 245–251. [Google Scholar]
- 21.Manel S, Holderegger R. 2013. Ten years of landscape genetics. Trends Ecol. Evol. 28, 614–621. ( 10.1016/j.tree.2013.05.012) [DOI] [PubMed] [Google Scholar]
- 22.Pysek P. 1998. Alien and native species in Central European urban floras. A quantitative comparison. J. Biogeogr. 25, 155–163. ( 10.1046/j.1365-2699.1998.251177.x) [DOI] [Google Scholar]
- 23.Brennan AC, Woodward G, Seehausen O, Muñoz-Fuentes V, Moritz C, Guelmami A, Abbott RJ, Edelaar P. 2014. Hybridization due to changing species distributions: adding problems or solutions to conservation of biodiversity during global change? Evol. Ecol. Res. 16, 475–491. [Google Scholar]
- 24.McKinney ML. 2006. Urbanization as a major cause of biotic homogenization. Biol. Conserv. 127, 247–260. ( 10.1016/j.biocon.2005.09.005) [DOI] [Google Scholar]
- 25.Aronson MFJ, et al. 2014. A global analysis of the impacts of urbanization on bird and plant diversity reveals key anthropogenic drivers. Proc. R. Soc. B 281, 20133330 ( 10.1098/rspb.2013.3330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinney ML. 2008. Effects of urbanization on species richness: a review of plants and animals. Urban Ecosyst. 11, 161–176. ( 10.1007/s11252-007-0045-4) [DOI] [Google Scholar]
- 27.Rhymer JM, Simberloff D. 1996. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27, 83–109. ( 10.1146/annurev.ecolsys.27.1.83) [DOI] [Google Scholar]
- 28.Helmus MR, Mahler DL, Losos JB. 2014. Island biogeography of the Anthropocene. Nature 513, 543–546. ( 10.1038/nature13739) [DOI] [PubMed] [Google Scholar]
- 29.Ashton GV, Davidson IC, Geller J, Ruiz GM. 2016. Disentangling the biogeography of ship biofouling: barnacles in the Northeast Pacific. Glob. Ecol. Biogeogr. 25, 739–750. ( 10.1111/geb.12450) [DOI] [Google Scholar]
- 30.Mooney HA, Cleland EE. 2001. The evolutionary impact of invasive species. Proc. Natl Acad. Sci. USA 98, 5446–5451. ( 10.1073/pnas.091093398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crispo E, Moore J-S, Lee-Yaw JA, Gray SM, Haller BC. 2011. Broken barriers. Human-induced changes to gene flow and introgression in animals. Bioessays 33, 508–518. ( 10.1002/bies.201000154) [DOI] [PubMed] [Google Scholar]
- 32.Olden JD, Leroy Poff N, Douglas MR, Douglas ME, Fausch KD. 2004. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol. Evol. 19, 18–24. ( 10.1016/j.tree.2003.09.010) [DOI] [PubMed] [Google Scholar]
- 33.Kolbe JJ, Larson A, Losos JB, de Queiroz K. 2008. Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol. Lett. 4, 434–437. ( 10.1098/rsbl.2008.0205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulte U, Veith M, Hochkirch A. 2012. Rapid genetic assimilation of native wall lizard populations (Podarcis muralis) through extensive hybridization with introduced lineages. Mol. Ecol. 21, 4313–4326. ( 10.1111/j.1365-294X.2012.05693.x) [DOI] [PubMed] [Google Scholar]
- 35.Abbott R, et al. 2013. Hybridization and speciation. J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 36.Michaelides SN, Goodman RM, Crombie RI, Kolbe JJ, Cowie R. 2017. Independent introductions and sequential founder events shape genetic differentiation and diversity of the invasive green anole (Anolis carolinensis) on Pacific Islands. Divers. Distrib. 13, 75. [Google Scholar]
- 37.Kolbe JJ, Glor RE, Rodríguez Schettino L, Lara AC, Larson A, Losos JB. 2004. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181. ( 10.1038/nature02807) [DOI] [PubMed] [Google Scholar]
- 38.Song Y, Endepols S, Klemann N, Richter D, Matuschka F-R, Shih C-H, Nachman MW, Kohn MH. 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr. Biol. 21, 1296–1301. ( 10.1016/j.cub.2011.06.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hewitt GM. 1988. Hybrid zones—natural laboratories for evolutionary studies. Trends Ecol. Evol. 3, 158–167. ( 10.1016/0169-5347(88)90033-X) [DOI] [PubMed] [Google Scholar]
- 40.Brumfield RT. 2010. Speciation genetics of biological invasions with hybridization. Mol. Ecol. 19, 5079–5083. ( 10.1111/j.1365-294X.2010.04896.x) [DOI] [PubMed] [Google Scholar]
- 41.Grabenstein KC, Taylor SA. 2018. Breaking barriers. Causes, consequences, and experimental utility of human-mediated hybridization. Trends Ecol. Evol. 33, 198–212. ( 10.1016/j.tree.2017.12.008) [DOI] [PubMed] [Google Scholar]
- 42.Allendorf FW, Leary RF, Spruell P, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16, 613–622. ( 10.1016/S0169-5347(01)02290-X) [DOI] [Google Scholar]
- 43.Frare CF, Matocq MD, Feldman CR, White AM, Manley PN, Jermstad KD, Hekkala ER. 2017. Landscape disturbance and sporadic hybridization complicate field identification of chipmunks. J. Wildl. Manage. 81, 248–258. ( 10.1002/jwmg.21193) [DOI] [Google Scholar]
- 44.Schulte U. 2008. Die Mauereidechse. erfolgreich im Schlepptau des Menschen. Bielefeld, Germany: Laurenti-Verlag. [Google Scholar]
- 45.Schulte U, Deichsel G. 2015. Eingeschleppte Mauereidechsen in Deutschland—ein Überblick mit Empfehlungen zum naturschutzfachlichen Umgang. In Verbreitung, Biologie und Schutz der Mauereidechse Podarcis muralis (LAURENTI, 1768) (ed. Laufer H, Schulte U), pp. 74–85. Mannheim, Germany: DGHT e. V. [Google Scholar]
- 46.Michaelides S, While GM, Bell C, Uller T. 2013. Human introductions create opportunities for intra-specific hybridization in an alien lizard. Biol. Invasions 15, 1101–1112. ( 10.1007/s10530-012-0353-3) [DOI] [Google Scholar]
- 47.While GM, et al. 2015. Sexual selection drives asymmetric introgression in wall lizards. Ecol. Lett. 18, 1366–1375. ( 10.1111/ele.12531) [DOI] [PubMed] [Google Scholar]
- 48.Schulte U, Veith M, Mingo V, Modica C, Hochkirch A. 2013. Strong genetic differentiation due to multiple founder events during a recent range expansion of an introduced wall lizard population. Biol. Invasions 15, 2639–2649. ( 10.1007/s10530-013-0480-5) [DOI] [Google Scholar]
- 49.Salvi D, Harris DJ, Kaliontzopoulou A, Carretero MA, Pinho C. 2013. Persistence across Pleistocene ice ages in Mediterranean and extra-Mediterranean refugia: phylogeographic insights from the common wall lizard. BMC Evol. Biol. 13, 147 ( 10.1186/1471-2148-13-147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gassert F, et al. 2013. From southern refugia to the northern range margin: genetic population structure of the common wall lizard, Podarcis muralis. J. Biogeogr. 40, 1475–1489. ( 10.1111/jbi.12109) [DOI] [Google Scholar]
- 51.Shirk AJ, Wallin DO, Cushman SA, Rice CG, Warheit KI. 2010. Inferring landscape effects on gene flow: a new model selection framework. Mol. Ecol. 19, 3603–3619. ( 10.1111/j.1365-294X.2010.04745.x) [DOI] [PubMed] [Google Scholar]
- 52.Prunier JG, Kaufmann B, Fenet S, Picard D, Pompanon F, Joly P, Lena JP. 2013. Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations. Towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol. Ecol. 22, 5516–5530. ( 10.1111/mec.12499) [DOI] [PubMed] [Google Scholar]
- 53.Landguth EL, Schwartz MK. 2014. Evaluating sample allocation and effort in detecting population differentiation for discrete and continuously distributed individuals. Conserv. Genet. 15, 981–992. ( 10.1007/s10592-014-0593-0) [DOI] [Google Scholar]
- 54.Schulte U, Gebhard F, Heinz L, Veith M, Hochkirch A. 2011. Buccal swabs as a reliable non-invasive tissue sampling method for DNA analysis in the lacertid lizard Podarcis muralis. North West. J. Zool. 7, 325–328. [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. ( 10.1007/s12686-011-9548-7) [DOI] [Google Scholar]
- 58.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. ( 10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 59.Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, Andrew RL. 2017. The K=2 conundrum. Mol. Ecol. 26, 3594–3602. ( 10.1111/mec.14187) [DOI] [PubMed] [Google Scholar]
- 60.Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. ( 10.1093/bioinformatics/btm233) [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, Kumar S. 2002. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 10, 1727–1736. ( 10.1093/oxfordjournals.molbev.a003995) [DOI] [PubMed] [Google Scholar]
- 62.Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. ( 10.1093/bioinformatics/bts460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goudet J.2001. FSTAT. A program to estimate and test gene diversities and fixation indices. Retrieved from http://www.unil.ch/izea/ .
- 64.Rohde K, Hau Y, Weyer J, Hochkirch A. 2015. Wide prevalence of hybridization in two sympatric grasshopper species may be shaped by their relative abundances. BMC Evol. Biol. 15, 83 ( 10.1186/s12862-015-0460-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. ( 10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 66.Koen EL, Garroway CJ, Wilson PJ, Bowman J, Bersier L-F. 2010. The effect of map boundary on estimates of landscape resistance to animal movement. PLoS ONE 5, e11785 ( 10.1371/journal.pone.0011785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peterman WE. 2018. ResistanceGA. An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 49, 340 ( 10.1111/2041-210X.12984) [DOI] [Google Scholar]
- 68.Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 19, 153–170. ( 10.1007/BF02300753) [DOI] [PubMed] [Google Scholar]
- 69.Miller MP. 2005. Alleles in space (AIS): computer software for the joint analysis of interindividual spatial and genetic information. J. Hered. 96, 722–724. ( 10.1093/jhered/esi119) [DOI] [PubMed] [Google Scholar]
- 70.Peterman WE, Connette GM, Semlitsch RD, Eggert LS. 2014. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 23, 2402–2413. ( 10.1111/mec.12747) [DOI] [PubMed] [Google Scholar]
- 71.Richardson JL, Brady SP, Wang IJ, Spear SF. 2016. Navigating the pitfalls and promise of landscape genetics. Mol. Ecol. 25, 849–863. ( 10.1111/mec.13527) [DOI] [PubMed] [Google Scholar]
- 72.van Etten J. 2015. R Package gdistance: distances and routes on geographical grids. J. Stat. Softw. 76(13), 1–21. ( 10.18637/jss.v076.i13) [DOI] [Google Scholar]
- 73.Shah VB, McRae BH. 2008. Circuitscape: A tool for landscape ecology. In Proceedings of the 7th Python in Science Conference, Pasadena, CA, 19–24 August 2008 (ed. Varoquaux G, Vaught T, Millman J), pp. 62–66. [Google Scholar]
- 74.Kolbe JJ, Glor RE, Schettino LR, Lara AC, Larson A, Losos JB. 2007. Multiple sources, admixture, and genetic variation in introduced Anolis lizard populations. Conserv. Biol. 21, 1612–1625. ( 10.1111/j.1523-1739.2007.00826.x) [DOI] [PubMed] [Google Scholar]
- 75.While GM, Williamson J, Prescott G, Horváthová T, Fresnillo B, Beeton NJ, Halliwell B, Michaelides S, Uller T. 2015. Adaptive responses to cool climate promotes persistence of a non-native lizard. Proc. R. Soc. B 282, 20142638 ( 10.1098/rspb.2014.2638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isaksson C, Hahs A. 2015. Urbanization, oxidative stress and inflammation. A question of evolving, acclimatizing or coping with urban environmental stress. Funct. Ecol. 29, 913–923. ( 10.1111/1365-2435.12477) [DOI] [Google Scholar]
- 77.Lowe WH, Muhlfeld CC, Allendorf FW. 2015. Spatial sorting promotes the spread of maladaptive hybridization. Trends Ecol. Evol. 30, 456–462. ( 10.1016/j.tree.2015.05.008) [DOI] [PubMed] [Google Scholar]
- 78.Beninde J, Feldmeier S, Veith M, Hochkirch A. 2018. Data from: Admixture of hybrid swarms of native and introduced lizards in cities is determined by the cityscape structure and invasion history Dryad Digital Repository. ( 10.5061/dryad.p564p61) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Beninde J, Feldmeier S, Veith M, Hochkirch A. 2018. Data from: Admixture of hybrid swarms of native and introduced lizards in cities is determined by the cityscape structure and invasion history Dryad Digital Repository. ( 10.5061/dryad.p564p61) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Input files for ResistanceGA and microsatellite genotypes are available on Dryad (http://dx.doi.org/10.5061/dryad.p564p61) [78].