Abstract
The initiation of DNA replication at replication origins in eukaryotic cells is tightly controlled to ensure that the genome is duplicated only once each cell cycle. We present evidence that in fission yeast, independent regulation of two essential components of the initiation complex, Cdc18 and Cdt1, contributes to the prevention of reinitiation of DNA replication. Cdc18 is negatively controlled by cyclin-dependent kinase (CDK) phosphorylation, but low level expression of a mutant form of Cdc18 lacking CDK phosphorylation sites (Cdc18CDK) is not sufficient to induce rereplication. Similar to Cdc18, Cdt1 is expressed periodically in the cell cycle, accumulating in the nucleus in G1 and declining in G2. When Cdt1 is expressed constitutively from an ectopic promoter, it accumulates in the nucleus throughout the cell cycle but does not promote reinitiation. However, constitutive expression of Cdt1, together with Cdc18CDK, is sufficient to induce extra rounds of DNA replication in the absence of mitosis. Significantly greater levels of rereplication can be induced by coexpression of Cdc18CDK and a Cdt1 mutant lacking a conserved C-terminal motif. In contrast, uncontrolled DNA replication does not occur when either mutant protein is expressed in the absence of the other. Constitutive expression of wild-type or mutant Cdt1 also leads to an increase in the levels of Cdc18CDK, possibly as a result of increased protein stability. Our data are consistent with the hypothesis that control of rereplication depends on a redundant mechanism in which negative regulation of Cdt1 functions in parallel with the negative regulation of Cdc18.
The initiation of DNA replication in eukaryotic cells is controlled precisely to ensure that the genome is duplicated exactly once each cell cycle. Many lines of evidence suggest that this control mechanism involves two sequential steps (for review see ref. 1). During the G1 phase, multiprotein complexes containing the origin recognition complex (ORC), Cdc6 (Cdc18 in Schizosaccharomyces pombe), Cdt1, and Mcm2–7, are formed at chromosomal origins of DNA replication. At the beginning of S phase these preformed initiation complexes are switched to an active state capable of supporting DNA synthesis, and at the same time the formation of new initiation complexes is blocked. Previous studies have established that an increase in cyclin-dependent kinase (CDK) activity at the G1/S transition plays a central role not only in triggering initiation but also in preventing the formation of new prereplication complexes (1–7). However, the precise machinery involved in this mechanism is not well understood.
Cdc6/Cdc18 is absolutely required for the assembly of prereplication complexes at origins of DNA replication. The protein is recruited to origins by interactions with ORC and is essential for the subsequent recruitment of the minichromosome maintenance (MCM) complex, which likely serves as the replicative helicase (8–13). In fission yeast it has been demonstrated that high level expression of Cdc18 can induce multiple rounds of DNA replication, indicating that it plays a key role in the onset of S phase (14, 15). We and others have shown that Cdc18 is a target of CDK-mediated phosphorylation at the G1/S transition and that phosphorylation causes the inactivation and degradation of the protein (6, 16, 17). Overexpression of a mutant form of Cdc18 lacking CDK phosphorylation sites leads to accumulation of the protein and a dramatic increase in rereplication. These observations suggest that inactivation of Cdc18 at the onset of S phase likely contributes to the inhibition of initiation complex formation, which limits DNA synthesis to once per cell cycle. However, inactivation of Cdc18, although important, cannot be the only mechanism involved in preventing reinitiation of DNA replication during the fission yeast cell cycle, because expression of the cdc18 mutant lacking CDK phosphorylation sites from the cdc18 promoter does not induce detectable rereplication. Thus, there must be additional proteins involved in the negative regulation of initiation of DNA replication.
One potential target of negative control is Cdt1. Initially identified in S. pombe, Cdt1 has now been described in Drosophila, Xenopus, and human cells and is likely conserved in most if not all eukaryotes (18–22). In fission yeast, Cdt1, similar to Cdc18, is expressed periodically in the cell cycle and is essential to load the MCM proteins onto chromatin during the assembly of the prereplication complex in G1 phase (18, 19). It has been shown that Cdt1 coimmunoprecipitates with Cdc18 and that the C-terminal half of Cdc18 is sufficient for this interaction (19). Overexpression of Cdt1 enhances the rereplication induced by high levels of Cdc18, indicating that Cdt1 cooperates with Cdc18 in the initiation of DNA replication in fission yeast (19). In Xenopus and other metazoans, Cdt1 interacts with geminin, an inhibitor of DNA replication, and it has been suggested that this interaction could play a role in preventing rereplication (22–24).
Here, we demonstrate that Cdt1 and Cdc18 act synergistically during DNA synthesis and that the regulation of both proteins is important in restricting DNA synthesis to once per cell cycle. Although overproduction of wild-type Cdt1 alone does not have a discernable effect on DNA synthesis, an increase in the DNA content of cells is observed upon coexpression of Cdt1 with a mutant Cdc18 protein lacking CDK phosphorylation sites. An even greater increase in DNA levels is observed upon coexpression of a mutant of Cdt1 (Cdt1S382A) together with the nonphosphorylatable Cdc18. Our results are consistent with the hypothesis that redundant regulatory mechanisms, targeting Cdc18 and Cdt1, operate within cells to ensure that the normal genome ploidy is maintained.
Materials and Methods
S. pombe Plasmids and Strains.
The plasmids pREP81X-cdt1, pREP41X-cdt1, and pREP3X-cdt1 encoding untagged Cdt1 were created by amplifying the cdt1+ ORF from S. pombe genomic DNA and inserting it into the BamHI site of pREP81X, REP41X, and pREP3X, respectively. The plasmid pREP41X-cdt1HA containing an epitope-tagged version of the cdt1+ gene under the control of the medium strength thiamine-repressible nmt1 promoter was constructed by inserting the Cdt1 coding sequence with a C-terminal triple-hemagglutinin (HA3) epitope tag into the SacI site of the expression vector pREP41X. The mutant allele of cdt1, cdt1S382A, was generated by using the QuikChange site-directed mutagenesis kit (Stratagene). The mutant Cdc18CDK, which has been described previously as cdc18ΔCDK1-5 (6), was recloned into the vector pUR18N for expression under its own promoter.
The plasmids pREP3X-cdt1, pREP81X-cdt1, pREP3X-cdt1(S382-A), and pREP81X-cdt1(S382A), expressing either the wild-type Cdt1 or the mutant Cdt1S382A under the control of nmt1 promoters were tested for their ability to rescue the viability of a strain carrying a deletion of the chromosomal cdt1+ gene. The diploid strain SP818 (h−/h+ ura4-D18/ura4-D18, leu1-32/leu1-32, ade6-M210/ade6-M216, cdt1+/cdt1∷ura4+; ref. 18) was transformed with plasmid DNA. Several independent transformants were sporulated, and the resulting haploid colonies were characterized. Ura+ colonies, which carry the chromosomal cdt1 deletion, were recovered at comparable frequencies from cells transformed with wild-type or mutant plasmids, and nearly all such colonies failed to grow on media containing thiamine.
The S. pombe strain VG234Y, expressing Cdt1 with a C-terminal HA3 epitope tag was constructed by transforming the strain VG55Y (h−, ura4-D18, leu1-32, his3-D1 ade6-M210) with the plasmid pKLG497-C-cdt1HA, linearized with NruI, which cleaves within the cdt1 ORF. To construct pKLG497-C-cdt1HA, a DNA fragment containing the C-terminal 1.1-kb of the cdt1 ORF together with the HA3 tag was cloned into the SalI site of the integrating vector pKLG497 (25).
The strain VG201Y was generated by introducing the plasmid pREP41X-cdt1HA into the diploid strain SP818 (see above). After sporulation, a haploid strain with the chromosomal cdt1 deletion covered by the pREP41X-cdt1HA plasmid was selected. The strain VG121Y was generated by the same method except that the covering plasmid was pREP41X-cdt1.
Immunofluorescence Assays.
Immunofluorescence studies were carried out as described in refs. 26 and 27. Regulated expression of Cdt1 under the control of the cdt1+ promoter was studied by using strain VG234Y, and constitutive expression of Cdt1 was studied by using strain VG201Y (28, 29).
Chromatin Binding Assay.
A cdc25-22 strain carrying an HA3 epitope-tagged copy of cdt1+ at its chromosomal locus was arrested in G2 by incubation at 35.5°C for 4 h. The cells were released from the block by incubation at 25°C and sampled at 20-min intervals. Cells in each sample were washed three times in CSE [1.2 M sorbitol, 50 mM sodium citrate, 50 mM sodium phosphate (dibasic), pH 5.6] and treated with 0.5 mg/ml Zymolyase 20T (Seikagaku Kogyo, Tokyo)/0.2 mg/ml Trichoderma lysing enzyme (Sigma) in CSE for 5 min at room temperature. The cells were lysed in a buffer containing 20 mM Tris, pH 7.5, 0.4 M sorbitol, 150 mM potassium acetate, 5 mM MgCl2, 5 mM MgSO4, 1% Triton X-100, 2 mM DTT, phosphatase inhibitors, and protease inhibitors. The resulting suspension was fractionated into detergent-insoluble (chromatin) and detergent-soluble (supernatent) fractions by centrifugation at 20,000 × g for 5 min at 4°C.
Coimmunoprecipitation Experiments.
Coimmunoprecipitation experiments were performed with extracts prepared from strains VG201Y or VG121Y. Extracts were prepared as described in ref. 30 with minor modifications. One gram of cells was resuspended in 1 ml of 2× LB buffer and disrupted by bead-beating five times for 45 seconds. NaCl and Tween 20 were added to a final concentration of 100 mM and 0.05%, respectively, and the lysates were clarified by stirring for 45 min followed by centrifugation in a microcentrifuge at 20,000 × g at 4°C. Extracts (1 mg) were incubated for 3 h at 4°C with 75 μg of either anti-HA (F7) (Santa Cruz Biotechnology) or anti-glutathione S-transferase monoclonal antibodies conjugated to agarose beads.
Results
Expression and Subcellular Localization of Cdt1.
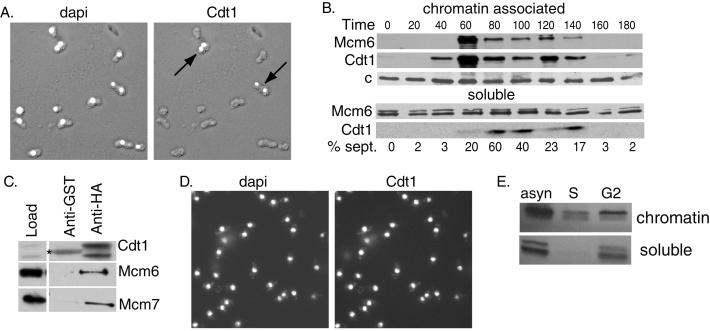
As a first step in assessing whether regulation of Cdt1 plays a role in limiting DNA replication in fission yeast, we used several methods to study the expression and localization of the protein during the cell cycle. For this purpose the chromosomal copy of the cdt1+ gene was tagged at its C terminus with an HA3 epitope. A population of asynchronous cells was fixed and stained with anti-HA antibody (Fig. 1A). Cdt1 was found to accumulate in the nucleus of a small subset of cells, consistent with previous observations (19). The strongest expression was observed in binucleated cells and a few small cells that had just completed division. In fission yeast such cells are postmitotic and in G1 or early S phase. Because it has been shown that other components of the initiation complex such as Cdc18 and the MCM complex are periodically associated with chromatin, we also analyzed the localization of Cdt1 by subcellular fractionation methods (refs. 19 and 31; Fig. 1B). A population of cdc25-22 cells, expressing Cdt1 with the HA3 epitope tag, were synchronized by release from a G2 block. At various times after release, cell extracts were prepared and separated into chromatin-enriched and soluble fractions. We observed that Cdt1 protein was absent in the starting G2 cell population and appeared first in the chromatin fraction at 40–60 min, 10–20 min before the peak of septation. Because initiation of DNA replication occurs immediately before septation, these data indicate that Cdt1 associates with chromatin in G1, consistent with the nuclear accumulation observed by indirect immunofluorescence. The amount of chromatin-associated Cdt1 declined rapidly from its peak level and reached very low levels in G2. In G1 phase nearly all the Cdt1 in the cell was associated with chromatin, although at later times a small amount of soluble protein was detectable. The initial association of Mcm6 occurred slightly later than that of Cdt1, but otherwise the pattern of chromatin association of the two proteins was similar. However, unlike Cdt1, Mcm6 was found consistently in the soluble fraction throughout the cell cycle, such that only a fraction of the total cellular Mcm6 protein is chromatin-bound in G1 and S phases. This observation is consistent with previous data (31).
Figure 1.
Regulation of Cdt1 expression in the cell cycle. (A) Cells expressing Cdt1-HA under the control of its endogenous promoter were fixed and stained with 4′,6-diamidino-2-phenylindole (dapi, left) or 16B12 anti-HA antibodies (right). The stained images are superimposed on Nomarski images of the fixed cells. (B) cdc25-22 cells were arrested in G2 by shifting to the nonpermissive temperature (35.5°C) for 4 h and released into synchronous culture at the permissive temperature (25°C). Cell extracts were prepared at the indicated times and fractionated into chromatin and soluble fractions. The Mcm6 and Cdt1 proteins were detected by Western blotting. A nonspecific band detected with anti-Mcm6 antibody (c) served as a loading control. (C) Extracts prepared from cells expressing Cdt1-HA under the control of the nmt1 promoter (pREP41X vector) were incubated with anti-HA antibody or nonspecific anti-glutathione S-transferase (anti-GST) antibody on agarose beads. The immunoprecipitates were analyzed by Western blotting to detect Cdt1, Mcm6, or Mcm7 proteins. A 5-fold excess of the immunoprecipitate was loaded compared with the starting material. The band marked with an asterisk is Ig heavy chain. (D) Cells expressing Cdt1-HA under the control of the 41X-nmt1 promoter were fixed and stained with 4′,6-diamidino-2-phenylindole (left) or anti-HA antibodies (right). (E) cdc25-22 cells expressing Cdt1 under the control of the 41X-nmt1 promoter were arrested in S phase (S) by incubation for 4 h in hydroxyurea (25 mM) or in G2 phase by incubation for 4 h at the nonpermissive temperature. Extracts prepared from the arrested cells and from a control population of asynchronous (asyn) cells were fractionated into detergent insoluble (chromatin) and soluble fractions. Cdt1 was detected by Western blotting with anti-Cdt1 antibodies.
It has been shown that Cdt1 is required for the association of MCMs with chromatin (19). Given this fact and our observation that Cdt1 associates with chromatin slightly before the Mcm6 protein, we asked whether Cdt1 associated with the MCMs in vivo. Cdt1 protein was immunoprecipitated from a strain overexpressing HA3-tagged Cdt1, and the immunoprecipitates were subjected to Western blot analysis with specific antibodies against the Cdt1, Mcm6, and Mcm7 proteins. As shown in Fig. 1C, both Mcm6 and Mcm7 proteins were coimmunoprecipitated by anti-Cdt1 antibody but not by control antibody. When the extracts were prepared from a strain expressing Cdt1 lacking the HA3 epitope tag, neither Cdt1 nor the MCM proteins were immunoprecipitated (data not shown). Thus, the observed association between Cdt1 and the Mcm6 and Mcm7 proteins is specific. Our data indicate that only a small fraction of the total pool of cellular MCM proteins is associated with Cdt1. One reasonable explanation for this finding is that the interaction between Cdt1 and MCM proteins is transient and occurs only during a narrow window of the cell cycle.
The cdt1+ gene is known to be under the control of Cdc10/Res1/Res2 transcription factor, which activates the transcription of target genes beginning in M phase and extending through the G1 phase of the cell cycle (17, 18). To assess the role of transcriptional control in the periodic expression of Cdt1, we placed the HA-tagged cdt1+ gene under the control of the repressible nmt1 promoter and carried out indirect immunofluorescence studies similar to those described above. When the nmt1 promoter was activated in an asynchronous population of cells, Cdt1 protein accumulated in the nuclei of all cells in the population, indicating that the expression of the protein is constitutive during the cell cycle under these conditions (Fig. 1D). Moreover, in contrast to the normal case (Fig. 1B), the constitutively expressed Cdt1 remained associated with chromatin in the G2 phase of the cell cycle (Fig. 1E). These data suggest that periodic expression and chromatin association of Cdt1 during the cell cycle depends, at least in part, on periodic activation of the cdt1+ promoter.
Simultaneous Dysregulation of Cdc18 and Cdt1 Results in Reinitiation of Replication.
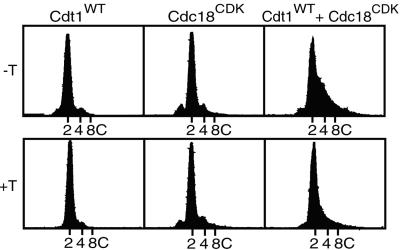
Because the normal periodicity of Cdt1 expression can be eliminated by expression from an ectopic promoter, we asked whether continuous expression of the protein altered the pattern of DNA replication in fission yeast cells. Cells with the cdt1+ gene under the control of the nmt1 promoter were derepressed by growth for 20 h in the absence of thiamine. This procedure resulted in a large increase in the expression of Cdt1 protein (Fig. 3C). However, we observed that constitutive high level expression of Cdt1 in an otherwise wild-type background had no discernable effect on DNA replication (Fig. 2, left panels). The cells maintained the normal 2C DNA content whether the nmt1 promoter was on or off. This result indicates that regulated expression of Cdt1 is either unimportant for preventing rereplication or that regulation of Cdt1 is redundant with other control mechanisms. Because our previous observations have suggested a role for CDK-dependent phosphorylation of Cdc18 in inhibiting reinitiation of DNA replication (6), we explored the effects of simultaneous dysregulation of both Cdt1 and Cdc18. A mutant form of Cdc18 lacking five CDK phosphorylation sites (Cdc18CDK) was expressed under the control of the cdc18+ promoter. As we reported previously, expression of Cdc18CDK alone failed to induce rereplication despite the fact that the mutant protein is resistant to negative control by CDK (Fig. 2, middle panels). In contrast, when Cdt1 was constitutively expressed at high levels in cells expressing the nonphosphorylatable Cdc18 mutant, nearly half the cells entered a second round of DNA replication and had DNA contents greater than 2C (Fig. 2, right panels). The observed rereplication depended on constitutive expression of Cdt1, because the fraction of cells with greater than 2C DNA content was reduced greatly when the nmt1 promoter was repressed by thiamine. Rereplication also depended on the mutations in the CDK phosphorylation sites of Cdc18, because no rereplication was observed when the wild-type Cdc18 protein, rather than the mutant, was coexpressed with Cdt1 (data not shown). We conclude from these observations that regulation of both Cdt1 and Cdc18 contributes to the limitation of replication to once per cell cycle.
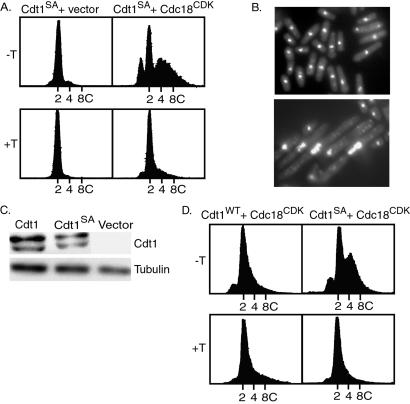
Figure 3.
Rereplication induced by coexpression of Cdt1 and Cdc18 mutants. (A) S. pombe strains expressing the Cdt1S382A mutant alone (left panels) or coexpressing Cdt1S382A and Cdc18CDK (right panels) were analyzed for DNA content by flow cytometry after 20 h under inducing (−T) or repressing (+T) conditions. As shown in Fig. 2, the Cdt1S382A mutant was expressed under the control of the nmt1 promoter by using a pREP3X vector, and the Cdc18CDK mutant was expressed under the control of the cdc18+ promoter on a separate plasmid. (B) Cells expressing Cdt1S382A (upper) or both Cdt1S382A and Cdc18CDK (lower) were fixed and stained with 4′,6-diamidino-2-phenylindole. (C) Extracts were prepared from cells transformed with plasmids encoding wild-type Cdt1 or the mutant Cdt1 (Cdt1S382A) or vector alone under the control of the 3X-nmt1 promoter after 20 h under inducing conditions. Cdt1 protein was detected by Western blotting. Tubulin represents a loading control. (D) Wild-type Cdt1 (left panels) or Cdt1S382A (right panels) under the control of the weak 81X-nmt1 promoter was coexpressed with Cdc18CDK under the control of the cdc18+ promoter. Cells were incubated for 20 h under inducing (−T) or repressing (+T) conditions. The DNA contents of the cells were determined by flow cytometry.
Figure 2.
Constitutive expression of Cdt1 induces rereplication in cells expressing nonphosphorylatable Cdc18. Three strains of S. pombe were constructed. The first strain (left panels) contained a plasmid expressing wild-type Cdt1 under the control of the nmt1 promoter (pREP3X vector). The second strain (center panels) contained a plasmid expressing a mutant form of Cdc18 (Cdc18CDK) under the control of the cdc18+ promoter. Cdc18CDK lacks five CDK phosphorylation sites in the N-terminal region of Cdc18. The third strain contained both plasmids (right panels). Cells were incubated for 20 h in the presence (+T) or absence (−T) of thiamine to repress or derepress the nmt1 promoter, and the DNA contents were measured by flow cytometry.
A Mutant Cdt1 Protein with Enhanced Rereplication Activity.
If, as the above results imply, negative control of Cdt1 may be an important cell cycle regulatory mechanism, we reasoned that it might be possible to obtain a mutant of cdt1 that, similar to Cdc18CDK, is less responsive to negative regulation. To search for such a mutant we began testing altered cdt1 genes for their ability to promote rereplication when coexpressed with the Cdc18CDK mutant. One mutation constructed in our initial experiments replaced a serine residue in a highly conserved SP motif at position 382 with alanine (Cdt1S382A). This SP motif, which is a potential CDK phosphorylation site, is conserved in Drosophila, Xenopus, and human Cdt1. We tested the ability of the Cdt1S382A mutant to rescue the viability of a S. pombe strain lacking the chromosomal copy of the cdt1 gene. A diploid S. pombe strain with one copy of the cdt1+ gene replaced with the ura4+ marker was transformed with a plasmid carrying the cdt1S382A gene under the control of the nmt1 promoter, and selected transformants were sporulated. We readily recovered haploid cells with both the chromosomal deletion of cdt1+ and the plasmid expressing Cdt1S382A. These cells failed to grow in the presence of thiamine, indicating that viability was maintained by the Cdt1S382A protein expressed under the control of the nmt1 promoter. Thus, the mutant protein is capable of fulfilling the normal function of Cdt1 in DNA replication.
As shown in Fig. 3A, coexpression of the Cdt1S382A mutant with Cdc18CDK greatly enhanced rereplication over that observed with the wild-type Cdt1 under the same conditions. A significant fraction of the cells displayed a DNA content greater than 4C. Expression of the Cdt1S382A mutant in a wild-type background had no effect on DNA replication, again indicating that rereplication requires dysregulation of both Cdt1 and Cdc18. The cells undergoing rereplication shown in Fig. 3A had swollen and distorted nuclei and were significantly elongated, indicating a failure of cell cycle progression (Fig. 3B). As might be expected, cells expressing both mutant proteins were nonviable (data not shown). To determine whether the mutation altered the steady-state levels of Cdt1, extracts prepared from cells overexpressing either the wild-type or mutant Cdt1 protein under the control of the 3X-nmt1 promoter were analyzed by Western blotting. As seen in Fig. 3C, there was no discernable difference between the steady-state levels of the two proteins.
Low Levels of Mutant Cdt1 Protein Are Sufficient to Promote Rereplication when Coexpressed with Cdc18CDK.
It could be argued that the rereplication observed upon coexpression of Cdc18CDK and Cdt1S382A represents an abnormal situation, because Cdt1S382A was expressed at high levels. Thus, we carried out an experiment similar to that described for Fig. 3A except that the expression of wild type or Cdt1S382A was under the control of a mutant nmt1 promoter (nmt1-81X) that drives very low level of expression in the absence of thiamine (less than 1% of the activity of the wild-type nmt1 promoter) (29, 32). With the weak promoter, very little if any rereplication was observed when wild-type Cdt1 was coexpressed with Cdc18CDK (Fig. 3D). However, significant rereplication was observed after coexpression of the Cdt1S382A mutant with Cdc18CDK. Thus, the Cdt1 mutant, even when expressed at low levels, can contribute to the bypass of the normal mechanism that restricts DNA replication to once per cell cycle.
Constitutive Expression of Cdt1 Results in an Increase in the Steady-State Levels of the Nonphosphorylatable Cdc18 Protein.
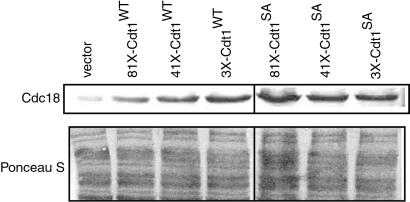
Given the genetic interaction between Cdt1 and Cdc18 described above, it was of interest to determine whether constitutive expression of Cdt1 affected the expression of Cdc18CDK. In these experiments wild-type or mutant Cdt1S382A protein was expressed under the control of the 81X-nmt1, 41X-nmt1, and 3X-nmt1 promoters, which drive increasing levels of expression of Cdt1 protein. The various promoters were derepressed for 20 h in a strain coexpressing the Cdc18CDK protein. Extracts prepared from these strains were analyzed by SDS/PAGE and Western blotting to detect Cdc18 CDK. As shown in Fig. 4, increased expression of wild-type Cdt1 caused an increase in the levels of Cdc18CDK over that observed with the vector control lacking the cdt1+ gene (compare lanes 2, 3, and 4 with lane 1). When the mutant Cdt1S382A protein was expressed from the same three promoters we also observed an elevated steady-state level of Cdc18CDK (Fig. 4, lanes 5–7). Although the mechanism responsible for the increased levels of Cdc18CDK is not yet clear, the most likely possibility, given the reported physical interaction between Cdt1 and Cdc18, is that Cdt1 stabilizes the Cdc18CDK protein (19). The increased levels of Cdc18CDK in the presence of Cdt1 or Cdt1S382A could contribute indirectly to the rereplication phenotype that we have observed, because very high expression of Cdc18 is sufficient to trigger rereplication in S. pombe (14, 15). However, the increase in Cdc18CDK expression in these experiments was relatively modest, and the increase was poorly correlated with the extent of rereplication. For example, the levels of Cdc18CDK were about the same when either wild-type Cdt1 or Cdt1S382A was expressed under the control of the 3X-nmt1 promoter (Fig. 4, lanes 4 and 7), but the extent of rereplication in the latter case was much greater (Figs. 2 and 3A).
Figure 4.
Stabilization of Cdc18CDK by overexpression of Cdt1. S. pombe strains expressing the Cdc18CDK alone (lane 1) or coexpressing wild-type (WT) Cdt1 and Cdc18CDK (lanes 2, 3, and 4) or coexpressing Cdt1S382A and Cdc18CDK (lanes 5, 6, and 7) were derepressed for 20 h. Extracts were prepared as described previously (15) and analyzed by SDS/PAGE followed by immunoblotting with 16B12 anti-HA monoclonal antibodies (Babco, Richmond, CA) to detect Cdc18CDK. The blot was stained with Ponceau S to confirm equal loading.
Discussion
The mechanisms responsible for ensuring that the genome is duplicated precisely once each cell cycle are incompletely understood. Our previous work, as well as that of others, has demonstrated that the activity of Cdc18 is negatively regulated by CDK phosphorylation and that this process likely plays a central role in the control of DNA replication during the cell cycle (1–7, 16, 17). Phosphorylation at the G1/S transition results in inactivation and rapid proteolysis of Cdc18 (6, 33). Given the absolute requirement for Cdc18 for the initiation of DNA replication, inactivation and/or elimination of Cdc18 is presumably sufficient to prevent a second round of DNA replication during the cell cycle. However, because low level expression of the Cdc18CDK mutant, which lacks CDK phosphorylation sites, does not result in uncontrolled DNA replication, we hypothesized that there must be other initiation proteins subject to negative regulation. The data presented in this paper suggest that control of Cdt1 is also important for preventing reinitiation of DNA synthesis within the cell cycle.
Cdt1 interacts with Cdc18 and MCM proteins and is essential for formation of the prereplication complex (19, 21). The Cdt1 protein is expressed only during a narrow window of the cell cycle before G1/S and is largely associated with chromatin. The periodic expression of Cdt1 is in part caused by regulated transcription of the cdt1+ gene under the control of the Cdc10/Res1/Res2 transcription factor. To assess the effects of unregulated expression of Cdt1 on the control of DNA replication, we placed the wild-type cdt1 gene under the control of an ectopic promoter. We observed that constitutive expression of Cdt1 led to nuclear accumulation of the protein throughout the cell cycle, but that this was not by itself sufficient to induce rereplication. On the other hand, constitutive high level expression of Cdt1 together with expression of the mutant Cdc18CDK protein lacking CDK phosphorylation sites induced a significant fraction of the cells to enter an additional round of DNA replication in the absence of mitosis. In these experiments the Cdc18CDK mutant was expressed at low levels under the control of its own promoter, a condition that, as noted above, does not induce rereplication by itself. These results indicate that uncontrolled expression of Cdt1 overcomes or interferes with a normal mechanism that acts to prevent rereplication. The simplest model is that inactivation of both Cdc18 and Cdt1 contribute independently to the prevention of rereplication. In a formal sense the mechanisms controlling the two proteins are redundant, because dysregulation of either protein by itself is not sufficient to induce uncontrolled DNA replication. However, by reducing the probability of reinitiation events, such apparent redundancy may be critical for maintaining the stability of the genome in the long term.
It has been demonstrated previously that high level overexpression of Cdc18 by itself is sufficient to drive rereplication in the absence of mitosis (14, 15). The extent of rereplication under these conditions can be enhanced by overexpression of Cdt1, suggesting that Cdt1 contributes to the efficiency of initiation (19). It is not clear why massive overexpression of Cdc18 can bypass all the normal restraints on DNA replication, and thus far this phenomenon is limited to fission yeast. Several lines of evidence indicate that the rereplication is not an indirect consequence of inhibition of CDK activity (6, 34). Moreover, as we have shown, low level expression of a form of Cdc18 that is resistant to negative control by CDKs does not cause rereplication. Thus, in addition to overcoming negative control of its own activity by CDKs, massive overexpression of Cdc18 must somehow block the negative regulation of other proteins involved in preventing reinitiation, including possibly Cdt1.
We have described a mutant form of Cdt1 (Cdt1S382A) that drives greatly enhanced rereplication when coexpressed with Cdc18CDK. This effect requires expression of only low levels of Cdt1S382A obtained by transcription from the weak 81X-nmt1 promoter. In contrast, coexpression of the wild-type Cdt1 under the control of the weak 81X-nmt1 promoter does not promote rereplication. There are several possible explanations for the phenotype of Cdt1S382A. One possibility is that the mutant protein is intrinsically more active in initiation than the wild-type protein. For example, the Cdt1 protein might contain an internal inhibitory domain, the effect of which is alleviated by the S382A mutation. A second possibility is that the Cdt1S382A protein is more resistant than wild-type Cdt1 to the effects of an inhibitory regulatory mechanism. For example, the activity or availability of Cdt1 might be limited by phosphorylation or other posttranslational modification and the S382A mutation might interfere with such modification. The SP motif at residue 382 is a potential phosphorylation site, but we do not yet know whether it is phosphorylated in vivo. Alternatively, the Cdt1S382A protein might have a lower affinity than wild-type Cdt1 for a putative inhibitory factor, perhaps analogous to geminin in Xenopus laevis (22–24). There is no obvious homologue of geminin in the S. pombe or Saccharomyces cerevisiae genome databases, but a similar function might be mediated by an apparently unrelated protein. All these potential explanations for the phenotype of Cdt1S382A imply the existence of additional mechanisms for the negative control of Cdt1 during the cell cycle. Further work will be required to determine which, if any, of these mechanisms are operative.
Our data indicate that the steady-state level of the Cdc18CDK protein is elevated when Cdt1 is overexpressed. This finding indicates that the amount of Cdc18 can be determined by mechanisms other than CDK phosphorylation. It has been reported that Cdt1 and Cdc18 interact in vivo, although it is not yet clear whether the interaction is direct or merely reflects the fact that the two proteins are part of the same prereplication complex (19). One simple explanation for the observed increase in Cdc18CDK protein level is that Cdt1 expression drives more Cdc18 into prereplication complexes where it may be less susceptible to proteolysis. At this point we cannot rule out the possibility that the increased level of Cdc18CDK contributes directly to the unregulated initiation of DNA replication that we have observed, although as noted above the correlation between Cdc18CDK level and the extent of rereplication is relatively weak.
Our data do not rule out the possibility that additional mechanisms besides control of Cdc18 and Cdt1 may contribute to the prevention of rereplication in eukaryotic cells. A recent study in S. cerevisiae has provided evidence that CDKs act to inhibit reinitiation of DNA replication by at least three independent and overlapping mechanisms (7). As in the case of S. pombe down-regulation of the activity of Cdc6, the budding yeast homologue of Cdc18, represents one important inhibitory mechanism. The other two mechanisms are exclusion of MCM proteins from the nucleus and inhibition of S. cerevisiae ORC function. There is no evidence at this point that either of the latter mechanisms are operative in fission yeast. Nuclear exclusion of MCM proteins is unlikely to play a role, because fission yeast MCM proteins, similar to those of most eukaryotic organisms, seem to be located in the nucleus throughout the cell cycle (35). However, it remains possible that regulation of the activity of ORC or other initiation proteins could play a role limiting replication in fission yeast to once per cell cycle (36). Multiple mechanisms to prevent rereplication may be a general phenomenon in eukaryotes, but the precise machinery may be different in different organisms.
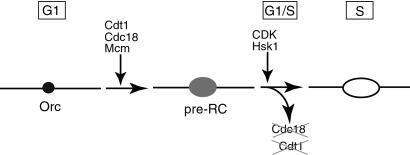
Our data are consistent with the following model for regulation of replication in fission yeast (Fig. 5). In the G1 phase of the cell cycle prereplication complexes (preRC) containing ORC, Cdc18, and Cdt1 are assembled at origins of replication. The Cdc18 and Cdt1 proteins are required for the loading of the MCM complex onto DNA but are dispensable thereafter. At the beginning of S phase, the initiation of DNA synthesis is triggered by the action of the CDK and Hsk1 protein kinases. Concurrent with this event, the key replication proteins, Cdt1 and Cdc18, are inactivated and degraded. Cdc18 is eliminated by CDK-dependent phosphorylation and ubiquitin-mediated degradation via the SCF pathway (33). The exact mechanism by which the activity of Cdt1 is eliminated is not known at present but could involve targeted degradation and/or a specific inhibitory factor. Rereplication is prevented until cells pass through mitosis into the next cell cycle and resynthesize Cdc18 and Cdt1. In this model the inactivation of either protein is sufficient to prevent reassembly of prereplication complexes and reinitiation of replication until the next cell cycle. The inherent redundancy in this model may provide a fail-safe mechanism to reduce the probability of reinitiation to a very low level over many generations.
Figure 5.
Model for control of rereplication in fission yeast. See Discussion for details.
Acknowledgments
This work was supported by grants from the National Institutes of Health and the Leukemia and Lymphoma Society.
Abbreviations
- ORC
origin recognition complex
- CDK
cyclin-dependent kinase
- MCM
minichromosome maintenance
- HA
hemagglutinin
References
- 1.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 2.Hayles J, Fisher D, Woollard A, Nurse P. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 3.Dahmann C, Diffley J F, Nasmyth K A. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 4.Jallepalli P V, Kelly T J. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 5.Jallepalli P V, Kelly T J. Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 6.Jallepalli P V, Brown G W, Muzi-Falconi M, Tien D, Kelly T J. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen V C, Co C, Li J J. Nature (London) 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 8.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 10.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 11.Ishimi Y. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima T, Takahashi N, Stillman B. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 13.Kearsey S E, Montgomery S, Labib K, Lindner K. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishitani H, Nurse P. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 15.Muzi-Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum B, Nishitani H, Yanow S, Nurse P. EMBO J. 1998;17:5689–5698. doi: 10.1093/emboj/17.19.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann J F, Beach D. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature (London) 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 20.Whittaker A J, Royzman I, Orr-Weaver T L. Genes Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- 21.Maiorano D, Moreau J, Mechali M. Nature (London) 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 22.Wohlschlegel J A, Dwyer B T, Dhar S K, Cvetic C, Walter J C, Dutta A. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 23.McGarry T J, Kirschner M W. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 24.Tada S, Li A, Maiorano D, Mechali M, Blow J J. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burke J D, Gould K L. Mol Gen Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- 26.Demeter J, Morphew M, Sazer S. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wente S R, Rout M P, Blobel G. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maundrell K. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 29.Forsburg S L. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown G W, Kelly T J. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa Y, Takahashi T, Masukata H. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forsburg S L, Sherman D A. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- 33.Jallepalli P V, Tien D, Kelly T J. Proc Natl Acad Sci USA. 1998;95:8159–8164. doi: 10.1073/pnas.95.14.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwood E, Nishitani H, Nurse P. J Cell Sci. 1998;111:3101–3108. doi: 10.1242/jcs.111.20.3101. [DOI] [PubMed] [Google Scholar]
- 35.Kearsey S E, Labib K. Biochim Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 36.Vas A, Mok W, Leatherwood J. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]