Abstract
Hoxa9 and Meis1a are homeodomain transcription factors that heterodimerize on DNA and are down-regulated during normal myeloid differentiation. Hoxa9 and Meis1a cooperate to induce acute myeloid leukemia (AML) in mice, and are coexpressed in human AML. Despite their cooperativity in leukemogenesis, we demonstrated previously that retroviral expression of Hoxa9 alone—in the absence of coexpressed retroviral Meis1 or of expression of endogenous Meis genes—blocks neutrophil and macrophage differentiation of primary myeloid progenitors cultured in granulocyte–macrophage colony-stimulating factor (GM-CSF). Expression of Meis1 alone did not immortalize any factor-dependent marrow progenitor. Because HoxA9-immortalized progenitors still execute granulocytic differentiation in response to granulocyte CSF (G-CSF) and monocyte differentiation in response to macrophage CSF (M-CSF), we tested the possibility that Meis1a cooperates with Hoxa9 by blocking viable differentiation pathways unaffected by Hoxa9 alone. Here we report that Meis1a suppresses G-CSF-induced granulocytic differentiation of Hoxa9-immortalized progenitors, permitting indefinite self-renewal in G-CSF. Meis1a also reprograms Hoxa9-immortalized progenitors to proliferate, rather than die, in response to stem cell factor (SCF) alone. We propose that Meis1a and Hoxa9 are part of a molecular switch that regulates progenitor abundance by suppressing differentiation and maintaining self-renewal in response to different subsets of cytokines during myelopoiesis. The independent differentiation pathways targeted by Hoxa9 and Meis1a prompt a “cooperative differentiation arrest” hypothesis for a subset of leukemia, in which cooperating transcription factor oncoproteins block complementary subsets of differentiation pathways, establishing a more complete differentiation block in vivo.
Oncoproteins cooperate in acute leukemias by deregulating combinations of cellular pathways that control proliferation, differentiation, and death (1). Hoxa9 and Meis1a are homeobox genes transcriptionally coactivated by proviral integration in spontaneous acute myeloid leukemia (AML) in BXH-2 mice (2–5), and Hoxa9 and Meis1 cooperate strongly in leukemogenesis, evidenced by the fact that retroviral coexpression of Hoxa9 plus Meis1 elicits rapid AML in marrow reconstitution experiments whereas expression of Meis1 alone fails to cause leukemia, and expression of Hoxa9 alone causes leukemia only after long latency (6). HoxA9 and Meis1 are also coexpressed in all but the promyelocytic subgroup of human AML (7, 8), suggesting that other human myeloid oncoproteins activate or maintain their transcription as a means to effect oncogenesis, emphasizing the importance of understanding how Hoxa9 and Meis1 cooperate to cause AML. The discovery that Hoxa9 and Meis1 interact in the absence of DNA (9) and heterodimerize on specific DNA elements (10) prompted early speculation that obligate Hoxa9:Meis1a heterodimers might target genes involved in leukemogenesis.
Despite the fact that coexpression of Meis1 is strongly correlated with leukemogenicity by Hoxa9, we demonstrated previously that Hoxa9 alone—in the absence of coexpressed retroviral Meis1 or of expressed endogenous Meis1, Meis2, or Meis3 genes—blocks neutrophil and macrophage differentiation of primary myeloid progenitors cultured in granulocyte–macrophage colony–stimulating factor (GM-CSF), but permits active granulocytic differentiation of this cell in response to granulocyte CSF (G-CSF) or monocytic differentiation in response to macrophage CSF (M-CSF; ref. 11). Similar to its ineffectual properties in marrow reconstitution experiments (6), retroviral expression of Meis1 alone also failed to immortalize any factor-dependent marrow progenitor in our studies (11). These observations demonstrated that Meis1 is dispensable for at least a subset of Hoxa9 transforming functions and suggested that Meis1 contributes a second independent function that cooperates with Hoxa9 in leukemogenesis. This second function would not necessarily require direct interaction with Hoxa9, because Pbx-Meis heterodimers and Meis monomers can control gene transcription in the absence of direct interaction with Hox proteins (12, 13), and because Meis1 also accelerates leukemogenesis by Hoxb3 (14), which falls in the category of Hox gene paralogues 1–8, which do not appear to bind Meis1 significantly in vitro (10).
Here we address the possibility that one cooperating function of Meis1 is to suppress myeloid differentiation pathways that are unaltered by Hoxa9 alone. We report that Meis1a alters the cellular response to G-CSF or to stem cell factor (SCF) in a manner that suppresses differentiation, and promotes proliferation and self-renewal. As Meis1 and Hoxa9 are expressed in early CD34+ but not later CD34− hematopoietic cells (7, 8), we propose that during normal myelopoiesis, Meis1a functions as a molecular switch that changes the response of a cell to both lineage-specific cytokines (e.g., G-CSF) and costimulatory cytokines (e.g., SCF), shifting that response from self-renewal when Meis1 is expressed in CD34+ cells to differentiation when Meis1 is down-regulated in CD34− cells. Through such a mechanism, extracellular factors could regulate the expansion and maintenance of hematopoietic progenitors by regulating transcription of Meis1. In like manner, the facts that Hoxa9 suppresses differentiation in the presence of GM-CSF or IL-3 (11, 15), and that deletion of Hoxa9 reduces levels of early myeloid and B-lymphoid progenitors (16) support the hypothesis that expression of HoxA9 also serves as a switch that controls cytokine-specific differentiation responses. We suggest that Meis1a and Hoxa9 cooperate in leukemogenesis by combining their abilities to promote progenitor self-renewal in response to different cytokines that activate complementary differentiation pathways. This forms the basis for a “cooperative differentiation arrest” hypothesis, which proposes that one basis for cooperativity between leukemia oncoproteins is their ability to block complementary differentiation pathways.
Methods
Construction of Recombinant Plasmids and Retroviral Vectors.
cDNAs encoding murine Meis1a, FLAG–Meis1a, Meis3, Hoxa9, and EE-tagged Hoxa9 were subcloned into the polylinker of the murine stem cell proviral vectors (MSCV), MSCVneo or MSCVpac. Helper-free virus was produced by cotransfection of 293T cells with MSCV vectors and a packaging-deficient murine leukemia virus provirus (17) and used for infection of primary murine marrow. EE-tagged Hoxa9 contained EEYMPEA (18) after the initiating methionine. FLAG-tagged Meis1a contained DYKDDDDK after the initiating methionine.
Marrow Infections.
Primary marrow progenitors were isolated and purified from the femurs and tibias of BALB/c mice as described (11). Ten thousand progenitors were transferred to each well of fibronectin-coated 24-well tissue culture plates, incubated with 1 ml of helper-free retrovirus containing 5 × 105 G418-resistance units, and 1 ml of marrow culture medium [MCM; RPMI medium 1640/10% FBS/1× antibiotics (penicillin, streptomycin)/1× glutamine/16 units/ml GM-CSF] containing Lipofectamine (GIBCO/BRL) at a concentration of 1 μl/ml. Cells were spun at 2,500 × g at 22°C for 1 h, incubated at 37°C under 5% CO2/95% air, and cultured in MCM medium thereafter. Nonadherent cells were transferred every 7 days to new plates.
Antiserum and Immunoblots.
Antiserum against the Hoxa9 homeodomain (HD) and C terminus has been described (11). Anti-Meis1a serum was raised in rabbits (Covance Research Products, Berkeley, CA) against the Meis1a HD and C terminus (Gly261–Arg340) fused to glutathione S-transferase sequences in PGEX2T (Amersham Pharmacia). The resulting 30-kDa fusion protein was purified by glutathione affinity column chromatography (Amersham Pharmacia) and dialyzed against PBS. For anti-Hoxa9 immunoblots, 5 × 104 cells boiled in Laemmli sample buffer were resolved by SDS/PAGE through 12.5% gels, transferred to a poly(vinylidene difluoride) membrane, detected by using polyclonal Hoxa9 antisera at a 1:1000 dilution, and visualized by using the Phototope-Star Chemiluminescent Detection kit (New England Biolabs). In the case of anti-Meis1a immunoblots, nuclear extract derived from 5 × 105 cells was analyzed in each lane, as described above.
Northern Blots.
Cytoplasmic RNA was purified from 2 × 108 myeloid progenitors (RNEasy, Qiagen). Cytoplasmic RNA (15 μg) was resolved by formaldehyde/agarose gels electrophoresis and transferred to positively charged nylon membrane (GeneScreenPlus, NEN). DNA probes labeled with [32P]dCTP were prepared from 100 ng of DNA that was subjected to random hexamer oligolabeling (Amersham Pharmacia). Hybridization in Ultrahyb (Ambion) and washing were carried out at 42°C according to the manufacturer's protocols. Probes were obtained from sources as described (11).
Electrophoretic Mobility-Shift Assay.
Oligonucleotides containing a consensus binding site for Pbx-Hoxa9 (bolded in tcacggTGATTTATgagcgactgctcgg) were synthesized (Genosys Biotechnologies, The Woodlands, TX) and labeled with [32P]ATP as described (11, 19). Nuclear extracts were prepared and electrophoretic mobility-shift assay analysis was performed as described (19).
Myeloid Differentiation Assays.
Myeloid progenitors (105) were washed twice in PBS. Cells were resuspended in RPMI medium 1640/10% FBS/1% antibiotics (penicillin and streptomycin) with addition of the appropriate cytokine (0.5 ng/ml G-CSF; 10 ng/ml M-CSF; 1 ng/ml IL-3, 1:200 dilution of SCF-containing supernatant). Neutrophil differentiation was examined by Wright–Giemsa staining after 72 h in G-CSF, and macrophage differentiation after 8 days in M-CSF.
Deconvolution Microscopy.
Images were captured with a DeltaVision deconvolution microscope system (Applied Precision, Issaquah, WA) and the data sets were deconvolved and analyzed by using SOFTWORX software (Applied Precision).
Results
Meis1a Promotes Self-Renewal and Suppresses Differentiation in Response to G-CSF.
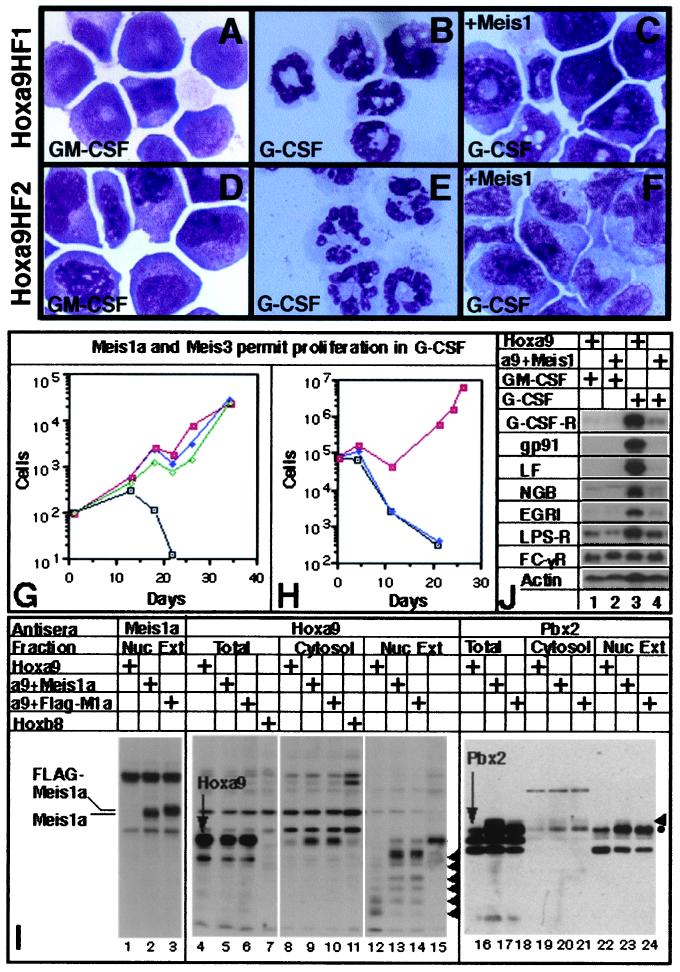
Hoxa9HF1 cells, a Hoxa9-immortalized, GM-CSF-dependent cell line that differentiates to neutrophils when GM-CSF is replaced with G-CSF (11), was infected with control MSCV retrovirus or MSCV encoding Meis1a, FLAG-Meis1a, or Meis3. After selection in puromycin, the ability of the Meis-expressing variants to proliferate in GM-CSF or to differentiate in G-CSF was compared with that of parental Hoxa9HF1 cells. Whereas G-CSF induced granulocytic differentiation of Hoxa9HF1 cells (Fig. 1 B vs. A), cells expressing Meis1a (Fig. 1C), FLAG-Meis1a, or Meis3 failed to exhibit terminal neutrophil differentiation and proliferated as cell lines (Fig. 1G) whose survival was strictly dependent on G-CSF. When G-CSF was withdrawn, these cells died. Meis1a also prevented G-CSF-induced differentiation of Hoxa9HF2 cells (Fig. 1 D–F; ref. 11) as well as the granulocytic differentiation of a third myeloid progenitor cell line immortalized by a version of Hoxa9 containing the EE tag at its N terminus (Fig. 1H). In no case did Meis1 prevent macrophage differentiation induced by M-CSF. A polyclonal antiserum against the Meis1 HD was generated in rabbits to confirm Meis1a production in the Hoxa9HF1 cells that proliferated in G-CSF. Both Meis1a and FLAG-Meis1a (≈1.0 kDa larger than Meis1a) were clearly present in nuclear extracts from Hoxa9HF1 cells infected by Meis1a and FLAG-Meis1a retrovirus, respectively (Fig. 1I, lanes 2 and 3) but not from uninfected cells (lane 1). These data suggested that Meis1a had a selective effect of suppressing granulocytic differentiation induced by G-CSF.
Figure 1.
Meis1a suppresses G-CSF-induced differentiation of Hoxa9-immortalized cells, increases Pbx2 abundance 3-fold, and does not alter the subcellular location of either Hoxa9 or Pbx2. Wright–Giemsa stains of Hoxa9HF1 and Hoxa9HF2 cells proliferating in GM-CSF (A and D) or 3 days after replacing GM-CSF with G-CSF (B and E). Hoxa9HF1 and Hoxa9HF2 cells expressing Meis1a 10 days after replacing GM-CSF with G-CSF (C and F, respectively). (G and H) Meis proteins permit immortalized proliferation of Hoxa9HF1 cells (G) and EE-tag-Hoxa9 cells (H) when shifted from GM-CSF to G-CSF. In G, tracing designations are uninfected cells (□), and cells infected with Meis1a virus (■), FLAG-Meis1a virus (♦), or Meis3 virus (⋄). In H, tracing designations are uninfected cells (□), and cells infected with control MSCV (♦) or Meis1a virus (■). (I) Western blot analysis demonstrating that expression of Meis1a increases Pbx2 abundance but does not alter the subcellular location of either Hoxa9 or Pbx2. Arrowheads indicate the positional of proteolytic fragments of Hoxa9 in lanes 12–14, and indicate a posttranslationally modified form of Pbx2 in lanes 16–24. The antiserum used and both the cell type and cell fraction examined are indicated across the top. (J) Northern blot analysis of total RNA from Hoxa9HF1 cells that do or do not coexpress Meis1a, and are proliferating in GM-CSF or shifted into medium with G-CSF for 11 days (indicated by + signs above lanes). The identity of each gene analyzed for expression is indicated at left.
Meis1a Induces a 3-Fold Up-Regulation of Pbx2, but Does Not Alter the Subcellular Localization of Pbx2 or Hoxa9.
Because HoxA9 was reported to reside principally in the cytosol of human AML cells that coexpress HoxA9 and Meis1a (7), and because Meis proteins bind Pbx proteins, increasing both their abundance and nuclear translocation (20–22), we addressed the possibility that, in addition to its direct function in the nucleus, Meis1a might also have the indirect function of either changing the abundance or altering the subcellular location of Hoxa9 or Pbx proteins. Immunoblots of total cell proteins demonstrated that Meis1a did not alter the abundance of HoxA9 (Fig. 1I, lanes 4–6), but did increase the abundance of Pbx2 ≈3-fold (lanes 16–18). Cell fractionation was used to investigate whether Meis1a altered the subcellular location of Pbx2 or HoxA9. In the presence or absence of Meis1, ≈70% of Pbx2 was nuclear (lanes 19–24). As reported earlier (11), significant proteolysis of Hoxa9 occurs during nuclear extraction procedures (Fig. 1I, lane 12–14) that yield other factors intact (e.g., Meis1, lanes 2 and 3). The proteolysis that we observed, which generated smaller HoxA9-DNA gel-shift complexes (11), is consistent with the appearance of smaller bands that stain with Hoxa9 antisera that are absent from parallel samples of nuclear extract from Hoxb8-immortalized progenitors (lane 15). Based on summation of these immunoreactive proteolytic fragments, the large majority of Hoxa9 was nuclear in the presence or absence of coexpressed Meis1a (compare Hoxa9 bands in lanes 12–14 with those in lanes 8–10). Nuclear extracts containing either Meis1a or FLAG-Meis1a suppressed the generation of smaller Hoxa9 fragments, suggesting that Hoxa9 is complexed to Meis1a and/or Pbx2 in vivo, and that such interactions prevent protect Hoxa9 from proteolysis in vitro (compare lanes 13 and 14 with lane 12).
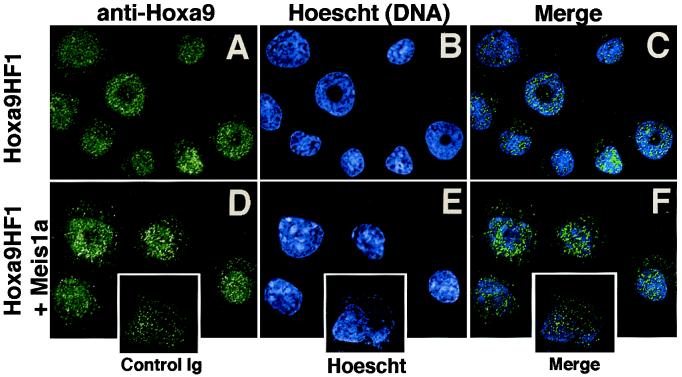
Because in vitro proteolysis of Hoxa9 could produce inaccurate conclusions regarding Hoxa9 localization by cell fractionation—especially if cleavage released the immunologically detected HD fragment into a different subcellular compartment—Hoxa9 was further localized by immunofluorescence, by using deconvolution microscopy (Fig. 2). In Hoxa9HF1 cells, Hoxa9 was totally nuclear (Fig. 2A), as confirmed by colocalization with Hoechst 33258 (blue), which stains DNA (Fig. 2 B and C). Cytosolic speckles resulted from nonspecific binding of rabbit Ig, as nonimmune pooled rabbit Ig yielded the same intensity of cytosolic staining without binding nuclear structures (Fig. 2, Insets). Expression of Meis1 had no effect on the nuclear location or subnuclear speckled pattern of Hoxa9 (Fig. 2 D–F).
Figure 2.
Meis1a does not alter Hoxa9 nuclear localization in Hoxa9-immortalized myeloid progenitors. Localization of Hoxa9 in Hoxa9HF1 cells (A–C) or Hoxa9HF1 cells expressing Meis1a (D–F), by using deconvolution microscopy and anti-Hoxa9 sera (green) (A, C, D, and F) and Hoescht 33258 (blue) as a nuclear stain (B, C, E, and F). (C and F) Merged images of anti-Hoxa9 plus Hoescht dye staining. (Inset) Hoxa9-immortalized cells stained with control rabbit Ig and Hoescht dye, demonstrating that cytosolic speckles arise from nonspecific staining.
Meis1a Suppresses Transcription of Terminal Differentiation Genes in Response to G-CSF in Hoxa9-Immortalized Progenitors.
In Hoxa9HF1 cells, Meis1a prevented G-CSF-induced transcriptional up-regulation of the genes encoding gp91phox (subunit of the respiratory burst oxidase), lactoferrin, Egr-1, neutrophil gelatinase B, and the lipopolysaccharide receptor (CD14), each of which is normally activated coincident with neutrophil differentiation (Fig. 1J). Meis1a also prevented up-regulation of the G-CSF receptor transcript accompanying neutrophil differentiation, but did not alter the basal level of this transcription. Therefore, Meis1 does not suppress G-CSF-induced differentiation by altering the initial abundance of the G-CSF, but could prevent differentiation events that specifically result from G-CSF-induced activation of G-CSF receptor gene transcription. Meis1a did not alter expression of Fc receptor 1 γ (CD64), which is expressed on both early and mature neutrophils. We conclude that Meis1a not only prevents phenotypic differentiation but also disrupts gene activation induced by signaling through the G-CSF receptor.
Simultaneous Expression of Meis1a and Hoxa9 in Primary Marrow Immortalizes a Myeloid Progenitor That Phenocopies the Effect of Sequential Expression of Meis1a in Hoxa9-Expressing Myeloid Progenitors.
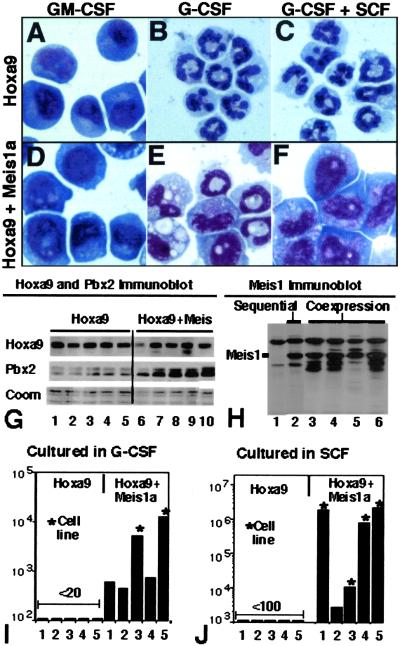
Coexpression of HoxA9 and Meis1 in human CD34+ cells and in human AML suggests that the most relevant context in which to study complementarity between Meis1a and Hoxa9 would be through their coexpression in primary marrow, as opposed to the sequential expression performed by transduction of Meis1a in Hoxa9-immortalized promyelocytes. We considered that simultaneous expression of Hoxa9 and Meis1a in primary marrow might generate a population of myeloid progenitors arrested at an earlier stage in differentiation—such as those immortalized by E2a-Pbx1, for example, which do not express the receptors for G-CSF or M-CSF (D.B.S., unpublished data). To explore this possibility, parallel infections of marrow progenitors were performed by using bicistronic retrovirus containing the neomycin-resistance cassette upstream of the Hoxa9 cDNA (NeoR__Hoxa9) or FLAG-Meis1a sequences upstream of Hoxa9 (FLAG-Meis1a__Hoxa9). Five independently derived, GM-CSF-dependent populations of myeloblasts immortalized by FLAG-Meis1a__Hoxa9 had an abundance of Hoxa9 comparable to five populations immortalized by NeoR__Hoxa9 (Fig. 3G, lanes 2–6 vs. lanes 7–11; compare Hoxa9 and Coomassie blue staining). The level of Meis1a expression from the bicistronic vector was comparable to that of Hoxa9HF1 cells selected for proliferation in G-CSF after infection with Meis1a retrovirus (Fig. 3H, lanes 3–6 vs. lane 2). As observed above, the abundance of Pbx2 in progenitors immortalized by coexpression of HoxA9 plus Meis1a was approximately 3 times that of progenitors immortalized by HoxA9 alone, suggesting that Meis1 directly stabilizes endogenous Pbx2. Consistent with our original report (10), greater than 96% of each population immortalized by Hoxa9 alone differentiated into neutrophils within 4 days of replacing GM-CSF with G-CSF (Fig. 3 A and B). By contrast, half of all cells in each culture coexpressing Hoxa9 plus Meis1a retained a progenitor morphology at day 4, and by day 11, cultures coexpressing Hoxa9 plus Meis1a contained 400 to 4,000 times the number of viable cells as cultures expressing Hoxa9 alone (Figs. 3 E and I). Two of the five populations continued to proliferate as G-CSF-dependent, immortalized cell lines (asterisks in Fig. 3I). All cell lines differentiated to macrophages in response to M-CSF (data not shown). These data indicated that coexpression of Meis1a and Hoxa9 immortalizes a late myeloid progenitor that is identical to that immortalized by Hoxa9 alone with regards to cytokine receptor expression (GM-CSF-R+, G-CSF-R+, M-CSF-R+), but that contains a selective defect in its ability to differentiate in response to G-CSF.
Figure 3.
Coexpression of Meis1a with Hoxa9 promotes self-renewal in response to G-CSF, self-renewal in response to SCF, and synergistic self-renewal in response to G-CSF plus SCF. Myeloid progenitors immortalized by Hoxa9 exhibit a progenitor morphology when cultured in GM-CSF (A), exhibit granulocytic differentiation when cultured in G-CSF (B), and exhibit granulocytic differentiation when cultured in G-CSF plus SCF (C) for 4 days. Myeloid progenitors immortalized by coexpression of Hoxa9 and FLAG-Meis1a exhibit a progenitor morphology when cultured in GM-CSF (D), significantly reduced granulocytic differentiation when cultured in G-CSF (E), and no granulocytic differentiation when cultured in G-CSF plus SCF (F) for 4 days. (G) Immunoblot analysis of Hoxa9 and Pbx2 in GM-CSF-dependent cell lines immortalized by Hoxa9 retrovirus (lanes 1–5) or by a bicistronic vector coexpressing Hoxa9 plus Meis1a (lanes 6–10). (H) Anti-Meis1 immunoblot of nuclear extracts derived from cultures 7–10 from G (lanes 3–6), from uninfected Hoxa9HF1 cells (lane 1), and from Hoxa9HF1 cells converted to G-CSF-dependent proliferation by expression of Meis1a (lane 2). (I and J) Abundance of five independent populations of myeloid progenitors immortalized from primary marrow by infection with Hoxa9 plus Neo retrovirus (first set of 5) or by Hoxa9 plus Meis1a retrovirus (second set of 5) 11 days after culturing 10,000 cells in G-CSF (I) or SCF (J). An asterisk signifies that the cells continued to proliferate as G-CSF- or SCF-dependent cell lines.
Meis1a Establishes a Strong Self-Renewal Response to SCF.
We tested the possibility that Meis1a might change proliferation and differentiation responses toward other cytokines. Meis1a had no effect on concentration-dependent proliferation rates in response to GM-CSF or IL-3 (data not shown). By contrast, Meis1a had a striking effect on responsiveness to SCF. By comparison to cells immortalized by Hoxa9 alone (Hoxa9HF1 and EE-Hoxa9 cells), which failed to proliferate in the presence of SCF, expression of Meis1a in these cells caused them to proliferate in response to SCF (data not shown). Analysis of the 10 cell lines generated with binary vectors demonstrated that >99% of cells in each of the five cultures immortalized by Hoxa9 alone stopped proliferating and either died or executed macrophage differentiation within 10 days of culturing in SCF alone, whereas each culture coexpressing Hoxa9 plus FLAG-Meis1a continued to proliferate (Fig. 3J), and four of five (denoted by asterisk) grew as immortal cell lines. SCF synergized with G-CSF in stimulating proliferation, as the total number of cells in the two G-CSF-dependent cell lines was augmented 2.5- and 4-fold by inclusion of SCF over a 72 h period, and those of the four SCF-dependent populations augmented 7- to 12-fold by inclusion of G-CSF over a 72-h period. SCF synergized with G-CSF to further suppress differentiation in Hoxa9- and Meis1-expressing cells (Fig. 3 F vs. E), but did not change G-CSF-induced neutrophil differentiation of Hoxa9-expressing cells (Fig. 3 C vs. B).
Flow cytometric analysis using antibodies against the SCF receptor (c-Kit; CD117), was performed to test the possibility that Meis1a up-regulates cell surface c-Kit, enabling SCF-responsiveness. The majority (78%) of Hoxa9-immortalized cells (proliferating in GM-CSF) expressed c-Kit with a relative mean fluorescence of 79, whereas those immortalized by Hoxa9 and Meis1a (also proliferating in GM-CSF) expressed c-Kit at a relative mean fluorescence of 58 (data not shown), indicating that Meis1a does not increase cell surface c-Kit. Consistent with this result, SCF was capable of synergizing with suboptimal concentrations of GM-CSF to increase the abundance of Hoxa9-immortalized progenitors 1.7-fold over a 72-h growth period. Therefore, although the synergistic effects of SCF and GM-CSF are apparent in the absence of Meis1a, the ability of cells to proliferate in response to SCF alone required Meis1a expression.
As a final test of the ability of Meis1a to suppress G-CSF-induced differentiation and permit SCF-induced proliferation, Meis1a was expressed in two populations of progenitors immortalized by the binary NeoR__Hoxa9 virus, and cells were assayed for their responses to GM-CSF, G-CSF, SCF, or G-CSF plus SCF. In both cases, Meis1a had no effect on proliferation rates in response to GM-CSF, whereas Meis1a induced a proliferative response to G-CSF, to SCF, and to G-CSF plus SCF (growth kinetics of one clone quantitated in Fig. 4).
Figure 4.
Meis1 induces rapid conversion of Neo-Hoxa9 cells to a phenotype that proliferates in response to G-CSF, SCF, or G-CSF plus SCF. The presence or absence of coexpressed Meis1 is designated adjacent to each tracing. Cytokines included in growth medium are indicated above each graph.
Discussion
Here we demonstrate that Meis1a suppresses G-CSF-induced differentiation in myeloid progenitors immortalized by Hoxa9. Furthermore, Meis1a endows Hoxa9-immortalized progenitors with the ability to proliferate in response to SCF alone. The independent functions of Hoxa9, in blocking differentiation in response to GM-CSF, and of Meis1a, in suppressing G-CSF-dependent differentiation and promoting SCF-dependent proliferation, establish a rational basis to propose that HoxA9 and Meis1a complement each other functionally to maintain progenitor self-renewal in human AML, and to explain how sequential activation of Hoxa9 and Meis1 by proviral integration could establish independent leukemogenic events leading to AML in BXH-2 mice. The facts that enforced Meis1a expression does not induce leukemia in vivo even after long latency (6) and that retroviral expressed Meis1 fail to either immortalize cultured marrow progenitors (11) or reestablish a differentiation block in myeloid progenitors conditionally immortalized by estrogen-regulated forms of E2a-Pbx1 (D.B.S., unpublished observation) suggests that the oncogenic function of Meis1 depends on preexisting mutations that up-regulate expression of Hoxa9 or Hoxa7, which is also coactivated with Meis1 in murine AML (4) and coexpressed with Meis1 in human AML (23). During the genesis of BXH-2 mouse AML, transcriptional activation of Hoxa9 may expand a pool of preleukemic progenitors whose differentiation continues to be induced by other cytokines, such as G-CSF (Fig. 5B). Secondary mutations that activate Meis1 transcription would accelerate leukemogenesis by (i) shifting the response to G-CSF from differentiation toward self-renewal, (ii) causing proliferation in response to SCF alone, and (iii) permitting SCF signaling to prevent differentiation induced by other cytokines. Genes whose activation selectively suppresses G-CSF-induced differentiation include cyclin D2 or D3 (24) and the CIS1/JAB family of JAK kinase modulators (25), and exemplify how Meis1 might alter gene expression to suppress G-CSF-induced differentiation. A somewhat different consequence of Hox oncoproteins on differentiation, combined with a consistent function of Meis1, could explain the observation of Thorsteinsdottir et al. (14), who found that Meis1 potentiates a leukemic phenotype that is defined by the Hox oncoprotein. Because neither Hoxa9-immortalized cells (11), nor their Meis-expressing derivatives (K.R.C., unpublished data) produce AML in sublethally irradiated BALB/c mice, generation of overt AML may require additional mutations (e.g., point mutants in Ras or Flt3, internal tandem duplications in Flt3, Bcr-Abl fusions); alternatively, the factor dependence and rapid (<3 months) outgrowth of AMLs induced by transplantation of mice with marrow coexpressing retroviral Hoxa9 and Meis1 (6) suggests that the impact of Hoxa9 and Meis1 on long-term reconstituting progenitors may be sufficient to induce leukemia, and that the cell lines we have characterized may have lost an intrinsic property retained by reconstituting progenitors that cooperates with Hoxa9 and Meis1 in leukemogenesis. The importance of genetic events that complement Hoxa9 and Meis1 in leukemogenesis, therefore, remains an open question.
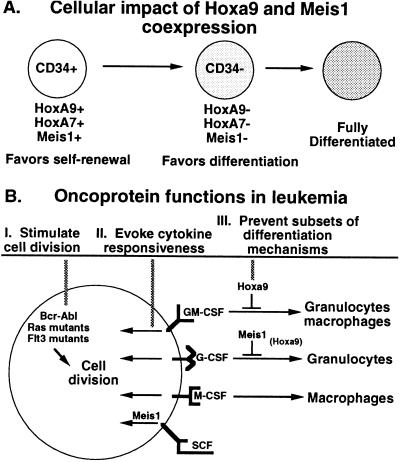
Figure 5.
Model for the function of Meis1a in AML.
The ability of Meis proteins to suppress G-CSF-induced differentiation has also recently been reported in the cell line 32Dcl3 (26), which executes granulocytic differentiation in response to G-CSF. While the 32Dcl3 cell system could not be used to demonstrate that Meis1 does not alter differentiation by M-CSF or GM-CSF, and was not used to show enhanced proliferation in response to SCF, it does confirm the ability of Meis1 to alter the differentiation response to G-CSF in another cell line that expresses Hox genes (32Dcl3 cells express Hoxa9 and Hoxb8).
If expression of HoxA9 and expression of Meis1 are essential events in the genesis of human AML, as their coexpression in all but acute promyelocytic leukemias suggests (7), a pivotal property of other myeloid transcription factor oncoproteins might be their ability to directly activate, or indirectly maintain activation of, Hoxa9 and Meis1 gene transcription. The fact that pre-B cell acute lymphocytic leukemia (ALL) specifically associated with mixed lineage leukemia (MLL) translocations coexpress Hoxa9 and Meis1, whereas other pre-B ALLs express neither gene (27) suggests strongly that Hoxa9 and Meis1 transcription is activated or maintained by MLL fusion oncoproteins. Recently we demonstrated that the human myeloid oncoprotein, Nup98-Hoxa9, immortalizes myeloid progenitors that proliferate in G-CSF or in SCF and, unlike progenitors immortalized by retroviral Hoxa9, maintain transcription of high levels of Hoxa9 and Meis1 at levels greater than those expressed in M1 cells, a myeloid leukemia initiated by retroviral coactivation of Hoxa9 and Meis1 (K.R.C., unpublished data). This finding implies that Hoxa9 and Meis1 are subordinate oncogenes of Nup98–HoxA9 and that the functions of Meis1 that we describe here may be operative in many human AMLs involving activated Meis1 transcription.
The biochemical mechanism by which Meis1 alters differentiation and proliferation responses to G-CSF and SCF, respectively, remains to be resolved. Meis1 can bind DNA as a monomer that cooperates in transcriptional activation with adjacent transcription factor complexes (13), it can bind DNA as a heterodimer with Pbx to promote gene transcription in a tissue-dependent manner (12), it can bind Pbx–Hox complexes independent of binding DNA directly (28), and it can heterodimerize with Hoxa9 on DNA (9), although this last function has not been observed in any cellular promoter analyzed to date. Each or all of these mechanisms may contribute to the functions of Meis1 that we report, as Meis1 up-regulates the abundance of Pbx2 and is coexpressed in cells with Hoxa9. A systematic mutational analysis of Meis1 that disrupts the isolated functions of binding DNA, binding Pbx cofactors through the cooperative functions of the M1 and M2 surfaces (29), and binding Hoxa9 through as-yet-unidentified surfaces remains an important goal to clarify how Meis1 alters differentiation and proliferation responses to G-CSF and SCF, respectively.
In normal hematopoiesis, amid a broad spectrum of cytokines, we propose that the combined functions of Meis1 and Hoxa9 control a genetic program that chooses between self-renewal or differentiation (Fig. 5A). Coexpression of Hoxa9 and Meis1 would favor strong self-renewal synergism and independent responsiveness to SCF, resulting in expansion of progenitors. Down-regulation of Hoxa9 and Meis1, which occurs during the early CD34+ to CD34− transition in the self-renewal of progenitors (7, 8), would curtail self-renewal to specific (i.e., G-CSF, GM-CSF) and costimulatory (i.e., SCF) cytokines, and permit terminal differentiation.
The impact of Hoxa9 and Meis1 on myelopoiesis suggest two hypotheses for how deregulation of cellular gene transcription can lead to leukemogenesis (Fig. 5B). The cytokine-specific differentiation blocks maintained by Hoxa9 and Meis1a prompt a hypothesis of “cooperative differentiation arrest” for the generation of a subset of acute leukemia. This hypothesis suggests that transcription factor oncoproteins may cooperate to establish a complete block to differentiation in vivo by blocking complementary subsets of differentiation pathways. This hypothesis further clarifies the general hypothesis that oncogenes that block differentiation (Fig. 5, mechanism III) cooperate with oncogenes that stimulate cell division (Fig. 5, mechanism I). The ability of Meis1 to reprogram gene transcription so as to permit a proliferative response to SCF prompts a “cytokine responsive” hypothesis, which states that genes involved in maintaining or establishing a proliferative response to environmental cytokines are targets for transcriptional activation in leukemia (Fig. 5, mechanism II).
Acknowledgments
We thank James Feramisco and Carolan Buckmaster for assistance with deconvolution microscopy; Marco Cassatella and Allen Seed for supplying the CD64 probe for Northern analysis; Christopher Glass and Gernot Walter for critical reading of this manuscript; and Michael David for pertinent discussions. This work was supported by National Institutes of Health Grant CA56876. K.R.C. is supported by National Institutes of Health Training Grant CA77109-01. D.B.S. is supported by Department of Defense Grant F4960-99-C-0054. M.P.K. is a scholar of the Leukemia and Lymphoma Society.
Abbreviations
- SCF
stem cell factor
- AML
acute myeloid leukemia
- MSCV
murine stem cell proviral vectors
- CSF
colony-stimulating factor
- GM-CSF
granulocyte-macrophage CSF
- G-CSF
granulocyte CSF
- M-CSF
macrophage CSF
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 2.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura T, Jenkins N A, Copeland N G. Oncogene. 1996;13:2235–2242. [PubMed] [Google Scholar]
- 4.Nakamura T, Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 5.Steelman S, Moskow J J, Muzynski K, North C, Druck T, Montgomery J C, Huebner K, Daar I O, Buchberg A M. Genome Res. 1997;7:142–156. doi: 10.1101/gr.7.2.142. [DOI] [PubMed] [Google Scholar]
- 6.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence H J, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, Buchberg A M, Largman C. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 8.Kawagoe H, Humphries R K, Blair A, Sutherland H J, Hogge D E. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 9.Shen W F, Rozenfeld S, Kwong A, Kömüves L G, Lawrence H J, Largman C. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen W F, Montgomery J C, Rozenfeld S, Moskow J J, Lawrence H J, Buchberg A M, Largman C. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo K R, Sykes D B, Pasillas M, Kamps M P. Mol Cell Biol. 2000;20:3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischof L J, Kagawa N, Moskow J J, Takahashi Y, Iwamatsu A, Buchberg A M, Waterman M R. J Biol Chem. 1998;273:7941–7948. doi: 10.1074/jbc.273.14.7941. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs Y, Schnabel C A, Cleary M L. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnabel C A, Jacobs Y, Cleary M L. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence H J, Helgason C D, Sauvageau G, Fong S, Izon D J, Humphries R K, Largman C. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- 17.Kamps M P, Wright D D. Oncogene. 1994;9:3159–3166. [PubMed] [Google Scholar]
- 18.Grussenmeyer T, Scheidtmann K H, Hutchinson M A, Eckhart W, Walter G. Proc Natl Acad Sci USA. 1985;82:7952–7954. doi: 10.1073/pnas.82.23.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoepfler P S, Kamps M P. Oncogene. 1997;14:2521–2531. doi: 10.1038/sj.onc.1201097. [DOI] [PubMed] [Google Scholar]
- 20.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 21.Pai C Y, Kuo T S, Jaw T J, Kurant E, Chen C T, Bessarab D A, Salzberg A, Sun Y H. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. Genes Dev. 1999;13:946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonja O, Smith J E, Cheng D M, Goldenberg A S, Amorosi E, Shimamoto T, Nakamura S, Ohyashiki K, Ohyashiki J, Toyama K, Takeshita K. Leuk Res. 2000;24:849–855. doi: 10.1016/s0145-2126(00)00059-x. [DOI] [PubMed] [Google Scholar]
- 24.Kato J Y, Sherr C J. Proc Natl Acad Sci USA. 1993;90:11513–11517. doi: 10.1073/pnas.90.24.11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki R, Sakamoto H, Yasukawa H, Masuhara M, Wakioka T, Sasaki A, Yuge K, Komiya S, Inoue A, Yoshimura A. Oncogene. 1998;17:2271–2278. doi: 10.1038/sj.onc.1202143. [DOI] [PubMed] [Google Scholar]
- 26.Fujino T, Yamazaki Y, Largaespada D, Jenkins N, Copeland N, Hirokawa K, Nakamura T. Exp Hematol. 2001;29:856–863. doi: 10.1016/s0301-472x(01)00655-5. [DOI] [PubMed] [Google Scholar]
- 27.Rozovskaia T, Feinstein E, Mor O, Roa R, Blechman J, Nakamura T, Croce C, Cimino G, Canaani E. Oncogene. 2001;15:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 28.Chang C P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoepfler P S, Calvo K R, Chen H, Antonarakis S E, Kamps M P. Proc Natl Acad Sci USA. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]