Abstract
The Notch signaling pathway has been implicated in homeostasis and disease, including cancer, in various tissues. Moreover, it has been involved both in stem cell maintenance and differentiation, in a context-dependent manner. Stem/progenitor cells, on the other hand, have long been suspected to be the cells of origin in various malignancies. In order to gain insight in the role of the Notch ligand Dll1 in mouse development, we generated a knock-in line expressing an inducible Cre recombinase. We have employed in vivo approaches in mice to genetically mark rare subpopulations of cells expressing Dll1 in various adult tissues. Moreover, we conditionally expressed a constitutively active Ras oncoprotein in these cells and showed that within days, mice develop squamous neoplasias in the skin, as well as in the stomach.
Introduction
The highly conserved Notch signaling pathway is involved in many processes in development and tissue homeostasis [1]. Its functions rely primarily, but not exclusively, upon the crosstalk of neighboring cells, which is achieved through the physical interaction of transmembrane receptors (Notch1–4 in mammals) and ligands (Dll1, 3, 4 and Jag1, 2). Although receptors and ligands can exhibit functional redundancy, it is widely accepted that the diverse outcomes of the Notch pathway in different cell contexts are in part due to specific ligand-receptor interactions [2], [3]. These interactions, which can act in trans (involving neighboring cells) or in cis (cell autonomously) [4], govern the role of the Notch signaling pathway in development and tissue homeostasis [5], stem cell maintenance and differentiation [6], as well as in disease [5]. Although it is implicated in the pathogenesis of various tissues, such as the nervous system [7], the heart [8], bones [9] and others, the Notch signaling pathway has been studied most extensively in the context of tumorigenesis [10], both as an oncogene and a tumor suppressor [11], [12], [13]. As effectors of Notch signaling and commonly mutated proteins in cancer, Notch receptors have monopolized researchers' interest in tumorigenesis studies. Less is known, however, about the individual Notch ligands, despite the fact that neither their expression pattern nor their mode of action is identical [14], [15], [16].
Several reports in the literature have identified the Notch ligand Dll1 as a putative stem/progenitor cell marker. In the mouse intestine, Dll1 marks a crypt subpopulation, which can functionally replace the bona fide Lgr5-expressing intestinal stem cells in crypts upon tissue damage [17]. In the central nervous system, Dll1 maintains quiescence of neural stem cells [18]. In the mammary gland, Dll1 marks cells with stem cell properties both in normal and neoplastic epithelium [19], [20]. Here, we present data indicating Dll1 expression in rare cells in various mouse tissues. Interestingly, Dll1-positive cells in the stomach and the skin respond to oncogenic Ras signaling, leading to tumor development within days. These findings imply that Dll1-positive cells could represent progenitors and/or cells of origin of cancer in the mouse.
Materials and Methods
Mice
Newly developed Dll1CreERT2-ires-EGFP heterozygotes were crossed to Gt(ROSA)26Sortm4(ACTB-tdTomato-EGFP)Luo (R26mTmG) to produce double heterozygotes Dll1CreERT2-ires-EGFP;R26mTmG/+. Male or female mice between 6 and 9 weeks of age were used for all experiments. They were injected intraperitoneally with 3.3 mg tamoxifen (Sigma) daily, for 5 consecutive days. Labeling of Dll1-positive cells was assessed 48 h after the last tamoxifen injection. Dll1CreERT2-ires-EGFP heterozygotes were crossed to KrasLSL-G12D mice [21] to produce double heterozygotes. Mice between 6 and 9 weeks of age were injected intraperitoneally with 2 mg tamoxifen daily, for 3 consecutive days. Animals were housed in individually ventilated cages under specific pathogen-free conditions, in full compliance with FELASA (Federation of Laboratory Animal Science Associations) recommendations in the Animal House Facility of the Biomedical Research Foundation of the Academy of Athens (BRFAA, Greece). All procedures for the care and treatment of animals were approved by the Institutional Committee on Ethics of Animal Experiments and the Greek Ministry of Agriculture.
Histology and Antibodies
Tissues derived from Dll1CreERT2-ires-EGFP;R26mTmG/+ were fixed in 4% paraformaldehyde at 4 °C for 2 h, thoroughly washed in 1x PBS, placed in 30% sucrose solution overnight and frozen in optimal cutting temperature (OCT) compound (Tissue Tek, Sakura). Frozen 10 um sections were obtained using a Leica (CM1950) cryostat. Sectioned tissues were washed for 5 min in 1x PBS and blocked in 1% bovine serum albumin, containing 0.1% Triton X-100 in 1x PBS (1x PBT), for 30 min. Primary antibodies were added at the appropriate dilutions in 1x PBT and tissues were incubated overnight at 4 °C in humidified chambers. Primary antibodies were washed out three times with 1x PBS, containing 0.1% Triton X-100 (1x PT), and tissues were further incubated with secondary antibodies in 1x PBT, for 2 h at room temperature. After three washes in 1x PT, sections were counterstained with 4,6-diamidino-2-phenylindole (Sigma) for 3 min and mounted with Vectashield (Vector Laboratories) [22]. Tissues from Dll1CreERT2-ires-EGFP;KrasLSL-G12D were fixed in formalin solution overnight, transferred to 70% EtOH and embedded in paraffin. Five-micrometer sections were cut and stained with hematoxylin and eosin, or with antibodies for immunofluorescence. Hematoxylin–eosin staining was performed using standard histology procedures [23]. Catalogue numbers and dilutions of the antibodies herein used are given as following: chicken anti-Krt14 (Biolegend #906001, 1:250), rabbit anti-Krt5 (Biolegend #905501, 1:1000), guinea pig anti-Krt10 (Progen Biotechnik #GP-K10, 1:200), rat anti-Troma I (DSHB #AB531826, 1:100), rabbit anti-Calbindin (Swant #CB38a, 1:500), sheep anti-Dll1 (R&D Systems #AF5026) and rabbit anti-E-cadherin (Cell Signaling Technology #3195S, 1:200). All secondary antibodies (Jackson Immunoresearch) were diluted 1:500.
Imaging
All images were captured on Leica TCS SP5 inverted confocal laser scanning microscope (CLSM), Leica HC, Leica DM IRE2, Leica DM LS2, or Nikon SM2800. Image processing and cell counts were performed using ImageJ and Adobe Photoshop CS3.
Flow Cytometry
Spleen was dissected from 7–8 week old Dll1CreERT2-ires-EGFP;R26mTmG/+ mice. Splenocyte suspensions were made by mechanical dissociation through a 70 μM strainer, in the presence of cold 1x PBS, containing 5% fetal bovine serum and 2 mM Ethylenediaminetetraacetic acid (EDTA) (1x PBF). Cells were treated with ACK (Ammonium Chloride Potassium) erythrocytes Lysis Buffer for 5 min and washed three times with 1x PBF. Cells were stained using directly conjugated antibodies obtained from Biolegend (anti-CD45BV510 #103137). Live cells were gated according to FSC/SSC profiles.
Reverse Transcription and Quantitative PCR
Total RNA from mGFP+ (mG+) and mGFP- (mG-) cells sorted from tamoxifen-injected Dll1CreERT2-ires-EGFP;R26mTmG mice was isolated using TRI-Reagent (Merck) protocol, according to manufacturer's instructions. Complementary DNA samples were prepared using PrimeScript RT Reagent Kit (Takara) and quantitative PCR reactions were performed using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems), in a Roche LightCycler 96. Three independent biological samples were quantified in technical duplicates and expression values were normalized to Glyceraldehyde 3-phosphate dehydrogenase (Gapdh). The following quantitative PCR oligonucleotide primers were used: Dll1_F: 5΄-CAT GAA CAA CCT AGC CAA TTG C-3΄; Dll1_R: 5΄-GCC CCA ATG CTA ACA GAA-3΄; Gapdh_F: 5΄-CCA GTA TGA CTC CAC TCA CG-3΄; Gapdh_R: 5΄-CTC CTG GAA GAT GGT GAT GG-3΄.
Data Availability
Data supporting the findings of this study are available within the article and from the corresponding author upon reasonable request.
Results
Dll1 is Expressed by a Small Subset of Cells in Various Mouse Tissues
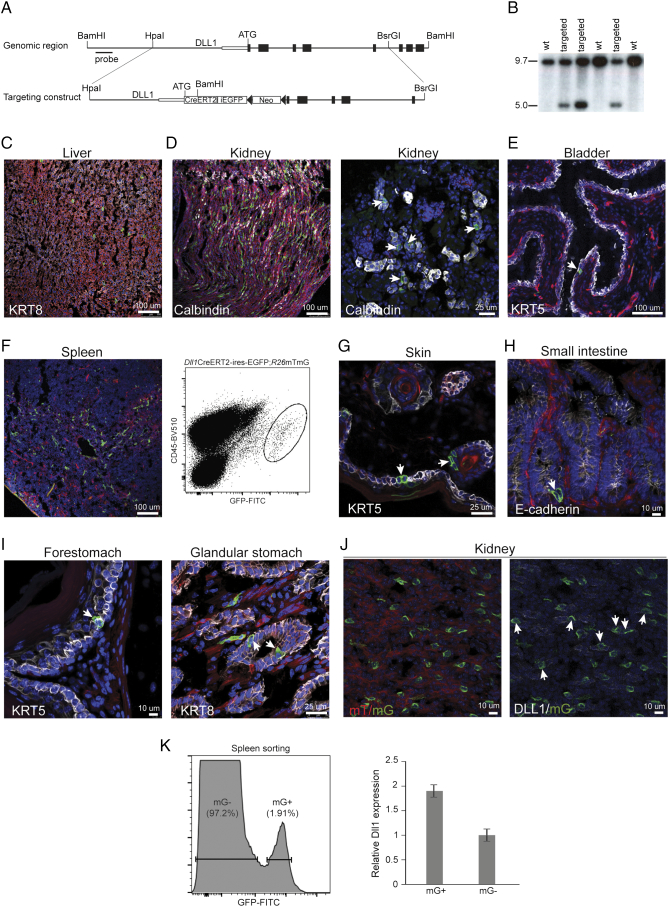
To identify cells expressing Dll1 and lineage-trace them in vivo, we herein employed a knock-in/knockout approach to generate a CreERT2 transgene by embedding the CreERT2 coding region into the endogenous Dll1 locus. To be able to identify Dll1-expressing cells without the need of antibody staining, we introduced an ires-EGFP cassette downstream of the CreERT2 open reading frame. An FRT-flanked PGKneo cassette was also introduced for clone selection (Figure 1A). The linearized targeting construct was electroporated into W9.5 wild-type 129SV embryonic stem (ES) cells (kind gift from Colin L. Stewart). Upon selection with G418, resistant cells were expanded and proper targeting events were identified by Southern blot analysis (Figure 1B). Surprisingly, 36/192 G418-resistant clones turned out to be correctly targeted. These targeted ES cells were injected into C57Bl/6 wild-type blastocysts to generate male chimeras, which were subsequently crossed with C57Bl/6 females to obtain germ line transmission of the allele Dll1CreERT2-ires-EGFP. Heterozygote mice were perfectly viable and indistinguishable from control littermates. Fluorescence microscopy on various tissues from Dll1CreERT2-ires-EGFP mice failed to detect native EGFP-driven fluorescence, possibly due to low level of expression (data not shown).
Figure 1.
Generation and characterization of the Dll1-CreERT2 mouse. (A) Schematic presentation of the genomic region and the targeting construct herein used. (B) Southern blot analysis of -candidate- targeted ES clones. (C) - (I) Confocal laser scanning microscope CLSM images of tissues derived from Dll1CreERT2-ires-EGFP;R26mTmG/+ bi-transgenic mice, counterstained with the indicated antibodies. Green cells indicate recombination events. For the spleen (F), a representative FACS profile is also included. (J) CLSM images of kidney sections derived from the same transgenic mouse, counterstained with anti-Dll1 antibodies. (K) Dll1 expression in sorted mG-positive (mG+) and mG-negative (mG-) subpopulations within spleen preparations from the same bi-transgenic mice. Data shown are representative of three independent experiments. In all fluorescence images DAPI was used as nuclear counterstain.
To assess recombinase activity, we crossed Dll1CreERT2-ires-EGFP mice with the Gt(ROSA)26Sortm4(ACTB-tdTomato-EGFP)Luo (R26mTmG) mouse strain, which constitutively expresses a membrane-tethered Tomato fluorescent protein (mT) and switches to membranous GFP (mG) expression upon Cre recombination [24]. Fluorescent microscopy on tissues of Dll1CreERT2-ires-EGFP;R26mTmG/+ bi-transgenic mice revealed sparse Dll1-positive cells in a variety of tissues. In the liver, Dll1 marks only a small fraction of both mono- and bi-nucleated Krt8-positive hepatocytes (Figure 1C); also it is expressed by a small portion of Calbindin-positive cells in the kidney (Figure 1D). Calbindin is a calcium binding protein marking distal tubules. It has been shown that Calbindin intensity is variable among epithelial cells even within the same tubule [25]. It is worth noting in this sense that Dll1 cells mostly stained weakly for Calbindin, a finding which merits further investigation. Further in the urogenital system, we observed very rare positive cells in the basal layer (Figure 1E), where stem cells reside [22], [26]. In the spleen, Dll1 marked CD45-positive leukocytes (Figure 1F), while it was absent from CD3-positive T cells, CD19-positive B cells and CD11b-positive macrophages (not shown). In agreement with previous reports, recombination was also observed in the epidermis [27] (Figure 1G) and the crypts of intestinal epithelium [17] (Figure 1H). Moreover, we observed recombination events in rare cells of both the KRT5-positive squamous forestomach and the KRT8-postive glandular stomach (Figure 1I). We should point out that for tissues such as the lung in which we did not detect Dll1 cells, we cannot exclude the possibility that Dll1 expression is too low to allow recombination and labelling of cells.
To directly mark Dll1 cells and validate the specificity of the Dll1CreERT2-ires-EGFP mouse line, we tried immunofluorescence with anti-Dll1 antibodies. After screening an exhaustive list of antibodies, we concluded that the best results were obtained from an R&D antibody (see Methods section). The best quality images came from the kidney and show that mG-positive cells largely overlap with antibody-stained Dll1-positive cells (Figure 1J). Moreover, mG-positive (recombined) and mG-negative (unrecombined) cells were sorted from Dll1CreERT2-ires-EGFP;R26mTmG/+ spleens, and the Dll1 transcript levels were assessed. As Figure 1K indicates, mG-positive cells showed twice as high Dll1 levels. Combined these findings indicate the Dll1CreERT2-ires-EGFP mouse indeed marks Dll1-expressing cells.
Skin Dll1 Cells Form Tumors in Response to Ras Signaling
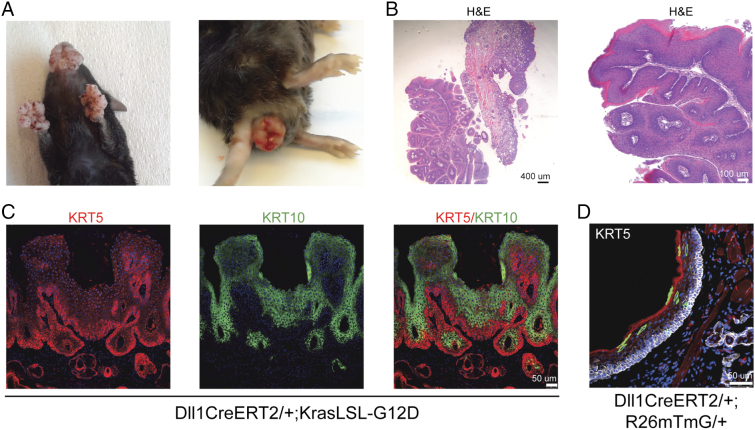
To investigate the functional role of Dll1-positive cells as cells of origin of cancer, we crossed Dll1CreERT2-ires-EGFP mice with a mouse line expressing the constitutively active KrasG12D oncoprotein under the control of the endogenous locus, in a Cre recombination-dependent manner (KrasLSL-G12D) [21]. Upon tamoxifen injection, Dll1CreERT2-ires-EGFP;KrasLSL-G12D mice developed visible skin lesions in various body sites, such as the lips, toes, back and anus (Figure 2A and data not shown), with an 100% percent penetrance. These neoplasms were visible as early as few days upon tamoxifen injection. Histological analysis of mouth/lip tumors showed squamous hyperplasias with extensive keratinization (Figure 2B). Tumors were positive for KRT5 and displayed extensive KRT10 staining, indicating keratinization (Figure 2C). Keratin profiling of normal lips skin epithelium from Dll1CreERT2-ires-EGFP;R26mTmG/+ mice indicated that the cells of origin are found within a small subpopulation of KRT5-positive cells (Figure 2D).
Figure 2.
Mutant Kras expression in Dll1-positive cells leads to skin lesions at various sites. (A) Images of Dll1CreERT2-ires-EGFP;KrasLSL-G12D bi-transgenic mice, following tamoxifen injection. (B) Low (left) and high (right) magnification images after Hematoxylin/Eosin staining of skin lesions from mouth/lips of the above transgenic mice. (C) CLSM images of mouth/lips lesions, indicating cytokeratin expression patterns. (D) CLSM images indicating the existence of rare cells in the skin of mouth/lips areas of transgenic mice. GFP-expressing cells, indicating Dll1CreERT2-ires-EGFP-dependent recombination, are located both within the basal KRT5-positive compartment, as well as in more superficial cells. In all fluorescence images DAPI was used as nuclear counterstain.
Dll1 Cells in the Forestomach are Cells of Origin of Squamous Hyperplasia
Dll1CreERT2-ires-EGFP;KrasLSL-G12D mice quickly lost weight and were either sacrificed or succumbed in less than a month (Figure 3A). Biopsy indicated the development of very large masses that filled the stomach of all mice (Figure 3B). These masses displayed squamous differentiation but not invasion and were always found in the forestomach, often adjacent to healthy glandular stomach (Figure 3C). As shown in Figure 1H, Dll1 is expressed in both the forestomach, co-localizing with KRT5, and the glandular stomach characterized by KRT8 cells. However, the fact that Kras-induced hyperplasias were exclusively detected in the forestomach, together with their cytokeratin profile showing KRT5, KRT14 and KRT10 expression both in diseased and normal tissue, indicate that the cells of origin are Dll1-positive cells within the KRT5 compartment of the squamous forestomach (Figure 3D-F). Of course, one cannot exclude the possibility that hyperplasias could originate from KRT8-expressing cells undergoing squamous metaplasia during the process. It is important to note, in this aspect, that similar phenotypes have been described with inducible expression of oncogenic Kras under the control of regulatory regions of KRT5 [28], or ubiquitously operating promoters [29]. Mutant Kras has been shown to induce tumors within the glandular stomach, albeit with a longer latency [30]. Hence, we suggest that Dll1CreERT2-ires-EGFP;KrasLSL-G12D mice do not survive long enough to develop such tumors.
Figure 3.
Mutant Kras expression in Dll1-positive cells in the mouse forestomach leads to squamous hyperplasia. (A) Kaplan–Meier curve indicating survival of tamoxifen-treated Dll1CreERT2-ires-EGFP;KrasLSL-G12D bi-transgenic mice. Time refers to days post last tamoxifen injection. (B) Images of dissected stomach from the above transgenic mice. (C) Low (left) and high (right) magnification images of Hematoxylin/Eosin (H&E) staining of the early-stage squamous lesions derived from a representative stomach from the above transgenic mice. g.s.: glandular stomach. Dotted lines separate the glandular regions from the squamous portion of the forestomach. (D) CLSM images of early-stage stomach lesions stained with indicated antibodies. (E) H&E and CLSM images of advanced stage stomach squamous lesions with indicated antibodies. (F) CLSM images of healthy stomach stained with indicated antibodies. In all fluorescence images DAPI was used as nuclear counterstain.
Discussion
Mutant Kras is probably the most powerful oncogene in human cancer. The highest percentage of Kras mutations are found in the pancreas, the lung and the colon [31]. While Kras mutations are not common in squamous cancers, the mouse squamous epithelium is particularly responsive to mutant Kras [28], [29], [32]. Expression of an oncogenic Kras in bitransgenic Krt5-rtTA;teto-KrasG12D mice leads to development of hyperplasias in various squamous epithelia including the skin, esophagus and the forestomach within 1 month [28]. In this model, Kras levels are likely artificially elevated and cannot be directly compared with the rest of the models, including the one reported here. Expression of oncogenic Kras from the endogenous locus in Krt5- or Krt14-expressing cells in the oral cavity leads to squamous hyperplasia after 5 months [32]. In another study, systemic activation of oncogenic Kras by means of a ubiquitous Cre recombinase driver led to development of gastric hyperplasia and metaplasia that appeared within 2–3 weeks [29]. Here, we show that expression of the KrasG12D mutant in a very small minority of Dll1-positive cells in skin epithelium at various sites, as well as in the forestomach, leads to rapid development of non-invasive squamous masses with latencies comparable, if not shorter, to those in previously published studies, in which expression of mutant Kras was induced in large cell compartments covering the whole organ, or even the whole mouse. Our data provide strong indication that the reported effect of Ras signaling on squamous epithelia in mice could be traced to a small subpopulation of Dll1-expressing cells within.
The Notch signaling pathway has been implicated in stem cell homeostasis, while the Dll1 ligand marks progenitors in the intestinal epithelium [17] and stem cell niches in normal and cancerous mammary epithelium [19], [20]. Little is known, however, about Dll1-expressing cells in other mouse tissues. In this study, we describe a knock-in/knockout mouse line that expresses CreERT2 under the direct control of the mouse Dll1 locus. Using inducible fluorescent reporter lines, we identified rare subpopulations of Dll1-carrying cells in the skin and the stomach epithelium. Cre recombinase-dependent expression of oncogenic KrasG12D in these cells leads to rapid development of hyperplasias, indicating that Dll1-positive cells might represent tissue progenitors that are sensitive to Ras activation. This work provides novel and powerful genetic tools to study stem cell and developmental biology in mouse squamous epithelia. Also, it offers novel mouse models for future basic, translational and preclinical research.
Acknowledgments
Acknowledgements
We would like to thank Dr. N. Paschalidis for help with FACS studies. This work was supported by a Horizon 2020 grant (BIOCDx), a Marie Curie Reintegration grant (MG stem), a Foundation Santé Grant in Biomedical Sciences and a Worldwide Cancer Research Grant (16/1217) to AK.
Author Contributions
AK and EV conceived the study and designed all experiments. ZK performed blastocyst injections for the Dll1CreERT2-ires-EGFP mouse. EV performed all experiments. DJS analyzed experimental data. AK wrote the manuscript.
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.del Alamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21:R40–R47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 5.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 6.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 7.Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Semin Cell Dev Biol. 2012;23:473–480. doi: 10.1016/j.semcdb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 12.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. Dynamic ligand discrimination in the notch signaling pathway. Cell. 2018;172:869–880.e819. doi: 10.1016/j.cell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preusse K, Tveriakhina L, Schuster-Gossler K, Gaspar C, Rosa AI, Henrique D, Gossler A, Stauber M. Context-dependent functional divergence of the notch ligands DLL1 and DLL4 in vivo. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu H, Okamoto R, Ito G, Fujii S, Nakata T, Suzuki K, Murano T, Mizutani T, Tsuchiya K, Nakamura T. Distinct expression patterns of Notch ligands, Dll1 and Dll4, in normal and inflamed mice intestine. PeerJ. 2014;2 doi: 10.7717/peerj.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi D, Furutachi S, Kawai H, Hozumi K, Gotoh Y. Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat Commun. 2013;4:1880. doi: 10.1038/ncomms2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti R, Celia-Terrassa T, Kumar S, Hang X, Wei Y, Choudhury A, Hwang J, Peng J, Nixon B, Grady JJ. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science. 2018;360(6396):1422–1433. doi: 10.1126/science.aan4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papafotiou G, Paraskevopoulou V, Vasilaki E, Kanaki Z, Paschalidis N, Klinakis A. KRT14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nat Commun. 2016;7:11914. doi: 10.1038/ncomms11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, Scorilas A, Klinakis A. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat Med. 2014;20:1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 24.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 25.Brandt LE, Bohn AA, Charles JB, Ehrhart EJ. Localization of canine, feline, and mouse renal membrane proteins. Vet Pathol. 2012;49:693–703. doi: 10.1177/0300985811410720. [DOI] [PubMed] [Google Scholar]
- 26.Shin K, Lee J, Guo N, Kim J, Lim A, Qu L, Mysorekar IU, Beachy PA. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature. 2011;472:110–114. doi: 10.1038/nature09851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM. Role of the Notch ligand Delta1 in embryonic and adult mouse epidermis. J Invest Dermatol. 2008;128:825–832. doi: 10.1038/sj.jid.5701113. [DOI] [PubMed] [Google Scholar]
- 28.Vitale-Cross L, Amornphimoltham P, Fisher G, Molinolo AA, Gutkind JS. Conditional expression of K-ras in an epithelial compartment that includes the stem cells is sufficient to promote squamous cell carcinogenesis. Cancer Res. 2004;64:8804–8807. doi: 10.1158/0008-5472.CAN-04-2623. [DOI] [PubMed] [Google Scholar]
- 29.Matkar SS, Durham A, Brice A, Wang TC, Rustgi AK, Hua X. Systemic activation of K-ras rapidly induces gastric hyperplasia and metaplasia in mice. Am J Cancer Res. 2011;1:432–445. [PMC free article] [PubMed] [Google Scholar]
- 30.Okumura T, Ericksen RE, Takaishi S, Wang SS, Dubeykovskiy Z, Shibata W, Betz KS, Muthupalani S, Rogers AB, Fox JG. K-ras mutation targeted to gastric tissue progenitor cells results in chronic inflammation, an altered microenvironment, and progression to intraepithelial neoplasia. Cancer Res. 2010;70:8435–8445. doi: 10.1158/0008-5472.CAN-10-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caulin C, Nguyen T, Longley MA, Zhou Z, Wang XJ, Roop DR. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. 2004;64:5054–5058. doi: 10.1158/0008-5472.CAN-04-1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are available within the article and from the corresponding author upon reasonable request.