Abstract
Introduction:
This prospective study aimed to study the composition and structure of the vaginal microbiota prior to Chlamydia trachomatis infection.
Methods:
A nested case control study was performed in 122 women, half of which acquired C. trachomatis within a year after of their first visit. At the first visit, the composition and structure of vaginal microbial communities were analyzed using 16S-rRNA-sequencing, in the context of the socio-demographic and sexual risk behaviour information using logistic regression.
Results:
Five vaginal community state types (CSTs) were identified. Four CSTs were dominated by Lactobacillus spp., of which L. crispatus (37%) and L. iners (33%) were the most common. One CST was characterized by the absence of Lactobacillus spp. (25%) and the presence of an array of strict and facultative anaerobes. Multivariate logistic regression analysis revealed that women with a L. iners dominated CST had an increased risk of C. trachomatis infection (P=0.04; OR: 2.6, 95%CI: 1.0–6.6).
Conclusions:
The distribution of CSTs dominated by Lactobacillus spp. agreed with previous studies. However, the frequency of dysbiosis amongst Caucasian women was relatively high (24%). Having vaginal microbiota dominated by L. iners was associated with an increased risk for C. trachomatis infection. Therefore, we hypothesize that specific signatures of vaginal microbiota are indicative of increased host predisposition to acquiring STIs.
Keywords: Vaginal microbiology, molecular epidemiology, Chlamydia infection, sexual health, women
Introduction
Chlamydia trachomatis genital infection is the most prevalent bacterial sexually transmitted infection (STI) in industrialized countries [1–2]. The infection often occurs asymptomatically thus contributing to transmission and high prevalence of chlamydia. C. trachomatis infections can increase the risk of HIV infection and lead to long-term complications in women such as pelvic inflammatory disease, ectopic pregnancy, and infertility [3]. Although the majority of C. trachomatis infections occur within transmission networks of specific risk groups, such as young adults and men having sex with men, infections with C. trachomatis are also endemic among the general population [4].
Microbial communities in the vagina are thought to play a protective role against colonization by pathogens responsible for bacterial vaginosis (BV), urinary tract infections, and STIs among others [5–9]. While the exact mechanism remains uncertain, it is believed that the acidic environment created by the production of copious amounts of lactic acid by Lactobacillus spp. is unfavorable to pathogen colonization [10–13]. Alterations in the composition and structure of vaginal bacterial communities may disturb the homeostasis of the vaginal environment, diminishing the defensive capacities thus increasing host susceptibility to vaginal infections.
In order to better understand the contribution of vaginal microbiota in STI susceptibility, this study aimed to compare the composition and structure of the vaginal microbiota prior to C. trachomatis infection in women who subsequently acquired C. trachomatis and in women who did not. Previous studies have demonstrated that the vaginal microbiota of women infected with C. trachomatis were depleted of Lactobacillus spp., and rather comprised a wide range of strict and facultative anaerobic bacterial species, many of which are also observed in BV [6,14].
Cross-sectional studies cannot determine whether BV-like vaginal microbiota often observed in C. trachomatis infected women have increased the risk of infection or are the consequence of C. trachomatis infection. Therefore, prospective longitudinal study designs that include characterizing the vaginal microbiota composition and structure prior to the infection are needed to better understand the contribution of the vaginal microbiota in the acquisition of C. trachomatis genital infection.
In this present study, we characterize the composition and structure of vaginal microbial communities prior to a C. trachomatis infection in a nested case control design that included 122 women, 61 of whom became infected with C. trachomatis within a year of the baseline visit. Vaginal swab samples were collected at baseline when all women were still negative of C. trachomatis genital infection, and then again 1 year later, at which time C. trachomatis positivity was evaluated. Vaginal community composition and structure was assessed using high-throughput sequencing of the V3-V4 hypervariable regions of the 16S rRNA gene. Vaginal microbiota composition and structure were evaluated in the context of extensive socio-demographic and sexual risk behaviour data collected during the 1-year visit at time of C. trachomatis testing.
Methods
Study population
A large trial to investigate the effectiveness of a population-based screening for C. trachomatis genital infection was conducted in the Netherlands from March 2008 to February 2012 [15]. In brief, yearly repeated chlamydia screenings were offered to all sexually active women and men aged between 16 and 29 years old listed in the municipal register. People were approached by postal invitations, which they could use to request a self-collection test kit online. Only those living in the Amsterdam area were included in this study. Vaginal swabs were self-collected, using the Aptima Vaginal Swab Specimen Collection kit (Hologic, Almere, the Netherlands). The self-collected samples were then sent to the Laboratory of Public Health in Amsterdam for C. trachomatis testing (Aptima single Ct test, Hologic, Almere, the Netherlands). After testing, the samples were stored at −80°C. If tested positive for C. trachomatis, treatment and partner notification were conducted by a general practitioner or at the local STI outpatient clinic. In addition to collecting vaginal samples for C. trachomatis testing, participants were also asked to fill out a standardized questionnaire about their sexual risk behaviour, medical history, and socio-demographic situation. Only women who provided vaginal swab samples and gave informed consent were included in this study. This study was approved by the Ethics Committee of the VU University of Amsterdam, METC number: 2007/239.
Study design
This study was set up as a nested case control study, in which we selected women who tested negative for C. trachomatis and then positive at the following year screening. Controls were women who tested negative for C. trachomatis at two consecutive yearly screenings. Cases were matched to controls according to age and ethnicity. The composition of the vaginal microbiota was profiled at baseline, prior to a possible acquisition of C. trachomatis, to evaluate whether its composition or structure were associated with C. trachomatis acquisition.
To calculate the sample size, the chance of having BV was taken as a proxy for having a vaginal microbiota associated with increased risk to STI because BV has been identified as a factor associated with increased risk for C. trachomatis infection (OR: 3.4; 95% CI: 1.5–7.8) [6]. Furthermore, BV is well characterized by a specific composition of the vaginal microbiota [8]. At a two sided confidence level of 95%, a power of 80%, assuming that cases and controls were evenly exposed to C. trachomatis, and an odds ratio of 3.4, a sample size was calculated of 48 cases and 48 controls. Another 13 cases and 13 controls were included to make sure the calculated sample size could be maintained, even when DNA isolation, amplification, and/or sequencing would fail in some of the samples. Therefore, 122 women in total were included in this study.
DNA extraction, amplification and sequencing of the V3-V4 region of bacterial 16S rRNA
Prior to extraction, the stored vaginal swabs were pre-treated by enzymatic and mechanical digestion as previously described by Fadrosh et al. [16]. Then, DNA was extracted from all 122 samples at baseline, using a QIAsymphony instrument as recommended by the manufacturer (Qiagen, Valencia, USA). DNA concentrations were determined using the Quant-iT Picogreen quantification system (Life technologies, Grand Island, USA) and diluted to a concentration of 5 ng/μl using a QIAgility instrument (Qiagen, Valencia, USA).
Dual indexed primers 319F and 806R were used for PCR amplification of the V3-V4 hypervariable regions of the 16S rRNA genes, as previously described by Fadrosh et al. [16]. Amplicon libraries were then sequenced using the 300 bp paired-end chemistry on a MiSeq platform, according to the manufacturer’s protocol (Illumina, San Diego, USA).
Sequence reads quality control, analysis, and taxonomic assignments
The index sequences of each paired-end sequence were combined and used to demultiplex samples as previously described by Fadrosh et al. [16]. Sequence read quality control was performed using strict criteria to ensure the high quality and full-length sequences of the entire amplicon region. In brief, reads with average quality greater than Q15 over a sliding window of 4 bp and with no ambiguous base calling were retained. Paired-end reads were assembled using FLASH [17] with a ~90bp overlapping region. Assembled reads were de-multiplexed by binning sequences with the same barcode and quality trimmed in QIIME (version 1.8.0) [18]. Both de novo and reference-based chimera detection was conducted in UCHIME (v5.1) [19] using the Greengenes database of 16S rRNA gene sequences (August 2013 version) as reference [20]. The processed 16S rRNA gene amplicon sequences were assigned to species and genera, using pplacer [21]. Samples were clustered in community state types (CSTs) using taxonomic composition and abundance and the Jensen-Shannon divergence metrics as previously described [8, 22].
Statistical analyses
The basic demographic characteristics of the women who acquired C. trachomatis within a year of baseline sampling were compared with the ones who did not, using the chi-square test. Median ages were compared among the two groups using the Mann-Whitney U-test.
Potential determinants for acquiring C. trachomatis were examined using logistic regression analyses. The following variables were evaluated based on preliminary tests: education, ethnicity, community state type, median age of the first sexual contact, number of sexual partners in the previous six months, and the type of relationship. Multivariate models were built by entering all variables that had a p-value <0.10 in the univariate analysis into the final model. The final model was checked for interactions between the variables present in the multivariate model. A p-value <0.05 was considered statistically significant. All analyses were performed using IBM SPSS Statistics version 21.
The linear discriminant analysis effect size (LEfSe) algorithm was used to identify bacterial taxa associated with acquiring C. trachomatis [23]. LEfSe uses the factorial Kruskal-Wallis rank-sum test to detect taxonomical classes with significant differential abundance with respect to the acquisition of C. trachomatis. Further, LEfSe uses linear discriminant analysis to estimate the effect size of each differentially abundant taxonomical class with respect to the acquisition of C. trachomatis.
Results
General characteristics
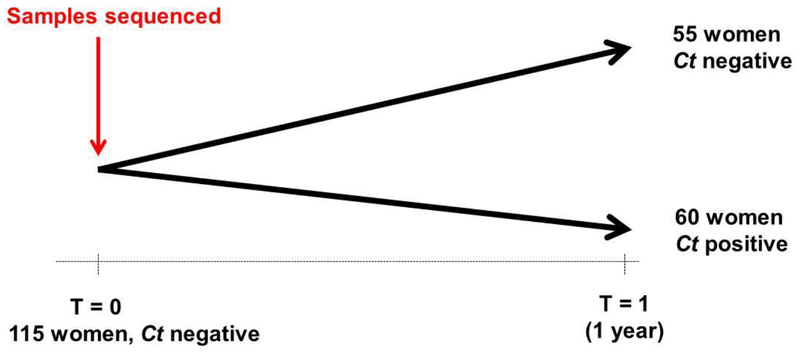
In total, 33,582 women living in the Amsterdam area participated in the Dutch Chlamydia screening project. A subset of 5,458 women (16%) participated at least twice and returned a self-collected vaginal swab and filled out the questionnaire. In this subset, 3,503 (64%) women had two yearly consecutive screenings and tested negative for C. trachomatis at the first visit and 61 tested positive for C. trachomatis at the subsequent yearly screening and had consented to use their samples for further research. These cases were then matched according to age and ethnicity to the control group. However, information on vaginal microbiota composition was available for 60 cases and 55 controls. The general characteristics at the baseline visit of these 115 are summarized in Table 1.
Table 1.
General characteristics of the women living in Amsterdam participating in the national Chlamydia screenings trial included in this study, at baseline
| Controls N=55 |
Cases N=60 |
P-value | |
|---|---|---|---|
| Median age (IQR) | 22 (19–25) | 23 (20–25) | 0.41 |
| Median age first sexual contact (IQR) | 16 (15–17) | 16 (15–17) | 0.52 |
| Born in the Netherlands | 46 (83.6%) | 50 (83.3%) | 0.96 |
| Ethnicity | 0.98 | ||
| Dutch | 39 (70.9%) | 41 (68.3%) | |
| Surinamese/Dutch Antilles | 11 (20.0% | 13 (21.7%) | |
| Asian | 3 (5.4.9%) | 3 (5.0%) | |
| Turkish | 1 (1.7%) | 1 (1.8%) | |
| Education | 0.28 | ||
| Low | 2 (3.6%) | - | |
| Middle | 23 (41.9%) | 22 (36.7%) | |
| High | 28 (50.9%) | 34 (56.7%) | |
| Sexual partners in the previous six months (IQR) | 1 (1–2) | 2 (1–3) | 0.07 |
| Previous infection with C. trachomatis* | 3 (5.5%) | 9 (15.0%) | 0.09 |
| Sexual health problems** | 6 (10.9%) | 13 (21.7%) | 0.27 |
| Relationship | 0.21 | ||
| Living together | 6 (10.9%) | 2 (3.3%) | |
| Living apart | 17 (30.9%) | 14 (23.3%) | |
| Single | 25 (45.5%) | 31 (51.7%) |
Self-reported
Self-reported complaints like having more vaginal discharge than normal, a burning sensation while urinating, abdominal pain, blood loss between menses or during or after sexual intercourse
IQR: InterQuartile Range
P-value: Probability value
Our study population mainly consisted of native Dutch Caucasian females (73%). The median age was 23 years (IQR: 20–25 years). There were no statistically significant differences between cases and controls at baseline.
None of the women in this study reported urogenital problems at the moment of inclusion. However, 6 controls and 13 cases reported urogenital problems in the past, such as more vaginal discharge than normal, abdominal pain, burning sensation while urinating, blood loss between menses, or during or after sexual intercourse. Before entering this study, 9 cases and 3 controls self-reported to have had an infection with C. trachomatis in the past.
Vaginal microbial community survey in the population
Amplification and high-throughput sequencing of the V3-V4 hypervariable regions of the 16S rRNA gene was performed on the samples at the baseline visit, when all women were negative for C. trachomatis. Sequence data could be obtained for 115 samples, 55 controls and 60 cases. After stringent quality control, 4,839,125 quality-filtered high-quality sequence reads were obtained, resulting in 42,448 sequence reads per sample.
By using hierarchical clustering based on microbiota composition and relative abundances, samples were grouped into five different vaginal microbiota clusters. Five clusters, so called community state types (CSTs) were observed in this dataset as depicted by the first sidebar in Figure 1. These were assigned according to Ravel et al. [8]. Four of these clusters were dominated by Lactobacillus spp. The two largest clusters were CST I (37%), dominated by L. crispatus, and CST III (33%). Within CST III, a larger diversity could be observed as compared to CST I. CST II was dominated by L. gasseri and CST V was dominated by L. jensenii, and they were observed in 3% and 2% of our cohort, respectively. CST IV was characterized by a depletion of Lactobacillus spp. and a wide array of strict and facultative anaerobic bacteria, like G. vaginalis, Megasphaera spp., Atopobium vaginae, and Prevotella spp. CST IV vaginal microbiota were found in 25% of the women included in this study.
Figure 1.
Hierarchical clustering of 115 vaginal samples from 60 women acquiring a C. trachomatis infection and 55 women who do not, within a year after entering the study. The dendrogram was generated using complete linkage clustering with Euclidean distance, based on the relative abundance of taxa in each sample. Relative abundance is illustrated by the colour key. The higher the abundance of a certain microbial species in a sample is, the more red it becomes. Sidebars depict the various community state types that could be assigned to the various clusters (CST I-V), acquiring C. trachomatis (in red), ethnicity, and the number of sexual partners in the past six months.
Ethnicities (Native Dutch Caucasian, Surinamese, Dutch Antilles, Turkish, and Asian) were evenly distributed over all CSTs . Noteworthy, 20 out of 84 (24%) native Dutch Caucasian women and 9 out of 24 (38%) Surinamese/Dutch Antilles women had CST IV vaginal microbiota (third sidebar, Figure 1). The total number of sexual partners in the past six months, both at baseline and a year later also did not show any relationship with CST (fourth sidebar, Figure 1).
Determinants for C. trachomatis acquisition in genital infection
To identify independent risk factors for acquiring a C. trachomatis infection within a year of sampling, a univariate and multivariate logistic regression analysis was performed (Table 2). A vaginal microbiota dominated by L. iners (CST III) was associated with an increased risk of acquiring C. trachomatis in our study population (OR: 2.58, 95% CI: 1.01–6.61). Being in a sexual relationship but living apart was also associated with an increased risk of acquiring C. trachomatis (OR: 14.51, 95% CI: 1.45–145.3).
Table 2.
Univariate and multivariate logistic regression analysis of determinants for the acquisition of C. trachomatis (N=115)
| Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|
| Acquiring Ct | OR | 95% CI | P | OR | 95% CI | P | |
| Median age first sexual contact | 16 | 1.03 | 0.89–1.19 | 0.71 | |||
| Education | |||||||
| Low | 2 (50%) | 1 | |||||
| Middle | 21 (50%) | 1.00 | 0.13–7.78 | 1.00 | |||
| High | 33 (53%) | 1.14 | 0.15–8.59 | 0.90 | |||
| Ethnicity | |||||||
| Dutch | 44 (52%) | 1 | |||||
| Surinamese/Dutch | 12 (50%) | 0.91 | 0.37–2.25 | 0.84 | |||
| Antilles | |||||||
| Asian | 3 (60%) | 1.36 | 0.22–8.58 | 0.74 | |||
| Turkish | 1 (50%) | 0.91 | 0.06–15.02 | 0.95 | |||
| CST before acquiring Ct | |||||||
| I (%) | 18 (30%) | 1 | 1 | ||||
| II (%) | 0 | 0 | 0 | 0.99 | 0 | 0 | 0.99 |
| III (%) | 25 (42%) | 2.67 | 1.08–6.59 | 0.03 | 2.58 | 1.01–6.61 | 0.04 |
| IV (%) | 15 (25%) | 1.49 | 0.58–3.84 | 0.41 | 1.42 | 0.53–3.81 | 0.49 |
| V (%) | 2 (3%) | 2.78 | 0.23–33.02 | 0.42 | 1.60 | 0.12–20.61 | 0.72 |
| Sexual partners in the previous six months | |||||||
| 0 | 2 (33%) | 1 | |||||
| 1 | 21 (45%) | 1.62 | 0.27–9.69 | 0.60 | |||
| 2–3 | 18 (56%) | 2.57 | 0.41–16.12 | 0.31 | |||
| >3 | 6 (60%) | 3.00 | 0.36–24.92 | 0.31 | |||
| Relationship | |||||||
| Living together | 1 (14%) | 1 | 1 | ||||
| Living apart | 20 (69%) | 13.33 | 1.39–127.6 | 0.03 | 14.51 | 1.45–145.3 | 0.02 |
| Single | 26 (47%) | 5.38 | 0.61–47.7 | 0.13 | 5.65 | 0.62–51.5 | 0.12 |
| Unknown | 13 (54%) | 7.09 | 0.74–68.2 | 0.09 | 7.54 | 0.76–74.9 | 0.09 |
Ct: C. trachomatis
CST: Community State Type
OR: Odds Ratio
95% CI: 95% Confidence Interval
P: Probability value
Median age, level of education, ethnicity, median age of the first sexual contact, and number of sexual partners in the previous six months were not significantly associated with acquiring a C. trachomatis infection (Table 2). To explore the effect of the various microbial taxa on acquiring C. trachomatis infection, the abundances of these taxa were incorporated in another logistic regression model. However, again only L. iners proved to be statistically significant (data not shown).
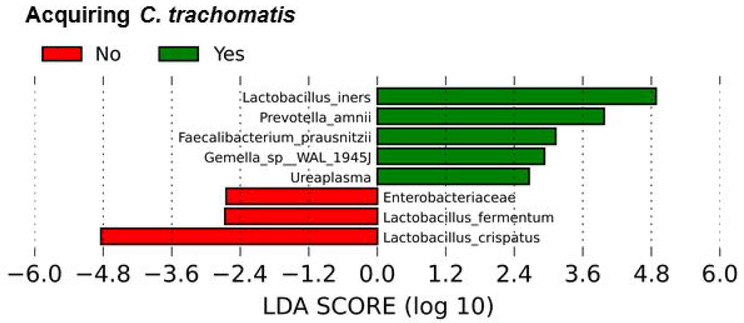
The linear discrimination analysis effect size (LEfSe) identified significantly abundant microbial taxa, when comparing samples collected from women who acquire C. trachomatis and women who do not with an alpha value <0.05. L. iners, P. amnii, and L. crispatus were the species that show statistically and biologically consistent differences between women acquiring C. trachomatis and women who do not (Figure 2). Furthermore, acquiring C. trachomatis infection was strongly associated with a higher abundance of L. iners, whereas not acquiring C. trachomatis infection was strongly associated with L. crispatus, both with an LDA score of 5 E05. Prevotella amnii was also associated with the acquisition of C. trachomatis. However, the abundance of this species was quite low in the various vaginal microbiota. Therefore, the meaning of this finding remains unclear.
Figure 2.
Association between microbial taxa and the acquisition of C. trachomatis. LEfSe analysis identified significantly abundant microbial taxa, when comparing samples collected from women who acquire C. trachomatis and women who do not with an alpha value <0.05. The taxa are ranked according to their effect size that are associated with different conditions with the highest median. The Linear Discriminant Analysis (LDA) score at the log10 scale is indicated at the bottom. The greater the LDA score is, the more significant the specific microbial taxa are in the comparison.
Discussion
This study focuses on the composition and structure of vaginal microbial communities prior to the acquisition of C. trachomatis infection in a relatively low risk population. In this cohort, the structure of the vaginal microbiota revealed five CSTs. Four of these CSTs were dominated by Lactobacillus species. One CST lacked Lactobacillus spp., and was heterogeneous in composition, comprising strict and facultative anaerobes. These CSTs have previously been described, albeit in different populations and in different frequencies [8]. In contrast to other studies, we observed a larger proportion of Caucasian women (24%) whose vaginal microbiota was not dominated by Lactobacillus spp. (CST IV). In a previous study, this dysbiotic state was present in 40% of African American and Hispanic women, just like our Surinamese/Dutch Antilles women, and only in 10% of Caucasian women [8].
Multivariate logistic regression analysis revealed that having a microbiota dominated by L. iners was an independent risk factor for acquiring C. trachomatis. This supported our hypothesis that aspects of the vaginal microbiota might create an environment associated with increased susceptibility to sexually transmitted pathogens, and that the environment either strongly favours or protects against C. trachomatis infection. Further, the analysis revealed that being in a relationship but living apart was also an independent risk factor for acquiring C. trachomatis, which might be explained by having casual sexual partners besides the steady sexual partner.
Epidemiological studies have shown an association between BV, either diagnosed by Amsel criteria or Nugent Score and increased risk of STIs including C. trachomatis [6,24,25]. High Nugent scores, which are indicative of the absence of Lactobacillus spp., and a high number of G. vaginalis and Mobiluncus spp., represents a diagnosis of BV. A large prospective study has shown that this high Nugent score was associated with an increased risk of acquiring STIs [14]. While no molecular definition of BV exists, it is characterized by a vaginal microbiota lacking Lactobacillus spp. and comprising a wide array of strict and facultative anaerobic bacteria [8,26,27]. This appears to be similar in composition to CST IV vaginal microbiota. Surprisingly, in our study, we did not observe an increased risk for C. trachomatis infection associated with women with CST IV vaginal microbiota. However, vaginal microbiota dominated by L. iners (CST III) at baseline was associated with an increased risk for C. trachomatis infection when assessed a year later. This result might be explained by the fact that vaginal microbiota dominated by L. iners has been shown to exhibit rapid change in composition in and out of community states similar to BV [21,28]. This might be explained by the larger diversity observed in the L. iners dominated CST as compared to the L. crispatus dominated CST. These findings might indicate that increased risk for STIs is actually not limited to women with BV at time of evaluation, but also women with vaginal microbiota comprising of L. iners. This is further supported by a study among Dutch women who were contact-traced by a chlamydia-positive partner. In this study, women who tested positive for C. trachomatis were more likely to have a vaginal microbiota dominated by L. iners as compared to women who tested negative [29].
The size of the study population was above the sample size needed to achieve 80%power and therefore well-powered to evaluate if the composition and structure of the vaginal microbiota is associated with susceptibility to C. trachomatis infection. However, this study should be repeated with a larger group of women to confirm our results and to give a more robust assessment on other possible determinants and confounders for the acquisition of C. trachomatis, such as ethnicity, sexual risk behavior, antibiotic and hormone contraceptive use. Another limitation of this study is the one year interval between assessing the composition of the vaginal microbiota and testing for C. trachomatis. The vaginal microbiota seems to be quite stable over time, especially in healthy women [22,30]. Some women undergo moderate or dramatic shifts just before and during menses. These fluctuations in vaginal microbiota composition often involve changes in the relative dominance of a limited number of different lactic acid producing bacterial species. Since our study population comprises healthy, low risk women, we do not expect to find major shifts in CST over time that could have influenced the outcome of this study. However, we do believe that these women should be followed over time, preferably with a shorter time interval between sampling, to see how their vaginal microbiota develops over time and what the influence of an infection with C. trachomatis is on the composition of the vaginal microbiota. Furthermore, serologic markers for C. trachomatis should be investigated to rule out any prior infection with C. trachomatis. In these follow-up studies, BV should also be studied, to see whether indeed only women with both BV and L. iners present in their vaginal microbiota are more susceptible for STIs.
This study demonstrates that vaginal microbiota dominated by Lactobacillus spp. does not necessarily protect against the acquisition of STIs like C. trachomatis. The specific species or possibly even strain of Lactobacillus is also of great importance to determine whether the vaginal microbiota can contribute to susceptibility to or protection against STIs. Having vaginal microbiota dominated by a L. iners-dominated community was associated with an increased the risk for acquiring C. trachomatis genital infection. Therefore, we hypothesize that the composition of the vaginal microbiota is indicative of increased host predisposition to acquiring STIs. The exact mechanism behind this still needs to be unraveled.
Key messages.
Having vaginal microbiota dominated by L. iners is associated with an increased risk for C. trachomatis infection
Specific signatures of vaginal microbiota could be indicative of increased host predisposition to acquiring STIs
The specific species or possibly even strain of Lactobacillus is of great importance to determine whether the vaginal microbiota can contribute to susceptibility to or protection against STIs
Acknowledgements
The authors would like to thank the Chlamydia Screening Intervention study group, JEAM van Bergen, IVF van den Broek, EEHG Brouwers, ELM op de Coul, HM Götz, CJPA Hoebe, JS Fennema, L Pars, and SM van Ravesteijn for supplying samples and data for this study. The authors also would like to thank Mike Humphrys and Hongqiu Yang from the Insitute for Genome Sciences in Baltimore for their help and support in processing and sequencing of the vaginal swabs.
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in STI and any other BMJPGL products and sub-licences such use and exploit all subsidiary rights, as set out in our licence http://group.bmj.com/products/journals/instructions-for-authors/licence-forms.
References
- 1.Centers for Disease Control and Prevention. Sexually transmitted diseases guidelines, 2015. MMWR Recomm Rep. 2015; 64(RR-03):1–137. [PMC free article] [PubMed] [Google Scholar]
- 2.Van Bergen JE, Götz HM, Richardus JH, et al. Prevalence of urogenital Chlamydia trachomatis increases significantly with level of urbanisation and suggests targeted screening approaches: results from the first national population based study in the Netherlands. Sex Transm Infect 2005;8:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Land JA, Van Bergen JE, Morré SA, et al. Epidemiology of Chlamydia trachomatis infection in women and the cost-effectiveness of screening. Hum Reprod Update 2010;16:189–204. [DOI] [PubMed] [Google Scholar]
- 4.Bebear C, de Barbeyrac B. Genital Chlamydia trachomatis infections. Clin Microbiol Infect 2009;15:4–10 [DOI] [PubMed] [Google Scholar]
- 5.Donders GG, Bosmans E, Dekeersmaecker A, et al. Pathogenesis of abnormal vaginal bacterial flora. Am J Obstet Gynecol 2000;182:872–8. [DOI] [PubMed] [Google Scholar]
- 6.Wiesenfeld HC, Hillier SL, Krohn MA, et al. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis 2003;36:663–8. [DOI] [PubMed] [Google Scholar]
- 7.Watts DH, Fazzari M, Minkoff H, et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J Infect Dis 2005;191:1129–39. [DOI] [PubMed] [Google Scholar]
- 8.Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 2011;108 Suppl 1 :4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buve A, Jespers V, Crucitti T, Fichorova RN. The vaginal microbiota and susceptibility to HIV. AIDS. 2014;28:2333–44. [DOI] [PubMed] [Google Scholar]
- 10.Witkin SS, Linhares IM, Giraldo P, Ledger WJ. An altered immunity hypothesis for the development of symptomatic bacterial vaginosis. Clin Infect Dis 2007;44:554–7. [DOI] [PubMed] [Google Scholar]
- 11.Mirmonsef P, Gilbert D, Zariffard MR, et al. The effects of commensal bacteria on innate immune responses in the female genital tract. Am J Reprod Immunol 2011;65:190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atassi F, Servin AL. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol Lett 2010;304:29–38. [DOI] [PubMed] [Google Scholar]
- 13.Gong Z, Luna Y, Yu P, et al. Lactobacilli inactivate Chlamydia trachomatis through lactic acid but not H2O2. PLoS One 2014;9:e107758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brotman RM, Klebanoff MA, Nansel TR, et al. Bacterial vaginosis assessed by gram stain and diminished colonization resistance to incident gonococcal, chlamydial, and trichomonal genital infection. J Infect Dis 2010;202:1907–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van den Broek IV, van Bergen JE, Brouwers EE, et al. Effectiveness of yearly, register based screening for chlamydia in the Netherlands: controlled trial with randomised stepped wedge implementation. BMJ 2012;345:e4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. Magoc T, Salzberg SL: FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011,27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC, Haas BJ, Clemente JC, et al. : UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011,27:2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal 2012,6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsen FA, Kodner RB, Armbrust EV. Pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 2010;11:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012;4:doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ness RB, Kip KE, Soper DE, et al. Bacterial vaginosis (BV) and the risk of incident gonococcal or chlamydial genital infection in a predominantly black population. Sex Transm Dis 2005;32:413–7. [DOI] [PubMed] [Google Scholar]
- 25.Allsworth JE, Peipert JF. Severity of bacterial vaginosis and the risk of sexually transmitted infection. Am J Obstet Gynecol 2011;205:e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005;353:1899–1911. [DOI] [PubMed] [Google Scholar]
- 27.Hardy L, Jespers V, Dahchour N, et al. Unravelling the Bacterial Vaginosis-Associated Biofilm: A Multiplex Gardnerella vaginalis and Atopobium vaginae Fluorescence In Situ Hybridization Assay Using Peptide Nucleic Acid Probes. PLoS One 2015;10:e0136658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravel J, Brotman RM, Gajer P, et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 2013;1:29. doi: 10.1186/2049-2618-1-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van der Veer C, Bruisten SM, Van der Helm JJ, et al. The cervicovaginal microbiota in women notified for Chlamydia trachomatis infection: A case-control study at the sexually transmitted infection outpatient clinic in Amsterdam, The Netherlands. Clin Infect Dis 2017;64:24–31. [DOI] [PubMed] [Google Scholar]
- 30.Chaban B, Links MG, Jayaprakash TP, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014. July 4;2:23. doi: 10.1186/2049-2618-2-23. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]