Abstract
Behavioral evidence suggests that beliefs about causal structure constrain associative learning, determining which stimuli can enter into association, as well as the functional form of that association. Bayesian learning theory provides one mechanism by which structural beliefs can be acquired from experience, but the neural basis of this mechanism is poorly understood. We studied this question with a combination of behavioral, computational, and neuroimaging techniques. Male and female human subjects learned to predict an outcome based on cue and context stimuli while being scanned using fMRI. Using a model-based analysis of the fMRI data, we show that structure learning signals are encoded in posterior parietal cortex, lateral prefrontal cortex, and the frontal pole. These structure learning signals are distinct from associative learning signals. Moreover, representational similarity analysis and information mapping revealed that the multivariate patterns of activity in posterior parietal cortex and anterior insula encode the full posterior distribution over causal structures. Variability in the encoding of the posterior across subjects predicted variability in their subsequent behavioral performance. These results provide evidence for a neural architecture in which structure learning guides the formation of associations.
SIGNIFICANCE STATEMENT Animals are able to infer the hidden structure behind causal relations between stimuli in the environment, allowing them to generalize this knowledge to stimuli they have never experienced before. A recently published computational model based on this idea provided a parsimonious account of a wide range of phenomena reported in the animal learning literature, suggesting a dedicated neural mechanism for learning this hidden structure. Here, we validate this model by measuring brain activity during a task that involves both structure learning and associative learning. We show that a distinct network of regions supports structure learning and that the neural signal corresponding to beliefs about structure predicts future behavioral performance.
Keywords: associative learning, Bayesian modeling, causal reasoning, context, fMRI, structure learning
Introduction
Classical learning theories posit that animals learn associations between sensory stimuli and rewarding outcomes (Rescorla and Wagner, 1972; Pearce and Bouton, 2001). These theories have achieved remarkable success in explaining a wide range of behaviors using simple mathematical rules, but numerous studies have challenged some of their foundational premises (Miller et al., 1995; Dunsmoor et al., 2015; Gershman et al., 2015). One particularly longstanding puzzle for these theories is the multifaceted role of contextual stimuli in associative learning. Some studies have shown that the context in which learning takes place is largely irrelevant (Bouton and King, 1983; Lovibond et al., 1984; Kaye et al., 1987; Bouton and Peck, 1989), whereas others have found that context plays the role of an “occasion setter,” modulating cue–outcome associations without itself acquiring associative strength (Swartzentruber and Bouton, 1986; Grahame et al., 1990; Bouton and Bolles, 1993; Swartzentruber, 1995). However, other studies suggest that context acts like another punctate cue, entering into summation and cue competition with other stimuli (Balaz et al., 1981; Grau and Rescorla, 1984). The multiplicity of such behavioral patterns defies explanation in terms of a single associative structure, suggesting instead that different structures may come into play depending on the task and training history.
Computational modeling has begun to unravel this puzzle using the idea that structure is a latent variable inferred from experience (Gershman, 2017). According to this theory, each structure corresponds to a causal model of the environment specifying the links among context, cues, and outcomes, as well as their functional form. The learner thus faces the joint problem of inferring both the structure and the strength of causal relationships, which can be implemented computationally using Bayesian learning (Griffiths and Tenenbaum, 2005; Körding et al., 2007; Meder et al., 2014). This theory can explain why different tasks and training histories produce different forms of context dependence: variations across tasks induce different probabilistic beliefs about causal structure. For example, Gershman (2017) showed that manipulations of context informativeness, outcome intensity, and number of training trials have predictable effects on the functional role of context in animal learning experiments (Odling-Smee, 1978; Preston et al., 1986).
If this account is correct, then we should expect to see separate neural signatures of structure learning and associative learning that are systematically related to behavioral performance. However, the direct neural evidence for structure learning is currently sparse (Collins et al., 2014; Madarasz et al., 2016; Tervo et al., 2016). In this study, we seek to address this gap using human fMRI and an associative learning paradigm adapted from Gershman (2017). On each block, subjects were trained on cue–context–outcome combinations that were consistent with a particular causal interpretation. Subjects were then asked to make predictions about novel cues and contexts without feedback, revealing the degree to which their beliefs conformed to a specific causal structure. We found that a variant of the structure learning framework developed by Gershman (2017) accounted for the subjects' predictive judgments, which led us to hypothesize a neural implementation of its computational components. We additionally found that an alternative structure learning model developed by Collins and Frank (2013) also accounts for the subjects' behavior, so we used both models to investigate the neural correlates of structure learning.
We found trial-by-trial signals tracking structure learning above and beyond associative learning. A whole-brain analysis revealed a univariate signature of Bayesian updating of the posterior distribution over causal structures in a frontoparietal network of regions, including the inferior part of posterior parietal cortex (PPC), lateral prefrontal cortex (lateral PFC), and rostrolateral PFC (RLPFC). Bayesian updating of structural beliefs according to the Collins and Frank (2013) model correlated with a network of regions that largely overlapped with the regions identified by our model, suggesting that both models tap into a generic structure learning mechanism in the brain. A multivariate analysis implicated some of those regions in the representation of the full posterior distribution over causal structures. Activity in two of those regions, the left inferior PPC and the right anterior insula, also predicted subsequent generalization on the test trials in accordance with the causal structure learning model. Our results provide new insight into the neural mechanisms of structure learning and how they constrain the acquisition of associations.
Materials and Methods
Subjects
Twenty-seven healthy subjects were enrolled in the fMRI portion of the study. Although we did not perform power analysis to estimate the sample size, it is consistent with the size of the pilot group of subjects that showed a robust behavioral effect (see Fig. 4, gray circles) Before data analysis, seven subjects were excluded due to technical issues, insufficient data, or excessive head motion. The remaining 20 subjects were used in the analysis (10 female, 10 male, 19–27 years of age, mean age 20 ± 2, all right handed with normal or corrected-to-normal vision). Additionally, 10 different subjects were recruited for a behavioral pilot version of the study that was conducted before the fMRI portion. All subjects received informed consent and the study was approved by the Harvard University Institutional Review Board. All subjects were paid for their participation.
Figure 4.
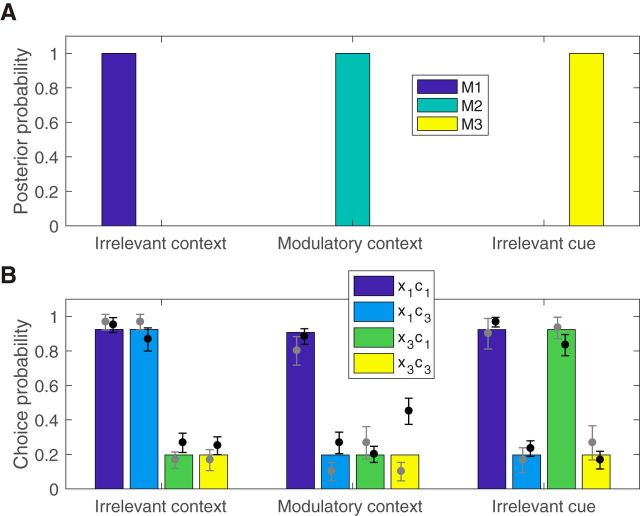
Generalization on the test trials. A, Posterior probability distribution over causal structures in each condition at the end of training. Each block was simulated independently and the posterior probabilities were averaged across blocks of the same condition. B, Choice probabilities on the test trials for subjects in the pilot (gray circles) and fMRI (black circles) portions of the study are shown overlaid over model choice probabilities (colored bars). Each color corresponds to a particular combination of an old (x1) or new (x3) cue in an old (c1) or new (c3) context. Error bars indicate within-subject SEM (Cousineau, 2005).
Experimental design and statistical analysis
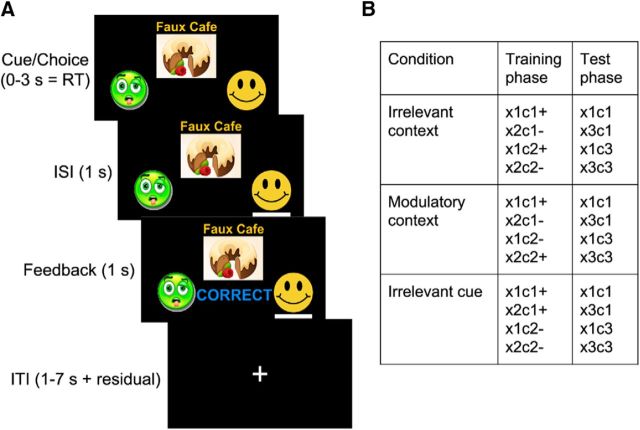
We adapted the task used in Gershman (2017) to a within-subjects design. Subjects were told that they would play the role of a health inspector trying to determine the cause of illness in different restaurants around the city. The experiment consisted of nine blocks. Each block consisted of 20 training trials followed by four test trials. On each training trial, subjects were shown a given cue (the food) in a given context (the restaurant) and asked to predict whether that cue–context combination would cause sickness. After making a prediction, they were informed whether their prediction was correct (Fig. 1A). On a given block, the assignment of stimuli to outcomes was deterministic such that the same cue–context pair always led to the same outcome. Even though the computational model could support stochastic and dynamically evolving stimulus–outcome contingencies, our goal was to provide a minimalist design that can assess the main predictions of the theory. There were four distinct training cue–context pairs (two foods × two restaurants) on each block such that two of the pairs always caused sickness and the other two never caused sickness. Each cue–context pair was shown five times in each block for a total of 20 randomly shuffled training trials.
Figure 1.
Experimental design. A, Timeline of events during a training trial. Subjects are shown a cue (food) and context (restaurant) and are asked to predict whether the food will make a customer sick. They then see a line under the chosen option, and feedback indicating a “correct” or “incorrect” response. B, Stimulus–outcome contingencies in each condition. Cues are denoted by x1, x2, and x3 and contexts by c1, c2, and c3. Outcome presentation is denoted by “+” and no outcome by “−.”
Crucially, the stimulus–outcome contingencies in each block were designed to promote a particular causal interpretation of the environment (Figs. 1B, 2). On “irrelevant context” blocks, one cue caused sickness in both contexts, whereas the other cue never caused sickness, thus rendering the contextual stimulus irrelevant for making correct predictions. On “modulatory context” blocks, the cue–outcome contingency was reversed across contexts such that the same cue caused sickness in one context but not the other and vice versa for the other cue. On these blocks, context thus acted like an “occasion setter” determining the sign of the cue–outcome association. Finally, on “irrelevant cue” blocks, both cues caused sickness in one context, but neither cue caused sickness in the other context, thus favoring an interpretation of context acting as a punctate cue. There were no explicit instructions or other signals that indicated the different block conditions other than the stimulus–outcome contingencies. We based our experimental design on the fact that a previously published model with similar structures could capture a wide array of behavioral phenomena (Gershman, 2017) and that the chosen stimuli–outcome contingencies establish a clear behavioral pattern that we can build upon to explore the neural correlates of structure learning.
Figure 2.
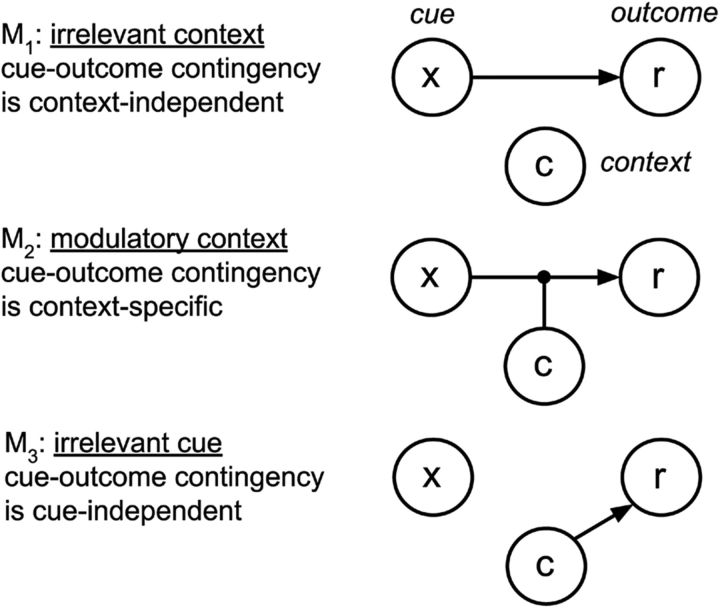
Hypothesis space of causal structures. Each causal structure is depicted as a network in which the nodes represent variables and the edges represent causal connections. In M2, the context modulates the causal relationship between the cue and the outcome. (Figure adapted with permission from Gershman, 2017.)
Behavior was evaluated on four test trials during which subjects were similarly asked to make predictions but this time without receiving feedback. Subjects were presented with one novel cue and one novel context, resulting in four (old cue vs new cue) × (old context vs new context) randomly shuffled test combinations (Fig. 1B). The old cue and the old context were always chosen such that their combination caused sickness during training. Importantly, different causal structures predict different patterns of generalization on the remaining three trials that contain a new cue and/or a new context. If context is deemed to be irrelevant, then the old cue should always predict sickness even when presented in a new context. If a modulatory role of context is preferred, then no inferences can be made about any of the three pairs that include a new cue or a new context. Finally, if context is interpreted as acting like a cue, then both the old cue and the new cue should predict sickness in the old context but not in the new context.
Each block was assigned to one of the three conditions (irrelevant context, modulatory context, or irrelevant cue) and each condition appeared three times for each subject for a total of nine blocks. The block order was randomized in groups of three such that the first three blocks covered all three conditions in a random order and so did the next three blocks and the final three blocks. We used nine sets of foods and restaurants corresponding to different cuisines (Chinese, Japanese, Indian, Mexican, Greek, French, Italian, fast food, and brunch). Each set consisted of three clipart food images (cues) and three restaurant names (contexts). For each subject, blocks were randomly matched with cuisines such that subjects had to learn and generalize for a new set of stimuli on each block. The assignment of cuisines was independent of the block condition. The valence of the stimuli was also randomized across subjects such that the same cue–context pair could predict sickness for some subjects but not others.
Before the experiment, the investigator read the task instructions aloud and subjects completed a single demonstration block of the task on a laptop outside of the scanner. Subjects completed nine blocks of the task in the scanner, with one block per scanner run. Each block had a duration of 200 s during which 100 volumes were acquired (TR = 2 s). At the start of each block, a fixation cross was shown for 10 s and the corresponding five volumes were subsequently discarded. This was followed by the training phase, which lasted 144 s. The event sequence within an example training trial is shown in Figure 1. At trial onset, subjects were shown a food and restaurant pair and instructed to make a prediction. Subjects reported their responses by pressing the left or the right button on a response box. After trial onset, subjects were given 3 s to make a response. A response was immediately followed by a 1 s interstimulus interval (ISI) during which their response was highlighted. The residual difference between 3 s and their reaction time was added to the subsequent intertrial interval (ITI). The ISI was followed by a 1 s feedback period during which they were informed whether their choice was correct. If subjects failed to respond within 3 s of trial onset, then no response was recorded and at feedback they were informed that they had timed out. During the ITIs, a fixation cross was shown. The trial order and the jittered ITIs for the training phase were generated using the optseq2 program (Greve, 2002) with ITIs between 1 and 12 s. The training phase was followed by a 4 s message informing the subjects that they were about to enter the test phase. The test phase lasted 36 s. Test trials had a similar structure as training trials, with the difference that subjects were given 6 s to respond instead of 3 and there was no ISI or feedback period. The ITIs after the first 3 test trials were 2, 4, and 6 s, randomly shuffled. The last training trial was followed by a 6 s fixation cross. The stimulus sequences and ITIs were pre-generated for all subjects. The task was implemented using the PsychoPy2 package (Peirce, 2007). The subjects in the behavioral pilot version of the study performed an identical version of the experiment except that it was conducted on a laptop.
Behavioral data were analyzed using t tests and computational modeling. Brain-imaging data were analyzed using general linear models. The modeling for behavioral and neural data is described in more detail below.
Causal structure learning model
We implemented the causal structure learning model presented in Gershman (2017), with the difference that the additive context structure was replaced by an irrelevant cue structure. This replacement was motivated by our observation that the model with an irrelevant cue structure had higher model evidence than the original model for our behavioral data. The key idea is that learners track the joint posterior over associative weights (w) and causal structures (M) computed using Bayes' rule as follows:
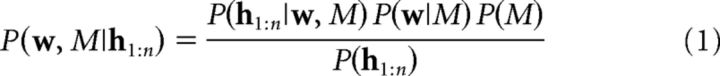 |
where h1:n = (x1:n, r1:n, c1:n) denotes the training history for trials 1 to n (cue–context–outcome combinations). The likelihood P(h1:n|w, M) encodes how well structure M predicts the training history, the prior P(w|M) specifies an inductive bias for the weight vector, and the prior over structures P(M) was taken to be uniform, reflecting the assumption that all structures are equally probable a priori.
Generative model.
Our model is based on the following assumptions about the dynamics that govern associations between stimuli and outcomes in the world. The training history is represented as h1:n = (x1:n, r1:n, c1:n) for trials 1 to n consisting of the following variables. The first variable, xn ∈ ℝD, is the set of D cues observed at time n, where xnd = 1 indicates that cue d is present and xnd = 0 indicates that it is absent. Therefore, each cue can be regarded as a “one-hot” D-dimensional vector and xn can be viewed as the sum of all cues present on trial n. In our simulations, we use D = 3 and we only have a single cue (the food) present on each trial. The second variable, cn ∈ {1, …, K}, is the context that can take on one of K discrete values. Although contexts could in principle be represented as vectors as well, we restrict the model to one context per trial for simplicity. In our simulations, we take K = 3. The third variable, rn ∈ ℝ, is the outcome. In our simulations, we use rn = 1 for “sick” and rn = 0 for “not sick”.
We consider three specific structures relating the above variables. All the structures have in common that the outcome is assumed to be drawn from a Gaussian with variance σr2 = 0.01 as follows:
where we have left the dependence on cn and xn implicit. The structures differ in how the mean r̄n is computed. For the irrelevant context (M1):
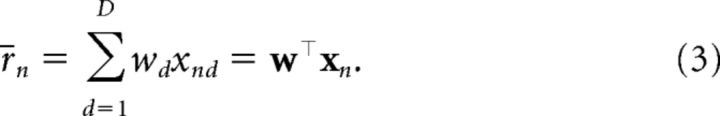 |
where d indexes the set of D cues. Under this structure, context cn plays no role in determining the expected outcome r̄n on trial n. Instead, a single set of weights w dictates the associative strength between each cue and the outcome such that the expected outcome on a given trial is the sum of the associative weights of all present cues. The idea that context is irrelevant for stimulus–outcome associations is consistent with number of behavioral studies (Bouton and King, 1983; Lovibond et al., 1984; Kaye et al., 1987; Bouton and Peck, 1989).
For the modulatory context (M2):
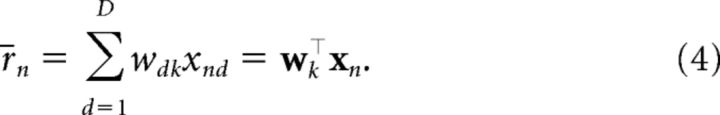 |
when cn = k. Under this structure, each context cn = k specifies its own weight vector wk. Therefore, the same cue can make completely different predictions in different contexts. The view that context modulates stimulus–outcome associations is also supported by previous behavioral findings (Swartzentruber and Bouton, 1986; Grahame et al., 1990; Bouton and Bolles, 1993; Swartzentruber, 1995).
For the irrelevant cue (M3):
where c̃nk = 1 if cn = k, and 0 otherwise. This structure is symmetric with respect to M1, in that we assume a one-hot context vector c̃n that encodes the context in the same way that xn encodes the cue in M1. The weight vector w thus contains entries for contexts only. Previous work also suggests that context sometimes acts like another cue (Balaz et al., 1981; Grau and Rescorla, 1984) and that cues are sometimes ignored when they are not predictive of outcomes (Mackintosh, 1975). Note that this is different from the additive structure used in Gershman (2017), in which the cue and the context summate together to predict the outcome. We chose this simpler structure because it more closely reflects the structure of the task and preliminary model comparisons revealed that it provides a better account of behavior (data not shown).
We assume that each weight is drawn independently from a Gaussian prior with mean w0 and variance σw2. Each weight can change slowly over time according to a Gaussian random walk with variance τ2. These free parameters were fit using data from the behavioral pilot version of the study.
In summary, each causal structure corresponds to an internal model of the world in which the relationship among cues, contexts, and outcomes can be described by a distinct linear-Gaussian dynamical system (LDS). Although the LDS assumptions might seem excessive given the deterministic nature of the task, they have been widely used in the classical conditioning studies (Dayan and Kakade, 2001; Kakade and Dayan, 2002; Kruschke, 2008; Gershman, 2015) to provide a parsimonious account for various learning phenomena. Here, we use them for the purposes of tractability and to remain consistent with the causal learning model that Gershman (2017) used to explain the seemingly contradictory roles of context reported in the animal learning literature. These causal structures were inspired by different theories that have been advanced in various forms in the literature, none of which has been able to capture the broad range of results on its own.
Probabilistic inference.
Assuming this generative model, a rational agent can use Bayesian inference to invert the model and use its training history h1:n to learn the underlying causal structure M and its associative weights w (Eq. 1). To achieve this, first, we can compute the posterior over the weights for a given model M using Bayes' rule as follows:
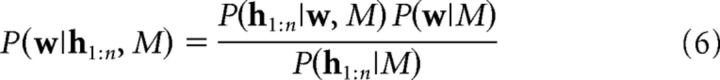 |
For M1, the posterior at time n is as follows:
with parameters updated recursively as follows:
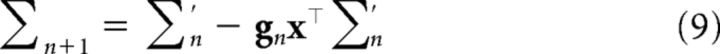 |
where ∑′n = ∑n + τ2I. These update equations are known as “Kalman filtering,” an important algorithm in engineering and signal processing that has recently been applied to animal learning (Dayan and Kakade, 2001; Kruschke, 2008; Gershman, 2015). The initial estimates are given by the parameters of the prior: ŵ0 = 0, ∑0 = σw2I. The Kalman gain gn (a vector of learning rates) is given by the following:
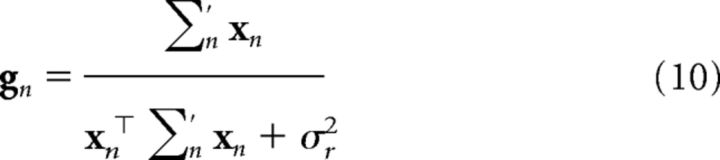 |
The same equations apply to M2, but the mean and covariance are context specific: ŵnk and ∑nk. Accordingly, the Kalman gain is modified as follows:
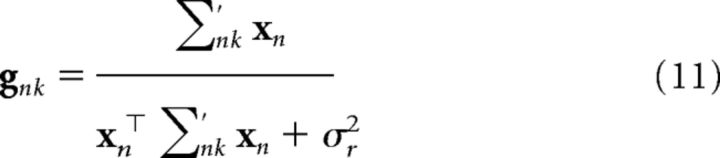 |
if cn = k and a vector of zeros otherwise. For M3, the same equations as M1 apply, but to the context vector c̃n.
To make predictions about future outcomes, we need to compute the posterior predictive expectation, which is also available in closed form as follows:
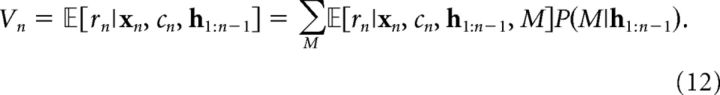 |
The first term in Equation 12 is the posterior predictive expectation conditional on model M as follows:
where, again, the variables are modified depending on what model is being considered. The second term in Equation 12 is the posterior probability of model M, which can be updated according to Bayes' rule as follows:
where the likelihood is given by the following:
To make predictions for the predictive learning experiment, we mapped the posterior predictive expectation onto choice probability (outcome vs no outcome) by a logistic sigmoid transformation as follows:
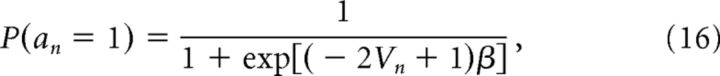 |
where an = 1 indicates a prediction that the outcome will occur and an = 0 indicates a prediction that the outcome will not occur. The free parameter β corresponds to the inverse softmax temperature and was fit based on data from the behavioral pilot portion of the study.
In summary, we use standard Kalman filtering to infer the parameters of the LDS corresponding to each causal structure. This yields a distribution over associative weights w for each causal structure M (Eq. 6), which we can use in turn to compute the posterior distribution over all three causal structures (Eq. 14). The joint posterior over weights and causal structures is then used to predict the expected outcome Vn (Eq. 12) and the corresponding decision an (Eq. 16). Our model thus makes predictions about computations at two levels of inference: at the level of causal structures (Eq. 14) and at the level of associative weights for each structure (Eq. 6).
Parameter estimation.
The model has four free parameters: the mean w0 and variance σw2 of the Gaussian prior from which the weights are assumed to be drawn, the variance of the process noise τ2, and the inverse temperature β used in the logistic transformation from predictive posterior expectation to choice probability. Intuitively, w0 corresponds to the initial weight given to a cue or context before observing any outcome, σw2 corresponds to the level of uncertainty in this initial estimate, τ2 reflects how much we expect the weights to change over time, and β reflects choice stochasticity. We estimated a single set of parameters based on choice data from the behavioral pilot version of the study using maximum log-likelihood estimation (see Fig. 4B, gray circles). We preferred this approach over estimating a separate set of parameters for each subject because it tends to avoid overfitting, produces more stable estimates, and has been frequently used in previous studies (Daw et al., 2006; Gläscher, 2009; Gershman et al., 2009; Gläscher et al., 2010). In addition, because none of these pilot subjects participated in the fMRI portion of the study, this procedure ensured that the parameters used in the final analysis were not overfit to the choices of the scanned subjects. For the purposes of fitting, the model was evaluated on the same stimulus sequences as the pilot subjects, including both training and test trials. Each block was simulated independently; that is, the parameters of the model were reset to their initial values before the start of training. The likelihood of the subject's response on a given trial was estimated according to the choice probability given by the model on that trial. Maximum likelihood estimation was computed using MATLAB's fmincon function with 25 random initializations. The bounds on the parameters were w0 ∈ [0, 1], σw2 ∈ [0, 10], τ2 ∈ [0, 1], and β ϵ [0, 10], all initialized with noninformative uniform priors.
The fitted values of the parameters are shown in Table 1. All other parameters were set to the same values as described in Gershman (2017). We used these parameter estimates to construct model-based regressors for the fMRI analysis. For the behavioral analysis, we trained and tested the model on each block separately and reported the choice probabilities on test trials averaged across conditions (see Fig. 4).
Table 1.
Model comparison favors the full causal structure learning model (M1, M2, and M3) and the clustering model (RL + clustering)
| Model | Free parameters | PXP | Pearson's r |
|---|---|---|---|
| M1, M2, M3 | σw2 = 0.0157, β = 2.6849, τ2 = 7.5724 × 10−5, w0 = 0.2382 | 0.8840 | r = 0.97, p < 10−7 |
| M1 | σw2 = 0.0079, β = 1.9441, τ2 = 1.3340 × 10−5, w0 = 0.2641 | 0.0013 | r = 0.61, p = 0.0347 |
| M2 | σw2 = 0.0570, β = 2.5302, τ2 = 1.3049 × 10−9, w0 = 0.3282 | 0.0017 | r = 0.73, p = 0.0076 |
| M3 | σw2 = 0.0111, β = 1.5085, τ2 = 3.8610 × 10−11, w0 = 0.1722 | 0.0003 | r = 0.59, p = 0.0447 |
| Simple RL | η = 0.8888, β = 2.3983, V 0 = 0.3188 | 0.0027 | r = 0.73, p = 0.0076 |
| RL + generalization | η = 0.5579, β = 2.3777, V 0 = 0.2175 | 0.0004 | r = 0.88, p = 0.0002 |
| RL + clustering | η = 0.8397, β = 2.4166, α = 1.3963, V 0 = 0.2624 | 0.1096 | r = 0.98, p < 10−7 |
Alternative models
Single causal structure.
We evaluated versions of the model that contain only a single causal structure (M1, M2, or M3). Theories corresponding to each of these structures have been advanced as potential explanations of the role of context in associative learning (Gershman, 2017), making them plausible candidates for explaining the data. We fit the four free parameters w0, σw2, τ2, and β separately for each of the three single structure models (Table 1, M1, M2, and M3).
Simple reinforcement learning.
We also evaluated a simple reinforcement learning (RL) model that learns a separate value Vn(x, c) for each cue–context pair (x, c). In particular, after observing the outcome rn on trial n, the expectation for the observed cue–context pair (xn, cn) is updated as follows:
where xn is the cue that was presented on trial n (i.e., xnxn = 1) and η is the learning rate. The values of all other cue–context pairs remain unchanged (i.e., Vn+1(i, j) = Vn(i, j) ∀ (i, j) ≠ (xn, cn)). Choices were modeled using the same logistic sigmoid transformation as before (Eq. 16). All values were initialized to V0.
This model has three free parameters: the learning rate η ϵ [0, 1], the inverse softmax temperature β ϵ [0, 10], and the initial values V 0 ϵ [0, 1], which were fit in the same way as the causal structure learning model (Table 1, simple RL).
Reinforcement learning with generalization.
Because the simple RL model treats each cue–context pair as a unique stimulus, it always predicts V0 for previously unseen cue–context pairs. To allow generalization to new cue–context pairs, we extended the simple RL model in the following way: if either the cue or the context is unknown, then the model takes the mean value over the unknown quantity as experienced in the current block. In particular, if a cue–context pair (xn, cn) has never been experienced, but either the cue xn or the context cn has been seen in other cue–context pairs, then the predicted value is computed as follows:
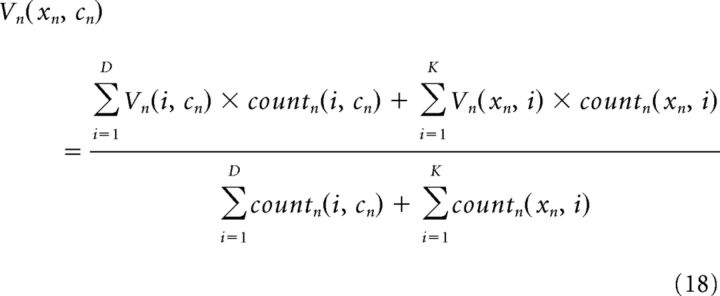 |
where countn(x, c) is the number of times a cue–context pair (x, c) has appeared in trials 1 … n. If neither the cue nor the context were seen before, then the predicted value is V0. Note that this extension pertains to predictions only; for learning, the value of new cue–context pairs is still initialized at V0. The free parameters η, β, and V0 were fit in the same way as the simple RL model (Table 1, RL + generalization).
Reinforcement learning with clustering.
We also implemented a structure learning model proposed by Collins and Frank (2013) that clusters cues and contexts into latent states, also referred to as “task sets.” Reinforcement learning is then performed over this clustered latent state space rather than the original space of cue–context pairs. Structure learning in this case refers to the process of building the latent state space, whereas in our model, we define structure learning as the process of arbitrating among an existing set of candidate causal structures.
Clustering is performed independently for cues and contexts such that cues are assigned to one set of clusters and contexts are assigned to a different set of clusters. Cluster membership is tracked probabilistically by P(zx|xn) and P(zc|cn) for cues and contexts, respectively. For a new cue xn on trial n, the cluster assignment probabilities are initialized as follows:
 |
where α is a concentration parameter and P(zx|i, h1:n−1) = 0 for unseen cues i. This is similar to a “Chinese restaurant process” (Gershman and Blei, 2012) and implements a “rich-get-richer” dynamic that favors popular clusters that already have many cues assigned to them. Note that a new cluster is created for each new cue. Cluster membership P(zc|cn, h1:n−1) for new contexts cn is initialized in the same way.
A prediction is generated by selecting the maximum a priori cue cluster zx′ = arg maxzx P(zx|xn, h1:n−1) and context cluster zc′ = arg maxzc P(zc|cn, h1:n−1) and using the value Vn(zx′, zc′) in the logistic sigmoid transformation (Eq. 16).
Once an outcome rn is observed, the posterior distributions over clusters are updated according to the following:
where the likelihood P(rn|zx, zc, h1:n−1) is estimated based on the cluster values Vn(zx, zc) and Equation 16.
Finally, the maximum a posteriori cue cluster zx″ = arg maxzx P(zx|xn, h1:n) and context cluster zc″ = arg maxzc P(zc|cn, h1:n) are selected based on the updated posterior distributions, and their value is updated according to the following:
This model has four free parameters: the learning rate η ϵ [0, 1], the inverse softmax temperature β ϵ [0, 10], the concentration parameter α ϵ [0, 10], and the initial values V 0 ϵ [0, 1], which were fit in the same way as the other models (Table 1, RL + clustering).
Model comparison
To select models for analyzing the neural data, we performed random-effects Bayesian model selection (Rigoux et al., 2014) based on the behavioral data from the fMRI session. Because we fit the free parameters using data from the pilot portion of the study, there was no need to penalize for overfitting, so we computed the model evidence as the probability of the subject's choices in the fMRI portion of the study (i.e., the model likelihood). This is equivalent to assuming that the probability density of the parameters is concentrated on the parameter settings obtained from the pilot data. The model evidences were then used to compute the protected exceedance probability (PXP) for each model, which indicates the probability that the given model is the most frequently occurring model in the population.
fMRI data acquisition
Scanning was carried out on a 3T Siemens Magnetom Prisma MRI scanner with the vendor 32-channel head coil at the Harvard University Center for Brain Science Neuroimaging. A T1-weighted high-resolution multi-echo magnetization-prepared rapid-acquisition gradient echo (ME-MPRAGE) anatomical scan (van der Kouwe et al., 2008) of the whole brain was acquired for each subject before any functional scanning (176 sagittal slices, voxel size = 1.0 × 1.0 × 1.0 mm, TR = 2530 ms, TE = 1.69 - 7.27 ms, TI = 1100 ms, flip angle = 7°, FOV = 256 mm). Functional images were acquired using a T2*-weighted echo-planar imaging (EPI) pulse sequence that employed multiband RF pulses and Simultaneous multi-slice (SMS) acquisition (Feinberg et al., 2010; Moeller et al., 2010; Xu et al., 2013). In total, 9 functional runs were collected per subject, with each run corresponding to a single task block (84 interleaved axial–oblique slices per whole-brain volume, voxel size = 1.5 × 1.5 × 1.5 mm, TR = 2000 ms, TE = 30 ms, flip angle = 80°, in-plane acceleration (GRAPPA) factor = 2, multiband acceleration factor = 3, FOV = 204 mm). The initial 5 TRs (10 s) were discarded as the scanner stabilized. Functional slices were oriented to a 25° tilt toward coronal from AC–PC alignment. The SMS–EPI acquisitions used the CMRR-MB pulse sequence from the University of Minnesota. Four subjects failed to complete all nine functional runs due to technical reasons and were excluded from the analyses. Three additional subjects were excluded due to excessive motion.
fMRI preprocessing
Functional images were preprocessed and analyzed using SPM12 (Wellcome Department of Imaging Neuroscience, London). Each functional scan was realigned to correct for small movements between scans, producing an aligned set of images and a mean image for each subject. The high-resolution T1-weighted ME-MPRAGE images were then coregistered to the mean realigned images and the gray matter was segmented out and normalized to the gray matter of a standard Montreal Neurological Institute (MNI) reference brain. The functional images were then normalized to the MNI template (resampled voxel size 2 mm isotropic), spatially smoothed with a 8 mm full-width at half-maximum Gaussian kernel, high-pass filtered at 1/128 Hz, and corrected for temporal autocorrelations using a first-order autoregressive model.
Univariate analysis
We defined two general linear models (GLMs) based on the causal structure learning model (GLM 1 and GLM 2) and two GLMs based on the clustering model (GLM 3 and GLM 4). Every GLM had two impulse regressors convolved with the canonical hemodynamic response function (HRF) on all training trials: a stimulus regressor at trial onset and a feedback regressor at feedback onset. For every GLM, the feedback regressor included two parametric modulators that differed across the GLMs. The parametric modulators were not orthogonalized. In addition, all GLMs included six motion regressors and a constant regressor for baseline activity.
For all group-level analyses, we report t-contrasts with single voxels thresholded at p < 0.001 and whole-brain cluster familywise error (FWE) correction applied at significance level α = 0.05. Anatomical regions of the peak voxels were labeled using the Automated Anatomical Labeling atlas (Tzourio-Mazoyer et al., 2002; Rolls et al., 2015), the SPM Anatomy Toolbox (Eickhoff et al., 2005), and the CMA Harvard-Oxford atlas (Desikan et al., 2006). All voxel coordinates are reported in Montreal Neurological Institute (MNI) space.
GLM 1.
The rationale behind GLM 1 was to look for brain regions that might be responsible for inferring the causal structure (i.e., structure learning, Eq. 14) versus inferring the associative weights (i.e., associative learning, Eq. 6).
At feedback onset on trial n, a region that updates the posterior over causal structures would exhibit a signal that is correlated with the magnitude of the discrepancy between the prior P(M|h1:n−1) and the posterior P(M|h1:n). We quantified this discrepancy by the Kullback–Leibler (KL) divergence:
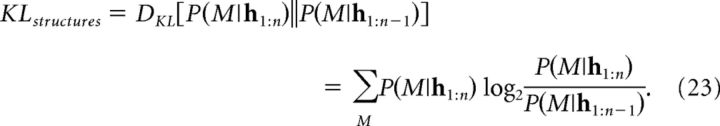 |
Similarly, a region involved in updating the associative weights would show activity correlated with the discrepancy between the weight prior and the weight posterior (i.e., the probability distribution over the weights before and after the update). Because the model keeps track of the weights for all structures, we reasoned that a region involved in associative weight updating would show activity that is correlated with the KL divergence between the joint prior and the joint posterior over the weights for all structures, which can be factored into the following:
where each summand represents the KL divergence between the posterior and the prior over the weights for the respective causal structure. The KL divergence for M1 is given by the following:
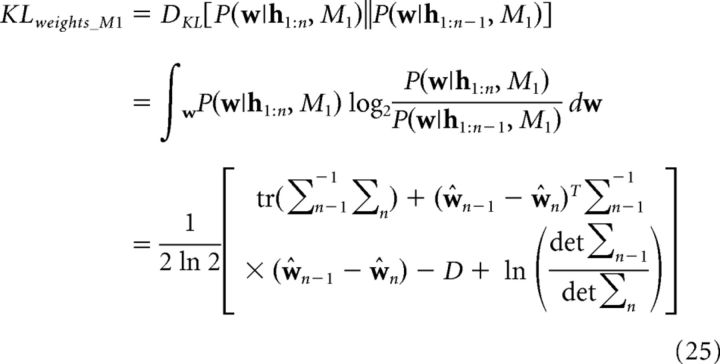 |
where D denotes the number of weights, σn denotes the posterior covariance on trial n, and dividing by ln 2 converts the result to bits. Equation 25 follows from the fact that the weights are normally distributed (Eq. 7). KLweights_M2 and KLweights_M3 were computed analogously.
We used KLstructures and KLweights as parametric modulators for the feedback regressor. We were primarily interested in KLstructures because it reflects the structure learning update. Previous work (Mumford et al., 2015) suggests that orthogonalizing it with respect to KLweights would not make a difference for the beta coefficients for KLstructures, whereas at the same time it would complicate the analysis of KLweights. Therefore, we did not orthogonalize the parametric modulators with respect to each other nor with respect to the feedback regressor. To look for signals specifically related to structure updating above and beyond associative weight updating, we computed the contrast KLstructures − KLweights.
GLM 2.
Another possibility is that only the weights of the most likely causal structure are updated. This approximation resembles the way in which the clustering model only updates the value of the maximum a posteriori clusters. GLM 2 is defined in the same way as GLM 1 except that only the weight update for the maximum a posteriori causal structure M′ = arg maxMP(M|h1:n) is included, as follows:
GLM 3. GLM 3 was based on the clustering model. Analogously to GLM 1, the purpose was to look for regions responsible for updating the cluster assignments (i.e., structure learning, in the sense used by Collins and Frank, 2013; Eq. 20) versus updating the cluster values (i.e., associative learning; Eq. 22).
Similarly to GLM 1, we quantified cluster updating as the KL divergence between the posterior (Eq. 20) and the prior (Eq. 19) over cluster assignments conditioned on the cue–context pair (xn, cn). Because clusterings for cues and contexts are independent, this can be factored as a sum of the KL divergences for cues and contexts as follows:
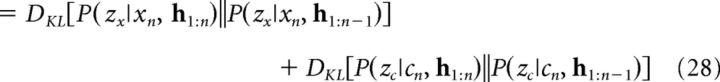 |
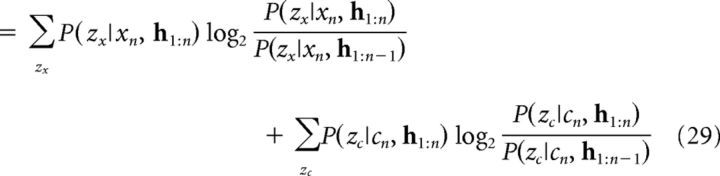 |
Associative updating was quantified by the (cluster) prediction error (Eq. 22) as follows:
We used KLclusters and cluster prediction error (CPE) as parametric modulators for the feedback regressor, not orthogonalized. As in GLM 1, we were primarily interested in KLclusters as a proxy for the structure learning update, so we computed the contrast KLclusters − CPE.
GLM 4.
As a control, we also included a GLM for the clustering model that was identical to the GLM used to analyze EEG data in Collins and Frank (2016). It had the clustering model prediction error CPE = rn − Vn(zx″, zc″) (Eq. 30) and the simple (or “flat”) RL prediction error FPE = rn − Vn(xn, cn) (Eq. 17) as parametric modulators at feedback onset, not orthogonalized. We then computed the contrast CPE − FPE to find brain regions that encode value updating specific to the clustering model.
GLM comparison.
We used random-effects Bayesian model selection (Rigoux et al., 2014) to compare GLMs based on how well they fit whole-brain neural activity. Although we did not expect our GLMs to account for the activity of all voxels, we did not select a priori regions of interest (ROIs) and therefore had no reason to exclude any particular voxels from the analysis. We approximated the log model evidence as −0.5 * BIC, where BIC is the Bayesian information criterion, which we computed using the residual variance of the GLM fits. The BIC quantifies how closely the GLM matches the neural activity of a given subject while adding a penalty proportional to the number of regressors in the GLM to account for overfitting. Bayesian model selection then produced a PXP for each GLM, which is the probability that this is the most frequently occurring GLM in the population.
Multivariate analysis
Representational similarity analysis (RSA).
We used RSA to identify candidate brain regions that might encode the full posterior distribution over causal structures (Eq. 14) in their multivariate activity patterns (Kriegeskorte et al., 2008). On a given trial, we expected Bayesian updating to occur when the outcome of the subject's prediction is presented at feedback onset (i.e., whether they were correct or incorrect). We therefore sought to identify brain regions that represent the posterior P(M|h1:n) at feedback onset.
To identify regions with a high representational similarity match for the posterior, we used an unbiased whole-brain “searchlight” approach. For each voxel of the entire volume, we defined a spherical ROI (searchlight) of 4 mm radius (Kriegeskorte et al., 2006) centered on that voxel, excluding voxels outside the brain (equivalently, radius = 2.6667 voxels, or up to 81 voxels in each searchlight). For each subject and each searchlight, we computed a 180 × 180 representational dissimilarity matrix R (the neural RDM) such that the entry in row i and column j is the cosine distance between the neural activity patterns on training trial i and training trial j as follows:
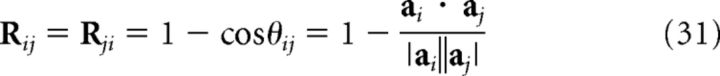 |
where θij is the angle between the 81-dimensional vectors ai and aj, which represent the instantaneous neural activity patterns at feedback onset on training trials i and j, respectively, in the given searchlight for the given subject. Neural activations entered into the RSA were obtained using a GLM with distinct impulse regressors convolved with the HRF at trial onset and feedback onset on each trial (test trials had regressors at trial onset only). The neural activity of a given voxel was thus simply its beta coefficient of the regressor for the corresponding trial and event. Since the matrix is symmetric and Rii = 0, we only considered entries above the diagonal (i.e., i < j). The cosine distance is equal to 1 minus the normalized correlation (i.e., the cosine of the angle between the two vectors), which has been preferred over other similarity measures as it better conforms to intuitions about similarity both for neural activity and for probability distributions (Chan et al., 2016).
Similarly, we computed an RDM (the model RDM) such that the entry in row i and column j is the cosine distance between the posterior on training trial i and training trial j, as computed by model simulations using the stimulus sequences experienced by the subject on the corresponding blocks.
If neural activity in the given searchlight encodes the posterior, then the neural RDM should resemble the model RDM: trials on which the posterior is similar should have similar neural representations (i.e., smaller cosine distances), whereas trials on which the posterior is dissimilar should have dissimilar neural representations (i.e., larger cosine distances). This intuition can be formalized using Spearman's rank correlation coefficient between the model RDM and the neural RDM (n = 180 × 179/2 = 16110 unique pairs of trials in each RDM). A high coefficient implies that pairs of trials with similar posteriors tend show similar neural patterns while pairs of trials with dissimilar posteriors tend to show dissimilar neural patterns. Spearman's rank correlation is a preferred method for comparing RDMs over other correlation measures because it does not assume a linear relationship between the RDMs (Kriegeskorte et al., 2008). Therefore, for each voxel and each subject, we obtained a single Spearman's ρ that reflects the representational similarity match between the posterior and the searchlight centered on that voxel.
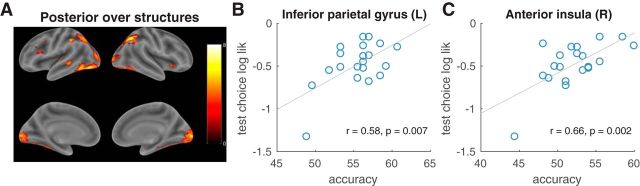
To aggregate these results across subjects, for each voxel, we Fisher z-transformed the resulting Spearman's ρ from all 20 subjects and performed a t test against 0. This yielded a group-level t-map in which the t-value of each voxel indicates whether the representational similarity match for that voxel is significant across subjects. We thresholded single voxels at p < 0.001 and corrected for multiple comparisons using whole-brain cluster FWE correction at significance level α = 0.05. We report the surviving clusters and the t-values of the corresponding voxels (see Fig. 6A).
Figure 6.
Neural signature of the posterior over causal structures. A, Statistical map showing regions with a high representational similarity match for the posterior over causal structures at feedback onset (p < 0.001, whole-brain cluster FWE corrected at α = 0.05). The color scales represent t-values. B, C, Between-subject correlation between peak classification accuracy on the training trials and the average log likelihood of the subject's choices on the test trials according to the causal structure learning model. Significant correlations were found for left inferior parietal gyrus (B) and right anterior insula (C) after Bonferroni correction with adjusted α = 0.05/6 = 0.0083. r, Pearson's correlation coefficient.
Because the posterior tends to be similar on trials that are temporally close to each other, as well as on trials from the same block, we computed two control RDMs: a “time RDM” in which the distance between trials i and j is |ti − tj|, where ti is the difference between the onset of trial i and the start of its corresponding block and a “block RDM” in which the distance between trials i and j is 0 if they belong to the same block, and 1 otherwise. Each Spearman's ρ was then computed as a partial rank correlation coefficient between the neural RDM and the model RDM, controlling for the time RDM and the block RDM, ruling out the possibility that our RSA results reflect within-block temporal autocorrelations that are unrelated to the posterior.
Temporal autocorrelation is a concern when performing RSA because it can bias the results (Diedrichsen et al., 2011; Alink et al., 2015; Cai et al., 2016). This concern is partially alleviated by using betas extracted from a GLM with a separate impulse regressor on each trial, in addition to controlling for the time RDM and the block RDM. Furthermore, most of the entries in the RDMs are for pairs of trials across different runs, so temporal autocorrelations are not an issue. Although this does not perfectly address the autocorrelation problem, the subsequent classification analysis and its link to behavior (described below) validate the ROIs identified by the RSA in a way that is not confounded by temporal autocorrelations in the BOLD signal.
We performed the same analysis for the clustering model. We looked for brain regions with a high representational similarity match with the joint posterior distribution over stimuli and clusters P(zx, x, zc, c), which we computed as follows:
The cluster assignments P(zx|x) and P(zc|c) were computed as in Equation 21. The priors P(x) and P(c) on a given trial were computed as the average number of times cue x and context c (respectively) have been encountered so far. Because this definition of the priors is somewhat ad hoc, we also performed the analysis assuming uniform P(x) and P(c), which makes the posterior equal to the conditional posterior over cluster assignments as follows:
Information mapping.
Performing RSA using the spatially smoothed functional images has the advantage of producing spatially continuous activation clusters that are consistent across subjects and easy to interpret. However, smoothing discards the fine-grained spatial structure of the signal (Kriegeskorte et al., 2006), which could contain rich information about variables involved in structure learning. Therefore, we chose to perform classification on the unsmoothed images but using ROIs selected from the smoothed images. This allows us to maximize the sensitivity of the classifier while accommodating between-subject variability in anatomical locations of the ROIs. Because the posterior closely tracks the block condition, we expect voxels that encode the posterior to be informative about the block condition. To identify such voxels, we used a whole-brain searchlight classification approach based on the unsmoothed neural data. We used the Searchmight toolbox (Pereira and Botvinick, 2011) on betas from a GLM identical to the GLM used for the RSA except that it was performed on functional images that did not undergo smoothing in the preprocessing step. As in the RSA, for each voxel in the whole-brain volume, we defined a 4 mm searchlight centered on that voxel. For each subject and each searchlight, we trained a separate linear discriminant analysis (LDA) classifier with a shrinkage estimator for the covariance matrix (Pereira and Botvinick, 2011) to predict the block condition (irrelevant context, modulatory context, or irrelevant cue) based on neural activity at feedback onset on the training trials. We only considered trials 6 to 20 because both subject performance and the posterior over causal structures plateaued around trial 6, so we did not expect trials 1–5 to be informative. Therefore, there were 15 × 9 = 135 data points, each consisting of up to 81 voxels. We trained and evaluated the classifier using stratified threefold cross-validation with whole blocks: there were three data partitions and each partition contained one block of each condition chosen at random (for a total of 15 × 3 = 45 data points per partition). Including entire blocks in the partitions was necessary due to the temporal autocorrelation of the fMRI signal within each block, which could overfit the classifier to individual blocks rather than block conditions. Because each block was part of one validation set, this allowed us to obtain performance for each data point by a classifier that had not seen that data point nor any other data points from the same block. Classification accuracy was computed based on the validation sets and was assigned to the center voxel of the searchlight. Therefore, for each subject, we obtained an accuracy map for the entire brain volume.
Correlating neural activity with behavior.
We sought to leverage the strengths of both the searchlight RSA and the searchlight classifier by combining the group-level ROIs identified by the RSA with the subject-specific accuracy maps identified by the classifier to predict subject behavior on the test trials. We conjectured that noise in the neural representation of the posterior might vary systematically across subjects. Subjects with noisier representations would produce test phase choices that are less consistent with the causal structure learning model. Furthermore, this noise would be reflected in the classifier performance, with noisier representations resulting in lower classification accuracy.
To test this prediction, we took the peak classification accuracy within each ROI identified by the RSA and correlated it with the log likelihood of the subject's test choices (averaged across blocks). We then applied Bonferroni correction to the resulting set of p-values. If a set of voxels encodes the posterior, then its classification accuracy should predict how well the subject's choices during the test phase conform to the predictions of the causal structure learning model. By restricting the analysis to ROIs identified by the RSA, this approach yields interpretable results on the group level while simultaneously taking into account idiosyncrasies in the precise locus of the neural representation of the posterior for each subject. Furthermore, because results from both the RSA and the classifier were based on training trials only, circularity in the analysis is avoided.
Results
Structure learning accounts for behavioral performance
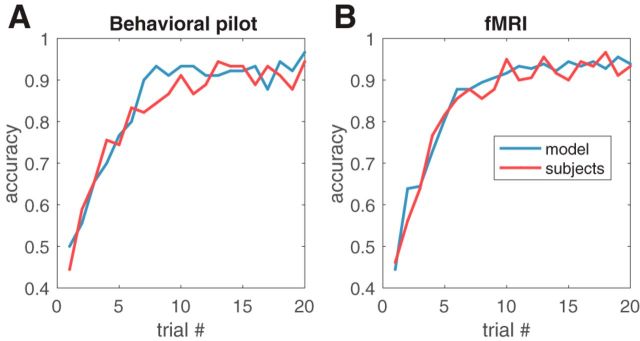
The behavioral results replicated the findings of Gershman (2017) using a within-subject design. Subjects from both the pilot and the fMRI portions of the study learned the correct stimulus–outcome associations relatively quickly, with average performance plateauing around the middle of training (Fig. 3). Average accuracy during the second half of training was 91.2 ± 2.5% (t9 = 16.8, p < 10−7, one-sample t test against 50%) for the pilot subjects and 92.7 ± 1.7% (t19 = 25.0, p < 10−15, one-sample t test against 50%) for the scanned subjects, well above chance.
Figure 3.
Learning curves during training. Performance during training is shown for behavioral pilot subjects (n = 10; A) and fMRI subjects (n = 20; B) averaged across subjects and blocks.
Importantly, both groups exhibited distinct patterns of generalization on the test trials across the different conditions, consistent with the results of Gershman (2017) (Fig. 4B). Without taking the computational model into account, these generalization patterns already suggest that subjects learned something beyond simple stimulus–response mappings. On blocks during which context was irrelevant (Fig. 4B, irrelevant context), subjects tended to predict that the old cue x1, which caused sickness in both c1 and c2, would also cause sickness in the new context c3 (circle for x1 c3) even though they had never experienced c3 before. Conversely, the new cue x3 was judged to be much less predictive of sickness in either context (t38 = 9.51, p < 10−10, paired t test). On modulatory context blocks, subjects appeared to treat each cue–context pair as a unique stimulus independent from the other pairs (Fig. 4B, modulatory context). On these blocks, subjects judged that the old cue is predictive of sickness in the old context significantly more compared with the remaining cue–context pairs (t38 = 9.01, p < 10−10, paired t test). On blocks during which the cue was irrelevant (Fig. 4B, irrelevant cue), subjects guessed that the old context c1, which caused sickness for both cues x1 and x2, would also cause sickness for the new cue x3 (circle for x3 c1), but that the new context c3 would not cause sickness (t38 = 11.1, p < 10−12, paired t test).
These observations were consistent with the predictions of the causal structure learning model. Using parameters fit with data from the behavioral pilot version of the study, the model quantitatively accounted for the generalization pattern on the test trials choices of subjects in the fMRI portion of the study (Fig. 4B; r = 0.97, p < 10−7). As expected, the stimulus–outcome contingencies induced the model to infer a different causal structure in each of the three conditions (Fig. 4A), leading to the distinct response patterns on the simulated test trials.
Of the alternative models, only the clustering model provided an equally compelling account of the generalization pattern on the test trials (Table 1, RL + clustering; r = 0.98, p < 10−7). Bayesian model comparison (Table 1) based on all of the subjects' choices favored both the causal structure learning model and the clustering model more strongly than the alternatives. For comparison, generalization was markedly worse when the hypothesis space was restricted to a single causal structure: the correlation coefficients were r = 0.61 for the irrelevant context structure (M1; p = 0.03), r = 0.73 for the modulatory context structure (M2; p = 0.008), and r = 0.59 for the irrelevant cue structure (M3; p = 0.04). As expected, performance of the simple RL model was comparable to M2 because they both treat each cue–context pair as a unique stimulus. RL with generalization showed an improvement in the generalization pattern (r = 0.88, p = 0.0002); however, it was not as good as the causal structure learning model nor the clustering model and its PXP indicated that it is unlikely to be the most prevalent model in the population. Therefore, we restricted our subsequent analysis of the neural data to the causal structure learning and clustering models.
Separate brain regions support structure learning and associative learning
We sought to identify brain regions in which the BOLD signal tracks beliefs about the underlying causal structure. To condense these multivariate distributions into scalars, we computed the KL divergence between the posterior and the prior distribution over causal structures on each training trial (KLstructures; Eq. 23), which measures the degree to which structural beliefs were revised after observing the outcome. Specifically, we analyzed the fMRI data using a GLM that included KLstructures as a parametric modulator at feedback onset. We reasoned that activity in regions involved in learning causal structure would correlate with the degree of belief revision.
Because we were interested in regions that correlate with learning on the level of causal structures rather than their associative weights, we included the KL divergence between the posterior and the prior distribution over associative weights (KLweights; Eq. 24). These weights encode the strength of causal relationships among cues, contexts, and outcomes separately for each causal structure. Including KLweights as an additional parametric modulator at feedback onset would capture any variability in the signal related to weight updating and allow us to isolate it from the signal related to structure updating.
Our Kalman filter implementation of structure learning assumes that the agent performs full Bayesian inference, which necessitates simultaneous updating of the weights for all causal structures regardless of the agent's beliefs about the causal structures. However, a biologically/cognitively plausible implementation might incorporate certain heuristics such as devoting less computational resources to updating the weights for causal structures that are less likely (Niv et al., 2015). To account for this possibility, we compared two GLMs that included both KLstructures and KLweights as parametric modulators and feedback onset, but differed in the way KLweights was computed. In GLM 1, KLweights was computed as the sum of the KL divergences for all causal structures (Eq. 24), consistent with our implementation that devotes the same amount of computational resources to updating the weights for all structures. In GLM 2, KLweights was computed only for the maximum a posteriori (MAP) structure on the current trial (Eq. 26). This is consistent with an implementation that only updates the weights for the most likely structure analogously to the clustering model, which only updates the value for the MAP cluster assignments.
We used on an analogous GLM (GLM 3) to identify brain regions that correlate with structural updates and associative updates based on the clustering model. The structure learned by the clustering model corresponds to the cluster assignments of the individual cues and contexts, so we reasoned that the structure learning update would elicit a signal proportional to the KL divergence between the posterior and the prior over cluster assignments (KLclusters; Eq 29). Associative learning in the clustering model corresponds to updating the value of the currently active cue cluster and context cluster, which can be quantified by the (cluster) prediction error (CPE, Eq. 30). As a control, we included another GLM (GLM 4) for the clustering model, which was based on the GLM used in Collins and Frank (2016). It had the CPE and the FPE as parametric modulators at feedback onset.
Bayesian model comparison favored GLM 1 and GLM 3 over the other GLMs (Table 2). The high PXP of GLM 1 compared with GLM 2 suggests that the most prevalent causal structure learning model in the population is the one that keeps updating the weights for all structures equally, as predicted by our Kalman filter implementation. The high PXP of GLM 3 compared with GLM 4 favors a model that performs RL over clusters alone rather than one which performs RL over clusters in addition to RL over individual cues and contexts. We therefore report group-level contrasts for GLM 1 and GLM 3 only.
Table 2.
GLM comparison cannot disambiguate between the causal structure learning model (GLM 1: M1, M2, and M3) and the clustering model (GLM 3: RL + clustering)
| GLM | Model | Parametric modulators | PXP |
|---|---|---|---|
| GLM 1 | M1, M2, M3 | KLstructures, KLweights (sum) | 0.4009 |
| GLM 2 | M1, M2, M3 | KLstructures, KLweights (MAP) | 0.0993 |
| GLM 3 | RL + clustering | KLclusters, CPE | 0.4006 |
| GLM 4 | RL + clustering, simple RL | CPE, FPE | 0.0992 |
PXPs were based on whole-brain activity from all trials.
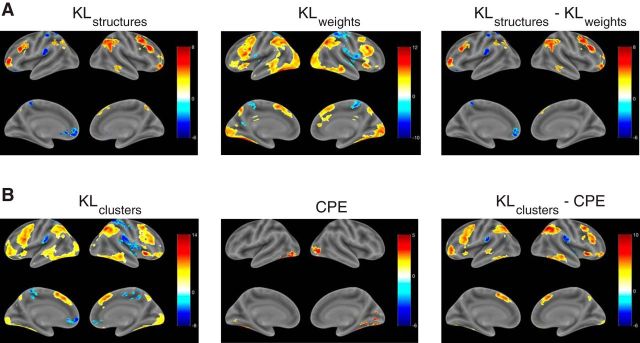
We were interested in identifying regions that track structure learning above and beyond associative learning. For GLM 1, this corresponds to the contrast KLstructures − KLweights (Fig. 5A, right, Table 3). We report clusters that show a significant positive effect (i.e., a stronger correlation with KLstructures than with KLweights) after thresholding single voxels at p < 0.001 and applying whole-brain cluster FWE correction at significance level α = 0.05 (minimum cluster extent = 211). The contrast highlighted a bilateral network of frontoparietal regions. We observed activations in inferior PPC, with a cluster in right angular gyrus and a smaller one spanning the left angular gyrus and left inferior parietal gyrus (IPG). We also found activations in lateral PFC, with a large cluster in right medial frontal gyrus (MFG), extending ventrally into inferior frontal gyrus (IFG) pars triangularis and dorsally into superior frontal gyrus (SFG), as well as a smaller cluster in IFG pars triangularis in the left hemisphere. We also found bilateral activations in rostrolateral PFC (RLPFC), extending into the orbital surface in the right hemisphere. Significant activations were also found on the medial surface of right SFG and in the occipitotemporal part of the right inferior temporal gyrus. Even though the regions that correlated with KLstructures (Fig. 5A, left) were highly overlapping with the regions that correlated with KLweights (Fig. 5A, middle), the fact that most of these regions survived in the contrast implies that the signal in these areas cannot be explained by associative learning alone, suggesting a dissociable network of regions that supports causal structure learning.
Figure 5.
Distinct neural signatures of structure learning and associative learning. Statistical maps for GLM 1 (A) and GLM 3 (B) using a threshold of p < 0.001, whole-brain cluster FWE corrected at α = 0.05. The color scales represent t-values. A, Regions tracking Bayesian updating of beliefs about causal structures (left), Bayesian updating of beliefs about associative weights for all structures (middle), and the contrast between the two (right). B, Regions tracking Bayesian updating of beliefs about cluster assignments (left), the prediction error for the currently active clusters (middle), and the contrast between the two (right).
Table 3.
GLM 1: KLstructures − KLweights
| Sign | Brain region | BA | Extent | t value | MNI coordinates |
|---|---|---|---|---|---|
| Positive | Middle frontal gyrus (R) | 9 | 2218 | 8.377 | 44 12 50 |
| Angular gyrus (R) | 39 | 1968 | 6.351 | 56 −60 30 | |
| IFG pars triangularis (L) | 48 | 698 | 6.341 | −38 26 26 | |
| Anterior orbital gyrus (R) | 11 | 1430 | 6.236 | 24 64 −14 | |
| Middle frontal gyrus (L) | 46 | 809 | 6.137 | −40 56 6 | |
| Cerebellum (L) | 423 | 5.584 | −40 −62 −46 | ||
| Superior frontal gyrus, dorsolateral (R) | 8 | 332 | 5.383 | 2 38 48 | |
| Inferior temporal gyrus (R) | 20 | 278 | 5.192 | 62 −44 −12 | |
| Inferior parietal gyrus (L) | 7 | 768 | 5.044 | −34 −66 46 | |
| Cerebellum (L) | 211 | 5.001 | −28 −72 −28 | ||
| Negative | Rolandic operculum (L) | 48 | 352 | −6.463 | −46 −28 20 |
| IFG pars orbitalis (L) | 11 | 308 | −6.207 | −24 32 −10 | |
| Superior parietal gyrus (L) | 5 | 554 | −5.831 | −18 −48 60 |
Brain regions in which the BOLD signal tracks Bayesian updating of causal structures above and beyond Bayesian updating of associative weights (corresponding to Figure 5A, right). The anatomical label and the MNI coordinates are based on the voxel with the maximum t-statistic from each cluster. Single voxels were thresholded at p < 0.001 and whole-brain cluster FWE correction was applied at significance level of α = 0.05. Regions were labeled using the Automated Anatomical Labeling atlas. BA, Brodmann's area.
For GLM 3, the contrast of interest was KLclusters − CPE (Fig. 5B, right, Table 4; minimum cluster extent = 195). This revealed a frontoparietal network of regions with a high degree of overlap with the KLstructures − KLweights contrast from GLM 1. We found bilateral clusters in inferior PPC (IPG and angular gyrus), lateral PFC (IFG and MFG), and RLPFC. Unlike GLM 1, there were also bilateral clusters in inferior temporal gyrus, middle temporal gyrus, anterior insula, and medial SFG. As in GLM 1, the regions that correlated with KLclusters (Fig. 5B, left) were largely present in the contrast as well, suggesting that their activity tracks a structure update signal that cannot be accounted for by associative updating alone.
Table 4.
GLM 3: KLclusters − CPE
| Sign | Brain region | BA | Extent | t-value | MNI coordinates |
|---|---|---|---|---|---|
| Positive | IFG pars triangularis (L) | 48 | 2996 | 10.548 | −42 14 30 |
| Angular gyrus (R) | 39 | 2433 | 10.541 | 32 −60 46 | |
| Anterior insula (R) | 47 | 506 | 10.275 | 30 22 −4 | |
| Superior frontal gyrus, dorsolateral (R) | 8 | 1625 | 9.032 | 2 28 44 | |
| IFG pars opercularis (R) | 44 | 2471 | 8.438 | 58 18 34 | |
| Middle frontal gyrus (L) | 10 | 1309 | 7.943 | −38 58 8 | |
| Cerebellum (L) | 2200 | 7.839 | −4 −80 −30 | ||
| Anterior orbital gyrus (R) | 11 | 1036 | 7.688 | 34 48 −16 | |
| Inferior parietal gyrus (L) | 7 | 2220 | 6.858 | −32 −56 48 | |
| Inferior temporal gyrus (R) | 37 | 903 | 6.792 | 44 −56 −10 | |
| Medial orbital gyrus (L) | 11 | 274 | 6.676 | −18 44 −18 | |
| Cerebellum (R) | 18 | 1112 | 6.172 | 24 −86 −22 | |
| Middle temporal gyrus (L) | 21 | 420 | 5.886 | −58 −28 −4 | |
| Precuneus (R) | 7 | 195 | 4.797 | 6 −68 48 | |
| Negative | Superior temporal gyrus (R) | 48 | 212 | −6.435 | 44 −12 −4 |
| Superior temporal gyrus (L) | 48 | 277 | −5.693 | −48 −28 18 | |
| Superior temporal gyrus (R) | 48 | 520 | −5.258 | 56 −32 22 |
Multivariate representations of the posterior over causal structures
If the brain performs Bayesian inference over causal structures, as our data suggest, then we should be able to identify regions that contain representations of the full posterior distribution over causal structures P(M|h1:n) (Eq. 14). We thus performed a whole-brain “searchlight” RSA (Kriegeskorte et al., 2008) using searchlights of 4 mm radius (Kriegeskorte et al., 2006). For each subject, we centered the spherical ROI on each voxel of the whole-brain volume and computed a representational dissimilarity matrix (RDM) using the cosine distance between neural activity patterns at feedback onset for all pairs of trials (see Materials and Methods). Intuitively, this RDM reflects which pairs of trials look similar and which pairs of trials look different according to the neural representations in the local neighborhood around the given voxel. We then used Spearman's rank correlation to compare this neural RDM with a model RDM based on the posterior over causal structures. If a given ROI encodes the posterior, then pairs of trials on which the posterior is similar would also show similar neural representations, whereas pairs of trials on which the posterior is different would show differing neural representations. This corresponds to a positive rank correlation between the model and the neural RDMs.
For each subject and each voxel, we thus obtained a Spearman's rank correlation coefficient, reflecting the similarity between variability in activity patterns around that voxel and variability in the posterior over causal structures (the representational similarity match). To aggregate these results at the group level for each voxel, we then performed a one-sample t test against 0 with the Fisher z-transformed Spearman's ρ from all subjects. The resulting t-values from all voxels were used to construct a whole-brain t-map, which was thresholded and corrected for multiple comparisons in the same way as the GLMs (Fig. 6A, Table 5; minimum cluster extent = 253). Each t-value in this map quantifies how likely it is that the given voxel exhibits a positive representational similarity match with the posterior across the population. This revealed some of the same frontoparietal regions identified by the structure learning contrast (Fig. 5A, right), including bilateral inferior PPC (angular gyrus and the neighboring IPG) and left IFG pars triangularis. We also found a large bilateral occipitotemporal cluster spanning the primary visual areas, fusiform gyrus, and inferior temporal gyrus. Additional matches were found in right anterior insula and left superior temporal gyrus.
Table 5.
Brain regions with a high representational similarity match between neural patterns at feedback onset and the posterior over causal structures (corresponding to Figure 6A)
| Brain region | BA | Extent | t-value | MNI coordinates | r | Unadjusted p-value | Adjusted p-value |
|---|---|---|---|---|---|---|---|
| Angular gyrus (R) | 7 | 1347 | 8.505 | 34 −58 42 | 0.03 | 0.908 | 1.000 |
| Inferior temporal gyrus, calcarine fissure, and surrounding cortex (L) | 37 | 6940 | 8.112 | −50 −58 −18 | 0.25 | 0.283 | 0.864 |
| Inferior parietal gyrus (L) | 7 | 1279 | 6.751 | −46 −38 44 | 0.58 | 0.007* | 0.042* |
| Superior temporal gyrus (L) | 48 | 270 | 6.162 | −46 −20 6 | 0.28 | 0.231 | 0.793 |
| Anterior insula (R) | 47 | 253 | 5.779 | 44 20 −6 | 0.66 | 0.002* | 0.010* |
| IFG pars triangularis (L) | 45 | 480 | 4.636 | −56 26 24 | 0.30 | 0.195 | 0.729 |
The voxel with the maximum t statistic from each cluster is also reported. All signs were positive. Single voxels were thresholded at p < 0.001 and whole-brain cluster FWE correction was applied at significance level α = 0.05. Notations are as in Table 3. r is the between-subject Pearson's correlation coefficient between peak classification accuracy within the ROI and test choice log likelihood.
*Significant after Bonferroni correction with adjusted α = 0.05/6 = 0.0083.
We then performed the same analysis for the clustering model using the posterior over clusters and stimuli P(zx, x, zc, c) (Eq. 32). We did not find any voxels that survived multiple comparisons correction. This was also true when we used the conditional posterior over cluster assignments P(zx, zc|x, c) (Eq. 33). Together, these results favor a causal structure learning account of the data and point to a network of regions for maintaining beliefs about causal structure, which get updated on a trial-by-trial basis by a distinct but overlapping network of frontoparietal regions.
Neural representations of the posterior predict subsequent choices
To confirm that ROIs identified by the RSA truly contain representations of the posterior over causal structures, we next sought to use the neural activity in those regions to predict subject behavior. We employed a whole-brain searchlight classification approach based on the unsmoothed functional images (see Materials and Methods). For each subject, this produced an accuracy map that quantifies the amount of information about the block condition contained in the local neighborhood of each voxel. To test the hypothesis that a particular ROI identified by the RSA encodes the posterior at the group level, we took the peak classification accuracy within that ROI and correlated it with the average log likelihood of the subject's responses during the test phase. Notice that because the RSA and the classifier results were based on training trials only, there is no circularity in this analysis. The resulting Pearson's correlation coefficients are shown in Table 5. After applying Bonferroni correction for all six RSA ROIs, we found a significant positive correlation in right anterior insula (adjusted p < 0.01, Fig. 6B) and left inferior PPC (adjusted p < 0.05, Fig. 6C).
We based this analysis on the assumption that there is some endogenous noise in the neural representation of the posterior (Legenstein and Maass, 2014; Haefner et al., 2016; Orbán et al., 2016). This noise would disrupt the close correspondence between the block condition and the posterior, resulting in lower classification accuracy. Therefore, the accuracy assigned to each voxel can be interpreted as the fidelity with which a particular subject represents the posterior in the searchlight around that voxel. The voxel with the highest accuracy within an ROI is also the best candidate for representing the posterior. At the same time, noise in the posterior would give rise to discrepancies between the subject's behavior and the model predictions, which are based on a noise-free representation of the posterior. Because this noise would likely be overshadowed by noise in the BOLD signal on any single trial, we turned to the group level, where any systematic variability in the noise of the posterior across subjects should be manifested as systematic variability in the both the classification accuracy and the likelihood of the subject's test phase choices. Although this analysis assumes subjects are using the structure learning model, subjects using a different model could show the same pattern as those having a noisy posterior: their classification accuracy would be low due to the incorrect representation and their test choice log likelihood would be low due to the discrepancy between their model and the structure learning model. That is, subjects should produce test phase choices in accordance with the posterior to the extent that they use the structure learning model and they have a less noisy neural representation of the posterior. Therefore, the fact that two of the ROIs matching the similarity pattern of the posterior also show this relationship with behavior provides strong evidence that these regions encode the full posterior distribution over causal structures in their multivariate patterns of activity.
Discussion
Behavioral evidence suggests that humans and animals infer both the structure and the strength of causal relationships (Griffiths and Tenenbaum, 2005; Körding et al., 2007; Meder et al., 2014; Gershman, 2017). Using functional brain imaging in humans, the current study provides neural evidence that the formation of stimulus–outcome associations is guided by the inferred structure of the environment. The neural data support the existence of a learning mechanism operating over structural representations that is distinct from the mechanism operating over associative representations, thus reifying the computationally hypothesized division of labor. Our univariate analysis identified areas that were sensitive to belief updates about structure, including inferior PPC, lateral PFC, and RLPFC. In addition, RSA revealed an overlapping network of brain areas that appear to represent the full posterior distribution over causal structures, with activity in two of those regions, the inferior PPC and anterior insula, showing a significant correlation with subsequent subject responses.
Our behavioral data were equally well explained by an alternative structure learning model put forward by Collins and Frank (2013), which implicated some of the same brain areas in relation to belief updates about structure. This is somewhat remarkable considering that their model offers a different interpretation of structure learning, namely that different stimulus dimensions (cues and contexts) are grouped into latent clusters and that associations are formed based on those latent clusters. In a sense, this offers greater flexibility than our model because it does not assume any preexisting knowledge of the relationships between different stimulus dimensions and it allows for a theoretically unbounded number of latent clusters. Indeed, their model and related latent cause models (Gershman et al., 2015) address the question of how structure might emerge in the first place. In contrast, our model endows the agent with an a priori set of relations between stimulus dimensions and outcomes, which are assumed to be innate or acquired through previous experience. This allows for more flexibility in the functional form of the associations such as the summation of values across different stimulus dimensions, something widely believed to be important for capturing classic animal learning phenomena such as blocking, overshadowing, and overexpectation (Rescorla and Wagner, 1972; Soto et al., 2014). The fact that a largely overlapping network of regions tracks belief updates about structure for both models despite their differences suggests a generic neural mechanism for discovering the latent structure of the world that is agnostic to the particular structure learning interpretation. The limitations of the current study preclude any strong conclusions favoring one model over the other, so further work will be required to disentangle the behavioral and neural predictions of the two models.
A notable feature of our data is that inferior PPC appears to encode the full posterior over structures, as well as its corresponding Bayesian update. Previous studies (Seghier, 2013) have linked this area with the integration of bottom-up multimodal input and top-down predictions from frontal areas. O'Reilly et al. (2013) found that angular gyrus encodes the discrepancy between the prior and the posterior distribution over outcomes in a statistical model based on task history. Gläscher et al. (2010) found a signature of the state prediction error in intraparietal sulcus and lateral PFC, implicating those regions in computing the discrepancy between the current model and the observed state transitions. Our results resonate with these findings and fit with the idea that inferior PPC acts as a crossmodal hub that integrates prior knowledge with incoming information.
One candidate region where such top-down predictions might originate is lateral PFC, an area with strong functional connectivity with the inferior parietal lobule (Vincent et al., 2008; Boorman et al., 2009). Previous studies on cognitive control (Koechlin et al., 2003; Badre and D'Esposito, 2007; Koechlin and Summerfield, 2007) have proposed the existence of a functional gradient in lateral PFC, with more anterior regions encoding representations of progressively higher levels of abstraction. Donoso et al. (2014) found evidence that RLPFC performs inference over multiple counterfactual strategies by tracking their reliability, whereas IFG pars triangularis is responsible for switching to one of those strategies if the current one is deemed unreliable. Work on hierarchical reinforcement learning (Badre et al., 2010; Frank and Badre, 2012) extends the notion of a functional hierarchy in lateral PFC to the acquisition of abstract latent rules that guide stimulus–outcome associations. If causal structures are likened to alternative strategies or latent rules, then these results may relate to our finding that RLPFC and IFG track structure updating and that IFG shows a representational similarity match with the posterior (although we were unable to link this representation with behavior, possibly due to the weak signal). Another region where top-down predictions might originate is orbitofrontal cortex (OFC), which has been linked with the representation of a posterior distribution over latent causes (Chan et al., 2016) and is thought to represent a cognitive map of task space (Wilson et al., 2014; Schuck et al., 2016). Consistent with this theory, we found a signature of the Bayesian update signal in right OFC, although our multivariate analysis did not implicate this region in the representation of the posterior.
One puzzling aspect of our results is that activity in anterior insula, a region traditionally implicated in affective processing, appears to encode the full posterior over structures and yet it does not correlate with the update signal. This might relate to previous work by Schapiro et al. (2013), who found that stimuli belonging to the same latent state elicit greater representational similarity in IFG and anterior insula, implicating these regions in some form of latent state inference. Further work will be required to investigate the functional role of anterior insula in relation to structure learning.
An important question that remains open is how structure learning might be implemented in biologically realistic neural circuits. Tervo et al. (2016) noted the parallels between the hierarchical architecture of cortical circuits and the hierarchical nature of structure learning, with empirical evidence suggesting that different layers of the hierarchy tend to be associated with separate cortical circuits. If the brain indeed performs Bayesian inference over causal structures, then this raises the more fundamental question of how ensembles of neurons could represent and perform operations on probability distributions. Different theories have been put forward, ranging from probabilistic population codes to Monte Carlo sampling (Pouget et al., 2013). Teasing apart the different possible mechanisms would require developing behavioral frameworks that lend themselves to computational modeling and quantitative predictions about the inferred probability distributions (Tervo et al., 2016). We believe our study is an important step in that direction.
In summary, we used a combination of behavioral, neural, and computational techniques to separate the neural substrates of structure learning from those of associative learning. Inference over the space of possible structures in the environment recruited frontoparietal regions that have been previously implicated in belief revision and latent state representations, such as inferior PPC, IFG, and RLPFC. Corresponding regions were activated regardless of whether we interpreted structure learning as arbitrating among a set of existing causal structures (Gershman, 2017) or as clustering stimuli into latent states (Collins and Frank, 2013). Additionally, our multivariate analysis found a representation of the posterior distribution over structures in inferior PPC and anterior insula that was predictive of subject responding. Together, these results provide strong support for the idea that the brain performs probabilistic inference over latent structures in the environment, enabling inductive leaps that go beyond the given observations.
Footnotes
This work was supported by the Office of Naval Research Science of Autonomy program (Grant N00014-17-1-2984) and the National Institutes of Health (Grant 1R01MH109177). This work involved the use of instrumentation supported by the NIH Shared Instrumentation Grant Program (Grant S10OD020039). We thank the University of Minnesota Center for Magnetic Resonance Research for the use of the multiband-EPI pulse sequences and Erik Kastman and Katie Insel for helping to design the experiment and collect the data.
The authors declare no competing financial interests.
References
- Alink A, Walther A, Krugliak A, van den Bosch JJ, Kriegeskorte N (2015) Mind the drift-improving sensitivity to fmri pattern information by accounting for temporal pattern drift. bioRxiv. Advance online publication. Retrieved December 4, 2015. doi: 10.1101/032391. [DOI] [Google Scholar]
- Badre D, D'Esposito M (2007) Functional magnetic resonance imaging evidence for a hierarchical organization of the prefrontal cortex. J Cogn Neurosci 19:2082–2099. 10.1162/jocn.2007.19.12.2082 [DOI] [PubMed] [Google Scholar]
- Badre D, Kayser AS, D'Esposito M (2010) Frontal cortex and the discovery of abstract action rules. Neuron 66:315–326. 10.1016/j.neuron.2010.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaz MA, Capra S, Hartl P, Miller RR (1981) Contextual potentiation of acquired behavior after devaluing direct context-us associations. Learning and Motivation 12:383–397. 10.1016/0023-9690(81)90001-1 [DOI] [Google Scholar]
- Boorman ED, Behrens TE, Woolrich MW, Rushworth MF (2009) How green is the grass on the other side? Frontopolar cortex and the evidence in favor of alternative courses of action. Neuron 62:733–743. 10.1016/j.neuron.2009.05.014 [DOI] [PubMed] [Google Scholar]
- Bouton ME, Bolles R (1993) Contextual control of the extinction of conditioned fear. Learning and Motivation 10:445–466. [Google Scholar]
- Bouton ME, King DA (1983) Contextual control of the extinction of conditioned fear: tests for the associative value of the context. J Exp Psychol Anim Behav Process 9:248–265. 10.1037/0097-7403.9.3.248 [DOI] [PubMed] [Google Scholar]
- Bouton ME, Peck CA (1989) Context effects on conditioning, extinction, and reinstatement in an appetitive conditioning preparation. Anim Learn Behav 17:188–198. 10.3758/BF03207634 [DOI] [Google Scholar]
- Cai MB, Schuck NW, Pillow JW, Niv Y (2016) A Bayesian method for reducing bias in neural representational similarity analysis. In Adv Neural Inf Process Syst, pp. 4951–4959. Available at https://papers.nips.cc/paper/6131-a-bayesian-method-for-reducing-bias-in-neural-representational-similarity-analysis.pdf. [DOI] [PMC free article] [PubMed]
- Chan SC, Niv Y, Norman KA (2016) A probability distribution over latent causes, in the orbitofrontal cortex. J Neurosci 36:7817–7828. 10.1523/JNEUROSCI.0659-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AG, Frank MJ (2013) Cognitive control over learning: creating, clustering, and generalizing task-set structure. Psychol Rev 120:190–229. 10.1037/a0030852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AGE, Frank MJ (2016) Neural signature of hierarchically structured expectations predicts clustering and transfer of rule sets in reinforcement learning. Cognition 152:160–169. 10.1016/j.cognition.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AG, Cavanagh JF, Frank MJ (2014) Human EEG uncovers latent generalizable rule structure during learning. J Neurosci 34:4677–4685. 10.1523/JNEUROSCI.3900-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau D. (2005) Confidence intervals in within-subject designs: a simpler solution to Loftus and Masson's method. Tutor Quant Methods Psychol 1:42–45. 10.20982/tqmp.01.1.p042 [DOI] [Google Scholar]
- Daw ND, O'Doherty JP, Dayan P, Seymour B, Dolan RJ (2006) Cortical substrates for exploratory decisions in humans. Nature 441:876–879. 10.1038/nature04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan P, Kakade S (2001) Explaining away in weight space. Adv Neural Inf Process Syst, pp. 451–457. Available at https://papers.nips.cc/paper/1852-explaining-away-in-weight-space.pdf.
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Diedrichsen J, Ridgway GR, Friston KJ, Wiestler T (2011) Comparing the similarity and spatial structure of neural representations: a pattern-component model. Neuroimage 55:1665–1678. 10.1016/j.neuroimage.2011.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso M, Collins AG, Koechlin E (2014) Foundations of human reasoning in the prefrontal cortex. Science 344:1481–1486. 10.1126/science.1252254 [DOI] [PubMed] [Google Scholar]
- Dunsmoor JE, Niv Y, Daw N, Phelps EA (2015) Rethinking extinction. Neuron 88:47–63. 10.1016/j.neuron.2015.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K (2005) A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 25:1325–1335. 10.1016/j.neuroimage.2004.12.034 [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E (2010) Multiplexed echo planar imaging for sub-second whole brain fMRI and fast diffusion imaging. PLoS One 5:e15710. 10.1371/journal.pone.0015710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Badre D (2012) Mechanisms of hierarchical reinforcement learning in corticostriatal circuits 1: computational analysis. Cereb Cortex 22:509–526. 10.1093/cercor/bhr114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman S, Blei D (2012) A tutorial on bayesian nonparametric models. J Math Psychol 56:1–12. 10.1016/j.jmp.2011.08.004 [DOI] [Google Scholar]
- Gershman SJ. (2015) A unifying probabilistic view of associative learning. PLoS Comput Biol 11:e1004567. 10.1371/journal.pcbi.1004567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman SJ. (2017) Context-dependent learning and causal structure. Psychon Bull Rev 24:557–565. 10.3758/s13423-016-1110-x [DOI] [PubMed] [Google Scholar]
- Gershman SJ, Norman KA, Niv Y (2015) Discovering latent causes in reinforcement learning. Curr Opin Behav Sci 5:43–50. 10.1016/j.cobeha.2015.07.007 [DOI] [Google Scholar]
- Gershman SJ, Pesaran B, Daw ND (2009) Human reinforcement learning subdivides structured action spaces by learning effector-specific values. J Neurosci 29:13524–13531. 10.1523/JNEUROSCI.2469-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläscher J. (2009) Visualization of group inference data in functional neuroimaging. Neuroinformatics 7:73–82. 10.1007/s12021-008-9042-x [DOI] [PubMed] [Google Scholar]
- Gläscher J, Daw N, Dayan P, O'Doherty JP (2010) States versus rewards: dissociable neural prediction error signals underlying model-based and model-free reinforcement learning. Neuron 66:585–595. 10.1016/j.neuron.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahame NJ, Hallam SC, Geier L, Miller RR (1990) Context as an occasion setter following either CS acquisition and extinction or CS acquisition alone. Learning and Motivation 21:237–265. 10.1016/0023-9690(90)90008-C [DOI] [Google Scholar]
- Grau JW, Rescorla RA (1984) Role of context in autoshaping. J Exp Psychol Anim Behav Process 10:324–332. 10.1037/0097-7403.10.3.324 [DOI] [Google Scholar]
- Greve DN. (2002) Optseq2 home page. Available online at http://surfer.nmr.mgh.harvard.edu/optseq (Accessed July 3, 2017).
- Griffiths TL, Tenenbaum JB (2005) Structure and strength in causal induction. Cogn Psychol 51:334–384. 10.1016/j.cogpsych.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Haefner RM, Berkes P, Fiser J (2016) Perceptual decision-making as probabilistic inference by neural sampling. Neuron 90:649–660. 10.1016/j.neuron.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Kakade S, Dayan P (2002) Acquisition and extinction in autoshaping. Psychol Rev 109:533–544. 10.1037/0033-295X.109.3.533 [DOI] [PubMed] [Google Scholar]
- Kaye H, Preston GC, Szabo L, Druiff H, Mackintosh NJ (1987) Context specificity of conditioning and latent inhibition: evidence for a dissociation of latent inhibition and associative interference. Q J Exp Psychol B 39:127–145. 10.1080/14640748708402258 [DOI] [Google Scholar]
- Koechlin E, Ody C, Kouneiher F (2003) The architecture of cognitive control in the human prefrontal cortex. Science 302:1181–1185. 10.1126/science.1088545 [DOI] [PubMed] [Google Scholar]
- Koechlin E, Summerfield C (2007) An information theoretical approach to prefrontal executive function. Trends Cogn Sci 11:229–235. 10.1016/j.tics.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Körding KP, Beierholm U, Ma WJ, Quartz S, Tenenbaum JB, Shams L (2007) Causal inference in multisensory perception. PLoS One 2:e943. 10.1371/journal.pone.0000943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P (2006) Information-based functional brain mapping. Proc Natl Acad Sci U S A 103:3863–3868. 10.1073/pnas.0600244103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Bandettini P (2008) Representational similarity analysis: connecting the branches of systems neuroscience. Front Syst Neurosci 2:4–10. 10.3389/neuro.01.016.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruschke JK. (2008) Bayesian approaches to associative learning: from passive to active learning. Learning and Behavior 36:210–226. 10.3758/LB.36.3.210 [DOI] [PubMed] [Google Scholar]
- Legenstein R, Maass W (2014) Ensembles of spiking neurons with noise support optimal probabilistic inference in a dynamically changing environment. PLoS Comput Biol 10:e1003859. 10.1371/journal.pcbi.1003859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovibond PF, Preston G, Mackintosh N (1984) Context specificity of conditioning, extinction, and latent inhibition. J Exp Psychol Anim Behav Process 10:360–375. 10.1037/0097-7403.10.3.360 [DOI] [Google Scholar]
- Mackintosh NJ. (1975) A theory of attention: variations in the associability of stimuli with reinforcement. Psychol Rev 82:276–298. 10.1037/h0076778 [DOI] [Google Scholar]
- Madarasz TJ, Diaz-Mataix L, Akhand O, Ycu EA, LeDoux JE, Johansen JP (2016) Evaluation of ambiguous associations in the amygdala by learning the structure of the environment. Nat Neurosci 19:965–972. 10.1038/nn.4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder B, Mayrhofer R, Waldmann MR (2014) Structure induction in diagnostic causal reasoning. Psychol Rev 121:277–301. 10.1037/a0035944 [DOI] [PubMed] [Google Scholar]
- Miller RR, Barnet RC, Grahame NJ (1995) Assessment of the Rescorla-Wagner model. Psychol Bull 117:363–386. 10.1037/0033-2909.117.3.363 [DOI] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Uğurbil K (2010) Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med 63:1144–1153. 10.1002/mrm.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford JA, Poline JB, Poldrack RA (2015) Orthogonalization of regressors in fMRI models. PLoS One 10:e0126255. 10.1371/journal.pone.0126255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niv Y, Daniel R, Geana A, Gershman SJ, Leong YC, Radulescu A, Wilson RC (2015) Reinforcement learning in multidimensional environments relies on attention mechanisms. J Neurosci 35:8145–8157. 10.1523/JNEUROSCI.2978-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odling-Smee FJ. (1978) The overshadowing of background stimuli: some effects of varying amounts of training and UCS intensity. Q J Exp Psychol 30:737–746. 10.1080/14640747808400698 [DOI] [PubMed] [Google Scholar]
- Orbán G, Berkes P, Fiser J, Lengyel M (2016) Neural variability and sampling-based probabilistic representations in the visual cortex. Neuron 92:530–543. 10.1016/j.neuron.2016.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly JX, Jbabdi S, Rushworth MF, Behrens TE (2013) Brain systems for probabilistic and dynamic prediction: computational specificity and integration. PLoS Biol 11:e1001662. 10.1371/journal.pbio.1001662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce JM, Bouton ME (2001) Theories of associative learning in animals. Annu Rev Psychol 52:111–139. 10.1146/annurev.psych.52.1.111 [DOI] [PubMed] [Google Scholar]
- Peirce JW. (2007) PsychoPy - psychophysics software in python. J Neurosci Methods 162:8–13. 10.1016/j.jneumeth.2006.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Botvinick M (2011) Information mapping with pattern classifiers: a comparative study. Neuroimage 56:476–496. 10.1016/j.neuroimage.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget A, Beck JM, Ma WJ, Latham PE (2013) Probabilistic brains: knowns and unknowns. Nat Neurosci 16:1170–1178. 10.1038/nn.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston G, Dickinson A, Mackintosh N (1986) Contextual conditional discriminations. Q J Exp Psychol 38:217–237. [Google Scholar]
- Rescorla RA, Wagner AR (1972) A theory of Pavlovian conditioning: variations on the effectiveness of reinforcement and non-reinforcement. In: Classical conditioning II: Current research and theory (Black AH, Prokasy WF, eds), pp. 64–99xs. New York, NY: Appleton-Century-Crofts. [Google Scholar]
- Rigoux L, Stephan KE, Friston KJ, Daunizeau J (2014) Bayesian model selection for group studies - revisited. Neuroimage 84:971–985. 10.1016/j.neuroimage.2013.08.065 [DOI] [PubMed] [Google Scholar]
- Rolls ET, Joliot M, Tzourio-Mazoyer N (2015) Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage 122:1–5. 10.1016/j.neuroimage.2015.07.075 [DOI] [PubMed] [Google Scholar]
- Schapiro AC, Rogers TT, Cordova NI, Turk-Browne NB, Botvinick MM (2013) Neural representations of events arise from temporal community structure. Nat Neurosci 16:486–492. 10.1038/nn.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck NW, Cai MB, Wilson RC, Niv Y (2016) Human orbitofrontal cortex represents a cognitive map of state space. Neuron 91:1402–1412. 10.1016/j.neuron.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghier ML. (2013) The angular gyrus. Neuroscientist 19:43–61. 10.1177/1073858412440596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto FA, Gershman SJ, Niv Y (2014) Explaining compound generalization in associative and causal learning through rational principles of dimensional generalization. Psychol Rev 121:526–558. 10.1037/a0037018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzentruber D. (1995) Modulatory mechanisms in Pavlovian conditioning. Anim Learn Behav 23:123–143. 10.3758/BF03199928 [DOI] [Google Scholar]
- Swartzentruber D, Bouton ME (1986) Analysis of the associative and occasion setting properties of contexts participating in a Pavlovian discrimination. J Exp Psychol Anim Behav Process 12:333–350. 10.1037/0097-7403.12.4.333 [DOI] [Google Scholar]
- Tervo DGR, Tenenbaum JB, Gershman SJ (2016) Toward the neural implementation of structure learning. Curr Opin Neurobiol 37:99–105. 10.1016/j.conb.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289. 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJW, Benner T, Salat DH, Fischl B (2008) Brain morphometry with multiecho MPRAGE. Neuroimage 40:559–569. 10.1016/j.neuroimage.2007.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL (2008) Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J Neurophysiol 100:3328–3342. 10.1152/jn.90355.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RR, Takahashi YK, Schoenbaum G, Niv Y (2014) Orbitofrontal cortex as a cognitive map of task space. Neuron 81:267–279. 10.1016/j.neuron.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, Uğurbil K (2013) Evaluation of slice accelerations using multiband echo planar imaging at 3 T. Neuroimage 83:991–1001. 10.1016/j.neuroimage.2013.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]