Abstract
Hepatocellular carcinoma (HCC) is a global health problem and one of the most common malignant tumors. Recent studies have shown that noncoding RNAs (ncRNAs) contribute to the pathogenesis of hepatocellular carcinoma (HCC). These RNAs may be involved in a variety of pathological processes such as cell proliferation, apoptosis, angiogenesis, invasion, and metastasis. In addition, abnormal expression of ncRNAs in HCC may provide potential prognostic or diagnostic biomarkers. This review provides an overview of the role and potential applications of ncRNAs, miRNAs, lncRNAs, circRNAs, and snoRNAs in liver cancer.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and a global health problem [1]. Like many other cancers, HCC is characterized by the involvement of multiple gene networks and the imbalance of signaling pathways [2, 3]. These genetic dysregulations involve protein-coding genes and noncoding RNA (ncRNA) genes [4]. Although the former has been the focus of research, the latter has only recently been recognized as playing a role in the pathological processes implicated in HCC [5]. Interestingly, the vast majority of the human genome is transcribed into ncRNA, while less than 2% of the genome directly encodes for proteins [6]. Noncoding RNA is a functional RNA that is not translated into protein [7].
NcRNAs include ribosomal RNA (rRNA), transfer RNA (tRNA), and small nuclear ribonucleic acids (snRNA) that process pre-mRNA, small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), microRNA (miRNA), long noncoding RNA (lncRNA), etc. [7]. The common characteristic of these RNA is their ability to exercise their biological functions at the level [8]. Prior work has generally focused on protein-coding genes, with little focus on ncRNA function, often dismissed as nothing more than transcriptional noise [9]. However, recent studies have discovered that ncRNAs play an important role in many biological processes. Research in the field of ncRNA has shown an explosive growth in recent years since their functional role has been recognized [10–12].
With the development of high-throughput sequencing technology, many ncRNAs have been characterized as functional molecules that play an important role in various biological processes and pathological states [13–15]. In the field of hepatocellular carcinoma, some key ncRNAs have been identified as participants in the pathophysiology of the disease [16, 17]. Thousands of universally transcribed ncRNAs have been identified, and these transcripts greatly outnumber those of protein-coding mRNAs [18]. In addition, some ncRNAs show significant evolutionary conservation, indirectly supporting their functional roles [19, 20]. For example, miRNA and lncRNAs regulate different biological and pathological processes, such as tumor occurrence [21–24].
2. Abnormal Expression miRNAs in HCCs and Serum of Patients with Hepatocellular Carcinoma
MicroRNA (miRNA) is a type of 20-24nt long biologically functional, small molecule that is highly conserved and provides negative regulation [25]. Since their discovery in 1993 in Caenorhabditis elegans, their important roles continue to be described [26, 27]. Their main functions are on the transcription of regulatory proteins encoded by gene expression [28–30]. Liver cancer is both common and lethal, mainly due to ineffective treatment options [31, 32]. MiRNAs regulate protein synthesis and could either be therapeutic agents or targets for intervention [33–35]. It is clear that miRNA plays a role in proliferation, persistence, invasion, metastasis, and prognostic indicators of hepatocellular carcinoma [36–39]. In liver cancer tissue, there are many abnormally expressed miRNAs, such as miR-224, miR-221, and miR-21 that influence hepatocellular carcinoma [40–42]. miR-224 has been shown to target HOXD10 RNA and enhance both PAK4-mediated phosphorylation and MMP-9 to promote the invasion and metastasis of cancer cells [43]. MiR-224 can inhibit SMAD4 expression and promote cell proliferation [44]. Also, miR-224 can target ppp2r1b leading to excessive activation of the AKT signaling pathway, increasing the risk of liver cancer [45]. MiR-221 can target cell cycle kinase inhibitory proteins p27 and p57, thereby promoting the progression of HCC cell cycle [46, 47]. At the same time, miR-221 can interfere with the mTOR signaling pathway by inhibiting another target DDIT4, thus promoting tumor development [48]. MiR-221 expression levels can be raised by HCV infection, a process that relies on the activation of NF-κB signaling [49]. MiR-221 can also target SOCS1 and SOCS3, enhancing the activation of the downstream interferon signaling pathway, thus enhancing the effectiveness of interferon against HCV [50].
In HCC cells, miR-21 can target MAP2K3 and promote the proliferation of cancer cells [51]. In parallel, miR-21 can inhibit PDCD4, thus activating the expression of downstream c-Jun, MMP-2, and MMP-9. Furthermore, AP-1 can modulate the transcription of miR-21 in a positive feedback loop, further increasing the risk of liver cancer invasion and metastasis [52]. The role of abnormally expressed miRNAs in HCCs and in the serum of HCC patients is listed in Table 1. Clinical data has shown that miR-21 expression in tumors of HCC patients is relatively high [85]. The response to postoperative IFN-Α/5-FU combined treatment is poor [86], suggesting that miR-21 may be a potential molecular marker for prognostic judgment and treatment of HCC. There are many abnormally expressed miRNAs in the serum of HCC patients. Among them, miR-16, let-7f, miR-21, miR-139, miR-101, miR-122, and miR-1 show reduced expression [87–90]. High expression of mir-17-5p may be indicative of HBV (Hepatitis B virus) and the recurrence of HCC [90]. In HCV-infected sera from HCC patients, the specific low expression of mir-30c-5p, mir-223-3p, mir-302c-5p, and mir-17-5p, as well as the specific high expression of miR-221, also suggest potential biomarkers indicating the recurrence of HCC [91, 92].
Table 1.
The function of abnormal expression miRNAs in HCC and in the serum of HCC patients.
| miRNAs | Target gene | Function | References |
|---|---|---|---|
| miR-224 | HOXD10 | Promoting the invasion and metastasis of cancer cells | [43] |
| miR-224 | SMAD4 | Promoting cell proliferation | [44] |
| miR-224 | ppp2r1b | Increasing the risk of liver cancer | [45] |
| miR-221 | p27 and p57 | Promoting the progression of hepatocellular carcinoma cell cycle | [46, 47] |
| miR-221 | DDIT4 | Promoting tumor development | [48] |
| miR-221 | SOCS1 and SOCS3 | Enhancing the effectiveness of interferon against HCV | [50] |
| miR-21 | map2k3 | Promoting the proliferation of cancer cells | [51] |
| miR-21 | PDCD4 | Increased the risk of liver cancer invasion and metastasis | [52] |
3. Roles of lncRNAs in HCC
Ten of thousands of lncRNAs are transcribed in humans [93]. Importantly, detecting their targets is challenging in contrast to miRNA [94]. Indeed, lncRNAs often need to form complex secondary and tertiary structures to predict targets [95].
With the development of large scale parallel sequencing technology, lncRNA has been shown to play an important role in the development of human HCC [96]. So far, many HCC-related lncRNA disorders, such as HULC, HOTAIR, MALAT1, and H19, have been used as predictive biomarkers for human disease diagnosis or prognosis [97, 98]. Furthermore, there is strong evidence that lncRNAs are associated with HCC via many signaling pathways. MyD88 levels were found to be elevated in multiple solid tumors, especially HCC [99]. Many ncRNAs, through a variety of mechanisms, regulate the location of binding sites of protein-coded genes. The abnormal increase in the expression of a new long noncoding Myd88 RNA (lnc-Myd88) is associated with HCC [66]. Chip analysis showed that lnc-Myd88 could increase Myd88 expression by enhancing the acetylation of H3K27 at the Myd88 gene promoter region, leading to the activation of NF-κB and PI3K/AKT signaling pathways [66]. Thus, lnc-Myd88 may be a new diagnostic and therapeutic target for HCC. Various examples of lncRNAs regulating signaling pathways in HCC are listed in Table 2. Hence, targeting these lncRNAs in combination with other therapeutic agents could have therapeutic potential for HCC.
Table 2.
Signaling pathways in HCC regulated by lncRNAs.
| lncRNAs | Pathways | Functions | References |
|---|---|---|---|
| URHC | ERK/MAPK pathway | Regulates cell proliferation and apoptosis | [53] |
| HULC | RXRA pathway | Modulates abnormal lipid metabolism | [54] |
| LINC00152 | mTOR pathway | Promotes proliferation in HCC | [55] |
| LINC01225 | EGFR-dependent pathway | Promotes occurrence and metastasis of HCC | [56] |
| CCHE1 | ERK/MAPK pathway | Promotes carcinogenesis of HCC | [57] |
| linc-cdh4-2 | R-cadherin pathway | Inhibits the migration and invasion of HCC cells | [58] |
| lncARSR | PTEN-PI3K/Akt Pathway. | Promotes doxorubicin resistance in HCC | [59] |
| TSLNC8 | Interleukin-6/STAT3 pathway | A tumor suppressor | [60] |
| PDIA3P1 | p53 pathway | Promotes cell proliferation, migration and invasion, and suppresses apoptosis | [61] |
| lncRNA00673 | Notch pathway | Promotes of proliferation and metastasis of HCC | [62] |
| Igf2as | ERK/MAPK pathway | Controls hepatocellular carcinoma progression | [63] |
| CCAL | Wnt/β-catenin pathway | Promotes HCC progression | [64] |
| LncDQ | EMT Pathway | Promotes HCC progression | [65] |
| lnc-Myd88 | NF-κB and PI3K/AKT pathways | Promotes HCC progression | [66] |
In addition, a new concept suggests that lncRNAs could be used as a protein scaffold close together to form ribose nuclear proteins [100]. However, only a few scaffolding lncRNAs have been identified and the broad extent of this function is unknown. LncRNAs participate in a variety of biological processes and play an important role in various human diseases, including fibrosis diseases, liver diseases, and rare human diseases [101–104]. With the rise of RNA sequencing (RNA-Seq) technology, the number of lncRNAs identified has increased rapidly [105]. However, most lncRNAs have not been well annotated, and their regulatory mechanisms remain elusive. Furthermore, many lncRNAs are not evolutionarily conserved [106]. Therefore, it is vital to study the key function of the conserved lncRNAs.
4. circRNA and HCC
In addition to miRNA and lncRNA, other ncRNAs also influence the development of HCC [67, 107]. Circular RNAs (circRNAs) were discovered in mouse testes in 1993 [108] and represent another class of endogenous, noncoding RNA. In addition to being a biomarker for HCC, circRNA has recently been found to be an important gene expression and pathological network regulatory factor [109–111].
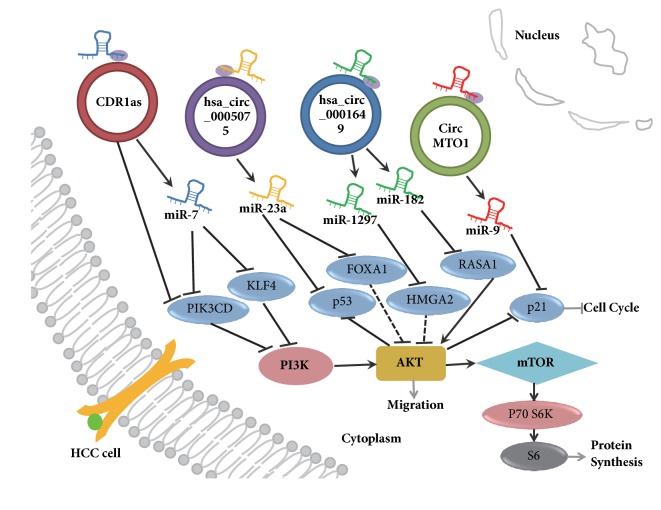
The interactions of hsa_circ_0005075-targeted miRNA genes, including miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p, and their corresponding mRNA, have been studied [70]. One circRNA, Cdr1as, inhibits and absorbs microRNA-7 (miR-7), a suppressor of HCC [69]. CircMTO1 (hsa_circRNA_0007874/ hsa_circRNA_ 104135) suppresses HCC progression by absorbing oncogenic miR-9, thus promoting p21 expression [68]. Recently, circrna_100338 was identified as a biomarker for HCC diagnosis and target for HCC therapeutics [67]. circRNAs function as microRNA sponges by targeting related genes shown in Figure 1 (modified from [108]) and Table 3. In addition, circ_0067934 directly inhibits miR-1324 that targets the 3′-UTR of FZD5 mRNA. Subsequently, the Wnt/β-catenin signaling pathway becomes downregulated in HCC, which enhances the proliferation, migration, and invasion of HCC [112].
Figure 1.
circRNAs function as microRNA sponges by targeting related genes.
Table 3.
circRNAs function as microRNA sponges in HCC.
| circRNA | microRNA sponges | Cells | References |
|---|---|---|---|
| circRNA_10033 | miR-141-3p | HCC and MHCC97H | [67] |
| circMTO1 | miR-9 | HCC | [68] |
| CDR1as | miR-7 | HCC cells, SMMC-7221, Hep3B, QGY-7703, and HepG2 | [69] |
| hsa_circ_0005075 | miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p |
HCC cells, HepG2, and SNU449 | [70] |
| hsa_circ_0001649 | miR-1297 | HCC cells, HepG2, and SMMC7721 | [71] |
5. snoRNA and Their Host Genes in HCC
Small nucleolar RNAs (snoRNAs) are a novel molecular species that may have significant influence on the development and progression of HCC [78]. The expression of snord113-1 in HCC is significantly downregulated [72]. The reduction of snord113-1 in HCC was clearly associated with decreasing patient. In essence, snord113-1 functionally inhibits the growth of HCC cells [72]. Small nucleolar RNA host gene 20 (SNHG20) expression in sk-hep-1 cells significantly inhibited cell proliferation, migration, and invasion. This suggests that SNHG20 could be used as an independent prognostic predictor for HCC patients [74]. The potential roles of snoRNA or their host genes in liver cancer are listed in Table 4.
Table 4.
Potential role of snoRNA or their host genes in liver cancer.
| SnoRNA/ their host genes | Potential role in liver cancer | Cells | References |
|---|---|---|---|
| SNORD113-1 | A tumor suppressor role in HCC and a potential diagnostic and therapeutic target for HCC. | HepG2 and Huh7 | [72] |
| SNHG3 | Associated with malignant status and poor prognosis in HCC patients. | HCC | [73] |
| SNHG20 | Up-regulated in patients with HCC, served as an independent prognostic predictor for HCC patients. | HCC and SK-Hep-1 | [74, 75] |
| SNHG1 | A prognostic biomarker and therapeutic target for HCC. | HCC and HepG2 | [76, 77] |
| SNORD78 | Associated with aggressive phenotype and poor prognosis of HCC | SK-Hep-1 | [78] |
| SNHG6 | Impacts HCC tumorigenesis by binding to up-frameshift protein 1 and regulating Smad7 expression. | HCC and L02 | [79] |
| SNORD126 | A therapeutic target | HepG2, LS174T and Huh7 | [80] |
| SNHG12 | A biomarker and a potential therapeutic target for HCC. | HCC | [81] |
| ACA11 | A promising prognostic biomarker and therapeutic target for patients with HCC. | HCCLM9 and SK-Hep1 | [82] |
| snoRA47 | A valuable biomarker and a potential therapeutic target for HCC. | HCC | [83] |
| SNORD76 | A promising prognostic biomarker in patients with HCC. | SK-Hep1 and Huh7 | [84] |
6. Conclusion
This review provides an overall view of ncRNA in hepatocellular carcinoma (HCC). In addition to the recent focus of research towards lncRNA and miRNA, other ncRNAs, including circRNA and snoRNA also influence liver cancer. Although the first miRNA was identified 20 years ago, other ncRNAs including lncRNAs, snoRNA, siRNA, and piRNAs were gradually discovered and proved to be important in the pathogenesis of cancer [113]. To fully grasp the complete picture of the role of ncRNAs in the pathogenesis of HCC, it is paramount to explore the regulatory networks including circRNA-mRNA, miRNA-lncRNA, lncRNA/snoRNA-piRNA, and other networks that have not been found. To fully understand the biological functions of ncRNAs, we must ascertain the functions of all the proteins and ncRNAs of each cell type and the interactions among them. This full understanding is still a long way off, far more difficult than the genome project.
Acknowledgments
The present study was supported by the Key Research Project of Science Technology Department of Zhejiang Province (no. 2015C03030), the Science Technology Department of Zhejiang Province (no. 2016C33116), the National Natural Science Foundation of China (nos. 81772575 and 81502463), the key project of Health Bureau of Zhejiang Province (no. 2018274734), the Natural Science Foundation of Zhejiang province (Y15H160158 and Q15H070012), and the CSCO Merck Serono Oncology Research Fund, SCORE (no. Y-MX2015-038).
Contributor Information
Dongsheng Huang, Email: dshuang@zju.edu.cn.
Liu Yang, Email: yangliuqq2003@163.com.
Conflicts of Interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Huaixiang Zhou, Qiuran Xu, Chao Ni, and Xiaoge Hu contributed equally to this work.
References
- 1.Ziogas I. A., Tsoulfas G. Advances and challenges in laparoscopic surgery in the management of hepatocellular carcinoma. World Journal of Gastrointestinal Surgery. 2017;9(12):233–245. doi: 10.4240/wjgs.v9.i12.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Huang J.-C., Cai K.-T., et al. Long non-coding RNA HOTTIP promotes hepatocellular carcinoma tumorigenesis and development: A comprehensive investigation based on bioinformatics, qRT-PCR and meta-Analysis of 393 cases. International Journal of Oncology. 2017;51(6):1705–1721. doi: 10.3892/ijo.2017.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Tang W., Chen G., et al. An encapsulation of gene signatures for hepatocellular carcinoma, microRNA-132 predicted target genes and the corresponding overlaps. PLoS ONE. 2016;11(7) doi: 10.1371/journal.pone.0159498.e0159498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto K., Suzuki A. M., Dos Santos A., et al. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Research. 2015;25(12):1812–1824. doi: 10.1101/gr.191031.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing W., Gao S., Zhu M., et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncology Reports. 2016;36:1085–1092. doi: 10.3892/or.2016.4859. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Li X. The role of noncoding RNA in hepatocellular carcinoma. Gland Surgery. 2013;2:25–29. doi: 10.3978/j.issn.2227-684X.2013.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht A.-S., Orom U. A. Bidirectional expression of long ncRNA/protein-coding gene pairs in cancer. Briefings in Functional Genomics. 2016;15(3):167–173. doi: 10.1093/bfgp/elv048. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S.-W., Fan X.-N. Computational methods for predicting ncRNA-protein interactions. Medicinal Chemistry. 2017;13(6):515–525. doi: 10.2174/1573406413666170510102405. [DOI] [PubMed] [Google Scholar]
- 9.Sarachana T., Dahiya N., Simhadri V. L., et al. Small ncRNA expression-profiling of blood from hemophilia a patients identifies mir-1246 as a potential regulator of Factor 8 gene. PLoS ONE. 2015;10(7) doi: 10.1371/journal.pone.0132433.e0132433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haberman Y., BenShoshan M., Di Segni A., et al. DOP015 Dysregulation of cell-type-specific long ncRNA in the ileum of treatment naïve early onset Crohn disease. Journal of Crohn's and Colitis. 2018;12(supplement_1):S040–S040. doi: 10.1093/ecco-jcc/jjx180.052. [DOI] [Google Scholar]
- 11.Zhang K., Yang L., Lu F., Wu X., Zhu J. Cancer Diagnosis: A Universal Upconversion Sensing Platform for the Sensitive Detection of Tumour-Related ncRNA through an Exo III-Assisted Cycling Amplification Strategy (Small 10/2018) Small. 2018;14(10):p. 1870044. doi: 10.1002/smll.201870044. [DOI] [PubMed] [Google Scholar]
- 12.Lackmann F., Belikov S., Burlacu E., Granneman S., Wieslander L. Maturation of the 90S pre-ribosome requires Mrd1 dependent U3 snoRNA and 35S pre-rRNA structural rearrangements. Nucleic Acids Research. 2018;46(7):3692–3706. doi: 10.1093/nar/gky036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra A., Bohra A. Non-coding RNAs and plant male sterility: current knowledge and future prospects. Plant Cell Reports. 2018;37(2):177–191. doi: 10.1007/s00299-018-2248-y. [DOI] [PubMed] [Google Scholar]
- 14.Jakobi T., Dieterich C. Circular RNAs. Vol. 1724. New York, NY: Springer New York; 2018. Deep Computational Circular RNA Analytics from RNA-seq Data; pp. 9–25. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 15.Qu S., Liu Z., Yang X., et al. The emerging functions and roles of circular RNAs in cancer. Cancer Letters. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Wilson C. L., Mann D. A., Borthwick L. A. Epigenetic reprogramming in liver fibrosis and cancer. Advanced Drug Delivery Reviews. 2017;121:124–132. doi: 10.1016/j.addr.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian F., Xu J., Xue F., Guan E., Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Bioscience Reports. 2017;37(4) doi: 10.1042/BSR20170301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S., Liu H., Huang P., et al. circlncRNAnet: An integrated web-based resource for mapping functional networks of long or circular forms of non-coding RNAs. GigaScience. 2017 doi: 10.1093/gigascience/gix118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oloomi M., Yardehnavi N., Bouzari S., Moazzezy N. Non-coding CK19 RNA in peripheral blood and tissue of breast cancer patients. Acta Medica Iranica. 2013;51(2):75–86. [PubMed] [Google Scholar]
- 20.Gebetsberger J., Wyss L., Mleczko A. M., Reuther J., Polacek N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biology. 2017;14(10):1364–1373. doi: 10.1080/15476286.2016.1257470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soengas M. S., Hernando E. TYRP1 mRNA goes fishing for miRNAs in melanoma. Nature Cell Biology. 2017;19(11):1311–1312. doi: 10.1038/ncb3637. [DOI] [PubMed] [Google Scholar]
- 22.Ghorbani S., Talebi F., Ghasemi S., Abad A. J. J., Vojgani M., Noorbakhsh F. MiR-181 interacts with signaling adaptor molecule DENN/MADD and enhances TNF-induced cell death. PLoS ONE. 2017;12(3) doi: 10.1371/journal.pone.0174368.e0174368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L., Lyu L., Wu W., et al. Genome-wide identification of long non-coding RNA and mRNA profiling using RNA sequencing in subjects with sensitive skin. Oncotarget . 2017;8(70):114894–114910. doi: 10.18632/oncotarget.23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Gao Y., Li D., Cao Y., Hao B. LncRNA-TP53TG1 Participated in the Stress Response Under Glucose Deprivation in Glioma. Journal of Cellular Biochemistry. 2017;118(12):4897–4904. doi: 10.1002/jcb.26175. [DOI] [PubMed] [Google Scholar]
- 25.Wang C., Ji B., Cheng B., Chen J., Bai B. Neuroprotection of microRNA in neurological disorders (Review) Biomedical Reports. 2014;2(5):611–619. doi: 10.3892/br.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee R. C., Feinbaum R. L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 27.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 28.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Yee D., Shah K. M., Coles M. C., Sharp T. V., Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. The Journal of Biological Chemistry. 2017;292(50):20683–20693. doi: 10.1074/jbc.M117.809053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma M., Wang X., Chen X., et al. MicroRNA-432 targeting E2F3 and P55PIK inhibits myogenesis through PI3K/AKT/mTOR signaling pathway. RNA Biology. 2017;14(3):347–360. doi: 10.1080/15476286.2017.1279786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H. Y., Yu X. P., Feng R., Hu H. J., Xiao W. W. Analysis of clinical prognosis and the correlation between bile duct injury after transcatheter arterial chemoembolization and the level of hepatic arterial embolization in patients with hepatocellular carcinoma. Zhonghua Zhong Liu Za Zhi. 2017;39(5):355–360. doi: 10.3760/cma.j.issn.0253-3766.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Hong D.-F., Liu Y.-B., Peng S.-Y., et al. Management of hepatocellular carcinoma rupture in the caudate lobe. World Journal of Gastroenterology. 2015;21(26):8163–8169. doi: 10.3748/wjg.v21.i26.8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X., Chang Y., Meng F., et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31(28):3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 34.Callegari E., Gramantieri L., Domenicali M., D'Abundo L., Sabbioni S., Negrini M. MicroRNAs in liver cancer: A model for investigating pathogenesis and novel therapeutic approaches. Cell Death & Differentiation. 2015;22(1):46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farra R., Grassi M., Grassi G., Dapas B. Therapeutic potential of small interfering RNAs/micro interfering RNA in hepatocellular carcinoma. World Journal of Gastroenterology. 2015;21(30):8994–9001. doi: 10.3748/wjg.v21.i30.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu F. F., Xie W. F., Zha G. Q., Chen H. W., Deng L. MiR-520f promotes cell aggressiveness by regulating fibroblast growth factor 16 in hepatocellular carcinoma. Oncotarget . 2017;8(65):109546–109558. doi: 10.18632/oncotarget.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan Y., Zheng N., Teng F., et al. MiR-199a/b-5p inhibits hepatocellular carcinoma progression by post-transcriptionally suppressing ROCK1. Oncotarget . 2017;8(40) doi: 10.18632/oncotarget.18052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lian J., Jing Y., Dong Q., et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget . 2016;7(3):2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X., Liang L., Zhang X.-F., et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58(1):158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y., Wei C., Guo C.-C., et al. Prognostic value of microRNAs in hepatocellular carcinoma: A meta-analysis. Oncotarget . 2017;8(63):107237–107257. doi: 10.18632/oncotarget.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi K.-Q., Lin Z., Chen X.-J., et al. Hepatocellular carcinoma associated microRNA expression signature: Integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget . 2015;6(28):25093–25108. doi: 10.18632/oncotarget.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon E. L., Yeon J. E., Ko E., et al. An explorative analysis for the role of serum mir-10b-3p levels in predicting response to sorafenib in patients with advanced hepatocellular carcinoma. Journal of Korean Medical Science. 2017;32(2):212–220. doi: 10.3346/jkms.2017.32.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q., Ding C., Chen C., et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. Journal of Gastroenterology and Hepatology. 2014;29(4):835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Ren J., Gao Y., et al. MicroRNA-224 targets sMAD family member 4 to promote cell proliferation and negatively influence patient survival. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0068744.e68744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma D., Tao X., Gao F., Fan C., Wu D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncology Letters. 2012;4(3):483–488. doi: 10.3892/ol.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fornari F., Gramantieri L., Ferracin M., et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Q., Loya K., Rani B., et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57(1):299–310. doi: 10.1002/hep.25984. [DOI] [PubMed] [Google Scholar]
- 48.Pineau P., Volinia S., McJunkin K., et al. miR-221 overexpression contributes to liver tumorigenesis. Proceedings of the National Acadamy of Sciences of the United States of America. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding C., Xu G., Ren H., et al. HCV infection induces the upregulation of miR-221 in NF-κB dependent manner. Virus Research. 2015;196:135–139. doi: 10.1016/j.virusres.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 50.Xu G., Yang F., Ding C. L., et al. MiR-221 accentuates IFNs anti-HCV effect by downregulating SOCS1 and SOCS3. Virology. 2014;462-463:343–350. doi: 10.1016/j.virol.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 51.Xu G., Zhang Y., Wei J., et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13(1, article 469) doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Q., Wang Z., Hu Y., et al. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncology Reports. 2012;27(5):1660–1668. doi: 10.3892/or.2012.1682. [DOI] [PubMed] [Google Scholar]
- 53.Xu W. H., Zhang J. B., Dang Z., et al. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. International Journal of Biological Sciences. 2014;10(7):664–676. doi: 10.7150/ijbs.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui M., Xiao Z., Wang Y., et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Research. 2015;75(5):846–857. doi: 10.1158/0008-5472.can-14-1192. [DOI] [PubMed] [Google Scholar]
- 55.Ji J., Tang J., Deng L., et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget . 2015;6(40):42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X., Zhang W., Tang J., et al. LINC01225 promotes occurrence and metastasis of hepatocellular carcinoma in an epidermal growth factor receptor-dependent pathway. Cell death & disease. 2016;7:p. e2130. doi: 10.1038/cddis.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng W., Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomedicine & Pharmacotherapy. 2016;83:450–455. doi: 10.1016/j.biopha.2016.06.056. [DOI] [PubMed] [Google Scholar]
- 58.Gao Y., Wang G., Zhang C., et al. Long non-coding RNA linc-cdh4-2 inhibits the migration and invasion of HCC cells by targeting R-cadherin pathway. Biochemical and Biophysical Research Communications. 2016;480(3):348–354. doi: 10.1016/j.bbrc.2016.10.048. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Ye Y., Feng B., Qi Y. Long Noncoding RNA lncARSR Promotes Doxorubicin Resistance in Hepatocellular Carcinoma via Modulating PTEN-PI3K/Akt Pathway. Journal of Cellular Biochemistry. 2017;118(12):4498–4507. doi: 10.1002/jcb.26107. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J., Li Z., Liu L., et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67(1):171–187. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 61.Kong Y., Zhang L., Huang Y., et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Letters. 2017;407:76–83. doi: 10.1016/j.canlet.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 62.Chen H., Liu JZ., GJ Hu., Shi LL., Lan T. Promotion of proliferation and metastasis of hepatocellular carcinoma by LncRNA00673 based on the targeted-regulation of notch signaling pathway. European Review for Medical and Pharmacological Sciences. 2017;21:3412–3420. [PubMed] [Google Scholar]
- 63.Bao H., Guo C.-G., Qiu P.-C., Zhang X.-L., Dong Q., Wang Y.-K. Long non-coding RNA Igf2as controls hepatocellular carcinoma progression through the ERK/MAPK signaling pathway. Oncology Letters. 2017;14(3):2831–2837. doi: 10.3892/ol.2017.6492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Liu Y., Yang Y., Wang T., et al. Long non-coding RNA CCAL promotes hepatocellular carcinoma progression by regulating AP-2α and Wnt/β-catenin pathway. International Journal of Biological Macromolecules. 2018;109:424–434. doi: 10.1016/j.ijbiomac.2017.12.110. [DOI] [PubMed] [Google Scholar]
- 65.Zeng B., Lin Z., Ye H., et al. Upregulation of LncDQ is Associated with Poor Prognosis and Promotes Tumor Progression via Epigenetic Regulation of the EMT Pathway in HCC. Cellular Physiology and Biochemistry. 2018;46(3):1122–1133. doi: 10.1159/000488841. [DOI] [PubMed] [Google Scholar]
- 66.Xu X., Yin Y., Tang J., et al. Long non-coding RNA Myd88 promotes growth & metastasis in hepatocellular carcinoma via regulating Myd88 expression through H3K27 modification. Cell Death & Disease. 2017;8(10) doi: 10.1038/cddis.2017.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang X., Huang Z., Xu Y., et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han D., Li J., Wang H., et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 69.Xu L., Zhang M., Zheng X., Yi P., Lan C., Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. Journal of Cancer Research and Clinical Oncology. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 70.Shang X., Li G., Liu H., et al. Comprehensive circular RNA profiling reveals that hsa-circ-0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95(22) doi: 10.1097/md.0000000000003811.e3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin M., Liu G., Huo X., et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16(1):161–169. doi: 10.3233/cbm-150552. [DOI] [PubMed] [Google Scholar]
- 72.Xu G., Yang F., Ding C.-L., et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Molecular Cancer. 2014;13(1, article no. 216) doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang T., Cao C., Wu D., Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumor Biology. 2016;37(2):2379–2385. doi: 10.1007/s13277-015-4052-4. [DOI] [PubMed] [Google Scholar]
- 74.Zhang D., Cao C., Liu L., Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. Journal of Cancer. 2016;7(5):608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu J., Lu C., Xiao M., Jiang F., Qu L., Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomedicine & Pharmacotherapy. 2017;89:857–863. doi: 10.1016/j.biopha.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 76.Zhang M., Wang W., Li T., et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomedicine & Pharmacotherapy. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H., Zhou D., Ying M., et al. Expression of long non-coding RNA (LncRNA) small nucleolar rna host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Medical Science Monitor. 2016;22:4820–4829. doi: 10.12659/MSM.898574.898574 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Ma P., Wang H., Han L., Jing W., Zhou X., Liu Z. Up-regulation of small nucleolar RNA 78 is correlated with aggressive phenotype and poor prognosis of hepatocellular carcinoma. Tumor Biology. 2016;37(12):15753–15761. doi: 10.1007/s13277-016-5366-6. [DOI] [PubMed] [Google Scholar]
- 79.Chang L., Yuan Y., Li C., et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Letters. 2016;383(2):183–194. doi: 10.1016/j.canlet.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 80.Fang X., Yang D., Luo H., et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. Journal of Molecular Cell Biology. 2017;9(3):243–255. doi: 10.1093/jmcb/mjw048. [DOI] [PubMed] [Google Scholar]
- 81.Lan T., Ma W., Hong Z., Wu L., Chen X., Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research. 2017;36(1, article no. 11) doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu L., Zheng J., Chen P., Liu Q., Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomedicine & Pharmacotherapy. 2017;90:705–712. doi: 10.1016/j.biopha.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 83.Li G., He Y., Liu X., et al. Small nucleolar RNA 47 promotes tumorigenesis by regulating EMT markers in hepatocellular carcinoma. Minerva Medica. 2017;108(5):396–404. doi: 10.23736/S0026-4806.17.05132-1. [DOI] [PubMed] [Google Scholar]
- 84.Wu L., Chang L., Wang H., Ma W., Peng Q., Yuan Y. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology. 2018;42(1):82–91. doi: 10.1016/j.clinre.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 85.Wang W.-Y., Zhang H.-F., Wang L., et al. MiR-21 expression predicts prognosis in hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology. 2014;38(6):715–719. doi: 10.1016/j.clinre.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 86.Tomimaru Y., Eguchi H., Nagano H., et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular carcinoma cells. British Journal of Cancer. 2010;103(10):1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ge W., Yu D.-C., Li Q.-G., Chen X., Zhang C.-Y., Ding Y.-T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clinical Laboratory. 2014;60(3):427–434. doi: 10.7754/Clin.Lab.2013.130133. [DOI] [PubMed] [Google Scholar]
- 88.Yang L., Yin D., Wang Y., Cao L. Inhibition of the growth of hepatocellular carcinoma cells through fibroblast growth factor 18 suppressed by miR-139. Oncology Reports. 2017;38(4):2565–2571. doi: 10.3892/or.2017.5869. [DOI] [PubMed] [Google Scholar]
- 89.Zhuang C., Jiang W., Huang D., et al. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology. 2016;40(4):386–396. doi: 10.1016/j.clinre.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 90.Lai Y., Ushio N., Rahman M. M., et al. Aberrant expression of microRNAs and the miR-1/MET pathway in canine hepatocellular carcinoma. Veterinary and Comparative Oncology. 2018;16(2):288–296. doi: 10.1111/vco.12379. [DOI] [PubMed] [Google Scholar]
- 91.Oksuz Z., Serin M. S., Kaplan E., et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Molecular Biology Reports. 2015;42(3):713–720. doi: 10.1007/s11033-014-3819-9. [DOI] [PubMed] [Google Scholar]
- 92.El-Garem H., Ammer A., Shehab H., et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World Journal of Hepatology. 2014;6(11):818–824. doi: 10.4254/wjh.v6.i11.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar M. M., Goyal R. LncRNA as a therapeutic target for angiogenesis. Current Topics in Medicinal Chemistry. 2017;17(15):1750–1757. doi: 10.2174/156802661766616111644744. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.León L. E., Calligaris S. D. MicroRNA Profiling. Vol. 1509. New York, NY: Springer New York; 2017. Visualization and Analysis of MiRNA–Targets Interactions Networks; pp. 209–220. (Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 95.Wu Q., Wu X., Ying X., et al. Suppression of endothelial cell migration by tumor associated macrophage-derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell International. 2017;17(1, article no. 62) doi: 10.1186/s12935-017-0430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin L., He Y., Tang S., Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. Journal of Cellular Physiology. 2017 doi: 10.1002/jcp.26257. [DOI] [PubMed] [Google Scholar]
- 97.Li C., Chen J., Zhang K., Feng B., Wang R., Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cellular Physiology and Biochemistry. 2015;36(2):423–434. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 98.Zhao J., Greene C. M., Gray S. G., Lawless M. W. Long noncoding RNAs in liver cancer: what we know in 2014. Expert Opinion on Therapeutic Targets. 2014;18(10):1207–1218. doi: 10.1517/14728222.2014.941285. [DOI] [PubMed] [Google Scholar]
- 99.Li Y.-J., Lu F.-M., Zhuang H. The role of MyD88 in inflammatory/infection-related hepatocellular carcinoma. Chinese Journal of Hepatology. 2010;18(3):232–234. doi: 10.3760/cma.j.issn.1007-3418.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 100.Jiang L., Shao C., Wu Q.-J., et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nature Structural & Molecular Biology. 2017;24(10):816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alvarez-Dominguez J. R., Lodish H. F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130(18):1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y., Luo G., Zhang M., Zhou J., Gao W. Critical effects of long non-coding RNA on fibrosis diseases. Experimental & Molecular Medicine. 2018;50:p. e428. doi: 10.1038/emm.2017.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pope C., Mishra S., Russell J., Zhou Q., Zhong X. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases. 2017;5(1):p. 11. doi: 10.3390/diseases5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.HE J., HAN Z., LI Y. Association between long non-coding RNA and human rare diseases (Review) Biomedical Reports. 2014;2(1):19–23. doi: 10.3892/br.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shumayla, Sharma S., Taneja M., Tyagi S., Singh K., Upadhyay S. K. Survey of high throughput RNA-seq data reveals potential roles for lncrnas during development and stress response in bread wheat. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li D., Yang M. Q. Identification and characterization of conserved lncRNAs in human and rat brain. BMC Bioinformatics. 2017;18 doi: 10.1186/s12859-017-1890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang B.-G., Li J.-S., Liu Y.-F., Xu Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumor Biology. 2017;39(7) doi: 10.1177/1010428317719577. [DOI] [PubMed] [Google Scholar]
- 108.Hu J., Li P., Song Y., et al. Progress and prospects of circular RNAs in Hepatocellular carcinoma: Novel insights into their function. Journal of Cellular Physiology. 2018;233(6):4408–4422. doi: 10.1002/jcp.26154. [DOI] [PubMed] [Google Scholar]
- 109.Meng S., Zhou H., Feng Z., et al. CircRNA: Functions and properties of a novel potential biomarker for cancer. Molecular Cancer. 2017;16(1, article no. 94) doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cui X., Niu W., Kong L., et al. Hsa-circRNA-103636: Potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomarkers in Medicine. 2016;10(9):943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 111.Zhang H.-D., Jiang L.-H., Sun D.-W., Hou J.-C., Ji Z.-L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1) doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 112.Zhu Q., Lu G., Luo Z., et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochemical and Biophysical Research Communications. 2018;497(2):626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 113.Weng W., Liu N., Toiyama Y., et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Molecular Cancer. 2018;17(1) doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]