Abstract
Background
Resistance to chemotherapy drugs (e.g. taxol) has been a major obstacle in successful cancer treatment. In A549 human lung adenocarcinoma, acquired resistance to the first-line chemotherapy taxol has been a critical problem in clinics. Sphingolipid (SPL) controls various aspects of cell growth, survival, adhesion, and motility in cancer, and has been gradually regarded as a key factor in drug resistance. To better understand the taxol-resistant mechanism, a comprehensive sphingolipidomic approach was carried out to investigate the sphingolipid metabolism in taxol-resistant strain of A549 cell (A549T).
Methods
A549 and A549T cells were extracted according to the procedure with optimal condition for SPLs. Sphingolipidomic analysis was carried out by using an UHPLC coupled with quadrupole time-of-flight (Q-TOF) MS system for qualitative profiling and an UHPLC coupled with triple quadrupole (QQQ) MS system for quantitative analysis. The differentially expressed sphingolipids between taxol-sensitive and -resistant cells were explored by using multivariate analysis.
Results
Based on accurate mass and characteristic fragment ions, 114 SPLs, including 4 new species, were clearly identified. Under the multiple reaction monitoring (MRM) mode of QQQ MS, 75 SPLs were further quantified in both A549 and A549T. Multivariate analysis explored that the levels of 57 sphingolipids significantly altered in A549T comparing to those of A549 (p < 0.001 and VIP > 1), including 35 sphingomyelins (SMs), 14 ceramides (Cers), 3 hexosylceramides (HexCers), 4 lactosylceramides (LacCers) and 1 sphingosine. A significant decrease of SM and Cer levels and overall increase of HexCer and LacCer represent the major SPL metabolic characteristic in A549T.
Conclusions
This study investigated sphingolipid profiles in human lung adenocarcinoma cell lines, which is the most comprehensive sphingolipidomic analysis of A549 and A549T. To some extent, the mechanism of taxol-resistance could be attributed to the aberrant sphingolipid metabolism, “inhibition of the de novo synthesis pathway” and “activation of glycosphingolipid pathway” may play the dominant role for taxol-resistance in A549T. This study provides insights into the strategy for clinical diagnosis and treatment of taxol resistant lung cancer.
Keywords: A549 human lung adenocarcinoma cell line - Taxol-resistant - LC-MS - Sphingolipids - Ceramide
Background
Lung cancer has been the leading cause of cancer mortality, and adenocarcinoma is its most prevalent form [1]. Paclitaxel (taxol) is commonly used as part of combination chemotherapy for the treatment of non-small cell lung cancer including adenocarcinoma A549. However, resistance to natural product chemotherapy drugs still constitutes a huge problem of successful cancer treatment, and the efficiency of chemotherapy is weakened because of paclitaxel resistance [2]. Potential mechanisms have been reported including multidrug resistance, β-tubulin alterations, detoxifying of paclitaxel, and apoptosis related genetic changes [3]. Although the extensive efforts have been made for understanding the underlying mechanisms, they are still elusive.
It has been recognized that the dysregulated metabolic profile of cancer is linked to the chemoresistance [4]. Cancer cells reprogram their metabolism to satisfy the demands of malignant phenotype, which decrease drug-induced apoptosis, conferring therapeutic resistance [5]. Since cellular SPLs appear to play a significant role in relation to cancer, their dysregulated synthesis and metabolism in drug-resistant cancer cells have been systematically studied [6]. Most previous studies focus on the biological effect of a kind of specific SPL like Cer [7] and S1P [8] on A549 cancer cell line. The sphingolipid profiles for A549 have been preliminary explored by using MALDI-TOF-MS, only two Cers have been defined as markers out of all the 9 SPLs detected in A549 [9]. The whole sphingolipidome in either A549 or A549T remains largely unrevealed. Recently, a versatile sphingolipidomic approach for both qualitative and quantitative analysis of up to 10 subclasses of SPLs has been established in our group [10]. In this study, the integrated LC-MS approach was employed to investigate the taxol resistance mechanism of A549T from the viewpoint of sphingolipidomic.
Methods
Chemicals and materials
The LIPID MAPS internal standard cocktail (internal standards mixture II, 25 μM each of 9 compounds in ethanol, catalog LM-6005) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). It was composed of uncommon SPLs which include: 17-carbon chain length sphingoid base analogs C17-sphingosine [So (d17:1)], C17-sphinganine [Sa (d17:0)], C17-sphingosine-1-phosphate [S1P (d17:1)], C17-sphinganine-1-phosphate [Sa1P (d17:0)], the C12-fatty acid analogs of the more complex SPLs C12-Ceramide [Cer (d18:1/12:0)], C12-ceramide-1-phosphate [C1P (d18:1/12:0)], C12-sphingomyelin [SM (d18:1/12:0)], C12-glucosylceramide [GlcCer (d18:1/12:0)], and C12-lactosylceramide [LacCer (d18:1/12:0)].
Acetic acid (CH3COOH, MS grade), formic acid (HCOOH, MS grade), ammonium acetate (NH4OAc, ACS grade) and potassium hydroxide (KOH, ACS grade) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The HPLC grade chloroform (CHCl3), isopropanol (IPA), as well as methanol (MeOH) were purchased from Merck (Darmstadt, Germany). Dulbecco’s Modified Eagle’s Medium (DMEM), Roswell Park Memorial Institute (RPMI) 1640 medium, Fetal Bovine Serum (FBS), Penicillin-Streptomycin (PS) were obtained from Gibco, New Zealand. Sodium dodecyl sulfate (SDS) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were acquired from Acros, USA. Ultrapure water (18.2 MΩ) was supplied with a Milli-Q system (Millipore, MA, USA).
Cell culture and SPLs extraction
A549 human lung adenocarcinoma cell line (Cat.No. KG007) and its taxol-resistant strain (A549T, Cat.No. KG124) were obtained from KeyGen Biotech Co., Ltd. (Nanjing, China). A549 was cultured in DMEM supplemented with 10% FBS and 1% PS in a humidified 5% CO2 atmosphere at 37 °C. A549T was cultured in RPMI 1640 medium supplemented with solution consisted of 10% FBS, 1% PS and 200 ng/mL taxol in a humidified 5% CO2 atmosphere at 37 °C. For lipid analysis, A549 and A549T cells were respectively seeded into 6-well plates at the density of 1.5 × 105 cells/well and incubated for 48 h. Lipids were extracted from the cells, when they were grown to 80% confluence. After rinsed twice by ice-cold PBS, the cells were scraped into a borosilicate glass tube, in which 0.5 mL of MeOH, 0.25 mL of CHCl3 and 10 μL of 2.5 μM internal standards cocktail were added. The extract procedure was carried out by incubation at 48 °C for 12 h after sonicated at ambient temperature for 30 s. After 75 μL of KOH in MeOH (1 M) was added, the mixture was placed into a shaking incubator at 37 °C for 2 h. Acetic acid was used to neutralize the mixture before the typical four-step extraction was carried out for the preparation of SPLs. Further details for extracting SPLs and sample preparation were the same as previously described [11]. MTT assay was employed to evaluate the sensitivity of A549 and A549T cells to taxol. The IC50s were 67.72 nM and 124.7 μM, respectively corresponding to A549 and A549T, showing almost 2000-fold difference in taxol sensitivity between these two cell lines.
LC-MS conditions
Sphingolipid analysis was performed by using our developed LC-MS method with minor optimization, just as described previously [10, 11]. Chromatographic separation was achieved by using an Agilent 1290 UHPLC system, and it was interfaced with an Agilent ultrahigh definition 6550 Q-TOF mass spectrometer and an Agilent 6460 triple-quadrupole mass spectrometer respectively for qualitative- and quantitative-analysis. The acquisition and data analysis were operated by using Agilent MassHunter Workstation Software.
Data analysis
Based on the Agilent Personal Compound Database and Library (PCDL) software and LIPID MAPS Lipidomics Gateway, a personal database has been established with the latest update of 32,622 SPLs until August 06 2016. The screening and identification of SPLs were carried out by searching against it.
In qualitative research, the sphingolipidomic approach was applied by analyzing QC samples equally pooled by A549 and A549T. In quantitative research, A549 cells (models, n = 10) and A549T cells (models, n = 10), as well as QC samples (n = 5), were analyzed in parallel. Multivariate statistical analysis, including principle component analysis (PCA) and partial least squares to latent structure-discriminant analysis (PLS-DA) methods, were performed to examine significant differences between A549 and A549T, using SIMCA-P+ software version 14.0 (Umetrics, Umea, Sweden). Variable Importance in the Project (VIP) value in PLS-DA model was used for selecting and identifying biomarkers. The altered SPL with a VIP value larger than 1.00 was considered as a biomarker.
Results
Comprehensive profiling of sphingolipids in A549 and A549T cells
QC samples were analyzed repeatedly to achieve comprehensive profiling of SPLs in A549 and A549T. In various subclasses of SPLs, the [M + H]+ ions exhibits highest intensities in positive ion mode. Totally 114 SPLs have been identified in the QC samples, among which Cer (d18:2/26:2), DHCer (d18:0/24:2), phytosphingosine (PTSo) t19:2, and PTSo t16:1 were new SPLs. Notably, 4 pairs of isobaric species (A1-A4 vs a1-a4) and 21 pairs of isomeric species (B1-B21 vs b1-b21) were clearly distinguished in this study. Respective qualitative test of A549 and A549T revealed that they share all the same species of SPLs. The full identification result was listed in Table 1.
Table 1.
Identification and quantification of SPLs in A549/A549T cells by using UHPLC-Q-TOF and UHPLC-QQQ MS
| Class | Name | [M + H]+ m/z | tR (min) | Molecular Formula | Measured Mass | Calculated Mass | Error (ppm) | MS/MS Fragments (m/z) | MRM transitions | |
|---|---|---|---|---|---|---|---|---|---|---|
| SM | d18:1/26:0 [B1] | 843.7314 | 18.483 | C49 H99 N2 O6 P | 842.7240 | 842.7241 | −0.12 | 264.2674, 184.0730 | 843.7 | 184.1 |
| d18:1/26:1 | 841.7096 | 17.136 | C49 H97 N2 O6 P | 840.7026 | 840.7084 | −6.90 | 264.2682, 184.0739 | 841.7 | 184.1 | |
| d18:1/25:0 [B2] | 829.7154 | 17.702 | C48 H97 N2 O6 P | 828.7080 | 828.7084 | −0.48 | 264.2683, 184.0732 | 829.7 | 184.1 | |
| d18:1/25:1 [B3] | 827.6993 | 16.288 | C48 H95 N2 O6 P | 826.6915 | 826.6928 | −1.57 | 264.2698, 184.0722 | 827.7 | 184.1 | |
| d18:1/24:0 [B4] | 815.6992 | 17.020 | C47 H95 N2 O6 P | 814.6925 | 814.6928 | −0.37 | 264.2697, 184.0732 | 815.7 | 184.1 | |
| d18:1/24:1 [B5] | 813.6841 | 15.507 | C47 H93 N2 O6 P | 812.6769 | 812.6771 | −0.25 | 264.2685, 184.0739 | 813.7 | 184.1 | |
| d18:1/24:2 | 811.6680 | 14.958 | C47 H91 N2 O6 P | 810.6607 | 810.6615 | −0.99 | 264.2688, 184.0732 | 811.7 | 184.1 | |
| d18:1/24:3 [B6] | 809.6526 | 14.243 | C47 H89 N2 O6 P | 808.6459 | 808.6458 | 0.12 | 264.2627, 184.0725 | 809.7 | 184.1 | |
| d18:1/23:0 [B7] | 801.6839 | 16.371 | C46 H93 N2 O6 P | 800.6769 | 800.6771 | −0.25 | 264.2645, 184.0727 | 801.7 | 184.1 | |
| d18:1/23:1 [B8] | 799.6683 | 15.241 | C46 H91 N2 O6 P | 798.6608 | 798.6615 | −0.88 | 264.2654, 184.0721 | 799.7 | 184.1 | |
| d18:1/23:2 | 797.6520 | 14.326 | C46 H89 N2 O6 P | 796.6443 | 796.6458 | −1.88 | 264.2398, 184.0727 | 797.7 | 184.1 | |
| d18:1/22:0 [B9] | 787.6679 | 15.723 | C45 H91 N2 O6 P | 786.6608 | 786.6615 | −0.89 | 264.2665, 184.0730 | 787.7 | 184.1 | |
| d18:1/22:1 [B10] | 785.6524 | 14.642 | C45 H89 N2 O6 P | 784.6452 | 784.6458 | −0.76 | 264.2606, 184.0730 | 785.7 | 184.1 | |
| d18:1/22:2 | 783.6362 | 13.728 | C45 H87 N2 O6 P | 782.6291 | 782.6302 | −1.41 | 264.2636, 184.0727 | 783.6 | 184.1 | |
| d18:1/21:0 | 773.6528 | 15.108 | C44 H89 N2 O6 P | 772.6455 | 772.6458 | −0.39 | 264.2651, 184.0721 | 773.7 | 184.1 | |
| d18:1/21:1 [B11] | 771.6361 | 14.010 | C44 H87 N2 O6 P | 770.6286 | 770.6302 | −2.08 | 264.2624, 184.0732 | 771.6 | 184.1 | |
| d18:1/20:0 | 759.6368 | 14.459 | C43 H87 N2 O6 P | 758.6295 | 758.6302 | −0.92 | 264.2658, 184.0730 | 759.6 | 184.1 | |
| d18:1/20:1 [B12] | 757.6201 | 13.428 | C43 H85 N2 O6 P | 756.6128 | 756.6145 | −2.25 | 264.2653, 184.0735 | 757.6 | 184.1 | |
| d18:1/19:0 | 745.6207 | 13.811 | C42 H85 N2 O6 P | 744.6134 | 744.6145 | −1.48 | 264.2677, 184.0725 | 745.6 | 184.1 | |
| d18:1/18:0 | 731.6054 | 13.195 | C41 H83 N2 O6 P | 730.5982 | 730.5989 | −0.96 | 264.2665, 184.0727 | 731.6 | 184.1 | |
| d18:1/18:1 [B13] | 729.5869 | 12.081 | C41 H81 N2 O6 P | 728.5786 | 728.5832 | −8.23 | 264.2659, 184.0732 | 729.6 | 184.1 | |
| d18:1/17:0 | 717.5896 | 12.613 | C40 H81 N2 O6 P | 716.5824 | 716.5832 | −1.12 | 264.2677, 184.0726 | 717.6 | 184.1 | |
| d18:1/16:0 | 703.5737 | 12.081 | C39 H79 N2 O6 P | 702.5666 | 702.5676 | −1.42 | 264.2680, 184.0748 | 703.6 | 184.1 | |
| d18:1/16:1 [B14] | 701.5583 | 11.366 | C39 H77 N2 O6 P | 700.5511 | 700.5519 | −1.14 | 264.2685, 184.0730 | 701.6 | 184.1 | |
| d18:1/15:0 | 689.5585 | 11.616 | C38 H77 N2 O6 P | 688.5512 | 688.5519 | −1.02 | 264.2627, 184.0726 | 689.6 | 184.1 | |
| d18:1/15:1 [B15] | 687.5420 | 10.752 | C38 H75 N2 O6 P | 686.5352 | 686.5363 | −1.60 | 264.2629, 184.0720 | 687.5 | 184.1 | |
| d18:1/14:0 | 675.5427 | 11.117 | C37 H75 N2 O6 P | 674.5355 | 674.5363 | −1.19 | 264.2686, 184.0734 | 675.5 | 184.1 | |
| d18:2/25:0 [b3] | 827.6988 | 16.504 | C48 H95 N2 O6 P | 826.6896 | 826.6928 | −3.87 | 262.2524, 184.0729 | |||
| d18:2/24:0 [b5] | 813.6763 | 15.873 | C47 H93 N2 O6 P | 812.6693 | 812.6771 | −9.60 | 262.2542, 184.0726 | |||
| d18:2/24:2 [b6] | 809.6503 | 15.723 | C47 H89 N2 O6 P | 808.6457 | 808.6458 | −0.12 | 262.2554, 184.0731 | |||
| d18:2/24:3 | 807.6343 | 14.659 | C47 H87 N2 O6 P | 806.6272 | 806.6302 | −3.71 | 184.0725 | |||
| d18:2/23:0 [b8] | 799.6684 | 15.474 | C46 H91 N2 O6 P | 798.6606 | 798.6615 | −1.13 | 184.0732 | 799.7 | 184.1 | |
| d18:2/22:0 [b10] | 785.6523 | 14.808 | C45 H89 N2 O6 P | 784.6451 | 784.6458 | −0.89 | 262.2440, 184.0732 | 785.7 | 184.1 | |
| d18:2/21:0 [b11] | 771.6357 | 14.193 | C44 H87 N2 O6 P | 770.6272 | 770.6302 | −3.89 | 184.0722 | |||
| d18:2/20:0 [b12] | 757.6200 | 13.545 | C43 H85 N2 O6 P | 756.6128 | 756.6145 | −2.25 | 262.2451, 184.0725 | 757.6 | 184.1 | |
| d18:2/18:0 [b13] | 729.5896 | 12.347 | C41 H81 N2 O6 P | 728.5822 | 728.5832 | −1.37 | 262.2554, 184.0728 | |||
| d18:2/16:0 [b14] | 701.5582 | 11.382 | C39 H77 N2 O6 P | 700.5510 | 700.5519 | −1.28 | 262.2504, 184.0728 | |||
| d18:2/15:0 [b15] | 687.5433 | 10.951 | C38 H75 N2 O6 P | 686.5355 | 686.5363 | −1.16 | 262.2503, 184.0716 | |||
| d18:1/12:0 [IS-1] | 647.5116 | 10.402 | C35 H71 N2 O6 P | 646.5042 | 646.5050 | −1.24 | 264.2699, 184.0732 | 647.5 | 184.1 | |
| DHSM | d18:0/26:0 | 845.7455 | 19.182 | C49 H101 N2 O6 P | 844.7382 | 844.7397 | −1.78 | 266.2711, 184.0730 | ||
| d18:0/26:1 [b1] | 843.7271 | 17.702 | C49 H99 N2 O6 P | 842.7202 | 842.7241 | −4.63 | 266.2787, 184.0727 | |||
| d18:0/25:0 | 831.7297 | 18.317 | C48 H99 N2 O6 P | 830.7224 | 830.7241 | −2.05 | 184.0731 | |||
| d18:0/25:1 [b2] | 829.7149 | 17.469 | C48 H97 N2 O6 P | 828.7071 | 828.7084 | −1.57 | 266.2729, 184.0729 | |||
| d18:0/24:0 | 817.7151 | 17.585 | C47 H97 N2 O6 P | 816.7081 | 816.7084 | −0.37 | 266.2696, 184.0732 | 817.7 | 184.1 | |
| d18:0/24:1 [b4] | 815.6992 | 16.405 | C47 H95 N2 O6 P | 814.6931 | 814.6928 | 0.37 | 266.2767, 184.0732 | |||
| d18:0/23:0 | 803.6995 | 16.937 | C46 H95 N2 O6 P | 802.6921 | 802.6928 | −0.87 | 184.0725 | 803.7 | 184.1 | |
| d18:0/23:1 [b7] | 801.6842 | 15.756 | C46 H93 N2 O6 P | 800.6767 | 800.6771 | −0.50 | 184.0728 | 801.7 | 184.1 | |
| d18:0/22:0 | 789.6838 | 16.272 | C45 H93 N2 O6 P | 788.6771 | 788.6771 | 0.00 | 184.0129 | 789.7 | 184.1 | |
| d18:0/22:1 [b9] | 787.6684 | 15.141 | C45 H91 N2 O6 P | 786.6612 | 786.6615 | −0.38 | 184.0731 | |||
| d18:0/21:0 | 775.6672 | 15.640 | C44 H91 N2 O6 P | 774.6598 | 774.6615 | −2.19 | 184.0728 | |||
| d18:0/20:0 | 761.6528 | 14.958 | C43 H89 N2 O6 P | 760.6452 | 760.6458 | −0.79 | 184.0728 | 761.7 | 184.1 | |
| d18:0/19:0 | 747.6355 | 14.326 | C42 H87 N2 O6 P | 746.6312 | 746.6302 | 1.34 | 184.0728 | 747.6 | 184.1 | |
| d18:0/18:0 | 733.6210 | 13.694 | C41 H85 N2 O6 P | 732.6140 | 732.6145 | −0.68 | 184.0730 | 733.6 | 184.1 | |
| d18:0/17:0 | 719.6056 | 13.096 | C40 H83 N2 O6 P | 718.5982 | 718.5989 | −0.97 | 266.2542, 184.0730 | 719.6 | 184.1 | |
| d18:0/16:0 | 705.5896 | 12.514 | C39 H81 N2 O6 P | 704.5823 | 704.5832 | −1.28 | 184.0739 | 705.6 | 184.1 | |
| d18:0/15:0 | 691.5737 | 11.982 | C38 H79 N2 O6 P | 690.5666 | 690.5676 | −1.45 | 184.0729 | 691.6 | 184.1 | |
| d18:0/14:0 | 677.5586 | 11.466 | C37 H77 N2 O6 P | 676.5512 | 676.5519 | −1.03 | 266.2797, 184.0729 | 677.6 | 184.1 | |
| Cer | d18:1/26:0 | 678.6745 | 20.495 | C44 H87 N O3 | 677.6679 | 677.6686 | −1.03 | 264.2681 | ||
| d18:1/25:0 | 664.6584 | 19.481 | C43 H85 N O3 | 663.6526 | 663.6529 | −0.45 | 264.2654 | |||
| d18:1/24:0 [B16] | 650.6436 | 18.566 | C42 H83 N O3 | 649.6365 | 649.6373 | −1.23 | 264.2652 | 650.6 | 264.3 | |
| d18:1/24:1 [B17] | 648.6280 | 17.219 | C42 H81 N O3 | 647.6208 | 647.6216 | −1.24 | 264.2681 | 648.6 | 264.3 | |
| d18:1/23:0 | 636.6272 | 17.070 | C41 H81 N O3 | 635.6251 | 635.6216 | 5.51 | 264.2681 | 636.6 | 264.3 | |
| d18:1/23:1 | 634.6120 | 16.588 | C41 H79 N O3 | 633.6045 | 633.6060 | −2.37 | 264.2685 | 634.6 | 264.3 | |
| d18:1/22:0 [B18] | 622.6124 | 17.086 | C40 H79 N O3 | 621.6050 | 621.6060 | −1.61 | 264.2680 | 622.6 | 264.3 | |
| d18:1/22:1 [B19] | 620.5964 | 15.956 | C40 H77 N O3 | 619.5891 | 619.5903 | −1.94 | 264.2681 | 620.6 | 264.3 | |
| d18:1/20:0 | 594.5805 | 15.706 | C38 H75 N O3 | 593.5732 | 593.5747 | −2.53 | 264.2686 | 594.6 | 264.3 | |
| d18:1/18:0 | 566.5497 | 14.376 | C36 H71 N O3 | 565.5423 | 565.5434 | −1.95 | 264.2673 | 566.5 | 264.3 | |
| d18:1/18:1 | 564.5302 | 13.478 | C36 H69 N O3 | 563.5233 | 563.5278 | 0.53 | 264.2669 | 563.5 | 264.3 | |
| d18:1/17:0 | 574.5155 | 13.744 | C35 H69 N O3 | 551.5259 | 551.5277 | −3.26 | 264.2670 | |||
| d18:1/16:0 | 538.5186 | 13.112 | C34 H67 N O3 | 537.5114 | 537.5121 | −1.30 | 264.2683 | 538.5 | 264.3 | |
| d18:1/16:1 [B20] | 536.5028 | 12.298 | C34 H65 N O3 | 535.4952 | 535.4964 | −2.24 | 264.2681 | 536.5 | 264.3 | |
| d18:1/15:0 | 524.5026 | 12.564 | C33 H65 N O3 | 523.4955 | 523.4964 | −1.72 | 264.2659 | 524.5 | 264.3 | |
| d18:1/14:0 | 510.4868 | 11.982 | C32 H63 N O3 | 509.4793 | 509.4808 | −2.94 | 264.2696 | |||
| d18:2/26:2 | 672.6258 | 18.566 | C44 H81 N O3 | 671.6185 | 671.6216 | −4.62 | 262.2530 | 672.6 | 262.3 | |
| d18:2/24:1 | 646.6121 | 16.288 | C42 H79 N O3 | 645.6047 | 645.6060 | −2.01 | 262.2528 | 646.6 | 262.3 | |
| d18:2/22:0 [b19] | 620.5960 | 16.139 | C40 H77 N O3 | 619.5886 | 619.5903 | −2.74 | 262.2528 | 620.6 | 262.3 | |
| d18:2/16:0 [b20] | 536.5029 | 12.554 | C34 H65 N O3 | 535.4953 | 535.4964 | −2.05 | 262.2532 | |||
| d18:1/12:0 [IS-2] | 482.4556 | 11.034 | C30 H59 N O3 | 481.4479 | 481.4495 | −3.32 | 264.2678 | 482.5 | 264.3 | |
| DHCer | d18:0/24:0 | 652.6587 | 19.198 | C42 H85 N O 3 | 651.6513 | 651.6529 | −2.46 | 266.2853 | 652.7 | 266.3 |
| d18:0/24:1 [b16] | 650.6434 | 17.735 | C42 H83 N O3 | 649.6361 | 649.6373 | −1.85 | 266.2837 | |||
| d18:0/24:2 [b17] | 648.6281 | 17.502 | C42 H81 N O3 | 647.6209 | 647.6216 | −1.08 | 266.2826 | |||
| d18:0/22:0 | 624.6277 | 17.569 | C40 H81 N O3 | 623.6202 | 623.6216 | −1.12 | 266.2859 | |||
| d18:0/22:1 [b18] | 622.6111 | 16.438 | C40 H79 N O3 | 621.6036 | 621.6060 | −3.86 | 266.2779 | |||
| d18:0/20:0 | 596.5956 | 16.222 | C38 H77 N O3 | 595.5878 | 595.5903 | −4.20 | 266.2811 | |||
| d18:0/18:0 | 568.5650 | 14.858 | C36 H73 N O3 | 567.5578 | 567.5590 | −2.11 | 266.2844 | |||
| d18:0/16:0 | 540.5343 | 13.561 | C34 H69 N O3 | 539.5272 | 539.5277 | −0.93 | 266.2822 | |||
| PTCer | t18:0/14:0 | 528.4981 | 11.333 | C32 H65 N O4 | 527.4908 | 527.4914 | −1.14 | 514.4823, 264.2687 | ||
| HexCer | d18:1/26:0 | 840.7273 | 18.467 | C50 H97 N O8 | 839.7189 | 839.7214 | − 2.98 | 264.2685 | 840.7 | 264.3 |
| d18:1/24:0 | 812.6974 | 17.020 | C48 H93 N O8 | 811.6893 | 811.6901 | −0.99 | 264.2685 | 812.7 | 264.3 | |
| d18:1/24:1 | 810.6809 | 16.654 | C48 H91 N O8 | 809.6732 | 809.6745 | −1.61 | 264.2677 | 810.7 | 264.3 | |
| d18:1/23:0 | 798.6804 | 16.388 | C47 H91 N O8 | 797.6725 | 797.6745 | −2.51 | 264.2676 | 798.7 | 264.3 | |
| d18:1/22:0 | 784.6656 | 15.740 | C46 H89 N O8 | 783.6579 | 783.6588 | −1.15 | 264.2689 | 784.7 | 264.3 | |
| d18:1/16:0 | 700.5717 | 12.115 | C40 H77 N O8 | 699.5621 | 699.5649 | −4.00 | 264.2689 | 700.6 | 264.3 | |
| d18:1/12:0 [IS-3] | 644.5101 | 10.435 | C36 H69 N O8 | 643.5007 | 643.5023 | −2.49 | 264.2684 | 644.5 | 264.3 | |
| LacCer | d18:1/24:0 | 974.7508 | 16.388 | C54 H103 N O13 | 973.7432 | 973.7429 | 0.31 | 264.2672 | 974.7 | 264.3 |
| d18:1/24:1 | 972.7329 | 15.257 | C54 H101 N O13 | 971.7253 | 971.7273 | −2.06 | 264.2650 | 972.7 | 264.3 | |
| d18:1/22:0 | 946.7190 | 15.124 | C52 H99 N O13 | 945.7112 | 945.7116 | −0.42 | 264.2679 | 946.7 | 264.3 | |
| d18:1/20:0 | 918.6866 | 13.894 | C50 H95 N O13 | 917.6780 | 917.6803 | −2.51 | 264.2688 | |||
| d18:1/18:0 | 890.6552 | 12.713 | C48 H91 N O13 | 889.6469 | 889.6490 | −2.36 | 264.2696 | 890.7 | 264.3 | |
| d18:1/16:0 | 862.6250 | 11.682 | C46 H87 N O13 | 861.6175 | 861.6177 | −0.23 | 264.2687 | 862.6 | 264.3 | |
| d18:1/12:0 [IS-4] | 806.5623 | 10.219 | C42 H79 N O13 | 805.5550 | 805.5551 | −0.13 | 264.2683 | 806.7 | 264.3 | |
| Sa | d19:0 [A1] | 316.3202 | 6.532 | C19 H41 N O2 | 315.3135 | 315.3137 | −0.63 | 298.3106, 272.2906 | 274.3 | 256.3 |
| d18:0 [B21] [A2] | 302.3050 | 6.993 | C18 H39 N O2 | 301.2972 | 301.2981 | −2.98 | 284.2921 | 302.3 | 284.3 | |
| d16:0 | 274.2739 | 4.881 | C16 H35 N O2 | 273.2666 | 273.2668 | 0.73 | 256.2627 | 274.3 | 256.3 | |
| PTSa | t17:0 | 304.2874 | 5.468 | C17 H37 N O3 | 303.2782 | 303.2773 | 2.68 | 286.2751 | ||
| d17:0 [A3] [IS-5] | 288.2901 | 6.632 | C17 H37 N O2 | 287.2829 | 287.2824 | 1.65 | 270.2794 | 288.3 | 270.3 | |
| So | d19:1 | 314.3051 | 10.668 | C19 H39 N O2 | 313.2978 | 313.2981 | −0.96 | 296.3320 | ||
| d18:1 | 300.2896 | 6.760 | C18 H37 N O2 | 299.2822 | 299.2824 | −0.67 | 282.2787, 264.2689 | 300.3 | 282.3 | |
| d17:1 [A4] | 286.2737 | 6.444 | C17 H35 N O2 | 285.2659 | 285.2668 | −3.15 | 270.2783 | |||
| d16:1 | 272.2582 | 5.264 | C16 H33 N O2 | 271.2505 | 271.2511 | −2.36 | 254.2833 | 272.3 | 254.3 | |
| d15:1 | 258.2425 | 6.711 | C15 H31 N O2 | 257.2347 | 257.2355 | −3.11 | 240.2319 | |||
| d15:2 | 256.2270 | 5.064 | C15 H29 N O2 | 255.2192 | 255.2198 | −2.35 | 238.2214 | |||
| PTSo | t19:1 | 330.3003 | 8.041 | C19 H39 N O3 | 329.2954 | 329.2930 | 7.29 | 312.3278 | ||
| t19:2 | 328.2846 | 6.245 | C19 H37 N O3 | 327.2771 | 327.2773 | −0.61 | 310.2996 | |||
| t18:1 [a1] | 316.2850 | 6.874 | C18 H37 N O3 | 315.2739 | 315.2773 | 10.8 | 298.2739, 280.2632, 262.2522 | 316.3 | 298.3 | |
| t17:1 [a2] | 302.2687 | 7.176 | C17 H35 N O3 | 301.2618 | 301.2617 | 0.33 | 284.2921, 266.2838 | 302.3 | 284.3 | |
| t16:1 [a3] | 288.2537 | 5.663 | C16 H33 N O3 | 287.2459 | 287.2460 | −0.35 | 270.2786 | |||
| d17:1 [a4] [IS-6] | 286.3106 | 6.558 | C18 H39 N O | 285.3034 | 285.3032 | 0.74 | 268.2643 | 286.3 | 268.3 | |
| SBA | Enigmol [b21] [A2] | 302.3052 | 5.081 | C18 H39 N O2 | 301.2974 | 301.2981 | −2.32 | 284.2930, 266.2090 | ||
| SBA | Xestoaminol C | 230.2477 | 5.131 | C14 H31 N O | 229.2404 | 229.2406 | −0.87 | 212.2355 | ||
| C1P | d18:1/12:0 [IS-7] | 562.4223 | 10.006 | C30 H60 N O6 P | 561.4149 | 561.4158 | −1.72 | 264.2688 | 562.5 | 264.3 |
| Sa1P | d17:0 [IS-8] | 368.2574 | 6.774 | C17 H38 N O5 P | 367.2504 | 367.2488 | 4.57 | 368.3 | 270.3 | |
| So1P | d17:1 [IS-9] | 366.2406 | 6.558 | C17 H36 N O5 P | 365.2331 | 365.2331 | 0.03 | 250.2510 | 366.2 | 250.3 |
The sphingolipids are classified according to “lipid classification system” (http://www.lipidmaps.org/)
SM sphingomyelin, DHSM dihydrosphingomyelin, Cer Ceramide, DHCer dihydroceramide, PTCer phytoceramides, HexCer hexosylceramide, LacCer lactosylceramide, Sa sphinganine, PTSa phytosphinganine, So sphingosine, PTSo phytosphingosine, SBA sphingoid base analog, C1P ceramide-1-phosphate, Sa1P sphinganine-1-phosphate, So1P sphingosine-1-phosphate
[A1-A4 vs a1-a4] 4 pairs of isomeric sphingolipids; [B1-B21 vs b1-b21], 21 pairs of isomeric sphingolipids; [IS], internal standard
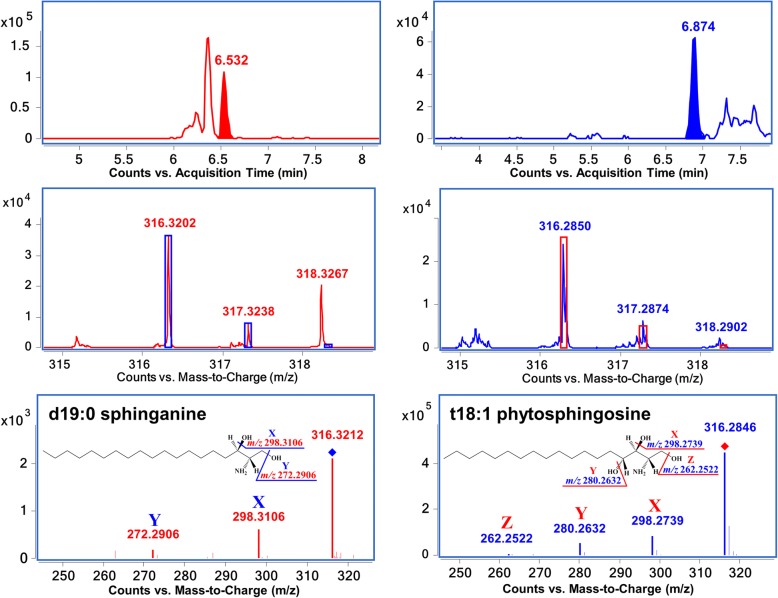
Interpretation of high resolution MS and MS/MS spectra of each identified ion, as well as searching against the latest database, allowed for the accurate identification of SPLs. For instance, isobaric lipids could be differentiated by the high-resolution mass spectrometry-based approaches. Two peaks yield m/z 316 ions, with accurate mass acquired by Q-TOF, m/z 316.3202 at 6.532 min and m/z 316.2850 at 6.874 min correspond to [C19H41NO2 + H]+ and [C18H37NO3 + H]+ respectively, facilitating assignment of sphinganine (Sa) d19:0 and phytosphingosine (PTSo) t18:1. Further fragmentation in MS/MS confirmed the identification, a consecutive loss of 3 hydroxy groups can be observed in the latter case, which is the characteristic cleavage of PTSo (Fig. 1).
Fig. 1.
Differentiation of isobaric SPLs by high resolution mass spectrometry. Accurate mass and isotope distribution can distinguish two m/z 316 compounds, corresponding to d19:0 sphinganine (C19H41NO2) and t18:1 phytosphingosine (C18H37NO3), respectively. Typical ion fragments in MS/MS confirmed the identification
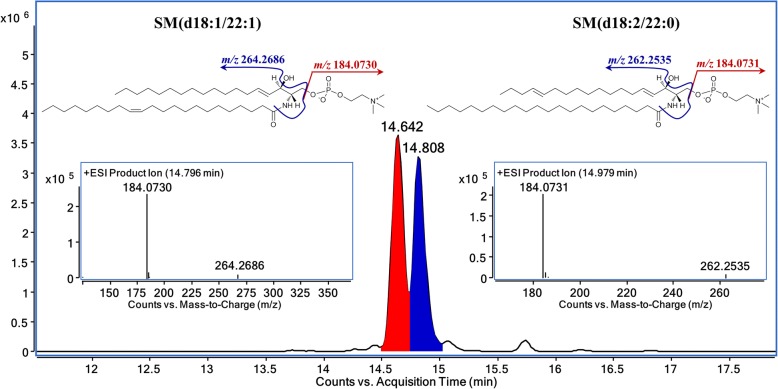
A more realistic interference in the identification of SPLs is the isomeric species that have same number of atoms of each element, thus MS/MS fragment data with the assistance of optimized separation are essential for distinguishing the isomers. Take SM (d18:1/22:1) and SM (d18:2/22:0) as example, there are 2 peaks corresponding to m/z 785.65 in extracted ion chromatogram of TOF MS. In accurate MS/MS data acquired by Q-TOF, two characteristic fragments (264.3 & 262.3) respectively corresponding to the sphingoid base chain of SM (d18:1/22:1) and SM (d18:2/22:0) were observed (Fig. 2). The targeted ion pairs as well as complete chromatographic separation make the accurate MRM quantification of isomers possible.
Fig. 2.
Differentiation of isomeric SPLs by accurate MS/MS. The extracted ion chromatogram of m/z 785.6423 at 5 ppm mass accuracy showed two peaks at 14.642 and 14.808 min. Targeted MS/MS of m/z 785.64 at respective time points gave distinct product ions corresponding to backbone of SM (d18:1/22:1) (m/z 264.3) and SM (d18:2/22:0) (m/z 262.3), providing evidence for the identification of these two species
Ceramides are prone to fragment into product ions corresponding to the sphingoid base backbone (e.g. m/z 262.25, 264.27, 266.28). In A549 QC samples, 29 Cers, including 20 dehydroceramides, 8 dihydroceramides (DHCers) and 1 phytoceramide (PTCer), were identified by comparing the MS information and retention time with those of SPLs in our previous study [10, 11]. Most Cers detected in the samples were with a d18:1 sphingoid backbone and the carbon number of N-acyl side chain varied from 14 to 26. A new dihydroceramide DHCer (d18:0/24:2), and Cer (d18:2/26:2), a dehydroceramide with high degree of unsaturation and long N-acyl chain, have been characterized for the first time to the best of our knowledge.
SM is the most multitudinous subclass of SPLs in A549 and A549T. Based on the exact mass in TOF MS and characteristic product ions obtained by Q-TOF MS/MS, a total of 56 SMs, including 38 dehydrosphingomyelins and 18 dihydrosphingomyelins (DHSMs), were unambiguously identified. All these SMs were characterized with a C18 sphingoid base chain, among which d18:1 type takes the largest proportion. In the N-acyl side chain, the number of carbon ranged between 14 and 26, with an unsaturation degree up to 5. Notably, all the DHSMs with 21 or less carbons in the N-acyl chain are fully saturated, while the others (with more than 21 carbons in the N-acyl chain) can be detected together with their corresponding de-hydrogen form. Three highly unsaturated SMs (total unsaturation degree no less than 4) including SM (d18:1/24:3), SM (d18:2/24:2) and SM (d18:2/24:3), have been detected in the QC sample of A549 & A549T cells.
Hexose-linked glycoceramide including galactosylceramide (GalCer) and glucosylceramide (GluCer) were represented as HexCer. All the 6 HexCers and 6 LacCers were found with d18:1 sphingoid base backbone. Only one HexCer with N-acyl chain in odd carbon number, HexCer (d18:1/23:0) was identified in A549. Notably, among all the HexCers and LacCers, only d18:1/24:1 species were identified as glycoceramides with unsaturated N-acyl fatty chain.
Seventeen sphingoid bases as well as the analogs were also successfully identified. The carbon number ranging from 14 to 19 and the degree of unsaturation falls between 0 and 2. Two PTSo with 3 hydroxyl groups, PTSo t19:2 and PTSo t16:1, have been discovered for the first time.
Quantitation of sphingolipids in A549 and A549T cells
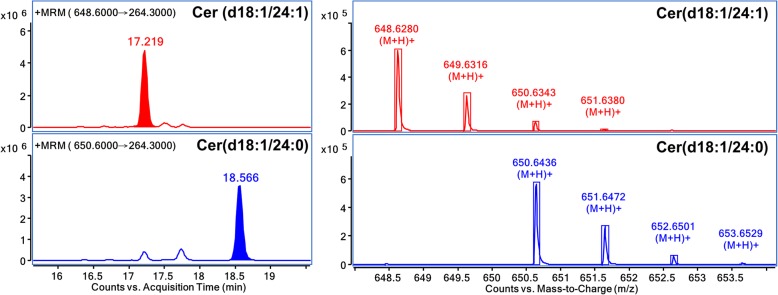
MRM mode of UHPLC-QQQ MS could provide accurate and sensitive approach under a wide range for quantitative analysis of SPLs. As the accuracy of triple-quadruple is about 0.1 Da, the quantification of SPLs cannot be accurately achieved merely with a QQQ analyzer, especially when suffering the isotopic interferences. Every unsaturated SPL could be recognized as an isotope of another one with the same characteristic backbone but less degree of unsaturation. For instance, if the LC separation is incomplete, the content of Cer (d18:1/24:0) will be artificially high due to the interference of Cer (d18:1/24:1) (Fig. 3). In this study, based on UHPLC complete separation and Q-TOF comprehensive profiling, accurate quantification was accomplished by eliminating the isotopic interference. By using the UHPLC-QQQ MS method with the optimized MRM parameters, a total of 75 species out of 114 identified SPLs were quantified in A549 and A549T cells, respectively. The amounts of these SPLs were quantified by comparing with the foregoing mentioned ISs.
Fig. 3.
Differentiation of isotopic SPLs by accurate MS together with complete separation. Cer (d18:1/24:1) (tR = 17.219 min) yields precursor ions at m/z 648.6280 and m/z 650.6343, the latter one is the [M + 2] isotopic ion which will interfere with the precursor ion of Cer (d18:1/24:0) (tR = 18.566 min) at m/z 650.6436. The mass differentiation cannot be distinguished by QQQ. If the two peaks cannot be completely separated by LC, the quantification result of Cer (d18:1/24:0) will be artificially high
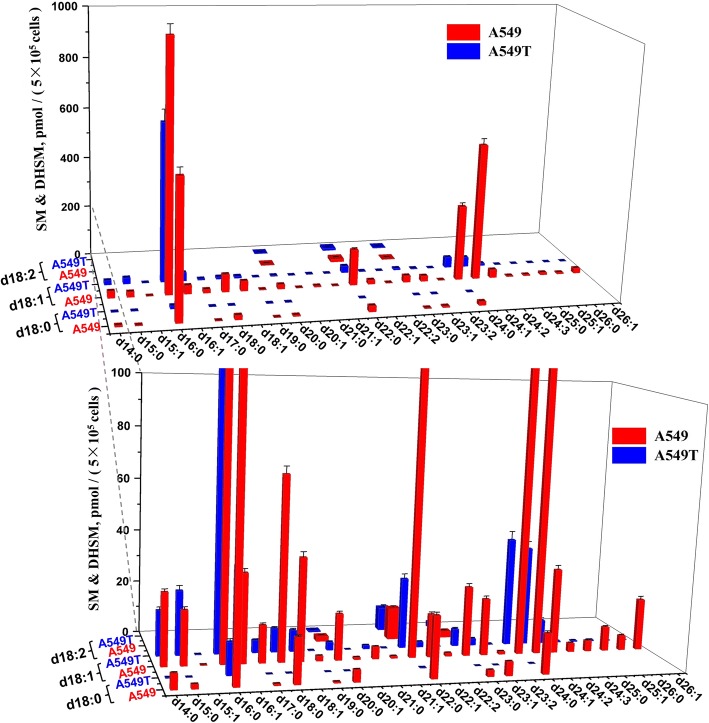
The quantitative results indicated that SMs account for the majority of all the SPLs in A549 and A549T, among which SMs with C16/C18/C22/C24 N-acyl side chain took the largest proportion of the total content. SMs with d18:1 sphingoid backbone are the most dominant species, which take 27 out of all the 41 quantified SMs (Fig. 4). For some SMs with high unsaturation degree or long N-acyl chain, the content is extremely low which cannot reach the limit of quantitation (LOQ). Figure 5 shows quantification data of 17 Cers. In general, the amounts of various Cers are significantly higher in A549 rather than those in A549T. Similar to SM, d18:1 Cers with C16/C18/C22/C24 N-acyl side chain showed relative high levels in both A549 and A549T, which take most proportion of Cer. LacCers and HexCers were only found with d18:1 sphingoid base backbone. All the 6 LacCers showed higher intensity in A549T than that in A549. But HexCer showed a species-dependent trend, HexCer d18:1/16:0, HexCer d18:1/22:0 and HexCer d18:1/23:0 increased in A549T, while HexCer d18:1/24:0, HexCer d18:1/24:1 and HexCer d18:1/26:0 decreased (Fig. 6). The overall content of sphingoid bases was similar in both cell types, Sa d16:0 was found with the highest intensity (Fig. 7). The relative abundance of each SPL varied greatly, but SPLs with N-acyl chain length of C16 and C24, respectively, are the most abundant species within each subclass.
Fig. 4.
Content of SM and DHSM in A549 and A549T. The X and Z axis represent the compose of fatty acid acyl chain and backbone chain, respectively. Comparisons were performed by the non-parametric Mann-Whitney test. Most SMs and DHSMs showed statistical significance between A549 and A549T (P < 0.0001), except for SM (d18:1/16:1) and SM (d18:2/23:0) (P > 0.05)
Fig. 5.
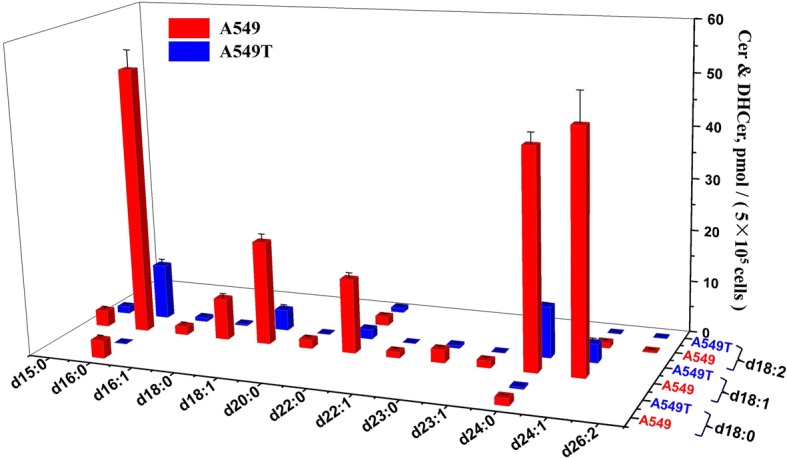
Content of Cer and DHCer in A549 and A549T. The X and Z axis represent the compose of fatty acid acyl chain and backbone chain, respectively. Comparisons were performed by the non-parametric Mann-Whitney test. All Cers and DHCers showed statistical significance between A549 and A549T (P < 0.0001)
Fig. 6.
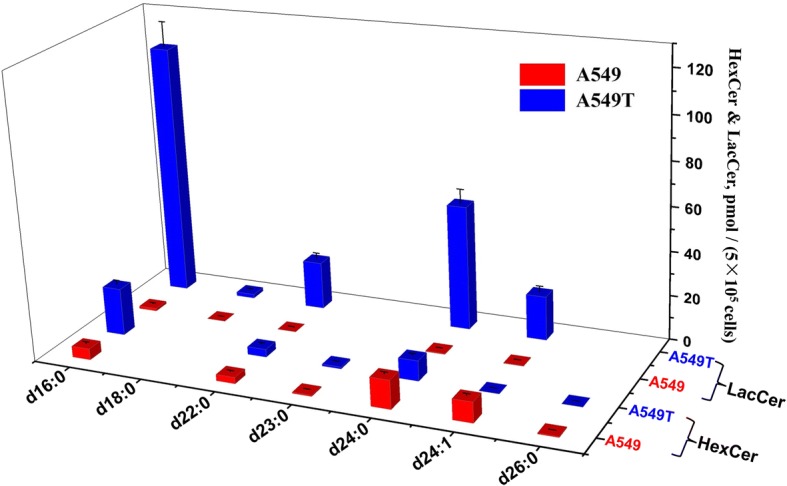
Content of HexCer and LacCer in A549 and A549T. The X axis represents the compose of fatty acid acyl chain of the d18:1 HexCer and d18:1 LacCer. Comparisons were performed by the non-parametric Mann-Whitney test. All HexCers and LacCers showed statistical significance between A549 and A549T (P < 0.0001), except for HexCer (d18:1/22:0) (P < 0.01) and HexCer (d18:1/23:0) (P < 0.001)
Fig. 7.
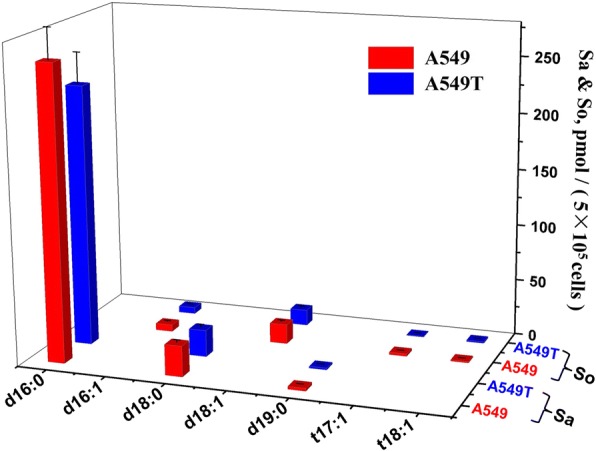
Content of sphingoid base in A549 and A549T. Comparisons were performed by the non-parametric Mann-Whitney test. All sphingoid bases showed statistical significance between A549 and A549T (P < 0.0001), except for Sa (d16:0) (P < 0.01), So (d16:1) (P > 0.05) and So (t18:1) (P > 0.05)
PCA was used for the overview of SPL dataset and the spotting of outliers, and thereby pick out trends of grouping or separation. It was performed to visualize general clustering among A549, A549T and QC groups [R2X (cum) = 0.874, Q2 (cum) = 0.845; Fig. 8a]. Supervised PLS-DA was used to further study the differences between A549 and A549T and to select potential biomarkers. In PLS-DA, the result of model showed the performance statistics of R2X (cum) = 0.880, R2Y (cum) = 0.999 with an excellent prediction parameter Q2 (cum) = 0.998, and the score plot showed good visual separation between A549 and A549T groups as well (Fig. 8b). A total of 57 potential biomarkers were identified according to scattering-plot and the VIP value (Table 2), among which most of them are SM and Cers. SM (d18:0/18:0) showed the largest decline in A549T, the content in decreased from 17.0 to 0.10 pmol/(5 × 105cells), that markedly contributes to the classification.
Fig. 8.
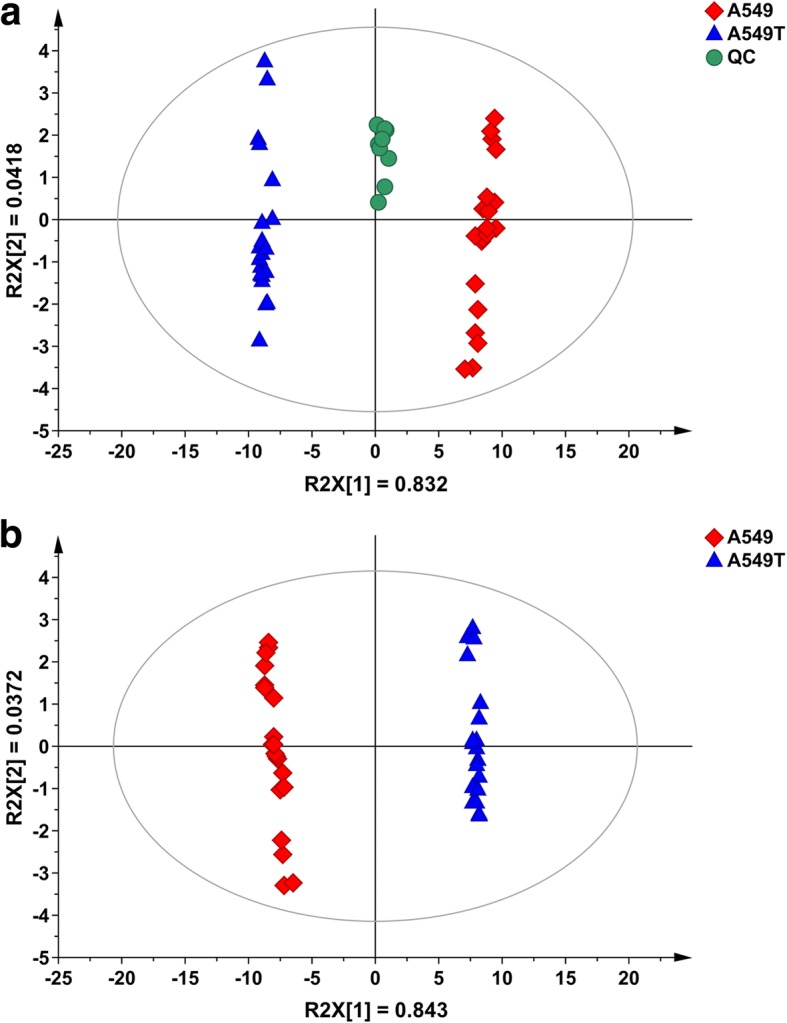
Principal component analysis and partial least squares discriminant analysis projecting scatter plots. a PCA [R2X (cum) = 0.874, Q2 (cum) = 0.845] score plots based on the content of SPLs obtained from A549 (red, diamond), A549T (blue, triangle), and QC (green, circle) groups; b PLS-DA [R2X (cum) = 0.880, R2Y (cum) = 0.999, Q2 (cum) = 0.998] score plots based on the content of SPLs obtained from A549 (red, diamond) and A549T (blue, triangle) groups
Table 2.
Quantification of SPLs (VIP > 1) in A549 and A549T
| SPLs | Content (pmol/5*105 cells) | ChangeA549T vs A549 | p value | VIP | |
|---|---|---|---|---|---|
| A549 (n = 20) | A549T (n = 20) | ||||
| SM (d18:2/20:0) | 1.96 ± 0.16 | 0.53 ± 0.07 | ↓ | < 0.001 | 1.07408 |
| SM (d18:1/26:1) | 19.8 ± 1.22 | 0.34 ± 0.04 | ↓ | < 0.001 | 1.08696 |
| SM (d18:1/26:0) | 5.67 ± 0.31 | 0.20 ± 0.02 | ↓ | < 0.001 | 1.08775 |
| SM (d18:1/25:1) | 9.28 ± 0.43 | 0.45 ± 0.05 | ↓ | < 0.001 | 1.08860 |
| SM (d18:1/25:0) | 4.09 ± 0.23 | 0.42 ± 0.04 | ↓ | < 0.001 | 1.08699 |
| SM (d18:1/24:3) | 3.34 ± 0.26 | 0.47 ± 0.07 | ↓ | < 0.001 | 1.08120 |
| SM (d18:1/24:2) | 32.5 ± 1.55 | 8.99 ± 0.87 | ↓ | < 0.001 | 1.08501 |
| SM (d18:1/24:1) | 542 ± 23.8 | 37.9 ± 2.59 | ↓ | < 0.001 | 1.08879 |
| SM (d18:1/24:0) | 298 ± 13.0 | 41.5 ± 3.16 | ↓ | < 0.001 | 1.08829 |
| SM (d18:1/23:2) | 1.12 ± 0.11 | 0.25 ± 0.04 | ↓ | < 0.001 | 1.06488 |
| SM (d18:1/23:1) | 21.6 ± 1.08 | 2.58 ± 0.30 | ↓ | < 0.001 | 1.08745 |
| SM (d18:1/23:0) | 26.4 ± 1.26 | 6.57 ± 0.51 | ↓ | < 0.001 | 1.08618 |
| SM (d18:1/22:2) | 1.25 ± 0.12 | 0.15 ± 0.02 | ↓ | < 0.001 | 1.07067 |
| SM (d18:1/22:1) | 16.4 ± 0.81 | 2.07 ± 0.18 | ↓ | < 0.001 | 1.08760 |
| SM (d18:1/22:0) | 142 ± 6.17 | 26.9 ± 1.78 | ↓ | < 0.001 | 1.08783 |
| SM (d18:1/21:1) | 0.82 ± 0.07 | 0.15 ± 0.02 | ↓ | < 0.001 | 1.07308 |
| SM (d18:1/21:0) | 4.66 ± 0.25 | 1.39 ± 0.12 | ↓ | < 0.001 | 1.08216 |
| SM (d18:1/20:1) | 0.87 ± 0.13 | 0.06 ± 0.00 | ↓ | < 0.001 | 1.06102 |
| SM (d18:1/20:0) | 17.6 ± 0.86 | 3.08 ± 0.31 | ↓ | < 0.001 | 1.08689 |
| SM (d18:1/19:0) | 1.94 ± 0.20 | 0.21 ± 0.03 | ↓ | < 0.001 | 1.07588 |
| SM (d18:1/18:1) | 39.0 ± 1.99 | 8.13 ± 0.58 | ↓ | < 0.001 | 1.08637 |
| SM (d18:1/18:0) | 69.5 ± 2.72 | 9.07 ± 0.54 | ↓ | < 0.001 | 1.08896 |
| SM (d18:1/17:0) | 14.1 ± 0.72 | 4.76 ± 0.39 | ↓ | < 0.001 | 1.08307 |
| SM (d18:1/16:0) | 982 ± 38.5 | 629 ± 23.5 | ↓ | < 0.001 | 1.06108 |
| SM (d18:1/14:0) | 27.3 ± 1.04 | 17.2 ± 0.65 | ↓ | < 0.001 | 1.06732 |
| SM (d18:0/24:0) | 15.3 ± 0.78 | 0.11 ± 0.02 | ↓ | < 0.001 | 1.08843 |
| SM (d18:0/23:0) | 2.45 ± 0.15 | 0.02 ± 0.00 | ↓ | < 0.001 | 1.08694 |
| SM (d18:0/22:0) | 23.3 ± 1.05 | 0.23 ± 0.04 | ↓ | < 0.001 | 1.08900 |
| SM (d18:0/20:0) | 4.59 ± 0.24 | 0.08 ± 0.01 | ↓ | < 0.001 | 1.08808 |
| SM (d18:0/19:0) | 0.49 ± 0.07 | 0.01 ± 0.00 | ↓ | < 0.001 | 1.05519 |
| SM (d18:0/18:0) | 17.0 ± 0.55 | 0.10 ± 0.03 | ↓ | < 0.001 | 1.09013 |
| SM (d18:0/17:0) | 0.65 ± 0.07 | 0.06 ± 0.01 | ↓ | < 0.001 | 1.07030 |
| SM (d18:0/16:0) | 545 ± 28.7 | 12.3 ± 0.94 | ↓ | < 0.001 | 1.08809 |
| SM (d18:0/15:0) | 1.74 ± 0.16 | 0.10 ± 0.02 | ↓ | < 0.001 | 1.08041 |
| SM (d18:0/14:0) | 5.83 ± 0.40 | 0.28 ± 0.03 | ↓ | < 0.001 | 1.08565 |
| Cer (d18:2/24:1) | 1.01 ± 0.21 | 0.03 ± 0.01 | ↓ | < 0.001 | 1.04397 |
| Cer (d18:1/24:1) | 46.1 ± 5.83 | 3.60 ± 0.44 | ↓ | < 0.001 | 1.07110 |
| Cer (d18:1/24:0) | 42.0 ± 2.08 | 9.83 ± 0.41 | ↓ | < 0.001 | 1.08653 |
| Cer (d18:1/23:1) | 1.37 ± 0.22 | 0.06 ± 0.01 | ↓ | < 0.001 | 1.06057 |
| Cer (d18:1/23:0) | 2.54 ± 0.27 | 0.53 ± 0.09 | ↓ | < 0.001 | 1.06941 |
| Cer (d18:1/22:1) | 1.20 ± 0.27 | 0.04 ± 0.01 | ↓ | < 0.001 | 1.03542 |
| Cer (d18:1/22:0) | 14.4 ± 0.94 | 1.90 ± 0.39 | ↓ | < 0.001 | 1.08443 |
| Cer (d18:1/20:0) | 1.69 ± 0.33 | 0.07 ± 0.02 | ↓ | < 0.001 | 1.04721 |
| Cer (d18:1/18:1) | 20.1 ± 1.33 | 4.03 ± 0.49 | ↓ | < 0.001 | 1.08187 |
| Cer (d18:1/18:0) | 8.03 ± 0.72 | 0.26 ± 0.04 | ↓ | < 0.001 | 1.08142 |
| Cer (d18:1/16:0) | 52.0 ± 3.59 | 10.7 ± 0.98 | ↓ | < 0.001 | 1.08243 |
| Cer (d18:1/15:0) | 3.14 ± 0.46 | 1.34 ± 0.30 | ↓ | < 0.001 | 1.00100 |
| Cer (d18:0/24:0) | 1.44 ± 0.11 | 0.34 ± 0.04 | ↓ | < 0.001 | 1.07587 |
| Cer (d18:0/16:0) | 3.46 ± 0.25 | 0.07 ± 0.01 | ↓ | < 0.001 | 1.05883 |
| HexCer (d18:1/26:0) | 0.41 ± 0.10 | 0.04 ± 0.02 | ↓ | < 0.001 | 1.00780 |
| HexCer (d18:1/24:1) | 9.25 ± 0.97 | 0.02 ± 0.00 | ↓ | < 0.001 | 1.07959 |
| HexCer (d18:1/16:0) | 5.06 ± 0.89 | 21.6 ± 2.10 | ↑ | < 0.001 | 1.07118 |
| LacCer (d18:1/24:1) | 0.22 ± 0.05 | 19.9 ± 2.17 | ↑ | < 0.001 | 1.07014 |
| LacCer (d18:1/24:0) | 0.37 ± 0.03 | 56.4 ± 3.16 | ↑ | < 0.001 | 1.07766 |
| LacCer (d18:1/22:0) | 0.06 ± 0.01 | 21.6 ± 2.85 | ↑ | < 0.001 | 1.07258 |
| LacCer (d18:1/16:0) | 1.15 ± 0.55 | 115 ± 11.6 | ↑ | < 0.001 | 1.08013 |
| So (t17:1) | 2.21 ± 0.17 | 0.62 ± 0.08 | ↓ | < 0.001 | 1.07483 |
Discussion
Using the sphingolipidomic approach, we obtained the detailed sphingolipid profiles for human lung adenocarcinoma cell A549 and its taxol resistant strain A549T, and then performed quantification. We found A549 and A549T share all the same species of SPLs, among which SM (dehydrosphingomyelin and DHSM), Cer (dehydroceramide, DHCer and PTCer), HexCer, LacCer, and sphingoid base were identified as the major SPLs. In contrast to normal A549, decreasing levels of Cer and SM concomitant with increasing of glycosphingolipids represent the main SPL metabolic profile of A549T. Totally 35 SMs, 14 Cers, 3 HexCers, 4 LacCers, and 1 sphingosine are recognized as metabolic pathway related biomarkers.
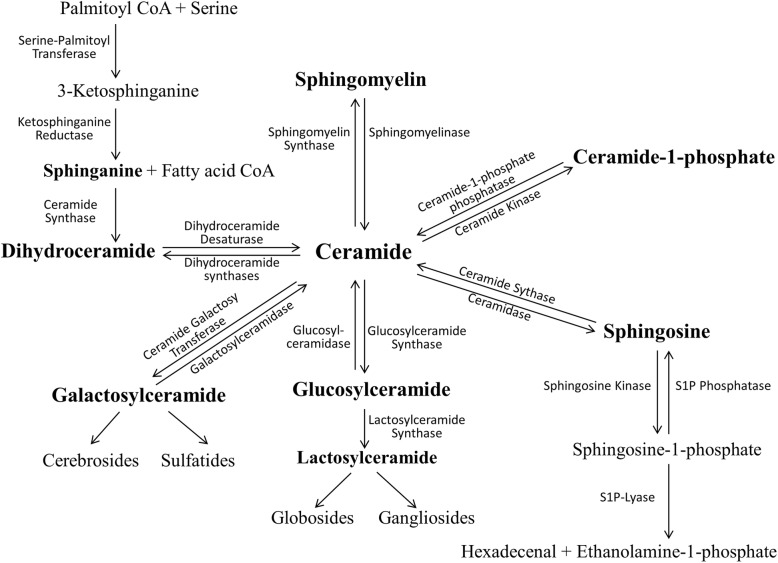
Cer is the basic SPL structural unit which balances cell growth and death by inducing apoptosis [12], and its definite efficacy in promoting apoptosis in A549 cells has been well studied [7]. It is noteworthy that Cers can be classified into SM-hydrolyzed and de novo-synthesized. The former is well known as triggering apoptotic death signaling in many cell types, while the specific role of the latter one seems important to tumor survival [13]. In human ovarian carcinoma cell line CABA I, anti-cancer drugs including taxol have been reported to activate SMase to generate Cer, which acts as a second messenger in triggering apoptosis [14]. While in lung carcinoma cells, the tumor tissues produce large amounts of both dihydroceramide and ceramide through the de novo synthesis pathway, but not through SM hydrolysis [13]. More relevantly, treatment of A549 cells with gemcitabine was demonstrated to increase Cer levels via the activation of de novo synthesis [15]. In the case of study of A549T in this paper, both Cer and SM levels were much lower than their levels in taxol-sensitive A549 cells, which indicates that the decrease of Cer may not attribute to “activating the SM pathway” as our previous study in A2780T [11]. Furthermore, DHCer was decreasing accompanied with Cer, which revealed the mechanism of taxol-resistance in A549T could be explained as “inhibiting the de novo synthesis pathway”. (Fig. 9).
Fig. 9.
Biosynthesis and metabolism pathway of sphingolipids
Both Cer and its catabolite sphingosine as negative regulators of cell proliferation could promote apoptosis, and the role of sphingosine as a messenger of apoptosis is of importance [16]. In small cell lung cancer (SCLC), multidrug-resistance-associated protein (MRP) contributes to the drug resistance, and pro-apoptotic SPLs (Cer and sphingosine) could further induce apoptosis overcome or bypass MRP-mediated drug resistance [17]. In non-small cell lung cancer (NSCLC) including A549, sphingosine kinase 2 (SphK2) is proposed to be the key regulator of sphingolipid signaling which may contribute to the apoptosis resistance [18]. Inhibition of SphK2 can enhance the apoptosis of NSCLC cells, and it will certainly result in an increase of the substrate sphingosine. In A549T, all sphingosines showed consistent trend of decrease comparing to A549. It’s known that sphingosine in mammalian cells is not synthesized de novo but it is generated from ceramides by ceramidases [19]. Thus, we can deduce that in A549T the concomitant decrease of sphingosine and Cer may be the result of activation of SphK2, which leads to the inhibition of apoptosis in taxol resistant strain.
Besides Cer and SM, glycosphingolipids including HexCers (GalCers & GluCers) and LacCers account for a large proportion of biomarkers in A549T. It has been observed that glucosylceramide synthase is up-regulated after drug intervention and suggests that glycolipids may be involved in chemotherapy resistance [2]. For decades, GluCer has been found to increase in the resistant cancer cells [20], suggesting that glycosylation plays an important role in evading Cer induced apoptosis. Glycosphingolipids have recently been reported as transactivating multidrug resistance 1/P-glycoprotein (MDR1) and multidrug resistance-associated protein 1 (MRP1) expression which further prevents accumulation of ceramide and stimulates drug efflux [21]. Specifically, GalCer and LacCer were characteristically increased in taxol-resistant human ovarian carcinoma-derived KF28TX cells [22]. Moreover, GalCer was demonstrated to be the apoptosis protector, and its upregulation was also thought to attenuate the Cer-mediated apoptotic signals [23]. Our findings revealed a significant overall increase of glycosphingolipids in A549T, among which all LacCers showed a consistent tendency of increase, while HexCers (including GalCer and GluCer) showed a species-dependent trend. It should be noted that GluCer could be converted into LacCer under the influence of lactosylceramide synthase. Therefore the decreased species of HexCer might be GluCer. For instance, the decrease of HexCer (d18:1/24:1) resulted in the concomitant increase of LacCer (d18:1/24:1).
Conclusions
Evidences suggest that tumor microenvironment including the sphingolipidome plays an important role in cancer drug resistance. So far to our knowledge, there is no sphingolipidomic study on taxol-resistant A549 human adenocarcinoma cell line. Based on the comprehensive identification and accurate quantification of SPLs, decreasing of Cer, SM and sphingosine concomitant with increasing of HexCer and LacCer have been characterized as the metabolic profile of A549T. It indicated that “inhibition of the de novo synthesis pathway” and “activation of glycosphingolipid pathway” played the dominant role for taxol-resistance, and the key enzymes related to the pathways may have been altered. These results provide evidence to unravel the mechanism of taxol resistance in A549T. The distinctive phenotype could facilitate clinical diagnosis of taxol-resistant adenocarcinoma and provide insights into targets for the development of new drug against taxol resistance.
Acknowledgments
Funding
This work was financially supported by Tertiary Education Services Office, Macau Special Administrative Region (GAES-17-001-SKL to Z.-H. Jiang); Macao Science and Technology Development Fund, Macau Special Administrative Region (015/2017/AFJ to Z.-H. Jiang and 023/2016/AFJ to J.-R Wang); and in part by Ph.D. start-up fund of Gannan Medical University (No.201304 granted to H. Huang). The funding bodies have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and analyzed during the current study were available from the corresponding author(s) on reasonable request.
Abbreviations
- A549T
Taxol-resistant strain of A549 cell
- C1P
Ceramide-1-phosphate
- Cer
Ceramide
- DHCer
Dihydroceramide
- DHSM
Dihydrosphingomyelin
- HexCer
Hexosylceramide
- LacCer
Lactosylceramide
- QC
Quality control
- SM
Sphingomyelin
- SPL
Sphingolipid
Authors’ contributions
ZHJ and JRW conceived the research; HH drafted the manuscript, ZHJ, JRW and LFY reviewed and revised it critically; TTT and LFY performed the cell experiments; HH, CCY and MJN carried out the sample preparation, LC-MS analysis and data interpretation; HH and TTT designed the figures. All authors read and approved the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hao Huang, Email: hhuang@must.edu.mo.
Tian-Tian Tong, Email: tttong@must.edu.mo.
Lee-Fong Yau, Email: lfyau@must.edu.mo.
Cheng-Yu Chen, Email: chychen@must.edu.mo.
Jia-Ning Mi, Email: mjnrhw@hotmail.com.
Jing-Rong Wang, Email: jrwang@must.edu.mo.
Zhi-Hong Jiang, Email: zhjiang@must.edu.mo.
References
- 1.Bender E. Epidemiology: the dominant malignancy. Nature. 2014;513(7517):S2–S3. doi: 10.1038/513S2a. [DOI] [PubMed] [Google Scholar]
- 2.Gouaze V, Yu JY, Bleicher RJ, Han TY, Liu YY, Wang H, Gottesman MM, Bitterman A, Giuliano AE, Cabot MC. Overexpression of glucosylceramide synthase and P-glycoprotein in cancer cells selected for resistance to natural product chemotherapy. Mol Cancer Ther. 2004;3(5):633–639. [PubMed] [Google Scholar]
- 3.Yusuf RZ, Duan Z, Lamendola DE, Penson RT, Seiden MV. Paclitaxel resistance: molecular mechanisms and pharmacologic manipulation. Curr Cancer Drug Targets. 2003;3(1):1–19. doi: 10.2174/1568009033333754. [DOI] [PubMed] [Google Scholar]
- 4.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giussani P, Tringali C, Riboni L, Viani P, Venerando B. Sphingolipids: key regulators of apoptosis and pivotal players in cancer drug resistance. Int J Mol Sci. 2014;15(3):4356–4392. doi: 10.3390/ijms15034356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurinna SM, Tsao CC, Nica AF, Jiffar T, Ruvolo PP. Ceramide promotes apoptosis in lung cancer-derived A549 cells by a mechanism involving c-Jun NH2-terminal kinase. Cancer Res. 2004;64(21):7852–7856. doi: 10.1158/0008-5472.CAN-04-1552. [DOI] [PubMed] [Google Scholar]
- 8.Pettus BJ, Bielawska A, Subramanian P, Wijesinghe DS, Maceyka M, Leslie CC, Evans JH, Freiberg J, Roddy P, Hannun YA, et al. Ceramide 1-phosphate is a direct activator of cytosolic phospholipase A2. J Biol Chem. 2004;279(12):11320–11326. doi: 10.1074/jbc.M309262200. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Sun G, Liu G, Wang Y, Shao Z, Chen Z, Yang J. Effects of mycoplasma pneumoniae infection on sphingolipid metabolism in human lung carcinoma A549 cells. Microb Pathog. 2009;46(2):63–72. doi: 10.1016/j.micpath.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Wang JR, Zhang H, Yau LF, Mi JN, Lee S, Lee KC, Hu P, Liu L, Jiang ZH. Improved sphingolipidomic approach based on ultra-high performance liquid chromatography and multiple mass spectrometries with application to cellular neurotoxicity. Anal Chem. 2014;86(12):5688–5696. doi: 10.1021/ac5009964. [DOI] [PubMed] [Google Scholar]
- 11.Huang H, Tong TT, Yau LF, Chen CY, Mi JN, Wang JR, Jiang ZH. LC-MS based Sphingolipidomic study on A2780 human ovarian Cancer cell line and its Taxol-resistant strain. Sci Rep. 2016;6:34684. doi: 10.1038/srep34684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravid T, Tsaba A, Gee P, Rasooly R, Medina EA, Goldkorn T. Ceramide accumulation precedes caspase-3 activation during apoptosis of A549 human lung adenocarcinoma cells. Am J Physiol Lung Cell Mol Physiol. 2003;284(6):L1082–L1092. doi: 10.1152/ajplung.00172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyanagi S, Kuga M, Soeda S, Hosoda Y, Yokomatsu T, Takechi H, Akiyama T, Shibuya S, Shimeno H. Elevation of de novo ceramide synthesis in tumor masses and the role of microsomal dihydroceramide synthase. Int J Cancer. 2003;105(1):1–6. doi: 10.1002/ijc.11024. [DOI] [PubMed] [Google Scholar]
- 14.Prinetti A, Millimaggi D, D'Ascenzo S, Clarkson M, Bettiga A, Chigorno V, Sonnino S, Pavan A, Dolo V. Lack of ceramide generation and altered sphingolipid composition are associated with drug resistance in human ovarian carcinoma cells. Biochem J. 2006;395(2):311–318. doi: 10.1042/BJ20051184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002;277(15):12587–12595. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 16.Cuvillier O. Sphingosine in apoptosis signaling. Biochim Biophys Acta. 2002;1585(2–3):153–162. doi: 10.1016/S1388-1981(02)00336-0. [DOI] [PubMed] [Google Scholar]
- 17.Khodadadian M, Leroux ME, Auzenne E, Ghosh SC, Farquhar D, Evans R, Spohn W, Zou Y, Klostergaard J. MRP- and BCL-2-mediated drug resistance in human SCLC: effects of apoptotic sphingolipids in vitro. Lung Cancer. 2009;66(1):48–57. doi: 10.1016/j.lungcan.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Yang C, Zhang S, Mei Z, Shi M, Sun S, Shi L, Wang Z, Wang Y, Li Z, et al. ABC294640, a sphingosine kinase 2 inhibitor, enhances the antitumor effects of TRAIL in non-small cell lung cancer. Cancer Biol Ther. 2015;16(8):1194–1204. doi: 10.1080/15384047.2015.1056944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta. 2008;1781(9):424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson KM, Quinn DM, Kellett GL, Warr JR. Preferential killing of multidrug-resistant KB cells by inhibitors of glucosylceramide synthase. Br J Cancer. 1999;81(3):423–430. doi: 10.1038/sj.bjc.6690711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyler A, Johansson A, Karlsson T, Gudey SK, Brannstrom T, Grankvist K, Behnam-Motlagh P. Targeting glucosylceramide synthase induction of cell surface globotriaosylceramide (Gb3) in acquired cisplatin-resistance of lung cancer and malignant pleural mesothelioma cells. Exp Cell Res. 2015;336(1):23–32. doi: 10.1016/j.yexcr.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Kiguchi K, Iwamori Y, Suzuki N, Kobayashi Y, Ishizuka B, Ishiwata I, Kita T, Kikuchi Y, Iwamori M. Characteristic expression of globotriaosyl ceramide in human ovarian carcinoma-derived cells with anticancer drug resistance. Cancer Sci. 2006;97(12):1321–1326. doi: 10.1111/j.1349-7006.2006.00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grazide S, Terrisse AD, Lerouge S, Laurent G, Jaffrezou JP. Cytoprotective effect of glucosylceramide synthase inhibition against daunorubicin-induced apoptosis in human leukemic cell lines. J Biol Chem. 2004;279(18):18256–18261. doi: 10.1074/jbc.M314105200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study were available from the corresponding author(s) on reasonable request.