Abstract
Cyclin E1 (formerly called cyclin E) and the recently described cyclin E2 belong to the family of E-type cyclins that operate during the G1/S phase progression in mammalian cells. The two E-cyclins share a catalytic partner, cyclin-dependent kinase 2 (CDK2), and activate their associated kinase activities at similar times during cell cycle progression. Despite these similarities, it is unknown whether the two proteins perform distinct functions, or, alternatively, they control S-phase entry of different cell types in a tissue-specific fashion. To start addressing in vivo functions of E-cyclins, we determined the expression pattern of cyclins E1 and E2 during normal mouse development. We found that the two E-cyclins showed very similar patterns of expression; both were expressed within the proliferating compartment during embryo development. Analyses of cells and tissues lacking members of the retinoblastoma (pRB) family of proteins revealed that the expression of both cyclins is controlled in a pRB-dependent, but p107- and p130-independent fashion, likely through the pRB-dependent E2F transcription factors. We also found that cyclins E1 and E2 are expressed at high levels in mouse breast tumors driven by the Myc oncogene. Last, we found that cyclin E2 is overexpressed in ≈24% of analyzed human mammary carcinomas. Collectively these findings suggest that the expression of cyclins E1 and E2 is governed by similar molecular circuitry.
Cyclins are key components of the core cell cycle machinery in mammalian cells. Two classes of cyclins operate during the G1 phase: D-type cyclins and cyclin E (1).
D-cyclins specify the family of three closely related proteins (cyclins D1, D2, and D3). The levels of D-cyclins are controlled by the extracellular environment. Once induced, D-cyclins associate with cyclin-dependent kinases CDK4 or CDK6 and phosphorylate the retinoblastoma tumor suppressor gene product (pRB). This phosphorylation leads to release of pRB-bound transcription factors, notably the E2F family of transactivators, and to derepression or activation of E2F-controlled genes, such as cyclin E. In addition, cyclin D–CDK complexes control the activity of cyclin E–CDK2 holoenzyme by titrating away the CDK inhibitors p27Kip1 and p21Cip1 from cyclin E–CDK2 complexes to cyclin D–CDK4/6 complexes, thereby triggering the kinase activity of cyclin E-CDK2 (1). Hence, D-type cyclins serve to couple the extracellular mitogenic stimulation with activation of cyclin E. While the three D-cyclins are biochemically indistinguishable, each of these cyclins shows a unique, tissue-specific pattern of expression during development and in adult tissues, and each is differentially controlled by distinct upstream activating pathways (2–5).
Cyclin E is believed to control G1/S phase progression. It associates with CDK2 and activates its kinase activity shortly before entry of cells into the S phase (6, 7). The targets for cyclin E–CDK2 kinase are largely unknown. Cyclin E is believed to control S phase entry by phosphorylating proteins involved in DNA replication (8). Cyclin E may also control centrosome duplication (8), histone gene transcription (9, 10), and pre-mRNA splicing (11).
Recently, another G1 cyclin was discovered (12–14). This protein shows substantial sequence homology to cyclin E and was hence named cyclin E2. Like cyclin E (now renamed cyclin E1), cyclin E2 associates with CDK2 and activates its kinase activity at the G1/S boundary (12–14). Despite these similarities, it is unknown whether these two cyclins perform redundant or distinct roles in controlling cell proliferation of various cellular compartments. To start elucidating in vivo functions of cyclins E1 and E2 in mouse development, we analyzed the expression of these two cyclins during normal mouse embryogenesis, in cells and tissues derived from various mouse mutants, and in mouse tumors driven by distinct oncogenic pathways. We hypothesized that, like D-type cyclins, cyclins E1 and E2 are expressed in a tissue-specific fashion and control S-phase entry in distinct cellular compartments.
Materials and Methods
Mice.
Mouse mammary tumor virus (MMTV)-v-Ha-Ras and MMTV-c-Myc mice were purchased from Charles River Breeding Laboratories. pRb−/−/p107−/−, and p130−/− mice were described previously (15, 16).
In Situ Hybridization.
Embryos were collected at days 9.5–13.5 post coitum (E9.5–E13.5) and processed as described (2). Sections were hybridized with α-[35S]thio-UTP-labeled riboprobes. Specimens were photographed by double exposure with the use of dark-field illumination with a red filter and Hoechst epifluorescence optics.
BrdUrd Staining.
Pregnant mothers were injected i.p. with BrdUrd (Sigma) at 100 μg/g of body weight. Embryos were collected after 2 h and fixed in 4% paraformaldehyde. Paraffin sections were stained with anti-BrdUrd antibody (Becton Dickinson) followed by detection with a Vectastain ABC kit (Vector Laboratories).
Northern Blot Analyses.
Twenty micrograms of total RNA (5 μg in the case of human breast tumor samples) were resolved with the use of 1% Mops-formaldehyde agarose gels, transferred to MagnaGraph membrane (Osmonics), and probed with 32P-labeled anti-cyclin E1 or anti-cyclin E2 riboprobes or with murine glyceraldehyde-3-phosphate dehydrogenase cDNA.
Analyses of Fibroblasts.
Fibroblasts were isolated from day 13.5 post coitum mouse embryos and serum starved in DMEM with 0.1% serum for 62–65 h before being restimulated by the addition of DMEM with 10% serum for the indicated times (16). Progression through the cell cycle was monitored by labeling cells with [3H]thymidine (16).
Human Tumors.
Samples of human mammary carcinomas were collected immediately after surgery by the Harvard SPORE tumor bank or by the Department of Pathology, Brigham and Women's Hospital. Tissue was snap frozen in Tissue-Tek optimal cutting temperature compound (Sakura). The sample included 49 ductal carcinomas, 9 lobular carcinomas, 8 mixed tumors, 7 metastatic adenocarcinomas, 2 other types, and 12 unknown. Fifty-eight samples were estrogen receptor (ER) positive, 20 were ER negative, and 9 were of unknown ER status.
Results
Expression of Cyclins E1 and E2 During Normal Mouse Embryogenesis.
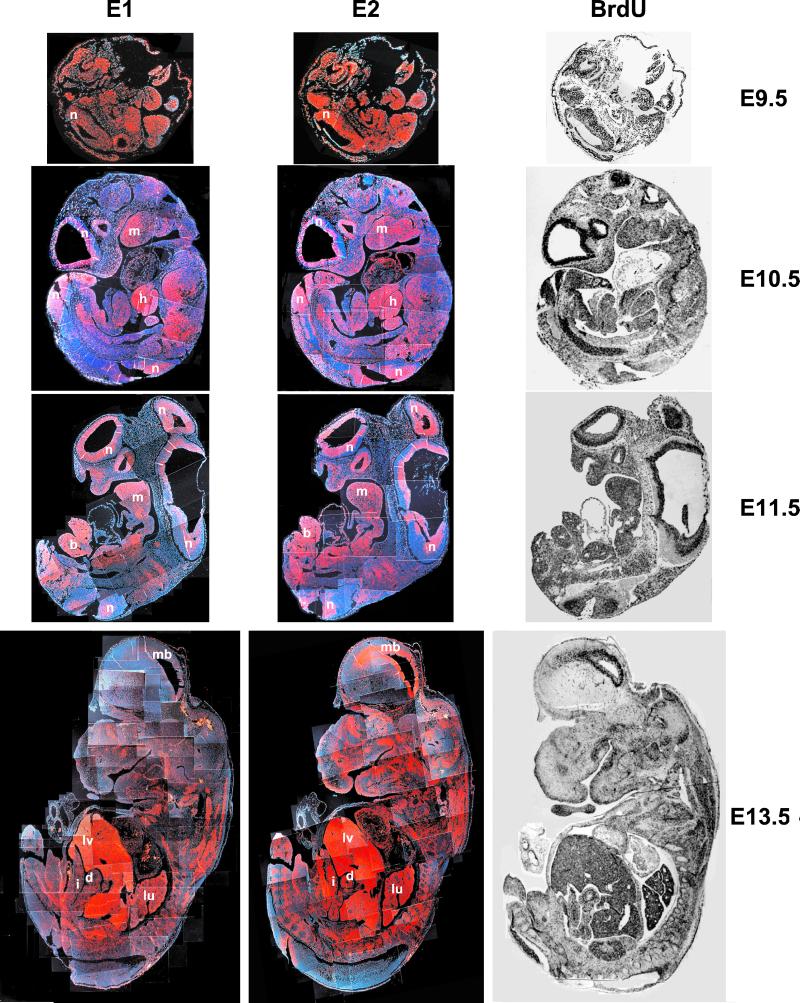
Contrary to our expectations, we found that the expression patterns of cyclin E1 and E2 mRNA during mouse development were very similar. Thus, at day E9.5 of gestation, both cyclins were expressed throughout the developing embryo, with high levels of transcripts in the neural tube. The ubiquitous expression of E-cyclins was consistent with the widespread staining of embryos with BrdUrd, indicating widespread proliferation at this stage (Fig. 1). At days E10.5 and E11.5 of embryo development, high levels of cyclins E1 and E2 were seen in the neuroepithelium, in the hepatic primordium, within the mandibular component of the first branchial arch, and in the hindlimb bud. Again, the expression patterns of cyclins E1 and E2 were very similar and closely mirrored the BrdUrd incorporation pattern (Fig. 1). This resemblance was also found at E13.5, when high levels of cyclins E1 and E2 were observed in the developing brain, liver, lungs, duodenum, and intestine (note that the lower level of cyclin E1 staining in the developing midbrain, as seen in Fig. 1, is not consistently observed). We concluded from these findings that, at least during our window of observation, cyclins E1 and E2 are coexpressed within the proliferating compartment. This coexpression is in stark contrast with the findings for D-type cyclins, which display a very distinct, tissue-specific pattern of expression during these stages of development (refs. 6 and 7 and M. Ciemerych and P.S., unpublished observations).
Figure 1.
Detection of cyclin E1 and E2 mRNA in developing mouse embryos, by in situ hybridization. Adjacent sections from embryos collected at indicated stages of development were hybridized with probes specific for cyclin E1 or cyclin E2. Red represents the hybridization signal. Blue represents the Hoechst 33258 staining of cell nuclei. The composite images of the whole embryo were obtained by assembling overlapping pictures taken at the same exposure times. (Right) Sections stained for BrdUrd. n, Neuroepithelium; m, mandibular component of first branchial arc; h, hepatic primordium; b, hindlimb bud; lv, liver; mb, midbrain, d, duodenum; lu, lung, i, intestine. (Magnifications: day E9.5, ×22; day E10.5, ×12; day E11.5, ×11; day E13.5, ×11.)
Cyclins E1 and E2 in Adult Tissues.
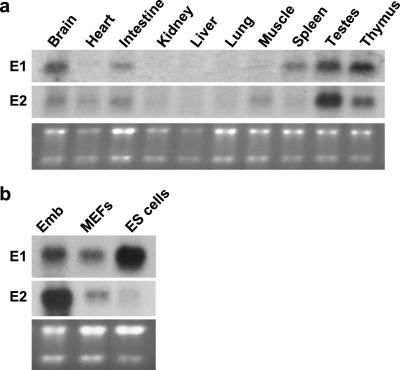
We next examined the expression of the two cyclins in adult tissues with the use of Northern blotting. Among the tissues analyzed, we found similar expression patterns of the two E-cyclins in brain, intestine, kidneys, liver, lung, testes, and thymus (Fig. 2a). However, we found that spleens expressed cyclin E1, but virtually no cyclin E2, whereas the converse was true for skeletal muscle and heart (Fig. 2a). Hence, unlike during embryo development, the expression patterns of the two cyclins in the tissues of adult mice are not identical.
Figure 2.
Expression of E-cyclin mRNAs in wild-type mice. (a) RNA was isolated from indicated organs of adult, 2- to 4-month-old, wild-type mice. Northern blots were probed with cyclin E1- and cyclin E2-specific probes. (b) RNA was isolated from wild-type embryos (Emb), mouse embryonal fibroblasts (MEFs), and embryonal stem cells (ES cells). Blots were probed with cyclin E1- and cyclin E2-specific probes. In both a and b, the bottom gels have been stained with ethidium bromide.
Cyclins E1 and E2 in Embryonal Stem (ES) Cells.
We also analyzed cyclin E1 and E2 levels in ES cells. As is shown in Fig. 2b, in vitro cultured ES cells expressed very high levels of cyclin E1, which clearly exceeded the level seen in the whole embryos. In marked contrast, cyclin E2 transcripts were barely detectable in ES cells. Hence, the ES cells seem to rely mainly on cyclin E1 for the G1/S phase progression.
The very high levels of cyclin E1 in ES cells may explain earlier observations that these cells are refractory to inhibitory effects of p16INK4a (17). Thus, in addition to extinguishing cyclin D-associated kinase, p16INK4a also needs to inhibit cyclin E–CDK2 complexes (by redistribution of p27Kip1) to arrest cell proliferation (1, 18). High levels of cyclin E1 in ES cells may exceed the threshold for p27Kip1 inhibition and may render these cells immune to p16INK4a. Indeed, ectopic overexpression of cyclin E1 was shown to bypass p16INK4a-induced cell cycle arrest in other systems (19, 20).
Regulation by the Retinoblastoma Family of Proteins.
Cyclin E1 is a downstream target of the E2F transcription factors (21, 22). Indeed, the cyclin E1 promoter contains several E2F-binding sites that confer cell cycle-specific regulation of the cyclin E1 transcription (23–25). The activity of E2Fs is negatively controlled by the retinoblastoma protein, pRB, and pRB-like proteins p107 and p130 (26). Consistent with this notion, embryos lacking pRB were shown to display elevated levels of cyclin E1 mRNA (15).
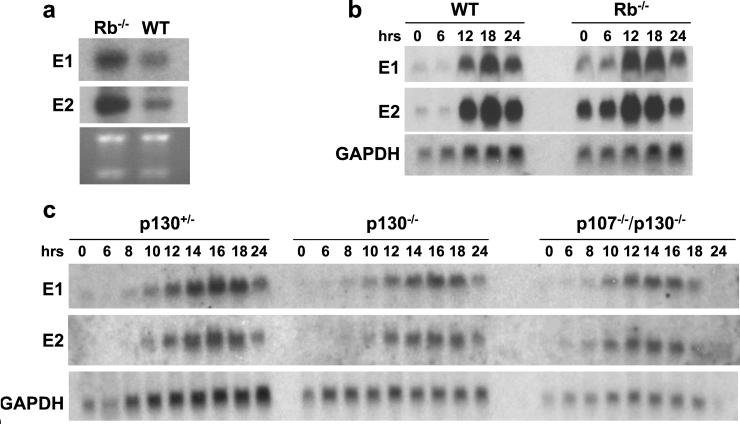
We asked whether the activity of cyclin E2 is also controlled in the same fashion. To this end, we determined the levels of cyclin E1 and E2 mRNA in brains derived from pRB−/− embryos. As reported (15), pRB−/− tissues contained increased amounts of cyclin E1 mRNA. Importantly, we found that the transcripts encoding cyclin E2 were equally up-regulated in pRB-deficient tissues (Fig. 3a).
Figure 3.
Expression of E-cyclin mRNAs in mutant mice. (a) RNA was isolated from brains derived from day E13.5 wild-type (WT) or pRB−/− embryos. Northern blots were hybridized with cyclin E1- and E2-specific probes. The bottom gel has been stained with ethidium bromide. (b) Wild-type or pRB−/− mouse embryonal fibroblasts (MEFs) were rendered quiescent by serum deprivation and then stimulated to enter the cell cycle by serum re-addition. Cells were harvested at indicated time points (in hours), and RNA was extracted and processed for Northern blot analyses. Blots were hybridized with probes specific for cyclin E1 or cyclin E2, or with a probe specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control. (c) Same type of experiment as in b, except that MEFs were prepared from p130+/− or p130-deficient or p130/p107-deficient embryos.
We extended these analyses by studying the expression of cyclins E1 and E2 during cell cycle progression of fibroblasts (mouse embryonal fibroblasts, MEFs) prepared from pRB−/− embryos. Wild-type and pRB−/− cells were rendered quiescent by serum starvation and stimulated to enter the cell cycle by the re-addition of serum. Cells were harvested at different time points, and RNA was isolated and processed for Northern blotting.
Hybridization of Northern blots with cyclin E1-specific probes revealed derepression of cyclin E1 mRNA during the G0 and G1 phases of the cell cycle, confirming the published reports (16, 27). Importantly, we also noted a strong derepression of cyclin E2 mRNA during the same cell cycle phases (Fig. 3b). We concluded that the expression of cyclins E1 and E2 is controlled in a pRB-dependent fashion, likely through the E2F transcription factors.
pRB and the pRB-related proteins p107 and p130 are known to regulate distinct members of the E2F family (26). Consistent with this notion are the observations that pRB and p107/p130 proteins control the activity of different E2F-responsive genes (16). We asked whether the expression of cyclin E2 was also influenced by these pRB-related proteins. To address this issue, MEFs were isolated from p130−/− and from double-knockout p107−/−/p130−/− embryos and analyzed for expression of cyclins E1 and E2 during cell cycle entry, as described above for pRB−/− MEFs.
We found that the loss of p130 or the combined loss of p107 and p130 proteins had no impact on the expression pattern of either cyclin (Fig. 3c). The slightly earlier induction of cyclins E1 and E2 during cell cycle progression of p107−/−/p130−/− MEFs can be accounted for by slightly accelerated entry of these cells into the S phase after serum stimulation (data not shown). We concluded that the expression of cyclins E1 and E2 in fibroblasts is governed by pRB-dependent but p107- and 130-independent molecular circuitry, likely through the pRB-dependent E2Fs.
Cyclins E1 and E2 in Ras- and Myc-Driven Breast Tumors.
To gain some insight into the regulation of cyclins E1 and E2 by distinct oncogenic pathways, we analyzed cyclin E1 and E2 mRNA levels in breast tumors arising in transgenic mice carrying MMTV-v-Ha-Ras or MMTV-c-Myc oncogenes. Female mice of these strains are highly prone to mammary carcinomas because of overexpression of Ras or Myc oncogenes in their mammary glands (28, 29). We chose to focus on these two particular oncogenes, because Ras is believed to signal in mammary epithelium through cyclin D1 (5, 30), whereas Myc signals through other targets, some of them possibly acting downstream of D-cyclins (20, 30, 31).
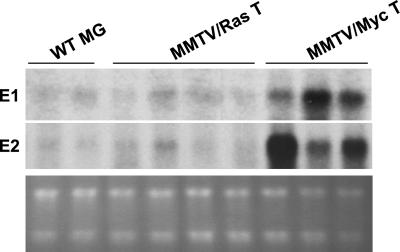
We found that all MMTV-v-Ha-Ras tumors analyzed (n = 8) expressed very low levels of cyclins E1 and E2, which were comparable to the levels seen in wild-type, nontransgenic mammary glands (Fig. 4). In marked contrast, a significant proportion of MMTV-c-Myc-driven tumors contained increased levels of cyclin E1 (five of six tumors) and E2 transcripts (four of six) (Fig. 4). These findings suggest that the expression of both cyclin E genes may be controlled, directly or indirectly, by the c-Myc-dependent pathway. They also strengthen our conclusion that the expression of the two cyclin genes is governed by similar molecular circuitry.
Figure 4.
Expression of E-cyclins in mouse breast tumors. RNA was isolated from mammary glands of nontransgenic, wild-type mice (WT MG), from breast tumors arising in transgenic MMTV-v-Ha-Ras female mice (MMTV/Ras T), or from breast tumors arising in transgenic MMTV-c-Myc female mice (MMTV/Myc T). Northern blots of representative samples were hybridized with probes specific for cyclin E1 or E2. (Bottom) Ethidium bromide-stained gel.
Cyclins E1 and E2 in Human Breast Cancers.
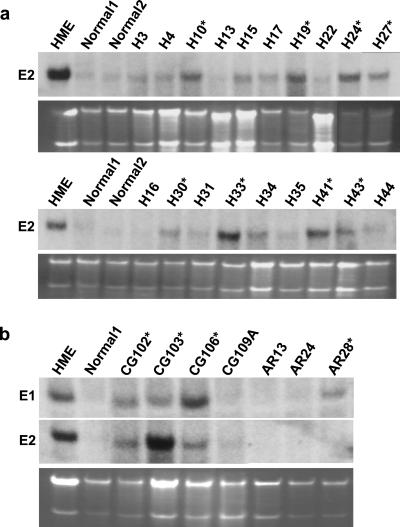
Last, we analyzed the expression of E-type cyclins in 87 samples of human breast cancers. Several independent samples of normal human mammary glands were used for comparison. Northern blot analyses revealed that 21 of 87 breast cancer samples (24.1%) contained elevated levels of cyclin E2 mRNA (Fig. 5a). We observed cyclin E2 overexpression in both ER-negative and ER-positive tumors (Table 1), with roughly similar frequency (14/58 in ER-positive and 5/20 in ER-negative; two cyclin E2-overexpressing tumor had unknown ER status). For technical reasons (the small amount of RNA used for Northern blots), we were unable to reprobe our blots with cyclin E1-specific probe. Instead, we prepared duplicate blots containing RNA extracted from 9 tumors overexpressing cyclin E2 and from 13 tumors with low cyclin E2 levels, and we probed these blots with cyclin E1-specific probe. We found that 5 of 9 cyclin E2-overexpressing tumors also contained elevated levels of cyclin E1 mRNA. In contrast, only 1 of 13 non-cyclin-E2-overexpressing mammary carcinomas displayed elevated levels of cyclin E1 (Fig. 5b). Hence, a significant fraction of cyclin-E2-overexpressing human mammary carcinomas may co-overexpress cyclin E1 mRNA. However, our sample is too small to draw statistically significant conclusions.
Figure 5.
Expression of E-cyclins in human breast cancers. (a) Examples of Northern blots of human breast cancer samples and normal mammary glands (normal) hybridized with cyclin E2-specific probe. The symbols of tumor samples are indicated above the respective lanes. Cyclin E2-overexpressing samples are marked with stars. RNA isolated from an immortalized human mammary epithelial cell line (HME) served as a positive control. (b) Selected tumor samples were rerun, and Northern blots were probed with cyclin E1- and E2-specific probes. RNA isolated from normal mammary glands (normal) and from the HME cell line was also included as in a. Cyclin E1-overexpressing samples are marked with stars. The bottom gels have been stained with ethidium bromide.
Table 1.
Histopathologic appearance of human breast cancers overexpressing cyclin E2
| Sample | Tumor type | ER status | E1 overexpression |
|---|---|---|---|
| AR3 | Ductal ca. | + | Yes |
| AR10 | Ductal ca. | + | No |
| AR17 | Ductal ca. | + | NA |
| AR18 | Ductal ca. | + | NA |
| AR27 | Ductal ca. | ± | No |
| AR30 | Mixed | ± | No |
| AR31 | Ductal ca. | + | NA |
| AR33 | Ductal ca. | − | NA |
| H10 | Metastatic adenoca. | − | NA |
| H17 | Ductal ca. | + | NA |
| H19 | Metastatic adenoca. | + | NA |
| H24 | Ductal ca. | + | NA |
| H27 | Ductal ca. | + | NA |
| H30 | Ductal ca. | ± | NA |
| H33 | Ductal ca. | + | NA |
| H41 | Ductal ca. | + | Yes |
| H43 | Metastatic adenoca. | − | NA |
| CG102 | Ductal ca. | − | Yes |
| CG103 | Ductal ca. | − | Yes |
| CG106 | Other | Unknown | Yes |
| CG107 | Ductal ca. | Unknown | No |
All samples overexpressing cyclin E2 are listed along with tumor type (ca., carcinoma) and ER status: +, ER-positive; ±, weakly positive; −, negative. Cyclin E1 overexpression status, if known, is also indicated: NA, not analyzed.
Discussion
The recently cloned cyclin E2 shares several features with the protein previously designated cyclin E and now renamed cyclin E1 (12–14). Because of these close similarities we speculated that the two E-type cyclins serve to control the S-phase entry of different cellular compartments, which was suggested by the tissue-specific expression of the D-type cyclins (2–4).
Contrary to our expectations, we found that the expression patterns of the two E-type cyclins were nearly identical during embryo development and similar in adult tissues. Moreover, we found that the expression of the two E-cyclins is negatively controlled by pRB, likely through pRB-dependent E2Fs, but is not influenced by genetic ablation of p107 and p130 proteins. Last, we observed up-regulation of the two cyclins in Myc- but not in Ras-driven mammary epithelial tumors. Collectively, these findings suggest that the expression of cyclin E1 and that of cyclin E2 are controlled in a similar fashion.
Why do the proliferating compartments coexpress the two E-cyclins? We note here that the degree of amino acid sequence similarity between the two E-cyclins is lower than that among D-type cyclins. For instance, human cyclins D1 and D2 display 60% overall amino acid identity (32), whereas the identity between cyclins E1 and E2 is 47% (12–14). We speculate that the two E-cyclins may control distinct, possibly partially overlapping, steps in G1/S-phase progression. Alternatively, the coexpression of two E-cyclins may represent a backup mechanism to ensure normal S-phase entry. Targeted deletion of the cyclin E1 and E2 genes may help to clarify this assumption.
E-Cyclins and Breast Cancer.
Our analyses of breast tumors arising in transgenic mice revealed that cyclins E1 and E2 are often co-overexpressed in tumors driven by the Myc oncogene. We note here that cyclin E1 was reported to represent a direct transcriptional target of c-Myc, but this notion remains controversial (33). Our observation that cyclin E2 mRNA is overexpressed in c-Myc-driven mouse breast tumors raises the possibility that cyclin E2 may be also regulated by c-Myc.
Cyclin E1 was reported to be overexpressed in a significant fraction of human mammary carcinomas. The overexpression of cyclin E1 correlates with increasing grade and stage of the tumor (34, 35). The work presented here revealed that approximately a quarter of human mammary carcinomas overexpress cyclin E2 mRNA. Moreover, our limited analyses suggest that both E-type cyclins may be co-overexpessed in the substantial proportion of cyclin E2-overexpressing mammary carcinomas. Anti-cyclin E2 antibodies, once they are available, will make it possible to precisely address the frequency of cyclin E2 abnormalities in human mammary carcinomas.
Acknowledgments
We thank B. Alpert, M. Ciemerych, L. Harris, J. D. Iglehart, K. Polyak, J. Rowinski, and E. Sicinska for help and advice. This work was supported by grants from the National Institutes of Health (CA83688-01) and by the Department of Defense Breast Cancer Idea Award (DAMD17–99–1–9164) to P.S. Y.G. and A.R. are partly supported by the National Cancer Institute Specialized Program of Research Excellence in Breast Cancer at the Dana–Farber Cancer Institute/Harvard Medical School. F.D. is a Fellow of the Leukemia and Lymphoma Society. K.Y.T. was supported by the Medical Scientist Training Program.
Abbreviations
- CDK
cyclin-dependent kinase
- pRB
retinoblastoma protein
- MMTV
mouse mammary tumor virus
- ER
estrogen receptor
- day En
embryonic day n
- ES cells
embryonal stem cells
- MEFs
mouse embryonal fibroblasts
References
- 1.Sherr C J, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 2.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 3.Sicinski P, Donaher J L, Geng Y, Parker S B, Gardner H, Park M Y, Robker R L, Richards J S, McGinnis L K, Biggers J D, et al. Nature (London) 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 4.Tam S W, Theodoras A M, Shay J W, Draetta G F, Pagano M. Oncogene. 1994;9:2663–2674. [PubMed] [Google Scholar]
- 5.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 6.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 7.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 8.Ewen M E. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma T, Van Tine B A, Wei Y, Garrett M D, Nelson D, Adams P D, Wang J, Qin J, Chow L T, Harper J W. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudas J M, Payton M, Thukral S, Chen E, Bass M, Robinson M O, Coats S. Mol Cell Biol. 1999;19:612–622. doi: 10.1128/mcb.19.1.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauper N, Beck A R, Cariou S, Richman L, Hofmann K, Reith W, Slingerland J M, Amati B. Oncogene. 1998;17:2637–2643. doi: 10.1038/sj.onc.1202477. [DOI] [PubMed] [Google Scholar]
- 14.Zariwala M, Liu J, Xiong Y. Oncogene. 1998;17:2787–2798. doi: 10.1038/sj.onc.1202505. [DOI] [PubMed] [Google Scholar]
- 15.Macleod K F, Hu Y, Jacks T. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 16.Hurford R K, Jr, Cobrinik D, Lee M H, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 17.Savatier P, Lapillonne H, van Grunsven L A, Rudkin B B, Samarut J. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- 18.Jiang H, Chou H S, Zhu L. Mol Cell Biol. 1998;18:5284–5290. doi: 10.1128/mcb.18.9.5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 20.Alevizopoulos K, Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duronio R J, O'Farrell P H. Genes Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- 22.Duronio R J, Brook A, Dyson N, O'Farrell P H. Genes Dev. 1996;10:2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- 23.Ohtani K, De Gregori J, Nevins J R. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Durr P. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 26.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 27.Herrera R E, Sah V P, Williams B O, Makela T P, Weinberg R A, Jacks T. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 29.Stewart T A, Pattengale P K, Leder P. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 30.Yu Q, Geng Y, Sicinski P. Nature (London) 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 31.Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Mol Cell Biol. 2000;20:3497–3509. doi: 10.1128/mcb.20.10.3497-3509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong Y, Menninger J, Beach D, Ward D C. Genomics. 1992;13:575–584. doi: 10.1016/0888-7543(92)90127-e. [DOI] [PubMed] [Google Scholar]
- 33.Amati B, Alevizopoulos K, Vlach J. Front Biosci. 1998;15:D250–D268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 34.Keyomarsi K, O'Leary N, Molnar G, Lees E, Fingert H J, Pardee A B. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 35.Nielsen N H, Arnerlov C, Emdin S O, Landberg G. Br J Cancer. 1996;74:874–880. doi: 10.1038/bjc.1996.451. [DOI] [PMC free article] [PubMed] [Google Scholar]