 A DNA/HDAC dual-targeting inhibitor containing nitrogen mustard and 2-aminobenzamide moieties was designed and evaluated as a promising candidate for cancer therapy.
A DNA/HDAC dual-targeting inhibitor containing nitrogen mustard and 2-aminobenzamide moieties was designed and evaluated as a promising candidate for cancer therapy.
Abstract
Histone deacetylases (HDACs) play a key role not only in gene expression but also in DNA repair. Herein, we report the rational design and characterization of a compound named chlordinaline containing nitrogen mustard and 2-aminobenzamide moieties as a DNA/HDAC dual-targeting inhibitor. Chlordinaline exhibited moderate total HDAC inhibitory activity. The HDAC isoform selectivity assay indicated that chlordinaline mostly inhibits HDAC3. Chlordinaline exhibited both DNA and HDAC inhibitory activities and showed potent antiproliferative activity against all the six test cancer cell lines with IC50 values of as low as 3.1–14.2 μM, which is significantly more potent than reference drugs chlorambucil and tacedinaline. Chlordinaline could induce the apoptosis and G2/M phase cell cycle arrest of A375 cancer cells. This study demonstrates that combining nitrogen mustard and 2-aminobenzamide moieties into one molecule is an effective method to obtain DNA/HDAC dual-targeting inhibitors as potent antitumor agents. Chlordinaline as the first example of such DNA/HDAC dual-targeting inhibitors could be a promising candidate for cancer therapy and could also be a lead compound for further optimization.
Introduction
Traditional genotoxic drugs targeting DNA are effective in killing cancer cells.1,2 However, the DNA damage caused by genotoxic drugs can be mitigated by cellular DNA repair machinery, thus enabling some cancer cells to survive and ultimately cause treatment failure.3–5 In the nucleus, DNA is noncovalently associated with histones to form the nucleosomes which make up chromatin subunits. Histone deacetylases (HDACs) are a class of enzymes that catalyze the removal of acetyl groups from histones, resulting in chromatin condensation.6–10 Modifications in chromatin conformation due to histone acetylation could expose DNA to DNA-damaging agents such as ultraviolet rays, ionising radiation, and genotoxic drugs, eventually leading to double strand breaks (DSB) in DNA.11 In addition to sensitizing DNA to exogenous genotoxic drugs, HDAC inhibitors could also down regulate the DNA damage repair machinery.12,13
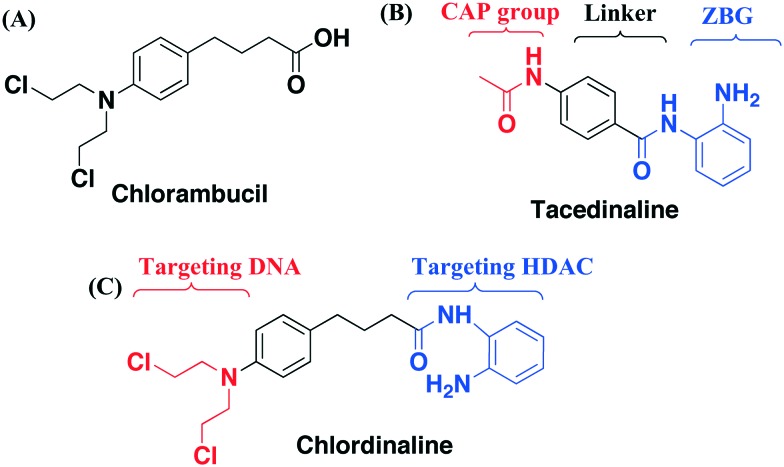
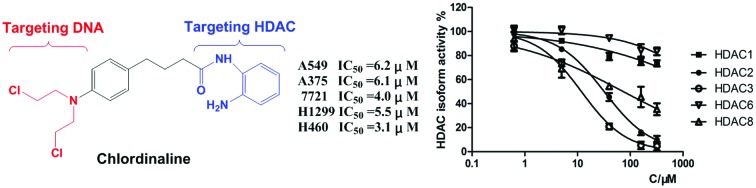
The emerging roles of HDACs in DNA repair provide new opportunities for improving traditional genotoxic drugs.14,15 Professor C. J. Marmion et al. designed and prepared a novel anti-cancer bifunctional platinum drug candidate with dual DNA binding and HDAC inhibitory activity.16–18 Professor J. Kasparkova et al. developed a photoactivatable platinum complex targeting DNA and HDAC.19 Herein, we report our effort in rational design and characterization of a compound named chlordinaline as a dual DNA/HDAC inhibitor by combining pharmacophores of two reference drugs, chlorambucil and tacedinaline. Nitrogen mustards represent an important branch of genotoxic drugs and one of them is in worldwide clinical use, namely, chlorambucil (Fig. 1A).20,21 HDAC inhibitors are characterized by a widely accepted pharmacophore model comprising a zinc binding group (ZBG) chelating with zinc at the bottom of the HDAC active site, a CAP group, recognizing and interacting with residues on the rim of the active site of HDACs, and a linker connecting the ZBG and the CAP groups (Fig. 1B).22–29 Tacedinaline is the first 2-aminobenzamide based HDAC inhibitor, which is under clinical trial II for the treatment of cancer.30 Nitrogen mustard is able to kill cancer cells by causing DNA damage. A HDAC inhibitor is able to down regulate the DNA damage repair machinery. Thus, we combined pharmacophores of nitrogen mustard drugs and HDAC inhibitors to obtain dual-targeting potent antitumor agents. Chlordinaline was designed to achieve such a dual functionality to target both DNA and HDACs (Fig. 1C).
Fig. 1. (A) Structure of chlorambucil; (B) structure of tacedinaline; (C) design of chlordinaline as a DNA/HDAC dual-targeting inhibitor.
Results and discussion
Chemistry
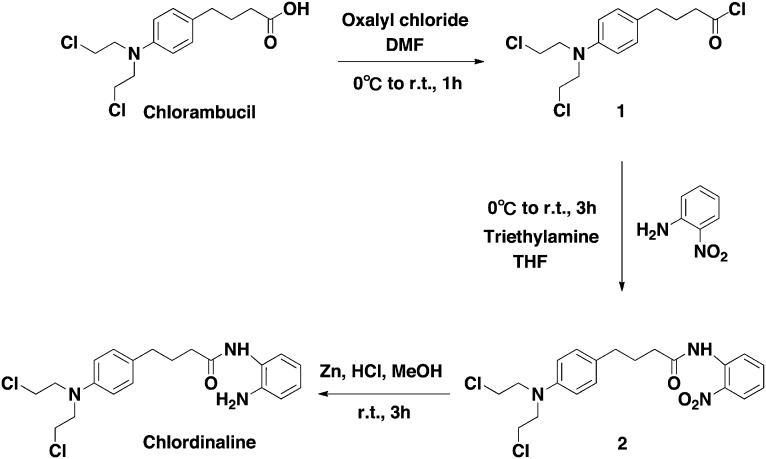
The synthetic route to obtaining chlordinaline is shown in Scheme 1. First, commercially available chlorambucil was treated with oxalyl chloride to provide intermediate 1. Then, intermediate 1 was directly coupled with 2-nitroaniline to obtain intermediate 2. Finally, intermediate 2 was reduced with zinc and hydrochloric acid to obtain chlordinaline.
Scheme 1. Synthetic route of chlordinaline.
HDAC inhibitory activity
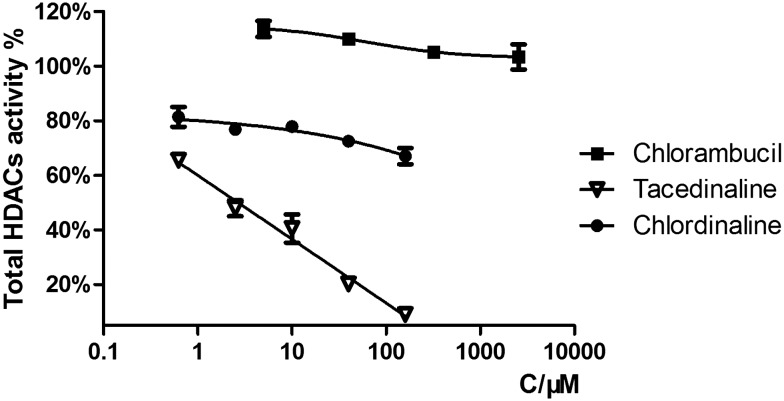
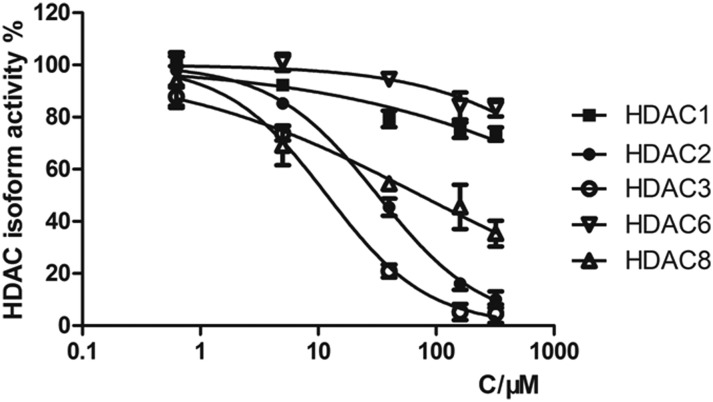
The HDAC family consists of 18 isoforms, which are divided into 4 classes or subfamilies according to their sequence homology and catalytic mechanism: class I (HDACs 1–3 and 8), class II (HDACs 4–7, 9 and 10), class IV (HDAC 11) and class III (sirtuins 1–7).31,32 It has been shown that the dysregulation of class I and II HDACs, especially class I isozymes, has been associated with the process of tumor cell proliferation and DNA repair.33–36 HDAC1, HDAC2, HDAC3, HDAC8 and HDAC6 are the most studied HDAC isoforms in tumor-related HDAC enzymes.37–40 Therefore, we selected HDAC1, HDAC2, HDAC3, HDAC8 and HDAC6 for selectivity investigation. We first tested the total HDAC inhibitory activity of chlordinaline using a HDAC Assay kit (BML-AK530, Enzo® Life Sciences) to investigate whether chlordinaline exhibited HDAC inhibitory activity or not. As expected (pharmacophoric hypothesis), chlordinaline displayed a moderate ability to inhibit HDACs (Fig. 2). Chlordinaline was then assessed for inhibitory activity against HDAC isoforms 1–3 and 8 (class I) and 6 (class II). As shown in Fig. 3, chlordinaline displayed the optimal inhibitory activity against HDAC3 (IC50 = 9.52 μM) and was inactive against HDAC1 and HDAC6.
Fig. 2. Total HDAC inhibitory activity of chlorambucil, tacedinaline and chlordinaline.
Fig. 3. HDAC isoform inhibitory activity of chlordinaline (IC50 > 160 μM against HDAC1, IC50 = 32.9 μM against HDAC2, IC50 = 9.52 μM against HDAC3, IC50 > 160 μM against HDAC6, IC50 = 75.2 μM against HDAC8).
Molecular modelling studies
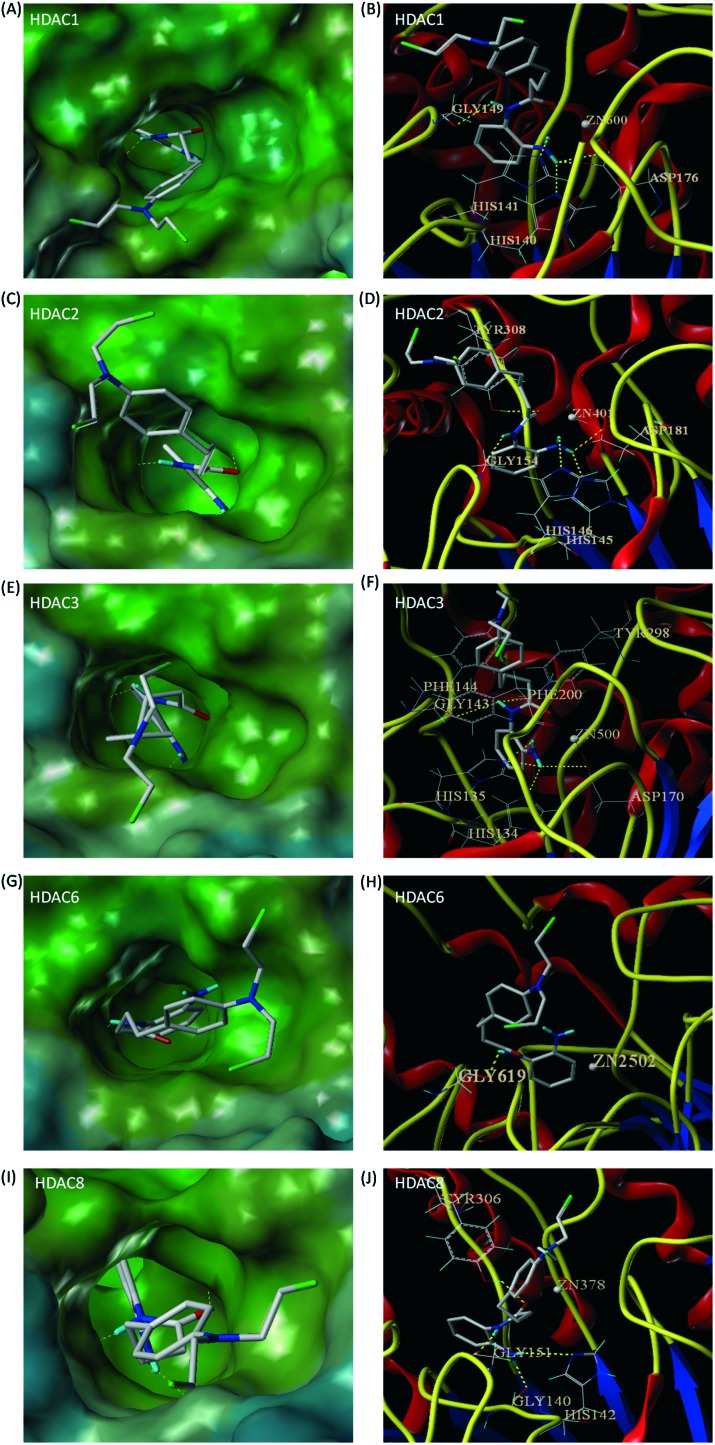
To better understand the HDAC3 isoform selective inhibitory activity of chlordinaline, we docked chlordinaline to the active sites of HDAC1 (PDB code: 4BKX),41 HDAC2 (PDB code: ; 4LXZ),42 HDAC3 (PDB code: ; 4A69),43 HDAC8 (PDB code: ; 1T69)44 and HDAC6 (PDB code: ; 5EDU)45 using Surflex-Dock. As shown in Fig. 4(E and F), chlordinaline could form six hydrogen bonds with GGLY143, TYR298, HIS135, HIS134, and ASP170 in the active site of HDAC3. In addition to these hydrogen bonding interactions, the phenyl ring of chlordinaline formed two π–π stacking interactions with PHE144 and PHE200 in the active site of HDAC3. The binding affinity of chlordinaline for HDAC3 was –9.9 kcal mol–1. As shown in Fig. 4(C and D), chlordinaline could also form five hydrogen bonds with TYR308, GLY154, ASP181, HIS145 and HIS146 in the active site of HDAC2. In addition to these hydrogen bonding interactions, the phenyl ring of chlordinaline formed one π–π stacking interaction with TYR308 in the active site of HDAC2. The binding affinity of chlordinaline for HDAC2 was –9.5 kcal mol–1. As shown in Fig. 4(A and B), chlordinaline could also form five hydrogen bonds with HIS140, HIS141, ASP176 and GLY149 in the active site of HDAC1, however, the phenyl ring of chlordinaline could not form a π–π stacking interaction with HDAC1. The binding affinity of chlordinaline for HDAC1 was –8.5 kcal mol–1. As shown in Fig. 4(I and J), chlordinaline could only form four hydrogen bonds with TYR306, GLY151, GLY140 and HIS142 in the active site of HDAC8, however, the phenyl ring of chlordinaline could not form a π–π stacking interaction with HDAC8. The binding affinity of chlordinaline for HDAC1 was –7.7 kcal mol–1. As shown in Fig. 4(G and H), chlordinaline could only form one hydrogen bond with GLY619 in the active site of HDAC6 and the binding affinity of chlordinaline for HDAC6 was only –3.6 kcal mol–1. Molecular docking results could well support the initial pharmacophoric hypothesis and rationalize the moderate potency and selectivity of chlordinaline against HDAC3.
Fig. 4. Predicted binding modes of chlordinaline on HDAC1 (PDB code: 4BKX), HDAC2 (PDB code: ; 4LXZ), HDAC3 (PDB code: ; 4A69), HDAC6 (PDB code: ; 5EDU) and HDAC8 (PDB code: ; 1T69). Interactions between the protein and the ligand are shown as yellow dotted lines. (A) Molecular surface of the HDAC1 binding pocket. (B) Chlordinaline interacted with the active site of HDAC1. (C) Molecular surface of the HDAC2 binding pocket. (D) Chlordinaline interacted with the active site of HDAC2. (E) Molecular surface of the HDAC3 binding pocket. (F) Chlordinaline interacted with the active site of HDAC3. (G) Molecular surface of the HDAC6 binding pocket. (H) Chlordinaline interacted with the active site of HDAC6. (I) Molecular surface of the HDAC8 binding pocket. (J) Chlordinaline interacted with the active site of HDAC8.
DNA-targeting activity
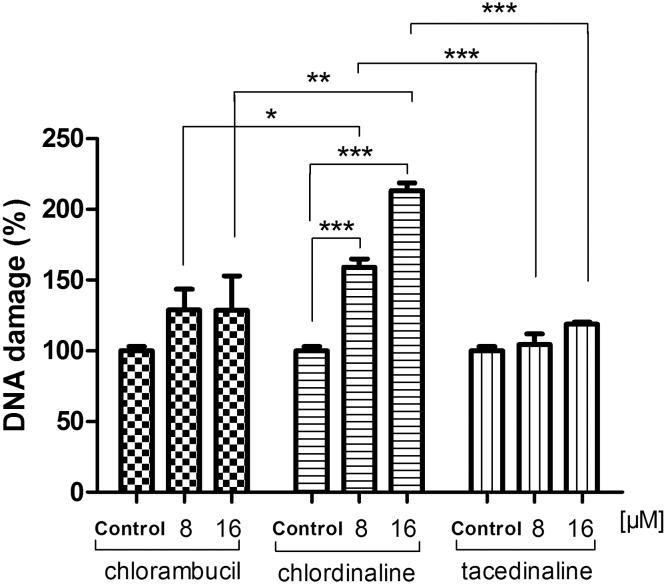
To examine whether chlordinaline causes DNA damage in cancer cells, we performed DNA damage determinations of chlordinaline against A375 cancer cells using a DNA damage assay kit (Epigentek, NY, USA). The assay is able to determine the phosphorylation of histone H2AX (γ-H2AX), which is a biomarker of DNA damage. As displayed in Fig. 5, after 48 h exposure, chlordinaline significantly induced more DNA damage (159.0 ± 5.9% at 8 μM, 213.3 ± 5.4% at 16 μM) compared with chlorambucil (128.9 ± 14.8% at 8 μM, 128.6 ± 24.3% at 16 μM) and tacedinaline (104.5 ± 7.5% at 8 μM, 118.8 ± 1.6% at 16 μM). These results indicate that chlordinaline exhibit both DNA and HDAC inhibitory activities as expected (pharmacophoric hypothesis).
Fig. 5. Chlorambucil, chlordinaline and tacedinaline induce DNA damage in A375 cells. A375 cells were treated with chlordinaline, chlorambucil and tacedinaline at 8 μM or 16 μM for 48 hours. Data were expressed as the percent of the control (no drug treatment), as mean ± SD (n = 3). P-Values were determined using a t-test (* indicates p < 0.05, ** indicates P < 0.01, *** indicates P < 0.001).
Antiproliferative activity
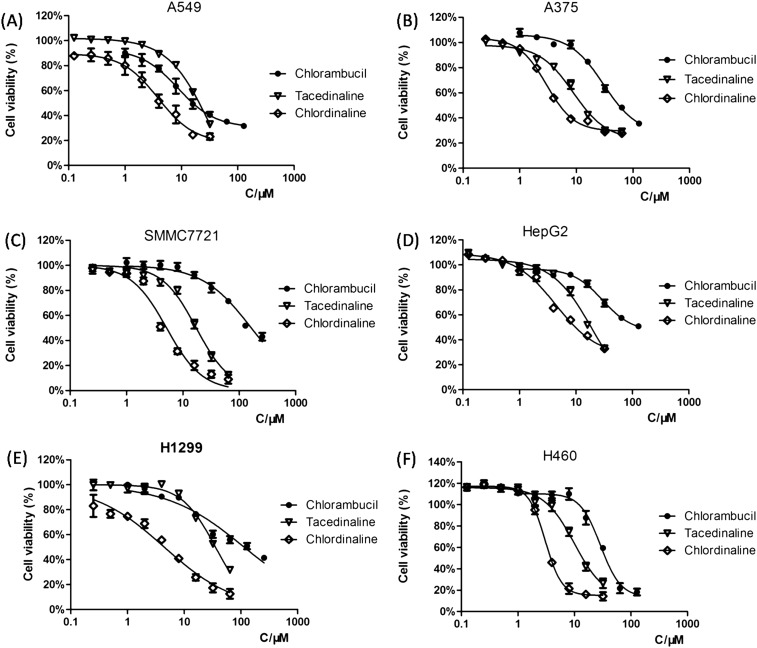
To investigate whether the DNA/HDAC dual-targeting inhibitory activity of chlordinaline is accompanied by enhanced anticancer activities or not, we investigated the growth-inhibitory activity of chlordinaline towards six cancer cell lines. As shown in Table 1, chlordinaline showed significantly enhanced anticancer potency with IC50 values of as low as 3.1–14.2 μM against six human cancer cell lines, more than 3.6–40.8-fold lower than those of chlorambucil and 1.3–6.1-fold lower than those of tacedinaline. The dose-dependent antiproliferative activities of chlordinaline against cancer cell lines are displayed in Fig. 6, demonstrating a clear dose-dependent inhibition of the anti-proliferative activity of chlordinaline.
Table 1. Antiproliferative activity data (IC50, μM) of chlordinaline against six cancer cell lines.
| Compound | IC50
a
(μM) |
|||||
| A549 | A375 | SMMC7721 | HepG2 | H1299 | H460 | |
| Chlorambucil | 22.2 ± 3.7 | 69.2 ± 9.9 | 163.0 ± 20.7 | 111.8 ± 17.0 | 116.3 ± 24.8 | 28.9 ± 3.5 |
| Tacedinaline | 20.1 ± 0.8 | 11.3 ± 1.1 | 16.9 ± 1.3 | 18.5 ± 1.8 | 33.8 ± 3.9 | 9.7 ± 1.1 |
| Chlordinaline | 6.2 ± 1.3 | 6.1 ± 0.9 | 4.0 ± 0.6 | 14.2 ± 4.0 | 5.5 ± 1.3 | 3.1 ± 0.2 |
aThe values represent the means of three experiments.
Fig. 6. Dose-dependent antiproliferative activities of chlordinaline against six cancer cell lines. The values represent the means of three experiments. (A) Treatment in A549 cells. (B) Treatment in A375 cells. (C) Treatment in SMMC7721 cells. (D) Treatment in HepG2 cells. (E) Treatment in H1299 cells. (F) Treatment in H460 cells.
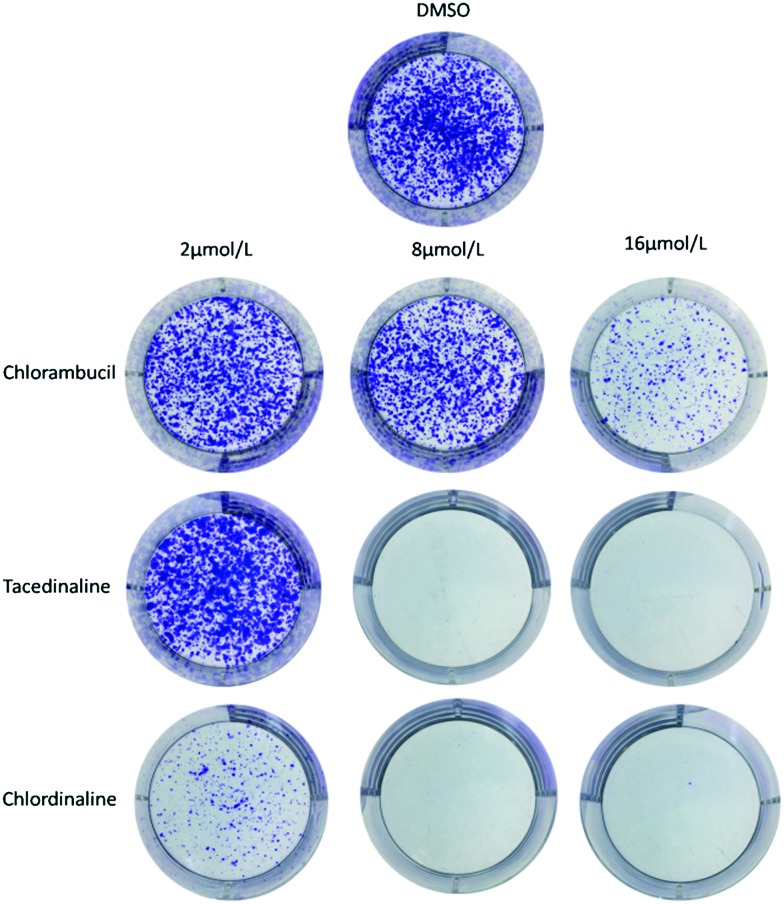
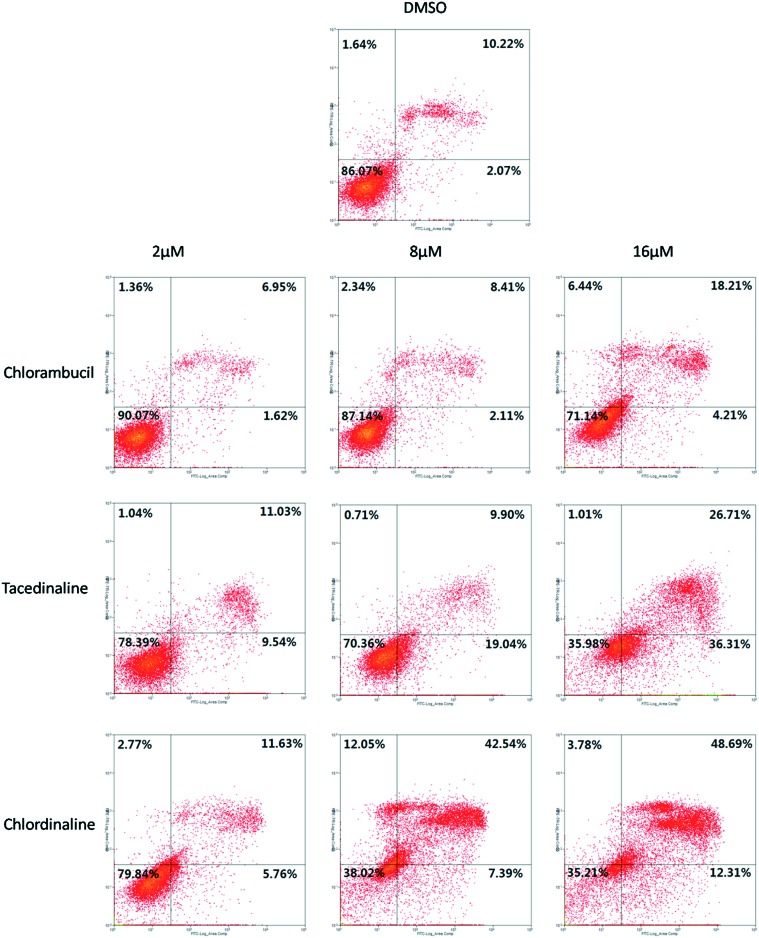
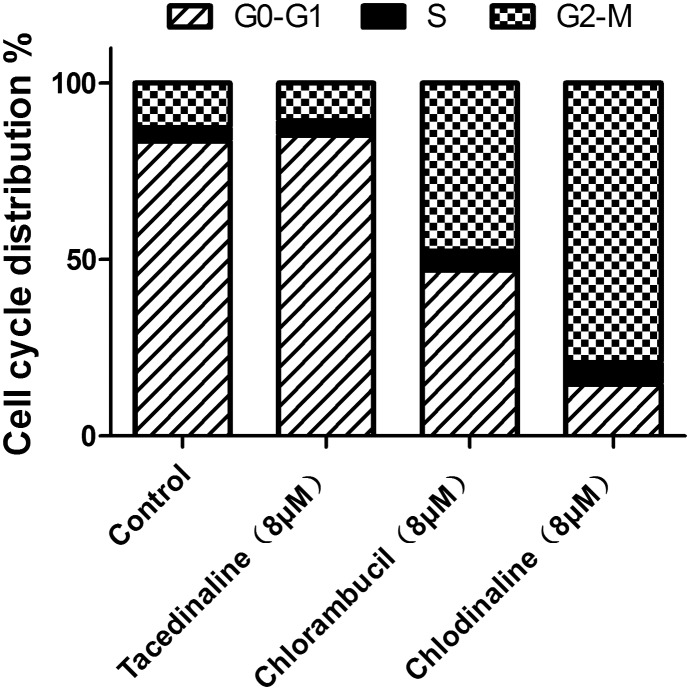
Since chlordinaline exhibited significantly improved antiproliferative activity and DNA damage activity, we further evaluated the effect of chlordinaline on the colony formation of A375 cancer cells with chlorambucil and tacedinaline as positive controls. The results are summarized in Fig. 7. Chlordinaline inhibited the colony formation of A375 cancer cells in a dose-dependent manner and is significantly more effective than chlorambucil and tacedinaline. Next, we also examined whether chlordinaline induces apoptosis using flow cytometry. As shown in Fig. 8, chlordinaline significantly induced the apoptosis of A375 cancer cells in a dose-dependent manner. Chlordinaline induced 20.16%, 61.98% and 64.78% apoptosis of A375 cancer cells at 2 μM, 8 μM and 16 μM, respectively. Meanwhile, chlorambucil only induced 9.93%, 12.85% and 28.86% apoptotic cells and tacedinaline only induced 21.61%, 29.64% and 64.02% apoptotic cells at the same concentration. Furthermore, cell cycle analysis showed that chlordinaline remarkably induced the accumulation of A375 cells at the G2/M phase (79.2% at 8 μM, Fig. 9), which demonstrates that it is obviously more potent than chlorambucil (47.6% at 8 μM, Fig. 9) and tacedinaline (10.8% at 8 μM, Fig. 9).
Fig. 7. Chlordinaline, chlorambucil and tacedinaline inhibit A375 cancer cell colony formation.
Fig. 8. Induction of apoptosis by chlordinaline, chlorambucil and tacedinaline in different concentrations in A375 cancer cells. The values represent the means of three experiments with SD less than 10%.
Fig. 9. Effects of chlordinaline, chlorambucil and tacedinaline on A375 cell cycle progression. The values represent the means of three experiments with SD less than 10%.
Conclusions
In summary, we successfully designed a DNA/HDAC dual-targeting inhibitor, chlordinaline, by combining pharmacophores of two reference drugs, chlorambucil and tacedinaline, for the first time. Chlordinaline exhibited significantly enhanced antiproliferative activity against all six tested cancer cell lines. Notably, chlordinaline exhibited excellent selective inhibition against HDAC3. The excellent selectivity of chlordinaline against HDAC3 was well rationalized by molecular docking results. Chlordinaline also significantly increased the expression of the DNA damage biomarker γ-H2AX. The above results demonstrate that a HDAC inhibitor plays a key role in the cytotoxicity of nitrogen mustard. Consequently, our study has highlighted the potential clinical values of DNA/HDAC dual-targeting drugs. Chlordinaline exhibits a simple structure, potent antitumor activity and special HDAC isoform inhibitory activity, and could be a promising candidate for cancer therapy and could also be a lead compound for further optimization to develop more potent DNA/HDAC dual-targeting inhibitors. Further structure optimization of chlordinaline and more detailed antitumor mechanism research are underway in our lab.
Experimental
General information
Materials and instruments
Chlorambucil and tacedinaline were purchased from Dalian Meilun Biotech Co., Ltd. The Cell Cycle Detection Kit (KGA512) was purchased from KeyGen Biotech (Nanjing, China). Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). The HDAC Inhibition Assay Kit was purchased from Enzo® Life Sciences. Other reagents and solvents were purchased from Beijing Chemical Works, Beijing Inno-Chem Co. Ltd. and other commercial sources. They were used without further purification. All cancer cell lines were purchased from the Cell Resource Center, Peking Union Medicinal College, Beijing, China. Melting points (uncorrected) were obtained using an XT5, manufactured by Beijing Keyiecopti Instrument Factory. Mass spectra were obtained with a Waters Xevo G2 Qtof mass spectrometer. 1H NMR and 13C NMR spectra were determined with a Bruker AV-400 spectrometer using tetramethylsilane (TMS) as an internal standard in DMSO-d6 solutions. Chemical shifts were reported in parts per million (ppm). All reactions were monitored by thin-layer chromatography (TLC) on pre-coated plates with silica gel F254, purchased from Qingdao Haiyang Chemical Co. Ltd. HPLC analysis was performed using a Diamonsil C18 (5 μm, 250 mm × 4.6 mm) with the solvent system consisting of methanol (mobile phase A) and water containing 0.1% phosphoric acid. The purity of chlordinaline was established to be 98.6% pure using HPLC.
Synthesis of chlordinaline
Step-a: A 100 mL, one-necked, round-bottomed flask was sequentially charged with chlorambucil (0.30 g, 1 mmol), 30 mL of tetrahydrofuran (THF) and 0.1 mL of N,N-dimethylformamide (DMF). The mixture was stirred at room temperature for 5 min, and then 2 ml of oxalyl chloride was added to the reaction mixture slowly and the temperature was maintained at 0 °C during the addition. When the addition was completed, the mixture was warmed to room temperature and stirred for another 1 h. The reaction mixture was concentrated in vacuo to afford intermediate 1 as colorless oil.
Step-b: 2-Nitroaniline (1 mmol) and triethylamine (1.05 mmol) were dissolved in anhydrous THF (10 mL), and then intermediate 1 (1 mmol) dissolved in dry THF (5 mL) was added dropwise at 0 °C. The mixture was stirred for 1 h at 0 °C and was stirred for another 2 hour at room temperature. TLC analysis, using ether/petroleum ether (2/1) as the eluent, indicated the complete reaction. The reaction mixture was poured into 400 mL of ice water with stirring leading to the precipitation of a solid. Stirring was continued for an additional 30 min and the resulting solid was then filtered off with suction through a sintered glass filter funnel to afford intermediate 2.
Step-c: A suspension of intermediate 2 (0.7 mmol) and zinc powder (1.4 mmol) in methanol (10 mL) and water (2 mL) was stirred at room temperature for 10 min, and then hydrochloric acid (3.5 ml, 1 mol L–1) was added dropwise. The reaction mixture was stirred at room temperature for another 3 h, and then the reaction mixture was poured into water and extracted with EtOAc (100 mL × 3). The combined organic layer was dried over anhydrous MgSO4, concentrated under vacuum and purified by silica gel chromatography to afford chlordinaline as a white solid, N-(2-aminophenyl)-4-{4-[bis(2-chloroethyl)amino]phenyl}butanamide (chlordinaline). Total yield 43%; HPLC purity: 98.6% (tR = 36.4 min); mp 140.8–141.9 °C; 1H NMR (400 MHz, DMSO-d6): δ = 9.12 (s, 1H, –CON = 9.12 (s, 1H, –CON̲H̲–), 7.17 (d, H = 9.12 (s, 1H, –CON̲H̲–), 7.17 (d, –), 7.17 (d, J = 7.8 Hz, 1H, ArH), 7.07 (d, J = 8.2 Hz, 2H, ArH), 6.91 (t, J = 7.5 Hz, 1H, ArH), 6.74 (d, J = 8.0 Hz, 1H, ArH), 6.69 (d, J = 8.2 Hz, 2H, ArH), 6.56 (t, J = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)H = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)2 = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)), 3.71 (m, 8H, (Cl–C = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)H = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)2 = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)C = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)H = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)2 = 7.5 Hz, 1H, ArH), 5.04 (s, 2H, –ArN̲H̲2̲), 3.71 (m, 8H, (Cl–C̲H̲2̲C̲H̲2̲–)–)2N–), 2.53 (t, J = 7.9 Hz, 2H, –ArC = 7.9 Hz, 2H, –ArC̲H̲2̲CH2CH2CO–), 2.33 (t, H = 7.9 Hz, 2H, –ArC̲H̲2̲CH2CH2CO–), 2.33 (t, 2 = 7.9 Hz, 2H, –ArC̲H̲2̲CH2CH2CO–), 2.33 (t, CH2CH2CO–), 2.33 (t, J = 7.2 Hz, 2H, –ArCH2CH2C = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); H = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); 2 = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); CO–), 1.85 (m, 2H, –ArCH2C = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); H = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); 2 = 7.2 Hz, 2H, –ArCH2CH2C̲H̲2̲CO–), 1.85 (m, 2H, –ArCH2C̲H̲2̲CH2CO–); CH2CO–); 13C NMR (400 MHz, DMSO-d6): δ = 27.3, 33.6, 35.2, 25.6, 52.2, 111.9, 115.9, 116.2, 123.6, 125.3, 125.6, 129.3, 129.9, 141.7, 144.4, 170.9 ppm; HRMS (ESI) m/z calcd for C20H26Cl2N3O [M + H]+ 394.1447, found: 394.1456.
HDAC inhibition assay
The total HDAC activity was determined using a Fluor de Lys®-Green HDAC Assay kit (BML-AK530, Enzo® Life Sciences). HDAC 1–3, 8 and 6 activity was determined using a Fluor de Lys® HDAC1 Assay kit (BML-AK511, Enzo® Life Sciences), a Fluor de Lys®-Green HDAC2 Assay kit (BML-AK512, Enzo® Life Sciences), a Fluor de Lys® HDAC3/NCOR1 Assay kit (BML-AK531, Enzo® Life Sciences), a Fluor de Lys® HDAC8 Assay kit (BML-AK518, Enzo® Life Sciences), and a Fluor de Lys® HDAC6 Assay kit (BML-AK516, Enzo® Life Sciences). All assays were performed according to the manufacturer's instructions. Briefly, 15 μL of HDAC was mixed with 10 μL of tested compounds at various concentrations in the corresponding microplate wells. The diluted Fluor de Lys®-Green substrate and the microtiter plate were allowed to equilibrate to the assay temperature (37 °C) for 5 minutes. The HDAC reaction was initiated by the addition of 25 μL diluted substrate to each well and the mixtures were mixed thoroughly. The plate was incubated at 37°C for 1 h and then 50 μL Fluor de Lys® developer was added to each well to stop the HDAC reaction. The plate was incubated at 25 °C for another 10 min and fluorescence measurements were obtained using an EnSpire multimode plate reader (PerkinElmer, USA) with excitation at 360 nm and emission at 460 nm. The HDAC activity was calculated as the percentage of activity compared with the control group. The 50% inhibition concentration (IC50) values for the test compounds were calculated using a regression analysis of the dose/inhibition data.
DNA damage assay
DNA damage was assessed using an EpiQuik™ in situ DNA Damage Assay Kit (Epigentek, NY, USA), which is a whole cell-based assay for the detection of DNA damage by measuring phosphorylation of H2AX at Ser139 (γH2AX). A total of 7000 cells per well were seeded in 96-well micro plates. The following day, A375 cancer cells were treated with chlordinaline, chlorambucil and tacedinaline at 8 μM or 16 μM for 48 hours. The assay was carried out according to the manufacturer's instructions. The absorbance signal was normalized to the cell number in each sample, and the samples were calculated relative to the untreated control.
Antiproliferative assay
Cell lines A549, SMMC77212, H460 and H1299 were cultured in RPMI1640 (Corning, USA) containing 10% fetal bovine serum (FBS) (Thermo Fisher, USA) and 1% penicillin/streptomycin. A375 and HepG2 cell lines were cultured in DMEM (Corning, USA) containing 10% fetal bovine serum (FBS) (Thermo Fisher, USA) and 1% penicillin/streptomycin. All cell lines were incubated at 37 °C in a humidified atmosphere of 5% CO2. Cells in the logarithmic phase were seeded in 96-well culture plates at a density of 3000–4000 cells per well. After 12 h, the cells were treated with various concentrations of compounds or solvent control. After 72 h of incubation, 10 μL of CCK-8 reagent was added to each well and the cells were incubated for an additional 1 h. Absorbance was measured at 450 nm using an EnSpire multimode plate reader (PerkinElmer, USA). IC50 values were calculated using the percentage of growth versus untreated control.
Colony formation assay
A375 cells were seeded in 6-well plates at a density of 3000 cells per well. After 24 h, the cells were treated with DMSO, chlordinaline, chlorambucil or tacedinaline. After 7 days, colonies were fixed with 3.7% formaldehyde and stained with 0.1% crystal violet.
Cell apoptosis analysis
Cell apoptosis was determined using a Vybrant Apoptosis Assay kit (Invitrogen). Briefly, A375 cancer cells (8 × 104 per well) were incubated in 6-well plates for 12 h and then treated with DMSO, chlordinaline, chlorambucil or tacedinaline. After 72 h, the cells were harvested and washed three times with pre-chilled PBS. 100 μL of 1X annexin-building buffer, 5 μL of Alexa Fluor 488 annexin and 1 μL of propidium iodide (100 μg ml–1) were added. The cells were incubated for 15 min in the dark. After staining, 400 μL of 1X annexin-building buffer was added, and the mixture was mixed gently and kept on ice. The samples were analyzed with a MoFlo XDP flow cytometer (Beckman Coulter, Inc.).
Cell cycle analysis
The cell cycle was analyzed using a Cell Cycle Detection Kit (KGA512, KeyGen Biotech, Nanjing, China). Briefly, A375 cancer cells (1.6 × 105 per well) were incubated in 6-well plates for 12 h and then treated with DMSO, chlordinaline, chlorambucil or tacedinaline. After 72 h, the cells were harvested and fixed with 70% ethanol in phosphate buffer at –20 °C overnight. The cells were incubated with 500 μL freshly prepared staining solution (Rnase : PI = 1 : 9) for 30 min at room temperature. The DNA content was measured using a MoFlo XDP flow cytometer (Beckman Coulter, Inc.).
Molecular docking
Molecular docking calculations were carried out with Surflex-Dock in Sybyl-X 2.0. (Tripos Inc.) The three-dimensional structures of HDAC1 (PDB code: 4BKX), HDAC2 (PDB code: ; 4LXZ), HDAC3 (PDB code: ; 4A69), HDAC8 (PDB code: ; 1T69) and HDAC6 (PDB code: ; 5EDU) were retrieved from the RCSB Protein Data Bank (; http://www.rcsb.org/pdb/home/home.do). For the protein preparation, all water molecules and co-crystallized ligands were removed and polar hydrogen was added. The structures of chlordinaline were optimized using the conjugated gradient method with the Tripos force field with the convergence criterion set at 0.001 kcal Å–1 mol–1. The active pocket was defined by selecting HDAC residues within 6 Å from the co-crystallized ligand. For other parameters the default values were accepted.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation of China (Grant No. 21636001).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00476a
References
- DeVita V. T., Chu E. Cancer Res. 2008;68:8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- Chabner B. A., Roberts T. G. Nat. Rev. Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- Jeggo P. A., Pearl L. H., Carr A. M. Nat. Rev. Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- Bouwman P., Jonkers J. Nat. Rev. Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- Lord C. J., Ashworth A. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Haberland M., Montgomery R. L., Olson E. N. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel M., Wascholowski V., Giannis A. Angew. Chem., Int. Ed. 2005;44:3186–3216. doi: 10.1002/anie.200461346. [DOI] [PubMed] [Google Scholar]
- Lane A. A., Chabner B. A. J. Clin. Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Venugopal B., Evans T. Curr. Med. Chem. 2011;18:1658–1671. doi: 10.2174/092986711795471284. [DOI] [PubMed] [Google Scholar]
- Johnstone R. W. Nat. Rev. Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- Kim M. S., Blake M., Baek J. H., Kohlhagen G., Pommier Y., Carrier F. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- Eot-Houllier G., Fulcrand G., Magnaghi-Jaulin L., Jaulin C. Cancer Lett. 2009;274:169–176. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Diyabalanage H. V., Granda M. L., Hooker J. M. Cancer Lett. 2013;329:1–8. doi: 10.1016/j.canlet.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith D., Morgan M. P., Marmion C. J. Chem. Commun. 2009:6735–6737. doi: 10.1039/b916715c. [DOI] [PubMed] [Google Scholar]
- Parker J. P., Nimir H., Griffith D. M., Duff B., Chubb A. J., Brennan M. P., Morgan M. P., Egan D. A., Marmion C. J. J. Inorg. Biochem. 2013;124:70–77. doi: 10.1016/j.jinorgbio.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Griffith D. M., Duff B., Suponitsky K. Y., Kavanagh K., Morgan M. P., Egan D., Marmion C. J. J. Inorg. Biochem. 2011;105:793–799. doi: 10.1016/j.jinorgbio.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Kasparkova J., Kostrhunova H., Novakova O., Křikavová R., Vančo J., Trávníček Z., Brabec V. Angew. Chem. 2015;127:14686–14690. doi: 10.1002/anie.201506533. [DOI] [PubMed] [Google Scholar]
- Kohn K. W., Hartley J. A., Mattes W. B. Nucleic Acids Res. 1987;15:10531–10549. doi: 10.1093/nar/15.24.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J., Lewensohn R., Ringborg U., Nilsson B. Cancer Res. 1987;47:2631–2637. [PubMed] [Google Scholar]
- Xie R., Yao Y., Tang P., Chen G., Liu X., Yun F., Chen C., Wu X., Yuan Q. Eur. J. Med. Chem. 2017;134:1–12. doi: 10.1016/j.ejmech.2017.03.038. [DOI] [PubMed] [Google Scholar]
- Xie R., Shi J. H., Qu Y., Tang P. W., Wu X. Y., Yang M., Yuan Q. P. Med. Chem. 2015;11:636–648. doi: 10.2174/1573406411666150429154107. [DOI] [PubMed] [Google Scholar]
- Xie R., Shi J., Cheng C., Yun F., Liu X., Tang P., Wu X., Yang M., Yuan Q. Med. Chem. 2016;12:767–774. doi: 10.2174/1573406412666160404125551. [DOI] [PubMed] [Google Scholar]
- Miller T. A., Witter D. J., Belvedere S. J. Med. Chem. 2003;46:5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- Zagni C., Floresta G., Monciino G., Rescifina A. Med. Res. Rev. 2017;37:1373–1428. doi: 10.1002/med.21437. [DOI] [PubMed] [Google Scholar]
- Mai A., Massa S., Rotili D., Cerbara I., Valente S., Pezzi R., Simeoni S., Ragno R. Med. Res. Rev. 2005;25:261–309. doi: 10.1002/med.20024. [DOI] [PubMed] [Google Scholar]
- Pontiki E., Hadjipavlou-Litina D. Med. Res. Rev. 2012;32:1–165. doi: 10.1002/med.20200. [DOI] [PubMed] [Google Scholar]
- Xie R., Li Y., Tang P., Yuan Q. Eur. J. Med. Chem. 2018;143:320–333. doi: 10.1016/j.ejmech.2017.08.041. [DOI] [PubMed] [Google Scholar]
- Gridelli C., Rossi A., Maione P. Crit. Rev. Oncol. Hematol. 2008;68:29–36. doi: 10.1016/j.critrevonc.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Bolden J. E., Peart M. J., Johnstone R. W. Nat. Rev. Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Yang X. J., Seto E. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W., Roske A., Gekeler V., Beckers T., Ebert M. P., Pross M., Dietel M., Denkert C., Rocken C. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- Weichert W., Röske A., Niesporek S., Noske A., Buckendahl A.-C., Dietel M., Gekeler V., Boehm M., Beckers T., Denkert C. Clin. Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- Fukuda T., Wu W., Okada M., Maeda I., Kojima Y., Hayami R., Miyoshi Y., Tsugawa K. i., Ohta T. Cancer Sci. 2015;106:1050–1056. doi: 10.1111/cas.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S., Chyla B. J., Amann J. M., Knutson S. K., Cortez D., Sun Z.-W., Hiebert S. W. Mol. Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams H., Fritzsche F. R., Dirnhofer S., Kristiansen G., Tzankov A. Expert Opin. Ther. Targets. 2010;14:577–584. doi: 10.1517/14728221003796609. [DOI] [PubMed] [Google Scholar]
- Weichert W., Röske A., Gekeler V., Beckers T., Stephan C., Jung K., Fritzsche F., Niesporek S., Denkert C., Dietel M. Br. J. Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W., Denkert C., Noske A., Darb-Esfahani S., Dietel M., Kalloger S. E., Huntsman D. G., Köbel M. Neoplasia. 2008;10:1021–1027. doi: 10.1593/neo.08474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Martin E., Mengwasser J., Schlag P., Janssen K.-P., Göttlicher M. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- Millard C. J., Watson P. J., Celardo I., Gordiyenko Y., Cowley S. M., Robinson C. V., Fairall L., Schwabe J. W. Mol. Cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer B. E., Mintzer R., Fong R., Mukund S., Tam C., Zilberleyb I., Flicke B., Ritscher A., Fedorowicz G., Vallero R. J. Biol. Chem. 2013;288:26926–26943. doi: 10.1074/jbc.M113.490706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P. J., Fairall L., Santos G. M., Schwabe J. W. Nature. 2012;481:335. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza J. R., Skene R. J., Katz B. A., Mol C., Ho J. D., Jennings A. J., Luong C., Arvai A., Buggy J. J., Chi E. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Hai Y., Christianson D. W. Nat. Chem. Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.