Summary
Defects in nucleocytoplasmic transport have been identified as a key pathogenic event in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) mediated by a GGGGCC hexanucleotide repeat expansion in C9ORF72, the most common genetic cause of ALS/FTD. Furthermore, nucleocytoplasmic transport disruption has also been implicated in other neurodegenerative diseases with protein aggregation, suggesting a shared mechanism by which protein stress disrupts nucleocytoplasmic transport. Here, we show that cellular stress disrupts nucleocytoplasmic transport by localizing critical nucleocytoplasmic transport factors into stress granules, RNA/protein complexes that play a crucial role in ALS pathogenesis. Importantly, inhibiting stress granule assembly, such as by knocking down Ataxin-2, suppresses nucleocytoplasmic transport defects as well as neurodegeneration in C9ORF72-mediated ALS/FTD. Our findings identify a link between stress granule assembly and nucleocytoplasmic transport, two fundamental cellular processes implicated in the pathogenesis of C9ORF72-xmediated ALS/FTD and other neurodegenerative diseases.
Graphical abstract
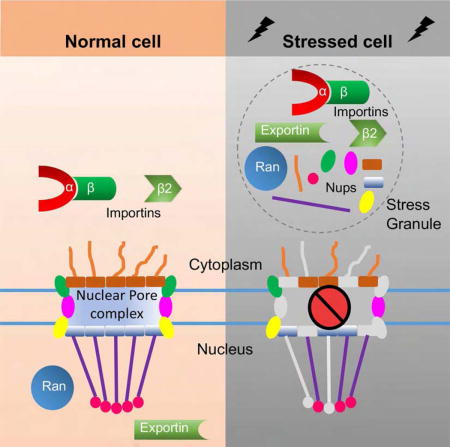
Introduction
A GGGGCC (G4C2) hexanucleotide repeat expansion in chromosome 9 open reading frame 72 (C9ORF72) is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). The driver of neurodegeneration in C9ORF72-mediated ALS/FTD (C9-ALS/FTD) is believed to be toxic gain-of-function derived from 1) the G4C2 mRNA and/or 2) its translation products, dipeptide repeat proteins generated via repeat associated, non-AUG translation (Ash et al., 2013; Donnelly et al., 2013; Mori et al., 2013). Arginine-containing dipeptide repeat proteins, poly-glycine-arginine(GR) and poly-proline-arginine(PR), are particularly cytotoxic when overexpressed (Kwon et al., 2014; Mizielinska et al., 2014).
Previous studies from our group and others have identified nucleocytoplasmic transport defects as a critical event in the pathogenesis of C9-ALS/FTD (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015), Huntington’s disease (Gasset-Rosa et al., 2017; Grima et al., 2017), and Alzheimer’s disease (unpublished). G4C2 mRNA binds and sequesters Ran GTPase activating protein (RanGAP1), leading to cytoplasmic mislocalization of its target, Ran GTPase (Ran) that is normally enriched in the nucleus (Zhang et al., 2015). Ran is a key nucleocytoplasmic transport regulator, and its nuclear-to-cytoplasmic gradient regulates the transport of proteins and RNAs through the nuclear pore complex (NPC) (Steggerda and Paschal, 2002). Since cytoplasmic mislocalization and aggregation of nuclear Tar-DNA binding protein 43 (TDP-43) is the pathological hallmark of ALS and most cases of FTD (Neumann et al., 2006), we postulate that disrupted nucleocytoplasmic transport may be a common pathogenic event in many types of ALS/FTD and other neurodegenerative diseases.
Stress granules (SGs) are membrane-less, RNA/protein condensates assembled upon diverse cellular stressors (Protter and Parker, 2016). Upon protein misfolding stress, polysomes disassemble, halting the translation of mRNAs that are subsequently embedded into SGs enriched in RNA binding proteins (Anderson and Kedersha, 2008). SG assembly involves liquid-liquid phase separation (LLPS) by proteins with low complexity sequence domains (LCDs), including TDP-43, FUS, and other heterogeneous nuclear ribonucleoproteins (hnRNPs) implicated in ALS (Molliex et al., 2015; Patel et al., 2015). SGs may play a role in ALS pathogenesis by inducing toxic deposition of these RNA-binding proteins, as most ALS-causing mutations in TDP-43 and some hnRNPs are found in their LCDs (Li et al., 2013). These mutant hnRNPs, as well as arginine-containing dipeptide repeat proteins, have also been shown to impair SG dynamics and function (Lee et al., 2016; Molliex et al., 2015). Furthermore, mutations in TIA-1 (Mackenzie et al., 2017) or Ataxin-2 (Elden et al., 2010), two crucial SG components (Gilks et al., 2004; Nonhoff et al., 2007), have been identified to either cause ALS or increase its risk. In addition, Ataxin-2 is a genetic modifier of neurodegeneration in ALS animal models, potentially through modulation of SG dynamics (Becker et al., 2017; Kim et al., 2014; Lee et al., 2016).
Although impaired nucleocytoplasmic transport and SG assembly can both lead to cytoplasmic mislocalization and deposition of TDP-43 or FUS, the connections between the two processes remain unclear. Importin α and β proteins have been previously identified as constituents of SGs (Chang and Tarn, 2009; Fujimura et al., 2010; Mahboubi et al., 2013), but their role in SG function is unknown. Here, we show that many nucleocytoplasmic transport factors are localized to SGs when exposed to stressors or mutant proteins implicated in ALS pathogenesis, leading to impaired nucleocytoplasmic transport. Importantly, SG inhibitors suppress nucleocytoplasmic transport defects as well as neurodegeneration in C9-ALS/FTD models. These findings link nucleocytoplasmic transport and SG assembly in a unified pathway contributing to the pathogenesis of C9-ALS/FTD.
Results
Cellular stress disrupts nucleocytoplasmic transport
Mislocalization of TDP-43 from the nucleus to cytoplasm is a common cytological event in ALS and believed to be a contributor to disease pathogenesis (Neumann et al., 2006). Given the important role of Ataxin-2 in modulating TDP-43 toxicity (Becker et al., 2017; Kim et al., 2014), we first analyzed whether Ataxin-2 regulates nucleocytoplasmic transport using a tdTomato protein tagged with a canonical nuclear localization signal (NLS) and a nuclear export sequence (NES) that shuttles between the cytoplasm and nucleus (Shuttle-tdTomato, or S-tdTomato) (Zhang et al., 2015). When expressed alone, S-tdTomato is enriched in the nucleus of HEK293T cells, whereas transient co-expression of Ataxin-2 leads to S-tdTomato cytoplasmic mislocalization (Figure S1A), suggesting that upregulating Ataxin-2 disrupts nuclear import. In addition, Ran GTPase (Ran) is a key regulator of nucleocytoplasmic transport that is normally enriched in the nucleus but mislocalized to the cytoplasm in C9-ALS (Zhang et al., 2015). As shown in Figure S1B, Ran is mislocalized to the cytoplasm in cells overexpressing Ataxin-2, further suggesting that upregulating Ataxin-2 disrupts nucleocytoplasmic transport.
Given that Ataxin-2 is an essential component of SGs (Nonhoff et al., 2007), we hypothesized that SG assembly could regulate nucleocytoplasmic transport. To test this hypothesis, we first treated HEK293T cells expressing S-tdTomato with 0.5 mM sodium arsenite, a commonly used to induce stress granules. Sodium arsenite causes oxidative stress and protein misfolding, leading to eIF2α phosphorylation, stalled translation, and SG assembly (Bernstam and Nriagu, 2000). As shown in Figure 1A, sodium arsenite causes cytoplasmic mislocalization of S-tdTomato in a time-dependent manner. In addition, we also treated S-tdTomato-expressing cells with 0.4 M sorbitol, a stressor that causes hyperosmosis and SG assembly independent of eIF2α (Bevilacqua et al., 2010; Kedersha et al., 2016). As shown in Figure 1A, sorbitol also causes cytoplasmic mislocalization of S-tdTomato. A one-hour treatment with sodium arsenite or sorbitol causes SG assembly, as indicated by cytoplasmic puncta of Ataxin-2 (Figure 1A). Thus, inducing SGs disrupts nucleocytoplasmic transport.
Figure 1. Stressors disrupt nucleocytoplasmic transport.
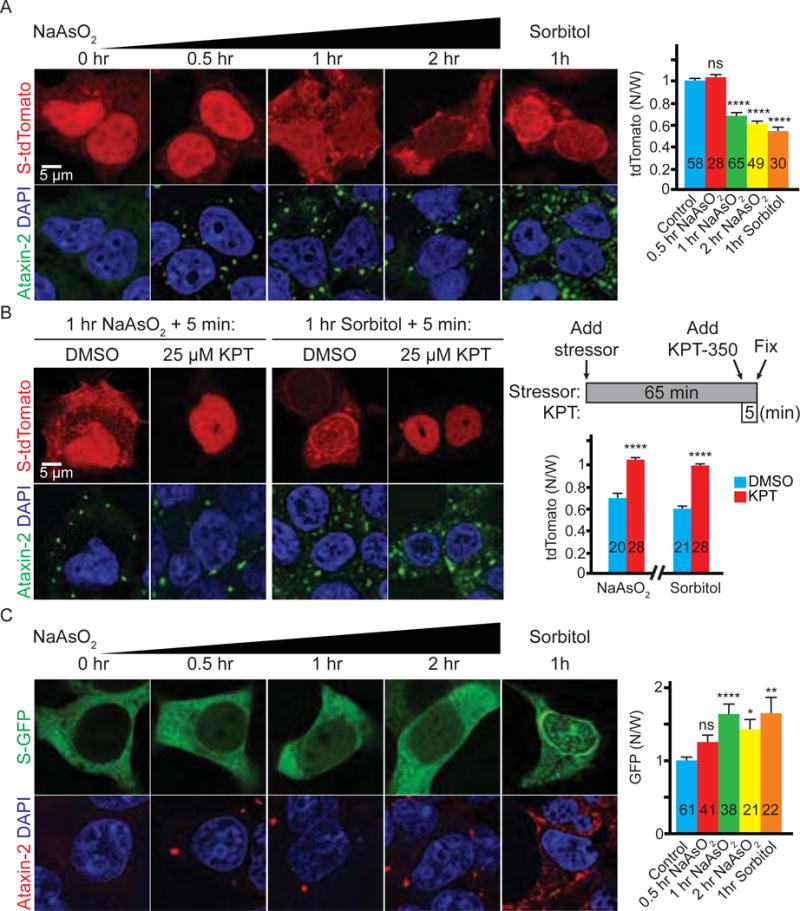
(A) HEK293T cells expressing S-tdTomato (red) stained with Ataxin-2 (green) and DAPI (blue). (B) Stressed HEK293T cells expressing S-tdTomato (red) treated with KPT-350 or DMSO and stained with Ataxin-2 (green) and DAPI (blue). (C) HEK293T cells expressing S-GFP stained with Ataxin-2 (red), GFP (green), and DAPI (blue). N: nuclear; W: whole cell. n numbers in graph. ns: not significant *: p<0.05; **: p<0.01; ****: p<0.0001. Data are represented as mean ± SEM.
To further test whether the cytoplasmic mislocalization of S-tdTomato is due to disrupted nucleocytoplasmic transport rather than other effects of these stressors, we treated stressed cells with KPT-350 (KPT), a selective inhibitor of Exportin-1-mediated nuclear export (Haines et al., 2015). As shown in Figure 1B, a five-minute KPT treatment completely reverses S-tdTomato mislocalization caused by one-hour arsenite or sorbitol stress, suggesting that this phenotype is indeed due to an imbalance between nuclear import and export. We also tested another reporter, S-GFP that is normally enriched in the cytoplasm (Woerner et al., 2016), and found that both arsenite and sorbitol cause nuclear mislocalization of S-GFP (Figure 1C). However, like S-tdTomato, S-GFP does not form immobile cytoplasmic aggregates under stress, as KPT also causes it to localize to the nucleus (Figure S1C). Together, our data suggest that acute cellular stress causes an imbalance between nuclear import and export.
Nucleocytoplasmic transport factors localize to stress granules
To understand how stressors disrupt nucleocytoplasmic transport, we analyzed the localization of Ran in arsenite- or sorbitol-treated HEK293T cells. As shown in Figure 2A, either arsenite or sorbitol causes cytoplasmic Ran accumulation that co-localizes with Ataxin-2 puncta, suggesting that Ran localizes to SGs. We further verified the localization of Ran in SGs using two additional SG markers, G3BP1 and TIA-1, in different cell types (Protter and Parker, 2016) (Figure S2A–S2B). Together, our data suggest that Ran localizes to SGs upon different stressors in multiple cell types.
Figure 2. Nucleocytoplasmic transport factors are localized to stress granules.
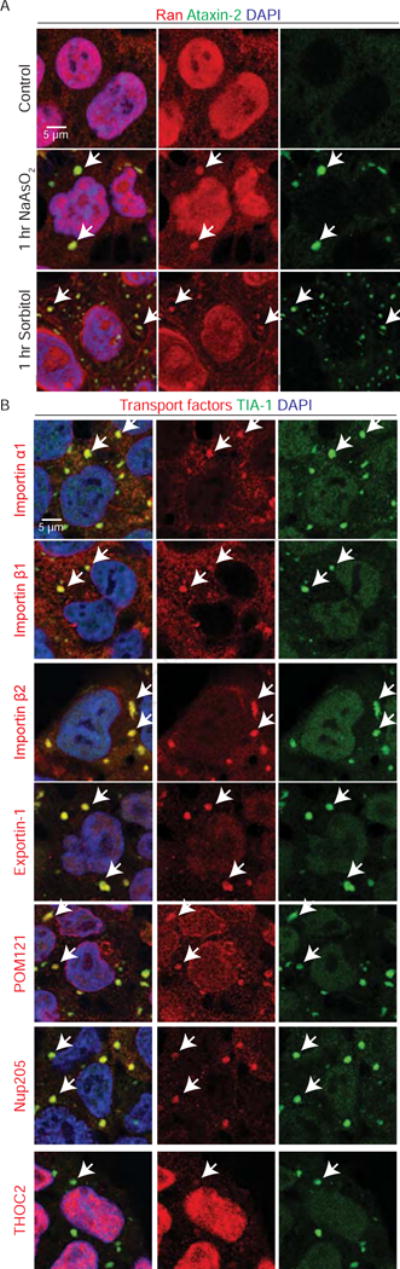
(A) Untreated (top row), arsenite- (middle row) or sorbitol- (bottom row) treated HEK293T cells stained with Ran (red), Ataxin-2 (green) and DAPI (blue). Arrows indicate co-localization. (B) Arsenite-treated HEK293T cells stained with transport factors (red), TIA-1 (green) and DAPI (blue). Arrows indicate co-localization.
Although sorbitol disrupts the nuclear versus cytoplasmic Ran gradient, this gradient is not changed upon arsenite treatment (Figure S2C). In addition, only around two percent of total cellular Ran is found in arsenite-induced SGs (Figure S2D), suggesting that other factors might contribute to nucleocytoplasmic transport disruption. Importins and Exportins are transport factors regulated by Ran that carry protein cargos with an NLS and/or NES to undergo nuclear import or export, respectively. Cytoplasmic proteins with a canonical NLS, including TDP-43, bind to Importin α1/β1 complex for nuclear import, whereas proteins with a noncanonical NLS, such as FUS and hnRNP A1, bind to Importin β2 for import (Fridell et al., 1997). In the nucleus, Exportin-1 binds to the NES of cargo proteins for their export. As shown in Figures 2B and S2D–E, Importin α1, β1, and β2 and Exportin-1 all co-localize with the SG marker TIA-1 upon arsenite or sorbitol treatment, consistent with previous findings (Chang and Tarn, 2009; Fujimura et al., 2010; Mahboubi et al., 2013).
The NPC consists of ~30 nucleoporins (Nups), through which macromolecules enter and exit the nucleus. Previous studies have shown that some Nups modulate C9ORF72-mediated toxicity and are accumulated in C9-ALS/FTD patients and animal models (Freibaum et al., 2015; Zhang et al., 2015; Zhang et al., 2016). As SGs may contribute to ALS pathogenesis by triggering the aggregation of its component proteins (Li et al., 2013), we hypothesized that some Nups may also localize to SGs. Indeed, as shown in Figures 2B, S2D–E, S3A, and Table S1, 14 out of 20 tested Nups co-localize with SG markers upon arsenite and/or sorbitol treatment, including Nup205, Nup50, and POM121 that are accumulated in C9-ALS/FTD patients and/or animal models. Together, our data suggest that some Nups also localize to SGs. Notably, Nups that localize to SGs (Table S1) do not belong to specific NPC subcomplexes, and include both Nups with and without phenylalanine-glycine (FG)-repeat motifs that have been shown to undergo LLPS (Shi et al., 2017).
In addition to Nups, previous studies have found that THOC2, a protein involved in mRNA export, mislocalizes and aggregates in cells overexpressing disease-related, LCD-containing proteins (Woerner et al., 2016). As seen with many Nups, either arsenite or sorbitol causes THOC2 to localize to SGs (Figure 2B and S2E), suggesting that SG assembly may also mediate THOC2 aggregation. In contrast, RanGAP1, Ran-Guanine nucleotide exchange factor (RanGEF) that opposes RanGAP1 in Ran regulation, and nuclear envelope protein Lamin B1 do not localize to SGs (Figure S3C). Therefore, our data suggest that many nucleocytoplasmic transport factors, including Ran, Importins, Exportin-1, multiple Nups, and THOC2, localize to SGs.
Nucleocytoplasmic transport factors are constituents of stress granules
SGs are sedimentable, protein/RNA condensates, which differ from both soluble complexes and insoluble aggregates (Banani et al., 2017). They consist of “cores” with high protein/RNA concentration and a surrounding “shell” with less concentrated material that weakly interacts with cores (Jain et al., 2016). Both the cores and shells can be enriched from the whole cell lysate by a series of centrifugations, and the cores can be further purified from the enriched fraction by immunoprecipitation (IP) of GFP-tagged G3BP1, a SG core protein. Furthermore, the same IP can co-purify both cores and shells if they are crosslinked by paraformaldehyde. Indeed, these approaches followed by mass spectrometry identified some nucleocytoplasmic transport factors, including Importins, Exportins, and several Nups, within purified SG shells (Jain et al., 2016), consistent with our staining data (Figures 2, S2 and S3).
As crosslinking by paraformaldehyde is not compatible with Western Blot analyses, we used alternative approaches to confirm that nucleocytoplasmic transport factors are recruited to SGs. As shown in Figure 3A, Ran, Importin α1, β1, and β2, and several Nups (Nup205, 88, and 50) are enriched in the sedimentable fraction containing SGs (P18000) of arsenite-stressed cells (Figure 3A), compared to non-stressed cells. As internal controls, two SG core proteins, Ataxin-2 and G3BP1, are also enriched in this fraction, in contrast to a previously known non-stress-granule protein, large ribosomal subunit RPL5 (Kimball et al., 2003). Together, these data suggest that arsenite causes nucleocytoplasmic transport factors to localize to sedimentable condensates consistent with SGs. However, we did not detect S-tdTomato in the stress-granule-enriched fraction (P18000, Figure 3A), consistent with our data that S-tdTomato is mobile under stress (Figure 1B). Therefore, our data suggest that nucleocytoplasmic transport factors are constituents of SGs.
Figure 3. Nucleocytoplasmic transport factors are constituents of stress granules.
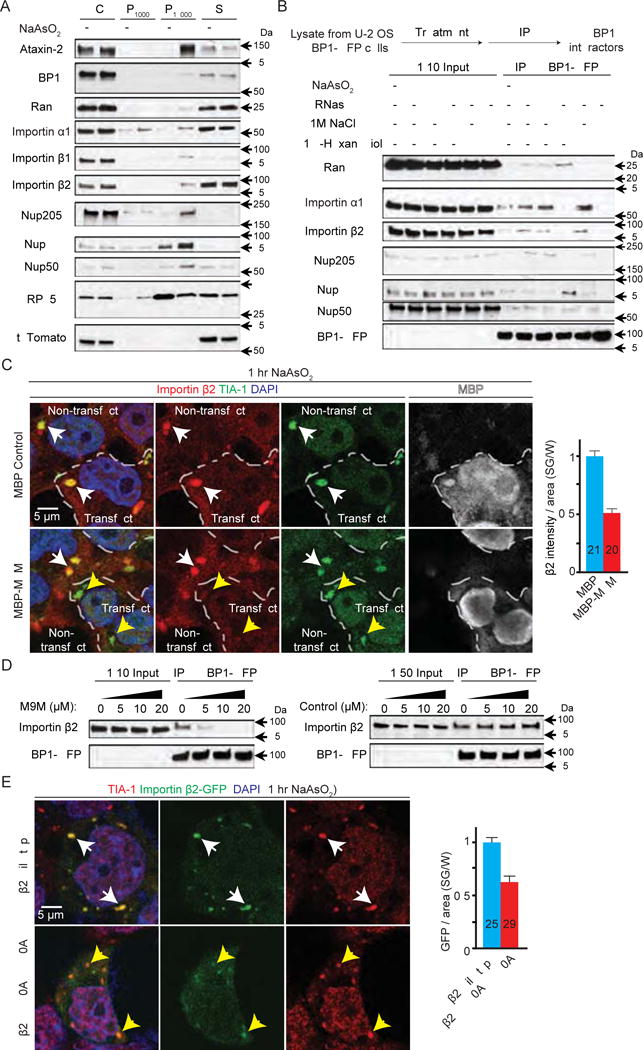
(A) Subcellular fractionation of HEK293T cells. P1000: pellet from 1,000 g; P18000: pellet from 18,000 g; S: supernatant after 18,000 g. (B) Co-IP of nucleocytoplasmic transport factors with G3BP1-GFP from U-2 OS cells expressing G3BP1-GFP. (C) HEK293T cells expressing MBP (control, top) or MBP-tagged M9M (bottom) were stained with Importin β2 (red), TIA-1 (green), DAPI (blue), and MBP (white). Dashed lines separate transfected versus non-transfected cells. White arrowheads indicate co-localization. Yellow arrowheads indicate TIA-1-positive puncta without Importin β2 co-localization. (D) Co-IP of Importin β2 and G3BP1-GFP with chemically synthesized M9M or control peptide. (E) HEK293T cells expressing GFP-tagged wild type (top) or mutant (bottom) Importin β2 (green) stained with TIA-1 (red) and DAPI (blue). Arrowheads indicate co-localization. W: whole cell, n numbers in graph. ****: p<0.0001. Data are represented as mean ± SEM.
To further investigate how nucleocytoplasmic transport factors are recruited to SGs, we performed co-IP of G3BP1 using a U-2 OS cell line stably expressing GFP-tagged G3BP1 (Figley et al., 2014) that has been widely used to study SG assembly (Boeynaems et al., 2017; Jain et al., 2016). As shown in Figure 3B, several nucleocytoplasmic transport factors interact with G3BP1 upon arsenite treatment, which is not disrupted by RNase, suggesting RNA-independent interactions. In addition, the interaction of G3BP1 with Importin α1, Importin β2, or Nup205 is inhibited by one molar sodium chloride (Figure 3B), suggesting an electrostatic interaction. Conversely, the interaction of G3BP1 with Ran, Nup88, or Nup50 is not inhibited by one molar sodium chloride alone but diminished by a mixture of sodium chloride and 6% 1,6-Hexanediol, an aliphatic alcohol that disrupts hydrophobic interactions (Shi et al., 2017). Together, these data suggest that both electrostatic and hydrophobic interactions mediate the recruitment of nucleocytoplasmic transport factors to SGs.
Because SG proteins FUS and hnRNPs are also cargos of Importin β2, we tested whether Importin β2 is recruited to SGs via its cargos by transiently expressing a peptide inhibitor M9M that competes with Importin β2 for NLSs (Cansizoglu et al., 2007). As shown in Figure 3C, transient expression of maltose binding protein (MBP)-tagged M9M (Bernis et al., 2014) prevents localization of Importin β2 to SGs, compared to the MBP control. In accord with these data, a chemically synthesized M9M, but not a control peptide, inhibits the interaction between Importin β2 and G3BP1-GFP in a dose-dependent manner in co-IP assays (Figure 3D), suggesting that the cargo-binding domain of Importin β2 mediates its recruitment to SGs. To further test this hypothesis, we expressed a GFP-tagged Importin β2 with mutations (W460A: W730A) that significantly decrease its NLS-binding activity (Lee et al., 2006). As shown in Figure 3E, the level of mutant Importin β2 in SGs upon arsenite treatment is significantly reduced, compared to the wild type control. Hence, our data suggest that Importin β2 is recruited to SGs via its cargo-binding domain.
Arsenite disrupts nucleocytoplasmic transport via stress granules
To determine whether SGs mediate the nucleocytoplasmic transport disruption under stress, we tested whether inhibiting SG assembly with Ataxin-2 knockdown (Becker et al., 2017) suppresses stressor-induced nucleocytoplasmic transport defects. We observed that Ran (Figure S1D) and other transport factors (data not shown) do not form cytoplasmic puncta in arsenite-treated HEK293T cells expressing Ataxin-2 RNAi. However, we were not able to analyze the localization of S-tdTomato or S-GFP, as knockdown of Ataxin-2 suppresses expression levels of these fluorescent proteins (data not shown). We therefore used two alternative approaches to inhibit arsenite-induced SGs assembly: 1) previously identified inhibitors GSK2606414 (GSK) (Axten et al., 2012) and integrated stress response inhibitor (ISRIB) (Sidrauski et al., 2015) and 2) genetic ablation of G3BP1 and 2 (Protter and Parker, 2016).
GSK is a selective inhibitor of PERK that phosphorylates eIF2α upon arsenite stress (Axten et al., 2012) and thus inhibits SG assembly, whereas ISRIB inhibits SG assembly downstream of eIF2α (Sidrauski et al., 2015). As shown in Figure 4A–C, a four-hour pretreatment with either GSK or ISRIB suppresses mislocalization of S-tdTomato or S-GFP caused by one-hour arsenite treatment in HEK 293T cells, suggesting that both inhibitors suppress arsenite-induced nucleocytoplasmic transport defects.
Figure 4. Stress granules mediate the nucleocytoplasmic transport defects caused by arsenite.
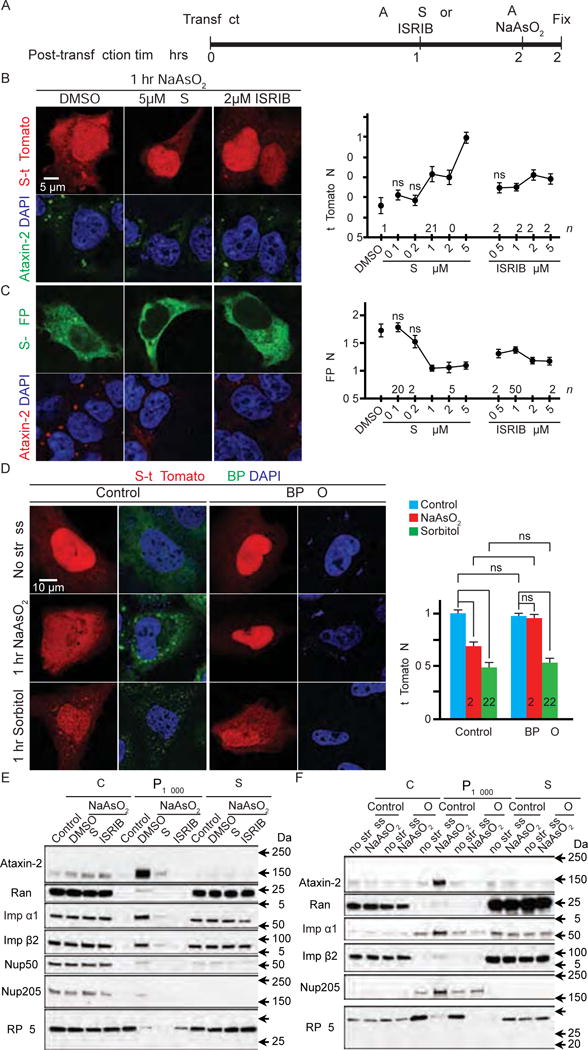
(A) Experimental design. (B and C) HEK293T cells expressing S-tdTomato (red) (B) or S-GFP (green) (C) were treated with arsenite and GSK or ISRIB and stained with Ataxin-2 (green in B and red in C) and DAPI (blue). (D) Control (left two columns) or G3BP1/2 double knockout (G3BP KO) U-2 OS cells (right two columns) expressing S-tdTomato (red) stained with G3BP (green) and (DAPI). N: nuclear; W: whole cell. (E and F) Subcellular fractionation of control and arsenite-treated HEK293T cells with or without GSK or ISRIB pre-treatment (E) or wild-type control or G3BP KO (KO) U-2 OS cells (F). WCL: whole cell lysate; P18000: pellet from 18,000 g; S: supernatant after 18,000 g. n numbers in graph. ns: not significant; *: p<0.05; **: p<0.01; ****: p<0.0001. Data are represented as mean ± SEM.
Besides inhibiting SG assembly, both GSK and ISRIB have also been shown to restore translation inhibited by arsenite stress (Sidrauski et al., 2015). Thus, it is possible that GSK and ISRIB suppress arsenite-induced S-tdTomato or S-GFP mislocalization by upregulating the translation of these fluorescent proteins and/or transport factors. However, as shown in Figure S4A, GSK and ISRIB do not affect the total levels of S-tdTomato, S-GFP, Ran, Importins, or Nup205. Indeed, neither arsenite nor sorbitol affect the levels of these proteins, suggesting that these stressors, as well as GSK and ISRIB, do not modulate nucleocytoplasmic transport via down- or up-regulating transport factors and S-tdTomato or S-GFP. Furthermore, mislocalization of S-tdTomato caused by siRNA-mediated loss of Ran or Importin α1 (Figure S1E) is not suppressed by GSK or ISRIB, and Ran or Importin α1 RNAi does not induce SG assembly (Figure S1F). Hence, our data suggest that GSK and ISRIB suppress arsenite-induced nucleocytoplasmic transport defects via inhibiting SG assembly.
Our second approach to inhibit SG assembly is to generate a U-2 OS cell line with double knockout (KO) of G3BP1 and 2 (G3BP) using CRISPR/Cas9. A prior study that independently generated this cell line showed that G3BP KO cells do not assemble SGs when treated with some stressors, including arsenite (Kedersha et al., 2016). Similarly, our G3BP KO cells do not assemble SGs upon arsenite treatment, as indicated by a lack of cytoplasmic puncta positive for Ataxin-2 or TIA-1 (Figure S4B). Importantly, in G3BP KO cells, arsenite treatment does not cause mislocalization of S-tdTomato (Figure 4D), demonstrating that arsenite-induced nucleocytoplasmic transport defects require SG assembly. In contrast, sorbitol induces SG assembly independent of G3BP (Figure S4B), and consistent with these data, sorbitol causes cytoplasmic mislocalization of S-tdTomato in G3BP KO cells to a similar degree as the control (Figure 4D). In contrast to GSK and ISRIB (Sidrauski et al., 2015), G3BP KO does not restore arsenite-inhibited translation, as indicated by eIF2α phosphorylation and decreased puromycin incorporation (Figure S4C–D). Indeed, S-tdTomato and nucleocytoplasmic transport factors are not upregulated in G3BP KO cells, compared to the control (Figure S4E). Thus, G3BP KO suppresses arsenite-induced nucleocytoplasmic transport defects via inhibiting SG assembly rather than upregulating transport factors.
To confirm that GSK, ISRIB, and G3BP KO prevent nucleocytoplasmic transport factors to localize to sedimentable condensates in arsenite-treated cells, we repeated our fractionation experiments as performed in Figure 3A. As shown in Figure 4E and F, GSK or ISRIB treatment, or G3BP KO, causes a strong reduction of SG core protein Ataxin-2 in the P18000 fractions in arsenite-treated cells (quantified in Figure S4F–G), consistent with an inhibition of SG assembly. Furthermore, Ran, Importin α1 and β2, and Nup205, but not RPL5, are also decreased in these fractions (Figure 4E–F, S4F–G). Together, our data suggest that arsenite disrupts nucleocytoplasmic transport by sequestering transport factors in SGs.
Dipeptide repeat proteins and mutant TDP-43 disrupt nucleocytoplasmic transport disruption via stress granule assembly
SGs have been suggested to contribute to ALS pathogenesis (Li et al., 2013). For C9-ALS, overexpression of poly-GR or poly-PR induces spontaneous assembly of poorly dynamic SGs (Boeynaems et al., 2017; Lee et al., 2016). As poly-GR/PR have also been shown to cause nucleocytoplasmic transport defects (Boeynaems et al., 2016; Jovicic et al., 2015), we hypothesize that they also disrupt nucleocytoplasmic transport via SGs. In addition, cytoplasmic mislocalization of TDP-43 is the pathological hallmark of almost all forms of ALS, including C9-ALS (Davidson et al., 2016), and transient overexpression of a cytoplasmically localized, truncated TDP-43 (TDP(cyto)) (Yang et al., 2010) has been shown to disrupt nucleocytoplasmic transport (Woerner et al., 2016). As TDP-43 is a constituent of SGs, and some ALS-linked TDP-43 mutations alter SG dynamics, we hypothesized that TDP(cyto) also disrupts nucleocytoplasmic transport via SGs.
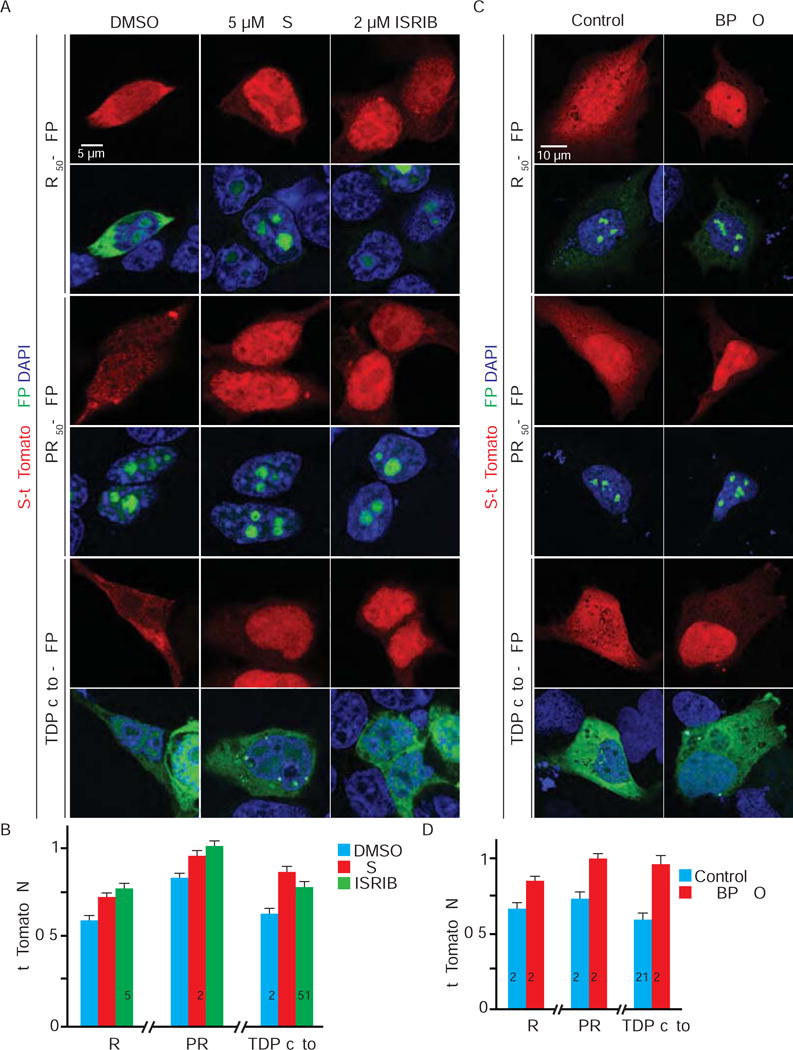
We co-expressed S-tdTomato with GFP-tagged poly-GR or PR (GFP-(GR)50 or (PR)50) (Wen et al., 2014) or GFP-tagged TDP(cyto) in HEK293T cells for 24 hours and analyzed S-tdTomato localization. As expected, S-tdTomato is mislocalized in (GR)50-, (PR)50-, or TDP(cyto)-expressing cells (Figure 5A), suggesting disrupted nucleocytoplasmic transport. In addition, (GR)50, (PR)50, or TDP(cyto) induces SG assembly in HEK293T cells, as indicated by cytoplasmic Ataxin-2 puncta (Figure 5B). Interestingly, like cells treated with arsenite or sorbitol, expression of these ALS proteins causes nucleocytoplasmic transport factors including Ran, Importins, Exportin-1, and POM121 to localize to SGs (Figure 5B and S5). Together, these data suggest that transient overexpression of (GR)50, (PR)50, or TDP(cyto) induces SG assembly and localization of nucleocytoplasmic transport factors into SGs.
Figure 5. Dipeptide repeat proteins and cytoplasmic TDP-43 cause nucleocytoplasmic transport defects.

(A) HEK293T cells co-expressing S-tdTomato (red) with GFP (left column), GFP-tagged 50 repeats of poly-GR (column 2) or poly-PR (column 3), or cytoplasmic TDP-43 (TDP(cyto)) (right column) stained with DAPI (blue). N: nuclear; W: whole cell. (B) HEK293T cells expressing GFP (top row), GFP-tagged (GR)50 (row 2) or (PR)50 (row 3), or TDP(cyto) (bottom row) were stained with Ran (red), Ataxin-2 (green) and DAPI (blue). GFP expression shown on right (white). Arrowheads indicate co-localization. Number of cells measured (n) for each condition indicated in graph. *: p<0.05; ****: p<0.0001. Data are represented as mean ± SEM.
To determine whether SGs mediate nucleocytoplasmic transport defects caused by (GR)50, (PR)50, or TDP(cyto), we again used GSK, ISRIB, or G3BP KO to inhibit SG assembly. We first transiently co-expressed S-tdTomato with GFP-(GR)50, GFP-(PR)50, or GFP-TDP(cyto) in HEK293T cells and treated them with GSK or ISRIB for five hours. As shown in Figure 6A–B and S6A, GSK or ISRIB partially suppresses S-tdTomato mislocalization in these cells. As expected, G3BP KO completely suppresses SG assembly and the S-tdTomato phenotype in response to GFP-(GR)50, GFP-(PR)50, or GFP-TDP(cyto) expression (Figure S6B and 6C and D). As GSK, ISRIB, and G3BP KO do not decrease levels of (GR)50, (PR)50, or TDP(cyto) (Figure S6C and D), our data suggest that SG assembly also mediates nucleocytoplasmic transport disruption caused by poly-GR, poly-PR, or TDP(cyto).
Figure 6. Stress granules contribute to nucleocytoplasmic transport defects caused by poly-GR, poly-PR or cytoplasmic TDP-43.
(A) HEK293T cells co-expressing S-tdTomato (red) and GFP-tagged (GR)50, (PR)50, or cytoplasmic TDP-43 (TDP(cyto)) (green) treated with DMSO, GSK or ISRIB and stained with DAPI (blue). Quantification in B. (C) Wild type control or G3BP1/2 double knockout (G3BP KO) U-2 OS cells co-expressing S-tdTomato (red) and GFP-tagged (GR)50, (PR)50, or TDP(cyto) (green) stained with DAPI (blue). Quantification in D. N: nuclear; W: whole cell. n numbers in graph. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. Data are represented as mean ± SEM.
Stress granule inhibitors suppress nucleocytoplasmic transport defects and neurodegeneration in C9-ALS models
We previously reported cytoplasmic mislocalization of Ran in iPS motor neurons (iPSNs) derived from C9-ALS patients (Zhang et al., 2015). Given that inhibiting SG assembly suppresses nucleocytoplasmic transport defects caused by poly-GR, poly-PR, or TDP(cyto), we tested whether it also suppresses Ran mislocalization in these neurons. We first treated four independent C9-ALS iPSNs (Figure S7A) with either GSK or ISRIB. As shown in Figure 7A and S7B, these inhibitors suppress Ran mislocalization in a dose-dependent manner, with 0.5 μM GSK or 2 μM ISRIB showing the strongest suppression. Next, we treated C9-ALS iPSNs with Ataxin-2 antisense oligonucleotides (ASOs) that also suppress arsenite-induced SG assembly (data not shown), as has been shown for Ataxin-2 siRNA (Figure S1D). Two exon-targeting ASOs (Figure S7C, #1 and 3) knock down Ataxin-2 and suppress Ran mislocalization in C9-ALS iPSN #1, whereas an intron-targeting ASO (Figure S7C, #2) is ineffective (Figure 7B). Furthermore, ASO #1 suppresses Ran mislocalization in three other C9-ALS iPSNs (Figure S7D). Together, these data suggest that inhibitors of SG assembly suppress Ran defects in C9-ALS iPSNs. Consistent with these data, we detected a mild increase in phospho-eIF2α levels in C9-ALS iPSNs (Figure S7E), suggesting that C9-ALS iPSNs are constitutively under low levels of stress.
Figure 7. GSK, ISRIB, and Ataxin-2 ASO suppress nucleocytoplasmic transport defects and neurodegeneration in C9-ALS models.
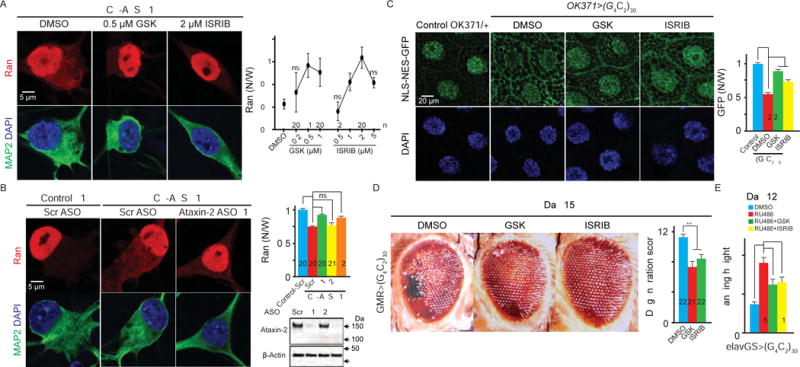
(A) C9-ALS iPSNs treated with DMSO, GSK or ISRIB were stained with Ran (red), MAP2 (green), and DAPI (blue). (B) Control or C9-ALS iPSNs treated with scrambled or Ataxin-2 ASOs were stained with Ran (red), MAP2 (green), and DAPI (blue). Bottom right: ASO-treated C9-ALS iPSNs immunoblotted for Ataxin-2 and β-Actin. (C) Fly salivary glands stained with GFP and DAPI. N: nuclear; W: whole cell. (D) Fly eye degeneration. (E) Flight assay. n numbers in the graph. ns: not significant; *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. Data are represented as mean ± SEM.
To study the effect of SG inhibitors in vivo, we employed a Drosophila model of C9-ALS/FTD (Xu et al., 2013). By expressing 30 G4C2 repeats using the UAS/GAL4 system, we previously showed that an NLS- and NES-tagged GFP reporter is mislocalized from the cytoplasm to the nucleus in salivary gland cells (Figure 7C and Zhang et al., 2015). Here, we show that feeding flies with 5 μM GSK or ISRIB suppresses these defects (Figure 7C), suggesting that SG inhibitors suppress nucleocytoplasmic transport defects caused by the G4C2 hexanucleotide repeat expansion in vivo. Furthermore, GSK and ISRIB do not decrease the amount of poly-GR protein (Figure S7F). Since poly-GR and PR are the only dipeptide repeat proteins toxic to flies among the five possibly expressed in C9-ALS/FTD (Mizielinska et al., 2014), and the structure of UAS/GAL4 construct does not allow poly-PR expression, our data suggest that the effect of GSK and ISRIB on nucleocytoplasmic transport is not via downregulating toxic dipeptide repeat proteins.
Nucleocytoplasmic transport defects are a critical pathogenic event in yeast, fly, and mouse models, as well as C9-ALS/FTD patients (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015; Zhang et al., 2016). Importantly, genetic and pharmacologic modulation of nucleocytoplasmic transport modify neurodegeneration in C9-ALS fly models. For example, feeding C9-ALS flies with KPT-276, an analogue of KPT-350, suppresses nucleocytoplasmic transport defects and neurodegeneration, whereas Nup50 RNAi or a dominant negative form of Ran (RanDN) enhances neurodegeneration. Given that SG inhibitors suppress nucleocytoplasmic transport defects in C9-ALS flies, we hypothesized that they might also suppress neurodegeneration. To test this hypothesis, we fed either GSK or ISRIB to flies expressing (G4C2)30 in the eye using GMR-GAL4. As shown in Figure 7D, both inhibitors significantly suppress eye degeneration in 15-day-old flies. Next, we fed either inhibitor to flies inducibly expressing (G4C2)30 in the nervous system, using elav gene-switch (elavGS), and analyzed their ability to fly. Induction of G4C2 repeat expression with RU486-feeding during adulthood causes progressive flight impairment with aging (Zhang et al., 2015) that is suppressed by GSK and ISRIB (Figure 7E). In contrast, GSK and ISRIB do not affect the flight defect induced by RanDN or Nup50 RNAi (Figure S7G). Together, our data suggest that inhibiting SG assembly suppresses neurodegeneration in a C9-ALS fly model by restoring nucleocytoplasmic transport.
Discussion
Defects in nucleocytoplasmic transport have recently been shown in C9-ALS/FTD (Freibaum et al., 2015; Jovicic et al., 2015; Zhang et al., 2015) and Huntington’s disease (Gasset-Rosa et al., 2017; Grima et al., 2017). In addition, overexpressing dipeptide repeat proteins or mutant TDP-43 or Huntingtin disrupts nucleocytoplasmic transport, suggesting a potentially common mechanism by which cytoplasmic protein aggregation impairs nucleocytoplasmic transport (Woerner et al., 2016). Here, we show that diverse stressors, including those that cause C9-ALS/FTD, disrupt nucleocytoplasmic transport by localizing key factors including Ran, karyopherins, and Nups into SGs. These data suggest a general mechanism by which cytoplasmic protein stress inhibits nucleocytoplasmic transport.
How transport factors are recruited to SGs is unclear. One possible mechanism is that karyopherins are recruited to SGs through binding to the NLS or NES of their SG component cargos, such as TDP-43, FUS, and other hnRNPs. Indeed, the NLS-binding domain is required for Importin β2 to localize to SGs (Figure 3). In this way, some Nups may also be recruited to SGs via their association with karyopherins (Otsuka et al., 2008). Alternatively, Nups containing FG-repeat motifs also have a propensity to phase separate (Shi et al., 2017), which may promote their recruitment to SGs.
Recruitment of TDP-43, FUS, and other disease-related, LCD-containing proteins to SGs triggers their cytoplasmic deposition in vitro (Li et al., 2013). However, many TDP-43 inclusions in ALS patients do not contain SG markers (Neumann et al., 2007), suggesting that its recruitment to SGs may precede aggregate formation. Similarly, in cells transiently expressing poly-GR, poly-PR or TDP(cyto), Importins are also localized to cytoplasmic puncta other than SGs (Figure S5A–B), possibly representing aggregates. Consistent with these data, Importins and Nups have been previously shown to aggregate in ALS patients and mouse models (Kinoshita et al., 2009; Zhang et al., 2006), suggesting that recruitment of these proteins to SGs may also trigger their aggregation.
As a common response to stress, cells halt their protein synthesis by inhibiting translation initiation via eIF2α phosphorylation (Anderson and Kedersha, 2008). Here, we show nucleocytoplasmic transport disruption upon stress, suggesting an alternative mechanism by which cells halt their protein synthesis. Indeed, a prior study has shown that stress suppresses the nuclear export of most mRNA (Saavedra et al., 1996). In contrast, since many stress-response proteins such as heat-shock proteins do not require eIF2α for their translation initiation, stress does not inhibit their translation (Thakor and Holcik, 2012). Furthermore, in accord with the cellular need for these proteins under stress, the export of their mRNAs is also selectively spared, due to specific nucleotide sequences that allow Ran-independent export. Hence, nucleocytoplasmic transport disruption is likely coupled with other cellular stress-response mechanisms.
While acute inhibition of nucleocytoplasmic transport may help cells cope with stress, chronic inhibition is likely detrimental. Indeed, loss of SG proteins Ataxin-2 or TIA-1 has been shown to suppress toxicity in yeast and animal models of ALS or tauopathies (Apicco et al., 2018; Elden et al., 2010; Kim et al., 2014). In addition, ASOs against Ataxin-2 have been shown to suppress SG assembly as well as neuronal toxicity in a TDP-43 transgenic ALS mouse model (Becker et al., 2017). In our study, SG inhibitors GSK, ISRIB or Ataxin-2 ASO suppress neurodegeneration in a C9-ALS fly model and iPSNs (Figure 7), further supporting critical roles for SG assembly and nucleocytoplasmic transport disruption in the pathogenesis of these diseases. Importantly, ISRIB has been shown to be neuroprotective in prion-diseased mice without deleterious side effects (Halliday et al., 2015), suggesting potential clinical translation. As SG assembly is a generic response to cytoplasmic protein misfolding, similar mechanisms may underlie the nucleocytoplasmic transport defects in other protein deposition diseases, including sporadic ALS and Huntington’s diseases, where mislocalization and aggregation of nucleoporins in the cytoplasm has been observed (Grima et al., 2017; Zhang et al., 2015). Hence, targeting SG assembly to prevent dysregulation of nucleocytoplasmic transport is a potential therapeutic approach for these diseases.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey D. Rothstein (jrothstein@jhmi.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Transformed human cells
HEK293T (of likely female origin due to lack of any trace of Y chromosome), SY5Y (of female origin), and U-2 OS cells (of female origin) were cultured in DMEM/F12 (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. For stressors, cells were cultured in the same media supplemented with 50 mM sodium arsenite or 0.4 M sorbitol. For KPT-350 experiments, 25 mM KPT-350 was dissolved in DMSO and added to cells to a final concentration of 25 μM. For KPT-350 treatment on S-GFP-expressing cells, after one-hour stress, the media was changed every 15 minutes together with fresh KPT-350 to wash out the stressors. All cells are maintained at 37°C in a hu midified incubator supplemented with 5% CO2. Transfection was performed using Lipofectamine 3000 (ThermoFisher). Cells were fixed in paraformaldehyde 24-48 hours post-transfection.
Human iPS cells
iPS cell generation and differentiation to motor neurons
iPS motor neurons were generated as previously described (Zhang et al., 2015). Briefly, patient fibroblasts were collected at Johns Hopkins Hospital with patient’s consent (IRB protocol: NA_00021979) as described previously. iPSC lines were created and initially characterized with an NIH-sponsored commercial agreement with iPierian (USA) using the four-vector method. Sox2, Oct4, Klf4 and c-Myc encoding vectors were transduced into human fibroblasts using retrovirus delivery. Selected colonies were evaluated for expression of multiple pluripotent markers by quantitative PCR (qPCR) and/or immunocytochemistry. In vitro pluripotency was further determined by three germ layer differentiation via embryoid body formation. iPSCs were maintained in mTeSR1 (StemCell Technology) and passed once a week using dispase (StemCell Technology) following the manufacturer’s instructions. Partially differentiated colonies were removed manually before differentiation analyses. The iPSCs were differentiated to neuroprogenitor cells, neurons and then motor neurons. At day 32 of differentiation, iPSC cells were treated with 20 μM Ara-C (Sigma) for 48 hours to eliminate iPS glial progenitor cells and enrich for iPSC neurons. iPSC neuronal cultures used for subsequent experiments were plated onto a confluent layer of mouse astrocytes. Media were changed every two days and cells were analyzed at day 50–60 of differentiation. Cells are maintained at 37°C in a humidified incubator supplemented with 5% CO2.
Sex, age, and the C9orf72 G4C2 expansion size of iPS cells
| Cell line ID | Line Name | Sex | Age | Estimated G4C2 repeat expansion |
|---|---|---|---|---|
| Control#1 | CS025 | Male | 76 | N/A |
| Control#2 | CS014 | Female | 52 | N/A |
| Control#3 | CS9XH7iCTR | Male | 57 | N/A |
| C9-ALS#1 | CS052 | Male | 49 | 6-8kb |
| C9-ALS#2 | JH034 | Female | 65 | >2.5kb |
| C9-ALS#3 | CS029 | Male | 47 | 6-8kb |
| C9-ALS#4 | CS030 | Female | 51 | 2.7kb |
Drosophila genetics
Flies were raised and maintained on yeast-cornmeal-syrup food at 25 °C unless otherwise indicated (see below). Stocks and crosses were transferred to new vials on a regular basis.
For subcellular localization of GFP, OK371-GAL4; UAS-(G4C2)30/TM6b, Tb, tub::GAL80 was crossed to UAS-NLS-NES(P12)/TM6b, Tb (III) and non-Tb offspring were selected for analysis (NES(P12) is referred to as NES).
For eye degeneration, GMR-GAL4, UAS-(G4C2)30/CyO, twi-GAL4, UAS-GFP were crossed to Canton-S flies, and GMR-GAL4, UAS-(G4C2)30/+ were selected from the offspring and aged at 25 °C for 15 days. Eye degeneration was quantified using a previously described method (Zhang et al., 2015). Briefly, points were added if there was complete loss of interommatidial bristles, necrotic patches, retinal collapse, loss of ommatidial structure, and/or depigmentation of the eye.
For the flight assay, elavGS (gene switch) was crossed to UAS-(G4C2)30, UAS-RanDN, or UAS-Nup50 RNAi. UAS/+; elavGS-GAL4/+ flies were selected and aged at 29 °C on regular fo od supplemented with DMSO, 300 μM RU486, or 300 μM RU486 and 5 μM GSK2606414 (GSK) or integrated stress response inhibitor (ISRIB). Flies were transferred to freshly made food every 2–3 days. After 15 days, individual female flies were dropped into a graduated cylinder through a hole at the center of its lid. The cylinder was graduated into 12 zones of 25 mm each (top: 0; bottom: 12). The landing height was noted as the zone number in which the fly landed.
For GR dot blot, hs-GAL4 were crossed to UAS-(G4C2)30; UAS-(G4C2)30/TM6, Tb, Hu. hs-GAL4/UAS-(G4C2)30; UAS-(G4C2)30/+ flies were selected and fed on food supplemented with DMSO, GSK, or ISRIB for 12 days at 25°C before moving to 37°C for one hour. The flies were then homogenized and whole tissue lysates were subjected to the dot blot.
METHOD DETAILS
Immunofluorescence staining and imaging
For cultured cells, cells were fixed in 4% paraformaldehyde for 20 min and then penetrated in PBS with 0.1% Triton X-100 for 10 min (for Exportin-1 antibody staining, this step is skipped), followed by blocking in wash buffer (PBS, 0.1% Tween-20, and 2mg/mL Heparin) supplemented with 3% donkey serum and 5% glycine for 1 h. After that, cells were incubated with primary antibodies in wash buffer with 3% donkey serum for 16 hours at 4 °C. Primary antibodies were used at 1:100 dilution. Next, cells were washed in wash buffer four times with 15 min each time at room temperature. Donkey secondary antibodies conjugated to Alexa Fluor 568, 488, and/or 633 (ThermoFisher) were used at 1:1,000 dilution in wash buffer with 3% donkey serum for 3 hours at room temperature. Cells were then washed in wash buffer four times for a total period of 1 h. For Drosophila salivary glands, tissues were dissected in pre-chilled PBS and fixed in 3.7% formaldehyde for 20 min, followed by penetration in PBS with 0.4% Triton X-100 (PBX) for 1 h. The tissues were then incubated with chicken anti-GFP (Abcam) at 1:1,000 dilution with 10% normal goat serum in PBX for 16 hours at 4 °C. After that, tissues were washed several times in PBX for a total period of 8 hours at room temperature and then incubated with goat secondary antibodies conjugated to Alexa Fluor 488 in PBX+10% NGS at 4 °C for 16 hours. Tissues were then washed several times in PBX for a total period of 6 hours at room temperature. Cells or tissues were mounted in ProLong Antifade Gold with DAPI and subjected to confocal microscopy analyses.
Fixed cells or tissues were analyzed under an LSM780 or LSM800 confocal microscope (Carl Zeiss) with their accompanying software using Plan Apochromat 63×, NA 1.4 objectives (Carl Zeiss) at room temperature. Images were captured by an AxioCam HRc camera (Carl Zeiss) and were processed using ImageJ/Fiji (National Institutes of Health). To quantify fluorescent or Western blot intensities, after opening the images in ImageJ/Fiji, certain areas/bands were circled and the intensities were measured. Experiments were repeated three to five times. Four to six fields of views were randomly selected for cell quantifications. Each field of view typically contains one to three cells.
Stress granule separation, co-IP assay, and immunoblot
Stress granules were separated as previously described with slight modifications (Wheeler et al., 2017). Cells were resuspended in lysis buffer (5 mM Tris-HCl pH 7.4, 100 mM KOAc, 2mM Mg(OAc)2, and 0.5% NP-40) supplemented with protease inhibitor cocktail (Complete, Roche), lysed by passing through a 25G needle 5 times, followed by centrifugation at 1,000 g for 5 min. The supernatant was further spun at 18,000 g for 20 min. After that, the pellet containing stress granules was washed and resuspended in lysis buffer.
For co-IP assays, cell lysate was precleared by protein A agarose beads for 30 min at room temperature and spun at 1,000 g for 5 min. The supernatant was incubated with GFP-TRAP A beads (ChromoTek) overnight at 4 °C. The beads were subsequently precipitated by centrifugation at 1,000 g for 5 min and washed in lysis buffer three times at 4 °C with 10 min each time. The beads were then resuspended in 50 μL lysis buffer and mixed with Laemmli buffer.
For immunoblot, the protein samples were heated in Laemmli buffer at 98 °C for 10 min. The protein samples were run on 4–15% SDS Mini-PROTEAN TGX Precast Gels (Bio-Rad) and transferred to nitrocellulose membranes. For dot blots, 2 μL protein samples were dotted on nitrocellulose membrane and then air-dried. TBST with 5% milk was used for blocking, except for the GR antibody that skipped this step. All primary antibodies were used at 1:1,000 dilution, except for mouse anti-β-Actin (Millipore) that was used at 1:5,000 dilution. The HRP-conjugated donkey secondary antibodies (Jackson ImmunoResearch) were used at 1:1,000 dilution, except for β-Actin and Importin α1, which was used at 1:5,000 dilution. All primary and secondary antibodies were diluted in TBST with 5% milk, except for the GR and phospho-eIF2α antibodies that are diluted in TBST.
Puromycin incorporation assay
Puromycin incorporation assay was performed as described (Kedersha et al., 2016). U-2 OS cells were treated with 10 μg/mL puromycin for 15 minutes prior to lysis. Whole cell lysates were analyzed by SDS-PAGE followed by Coomassie staining or Western Blot using a puromycin antibody (Millipore).
Drug treatment and/or feeding
GSK and ISRIB were added to cell culture media or fly food at indicated concentrations. For HEK293T cells, the chemicals were added 4 hours prior to fixation. For iPSNs, the chemicals were added 5 days prior to fixation and replenished every two days when changing the media. For flies, OK371-GAL4; UAS-(G4C2)30/TM6b, Tb, tub::GAL80 and UAS-NLS-NES(P12)/TM6b, Tb (III) were crossed on cornmeal-molasses-yeast fly food supplemented with the chemicals, and their offspring raised on the same food until third instar larval stage for GFP analysis. For adult flies used in eye degeneration and flight assays, newly-eclosed flies were raised and aged on food supplemented with the chemicals. Flies were transferred to freshly-made food every two days.
ASOs were added to iPSN media only once with a final concentration of 5 μM, 5 days prior to fixation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Student’s t-tests were used for comparisons between two samples. For multiple comparisons to the same sample, one-way ANOVA was used followed by Dunnett’s tests. For comparisons in the G3BP KO experiments with stressors, two-way ANOVA was used followed by Sidak’s tests (Figure 4D, S4C, D, E and G). Statistical analyses were performed using GraphPad Prism 7 software.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human Ataxin-2 | BD biosciences | Cat#611378; RRID: AB_398900 |
| Rabbit anti-human Ataxin-2 | Bethyl Laboratories | Cat#A301-118A; RRID: AB_2274451 |
| Mouse anti-human Ran | BD biosciences | Cat#610341; RRID: AB_397731 |
| Rabbit anti-human G3BP | Abcam | Cat#ab181149; |
| Mouse anti-Importin alpha 1 | BD biosciences | Cat#610485; RRID: AB_397855 |
| Mouse anti-human NTF97/Importin beta 1 | Abcam | Cat#ab2811; RRID: AB_2133989 |
| Mouse anti-Importin β2/Transportin 1 | Abcam | Cat#ab10303; RRID: AB_2206878 |
| Goat anti-Nup96 | Santa Cruz | Cat#sc-27400; RRID: AB_677310 |
| Rabbit anti-Nup50 | Abcam | Cat#ab137092; |
| Rabbit anti-Nup153 | Abcam | Cat#ab84872; RRID: AB_1859766 |
| Rabbit anti-Nup155 | Novus Biologicals | Cat#NBP-1-82959; RRID: AB_11021937 |
| Rabbit anti-Nup93 | Novus Biologicals | Cat#NBP-1-81546; RRID: AB_11037525 |
| Rat anti-Nup98 | Abcam | Cat#ab50610; RRID: AB_881769 |
| Mouse anti-Nup205 (for immunofluorescent staining) | Santa Cruz | Cat#sc-377047; |
| Rabbit anti-Nup205 (for Western Blot) | Abcam | Cat#ab157090; |
| Rabbit anti-Nup88 | Abcam | Cat#ab79785; RRID: AB_2042496 |
| Rabbit anti-Nup54 | Sigma | Cat#HPA-035929; RRID: AB_10671236 |
| Rabbit anti-Nup85 | Protein tech | Cat#19370-1-ap; RRID: AB_10859826 |
| Rabbit anti-Nup214 | Bethyl | Cat#A300-716A; RRID: AB_533409 |
| Rabbit anti-Nup358 | Bethyl | Cat#A301-796A; RRID: AB_1211503 |
| Rabbit anti-GP210 | Abcam | Cat#ab15601; RRID: AB_2236461 |
| Rabbit anti-NupL2 | Novus Biologicals | Cat#NBP2-31884; |
| Rabbit anti-POM121 | Thermo Fisher | Cat#PA5-36498; RRID: AB_2553555 |
| Rabbit anti-CRM1 | Novus Biologicals | Cat#NBP-2-16014; |
| Mouse anti-NUPL1 | Novus Biologicals | Cat#H00009818-M01; RRID: AB_1200057 |
| Rabbit anti-THOC2 | Sigma | Cat#HPA047921; RRID: AB_10960388 |
| Mouse anti-Gle1 | Santa Cruz | Cat#sc-514796; |
| Rabbit anti-TPR | Bethyl | Cat#IHC00099; RRID: AB_2206159 |
| Rabbit anti-Rae1 | MyBioSource | Cat#MBS9125380; |
| Rabbit anti-RanGEF/RCC1 | Sigma | Cat#HPA027574; RRID: AB_10601236 |
| Rat anti-Nup62 | Millipore | Cat#MABE1043; |
| Chicken anti-GFP | Abcam | Cat#ab13970; RRID: AB_300798 |
| Rabbit anti-RFP | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Guinea pig anti-MAP2 | Synaptic systems | Cat#188-004; RRID: AB_2138181 |
| Goat anti-human TIA-1 | Santa Cruz | Cat#sc-1751; RRID: AB_2201433 |
| Rabbit anti-human RanGAP1 | Santa Cruz | Cat#sc-25630; RRID: AB_2176978 |
| Mouse anti-β-Actin | Millipore | Cat#Mab1501; RRID: AB_2223041 |
| Rabbit anti-eIF2α | Cell signaling | Cat#9722; RRID: AB_2230924 |
| Rabbit anti-phospho-eIF2α (S51) | Cell signaling | Cat#9721; RRID: AB_330951 |
| Rabbit anti-human Lamin B1 | Abcam | Cat#ab16048; RRID: AB_443298 |
| Mouse anti-puromycin | Millipore | Cat#MABE343; RRID: AB_2566826 |
| Rabbit anti-GR | Proteintech | Cat#23978-1-AP; |
| GFP-TRAP® A | ChromoTek | Cat#gta-10; |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor | Roche | Cat#11873580001 |
| Lipofectamine 3000 | ThermoFisher | Cat#L3000015 |
| Sodium arsenite | Sigma | Cat#1062771000 |
| Sorbitol | Sigma | Cat#1617000 |
| PERK inhibitor GSK2606414 | Sigma | Cat#516535 |
| ISRIB | Sigma | Cat#SML0843 |
| M9M peptide | Peptide 2.0, Cansizoglu et al., 2007 | N/A |
| Control peptide: AHGDKVIVVDQSSNPKGFQFY ATAARTGKG | Peptide 2.0 | N/A |
| KPT-350 | Karyopharm Therapeutics/Sharon Tamir | https://www.karyopharm.com/ |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | Cat#CRL-3216; RRID: CVCL_0063 |
| Human: SY5Y | ATCC | Cat#CRL-2266; RRID: CVCL_0019 |
| Human: U-2 OS | ATCC | Cat#HTB-96; RRID: CVCL_0042 |
| Human: U-2 OS with G3BP1 and 2 double KO | This paper | N/A |
| Human: U-2 OS stably expressing G3BP1-GFP | Figley et al., 2014 | N/A |
| Human: iPS cell CS025, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Human: iPS cell CS014, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Human: iPS cell CS9XH7iCTR, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Human: iPS cell CS052, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Human: iPS cell JH034, see Figure S7 | Johns Hopkins Hospital | https://www.hopkinsmedicine.org/institute_cell_engineering/news/iPS_cells_Nobel.html |
| Human: iPS cell CS029, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Human: iPS cell CS030, see Figure S7 | Cedars-Sinai Induced Pluripotent Stem Cell (iPSC) Core | https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/ |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: OK371-GAL4; UAS-(G4C2)30/TM6b, Tb, tub::GAL80 | Zhang et al., 2015 | N/A |
| D. melanogaster: UAS-NLS-NES(P12)/TM6b, Tb (III) | Bloomington Drosophila Stock Center | BDSC: 7033; Flybase: FBst0007033 |
| D. melanogaster: GMR-Gal4, UAS-(G4C2)30/CyO, twi-GAL4, UAS-GFP | Zhang et al., 2015 | N/A |
| D. melanogaster. elavGS-GAL4 | Mizielinska et al., 2014 | N/A |
| D. melanogaster: UAS-(G4C2)30 | Xu et al., 2013 | N/A |
| D. melanogaster: UAS-RanDN: UAS-RanT24N | Cesario and McKim, 2011 | N/A |
| D. melanogaster. UAS-Nup50 RNAi | Bloomington Drosophila Stock Center | BDSC: 34580; Flybase: FBst0034580 |
| Oligonucleotides | ||
| Ataxin-2 siRNA | Qiagen | Cat#1027416 |
| Ataxin-2 ASOs | IONIS, Scoles et al., 2017 | N/A |
| Ran siRNA | Qiagen | Cat#GS5901 |
| Importin α1 siRNA | Qiagen | Cat#GS3838 |
| Importin β2 siRNA | Qiagen | Cat#GS3842 |
| Control siRNA | Qiagen | Cat#1027310 |
| Recombinant DNA | ||
| Plasmid: pcDNA6-His-V5-Ataxin-2 cDNA | Elden et al., 2010 | N/A |
| Plasmid: pEGFP-(GR or PR)50 | Wen et al., 2014 | N/A |
| Plasmid: pEGFP-TDP(cyto) a.k.a., tdp43-EGFP construct 4 | Yang et al., 2010 | Addgene plasmid #28197 |
| Plasmid: pLenti-NLS-tdTomato-NES | Zhang et al., 2015 | N/A |
| Plasmid: pNLS-NES-EGFP | Woerner et al., 2016 | N/A |
| Plasmid: pCS2-myc-MBP | Bernis et al., 2014 | N/A |
| Plasmid: pCS2-myc-MBP-M9M | Bernis et al., 2014 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Supplementary Material
Figure S1: Stress disrupts nucleocytoplasmic transport (related to Figure 1). (A) HEK293T cells co-transfected with S-tdTomato (red) and Ataxin-2 cDNA (bottom row) were stained with Ataxin-2 (green) and DAPI (blue), compared to cells transfected with S-tdTomato only (top row). Nuclear S-tdTomato was quantified on the right. (B) Cells transfected with Ataxin-2 cDNA (bottom row) were stained with Ran (red), Ataxin-2 (green), and DAPI (blue), compared to non-transfected cells (top row). Nuclear Ran was quantified on the right. In A and B, white lines outline the nuclei and stars indicate cytoplasmic mislocalization of tdTomato or Ran. (C) HEK293T cells expressing S-GFP were treated with 0.5 mM sodium arsenite or 0.4 M sorbitol for an hour, and then with 25 μM KPT-350 in fresh media without stressors for an hour (column 2 and 4), and stained with Ataxin-2 (red), GFP (green), and DAPI (blue), compared to no KPT-350 treatment (column 1 and 3). Nuclear S-GFP was quantified on the right. Experimental design summarized at top. (D) Arsenite-treated HEK293T cells expressing control (left column) or Ataxin-2 siRNA (right column) were stained with Ran (red), Ataxin-2 (green) and DAPI (blue). (E) Immunoblots showing the efficiencies of siRNA against Ran or Importin α1 or β2. (F) HEK293T cells co-expressing S-tdTomato (red) with control (left column), Ran (middle column), or Importin α1 (right column) siRNA were stained with Ataxin-2 (green) and DAPI (blue). Nuclear S-tdTomato was quantified at the bottom. Ctrl: control; N: nuclear; W: whole cell. Number of cells measured (n) for each condition indicated in graph. ns: not significant; *: p<0.05; **: p<0.01; ****: p<0.0001. Data are represented as mean ± SEM.
Figure S2: Nucleocytoplasmic transport factors localize to stress granules (related to Figure 2). (A) Arsenite- (top row) or sorbitol-treated (bottom row) HEK293T cells were stained with Ran (red), G3BP (left two columns) or TIA-1 (right two columns) (green), and DAPI (blue). Arrowheads indicate co-localization of G3BP or TIA-1 with Ran. (B) Arsenite-treated SY5Y (top row) and U-2 OS (bottom row) cells were stained with Ran (red), G3BP1 (green) and DAPI (blue). Arrowheads indicate co-localization of G3BP1 with Ran. (C) Quantification of nuclear Ran in Figure 2A. (D) Quantification of transport factors localized in arsenite-induced stress granules. N: nuclear; W: whole cell; SG: stress granule. (E) Control (left three columns) or sorbitol-stressed (right three columns) HEK293T cells were stained with nucleocytoplasmic transport factors (red), including Importin α1 (top row), β1 (row 2), β2 (row 3), Exportin-1 (row 4), POM121 (row 5), Nup205 (row 6), and THOC2 (bottom row), together with TIA-1 (green) and DAPI (blue). Arrowheads indicate co-localization of TIA-1 and nucleocytoplasmic transport factors. Ctrl: control; SG: stress granule; W: whole cell. Number of cells measured (n) for each condition indicated in graph and table. ns: not significant, ****: p<0.0001. Data are represented as mean ± SEM.
Figure S3: Nucleoporins (Nups) localize to stress granules (related to Figure 2). Arsenite-(top two rows) or sorbitol-treated (bottom two rows) HEK293T cells were stained with Nups (A and C), RanGAP1, RanGEF, or Lamin B1 (C) (red), together with TIA-1 (or Ataxin-2 for Nup96 and Lamin B1, green), and DAPI (blue). Arrowheads in (A) indicate co-localization of TIA-1 (or Ataxin-2) and Nups. (B) Arsenite-treated HEK293T cells were stained with transport factors (red) and DAPI (blue) without counter-staining of stress granule markers, as a control to exclude possibility of fluorescence bleed-through.
Figure S4: Stress granule assembly mediates nucleocytoplasmic transport defects caused by arsenite treatment (related to Figure 4). (A) Immunoblots of S-tdTomato, S-GFP, Ran, Importin α1, β1, and β2, Nup205, and β-actin in control, arsenite- or sorbitol-treated cells, and arsenite-treated cells pre-treated with 5 μM GSK or 2 μM ISRIB, quantified on the right. (B) Control (row 1 and 3) or G3BP1/2 double knockout (G3BP KO, row 2 and 4) U-2 OS cells treated with arsenite (top two rows) or sorbitol (bottom two rows) were stained with TIA-1 (left column), Ataxin-2 (middle column), and G3BP (right column). (C) Non-stressed or arsenite-treated control or G3BP KO cells were immunoblotted for phospho-eIF2α or eIF2α, quantified on the right. (D) Non-stressed or arsenite-treated control or G3BP KO cells were treated with 10 μM puromycin for 15 minutes and immunoblotted for puromycin or subjected to Coomassie staining, quantified on the right. (E) Immunoblots of S-tdTomato, Ran, Importin α1 and β2, Nup205, and β-actin in non-stressed or arsenite-treated control or G3BP KO cells, quantified on the right. (F and G): Quantification of Figure 4E and F, respectively. Ctrl: control; KO: G3BP KO. Number of cells measured (n) for each condition indicated in graph. ns: not significant, *: p<0.05; **: p<0.01; ***: p<0.001, ****: p<0.0001. Data are represented as mean ± SEM.
Figure S5: Dipeptide repeat proteins and cytoplasmic TDP-43 cause nucleocytoplasmic transport factors to localize to stress granules (related to Figure 5). HEK293T Cells expressing GFP-tagged 50 repeats of poly-GR (top row) or poly-PR (middle row), or cytoplasmic TDP-43 (TDP(cyto)) (bottom row) were stained with nucleocytoplasmic transport factors (red), including Importin α1 (A), β1 (B), or β2 (C), Exportin-1 (D), or POM121 (E), together with Ataxin-2 (green) and DAPI (blue). White arrowheads indicate co-localization of Ataxin-2 with nucleocytoplasmic transport factors. Yellow arrowheads in (E) indicate co-localization of POM121 with nucleolar GR or PR.
Figure S6: Stress granules mediate the nucleocytoplasmic transport defects caused by poly-GR or PR or TDP(cyto) (related to Figure 6). (A) Additional cells to Figure 6A. (B) Control or G3BP1/2 double knockout (G3BP KO) U-2 OS cells expressing GFP-tagged 50 repeats of GR (column 1 and 2) or PR (column 3 and 4), or cytoplasmic TDP-43 (TDP(cyto)) (column 5 and 6) were stained with TIA-1 (top row), Ataxin-2 (mid row) and DAPI (blue, bottom row). GFP were shown in green (bottom row). (C and D) Dot blots of the levels of GFP-tagged 50 repeats of GR or PR or TDP(cyto) in HEK293T cells upon GSK or ISRIB treatment (C), or in control and G3BP KO cells (D), quantified below. Ctrl: control; KO: G3BP KO. Experiments repeated n=3 times; ns: not significant, **: p<0.01. Data are represented as mean ± SEM.
Figure S7: GSK, ISRIB, and Ataxin-2 ASO suppresses Ran defects in C9-ALS (related to Figure 7). (A) iPSN lines used in the study. (B) Nuclear Ran quantification of C9-ALS lines (#2-4) treated with DMSO, 5 μM GSK, or 2 μM ISRIB. (C) Ataxin-2 ASOs used in this study. (D) Nuclear Ran quantification of C9-ALS lines (#2-4) treated with scrambled (Scr) or Ataxin-2 ASO #1. N: nuclear; W: whole cell. (E) Immunoblot of phospho-eIF2α and eIF2α in control and C9-ALS iPSNs, quantified below (n is number of experiments; m is number of measurements). (F) Dot blot of GR in control and C9-ALS flies fed with DMSO, GSK, or ISRIB, quantified below. (G) Flight assays for flies expressing RanDN or Nup205 RNAi. Ctrl: control Number of cells measured (n) for each condition indicated in graphs. (H–M) Original Western blots. ns: not significant, *: p<0.05; **: p<0.01; ***: p<0.001, ****: p<0.0001. Data are represented as mean ± SEM.
Highlights.
Stress granule inducers disrupts nucleocytoplasmic transport
Nucleocytoplasmic transport factors localize to stress granules
Dipeptide repeat proteins disrupt nucleocytoplasmic transport via stress granules
Inhibiting stress granule assembly suppresses neurodegeneration in C9-ALS/FTD
Acknowledgments
We thank Xiaopei Tang, Weibo Zhou, and Lin Xue for their technical support, Joshua Wheeler for insightful discussions and the following individuals for various constructs: Mark Hipp (S-GFP construct), Yuh Min Chook (MBP-M9M and MBP constructs), James Shorter (GFP-Importin β2 constructs), Xinmei Wen and Davide Trotti (dipeptide repeat proteins constructs). KPT-350 is from Karyopharm Therapeutics. We thank Adrian Isaacs, Kim McKim, and the Bloomington Drosophila Stock Center for fly stocks. K.Z. is a recipient of Milton Safenowitz Postdoctoral Fellowship. K.M.C. and J.C.G are recipients of F31 NRSA Awards. J.C.G is a recipient of NSF GRFP, Thomas Shortman, and Axol Science Scholarships. J.D.R. is supported by NIH (R01NS085207; R01NS099320; P01NS09911; R01NS094239; U24NS078736), Target ALS, Muscular Dystrophy Association, ALS Association, Answer ALS and The Robert Packard Center for ALS Research. T.E.L. is supported by NIH (R01 NS082563 and NS094239 and P30 NS050274), ALSA, Target ALS, and the Robert Packard Center for ALS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Conceptualization, K.Z., J.G.D., A.D.G., J.D.R., and T.E.L.; Methodology, K.Z., K.M.C., K.R., A.N.C., J.C.G., K.E.B., H.W., P.Y., and F.R.; Investigation, K.Z., K.E.B., J.P.T., J.D.R., and T.E.L.; Writing: Original Draft, K.Z., J.D.R., and T.E.L.; Writing: Review & Editing, K.Z., A.D.G., J.D.R., and T.E.L.; Funding acquisition: J.D.R. and T.E.L.; Supervision, J.D.R. and T.E.L.
Declaration of interests: J.P.T. received support from Inception Sciences. The remaining authors declare no competing interests.
Reference List
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Apicco DJ, Ash PEA, Maziuk B, LeBlang C, Medalla M, Al AA, Ferragud A, Botelho E, Ballance HI, Dhawan U, et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci. 2018;21:72–80. doi: 10.1038/s41593-017-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, III, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) J Med Chem. 2012;55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernis C, Swift-Taylor B, Nord M, Carmona S, Chook YM, Forbes DJ. Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol Biol Cell. 2014;25:992–1009. doi: 10.1091/mbc.E13-08-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstam L, Nriagu J. Molecular aspects of arsenic stress. J Toxicol Environ Health B Crit Rev. 2000;3:293–322. doi: 10.1080/109374000436355. [DOI] [PubMed] [Google Scholar]
- Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, Zhu X, Jordan LE, Scheuner D, Kaufman RJ, et al. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De DM, Jaspers T, Ryan VH, et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol Cell. 2017;65:1044–1055. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, De BG, Scheveneels W, Steyaert J, Cuijt I, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario J, McKim KS. RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J Cell Sci. 2011;124:3797–3810. doi: 10.1242/jcs.084855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Tarn WY. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson Y, Robinson AC, Liu X, Wu D, Troakes C, Rollinson S, Masuda-Suzukake M, Suzuki G, Nonaka T, Shi J, et al. Neurodegeneration in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9orf72 is linked to TDP-43 pathology and not associated with aggregated forms of dipeptide repeat proteins. Neuropathol Appl Neurobiol. 2016;42:242–254. doi: 10.1111/nan.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD. Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110(Pt 11):1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Suzuki T, Yasuda Y, Murata M, Katahira J, Yoneda Y. Identification of importin alpha1 as a novel constituent of RNA stress granules. Biochim Biophys Acta. 2010;1803:865–871. doi: 10.1016/j.bbamcr.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, Lagier-Tourenne C. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron. 2017;94:48–57. doi: 10.1016/j.neuron.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron. 2017;94:93–107. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JD, Herbin O, de la Hera B, Vidaurre OG, Moy GA, Sun Q, Fung HY, Albrecht S, Alexandropoulos K, McCauley D, et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nat Neurosci. 2015;18:511–520. doi: 10.1038/nn.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Radford H, Sekine Y, Moreno J, Verity N, le QJ, Ortori CA, Barrett DA, Fromont C, Fischer PM, et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015;6:e1672. doi: 10.1038/cddis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, III, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, Bonini NM. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Ito H, Hirano A, Fujita K, Wate R, Nakamura M, Kaneko S, Nakano S, Kusaka H. Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2009;68:1184–1192. doi: 10.1097/NEN.0b013e3181bc3bec. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron. 2017;95:808–816. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, Seganathy E, Kong D, Stochaj U. Identification of Novel Stress Granule Components That Are Involved in Nuclear Transport. PLoS One. 2013;8:e68356. doi: 10.1371/journal.pone.0068356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van BC, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Neumann M, Igaz LM, Kwong LK, Nakashima-Yasuda H, Kolb SJ, Dreyfuss G, Kretzschmar HA, Trojanowski JQ, Lee VM. Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta Neuropathol. 2007;113:543–548. doi: 10.1007/s00401-007-0221-x. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Iwasaka S, Yoneda Y, Takeyasu K, Yoshimura SH. Individual binding pockets of importin-beta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc Natl Acad Sci U S A. 2008;105:16101–16106. doi: 10.1073/pnas.0802647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Protter DS, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife. 2015;4:e05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steggerda SM, Paschal BM. Regulation of nuclear import and export by the GTPase Ran. Int Rev Cytol. 2002;217:41–91. doi: 10.1016/s0074-7696(02)17012-4. [DOI] [PubMed] [Google Scholar]
- Thakor N, Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2alpha-independent manner during stress. Nucleic Acids Res. 2012;40:541–552. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tan W, Whittle C, Qiu L, Cao L, Akbarian S, Xu Z. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ito H, Wate R, Ohnishi S, Nakano S, Kusaka H. Altered distributions of nucleocytoplasmic transport-related proteins in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Acta Neuropathol. 2006;112:673–680. doi: 10.1007/s00401-006-0130-4. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Stress disrupts nucleocytoplasmic transport (related to Figure 1). (A) HEK293T cells co-transfected with S-tdTomato (red) and Ataxin-2 cDNA (bottom row) were stained with Ataxin-2 (green) and DAPI (blue), compared to cells transfected with S-tdTomato only (top row). Nuclear S-tdTomato was quantified on the right. (B) Cells transfected with Ataxin-2 cDNA (bottom row) were stained with Ran (red), Ataxin-2 (green), and DAPI (blue), compared to non-transfected cells (top row). Nuclear Ran was quantified on the right. In A and B, white lines outline the nuclei and stars indicate cytoplasmic mislocalization of tdTomato or Ran. (C) HEK293T cells expressing S-GFP were treated with 0.5 mM sodium arsenite or 0.4 M sorbitol for an hour, and then with 25 μM KPT-350 in fresh media without stressors for an hour (column 2 and 4), and stained with Ataxin-2 (red), GFP (green), and DAPI (blue), compared to no KPT-350 treatment (column 1 and 3). Nuclear S-GFP was quantified on the right. Experimental design summarized at top. (D) Arsenite-treated HEK293T cells expressing control (left column) or Ataxin-2 siRNA (right column) were stained with Ran (red), Ataxin-2 (green) and DAPI (blue). (E) Immunoblots showing the efficiencies of siRNA against Ran or Importin α1 or β2. (F) HEK293T cells co-expressing S-tdTomato (red) with control (left column), Ran (middle column), or Importin α1 (right column) siRNA were stained with Ataxin-2 (green) and DAPI (blue). Nuclear S-tdTomato was quantified at the bottom. Ctrl: control; N: nuclear; W: whole cell. Number of cells measured (n) for each condition indicated in graph. ns: not significant; *: p<0.05; **: p<0.01; ****: p<0.0001. Data are represented as mean ± SEM.
Figure S2: Nucleocytoplasmic transport factors localize to stress granules (related to Figure 2). (A) Arsenite- (top row) or sorbitol-treated (bottom row) HEK293T cells were stained with Ran (red), G3BP (left two columns) or TIA-1 (right two columns) (green), and DAPI (blue). Arrowheads indicate co-localization of G3BP or TIA-1 with Ran. (B) Arsenite-treated SY5Y (top row) and U-2 OS (bottom row) cells were stained with Ran (red), G3BP1 (green) and DAPI (blue). Arrowheads indicate co-localization of G3BP1 with Ran. (C) Quantification of nuclear Ran in Figure 2A. (D) Quantification of transport factors localized in arsenite-induced stress granules. N: nuclear; W: whole cell; SG: stress granule. (E) Control (left three columns) or sorbitol-stressed (right three columns) HEK293T cells were stained with nucleocytoplasmic transport factors (red), including Importin α1 (top row), β1 (row 2), β2 (row 3), Exportin-1 (row 4), POM121 (row 5), Nup205 (row 6), and THOC2 (bottom row), together with TIA-1 (green) and DAPI (blue). Arrowheads indicate co-localization of TIA-1 and nucleocytoplasmic transport factors. Ctrl: control; SG: stress granule; W: whole cell. Number of cells measured (n) for each condition indicated in graph and table. ns: not significant, ****: p<0.0001. Data are represented as mean ± SEM.
Figure S3: Nucleoporins (Nups) localize to stress granules (related to Figure 2). Arsenite-(top two rows) or sorbitol-treated (bottom two rows) HEK293T cells were stained with Nups (A and C), RanGAP1, RanGEF, or Lamin B1 (C) (red), together with TIA-1 (or Ataxin-2 for Nup96 and Lamin B1, green), and DAPI (blue). Arrowheads in (A) indicate co-localization of TIA-1 (or Ataxin-2) and Nups. (B) Arsenite-treated HEK293T cells were stained with transport factors (red) and DAPI (blue) without counter-staining of stress granule markers, as a control to exclude possibility of fluorescence bleed-through.
Figure S4: Stress granule assembly mediates nucleocytoplasmic transport defects caused by arsenite treatment (related to Figure 4). (A) Immunoblots of S-tdTomato, S-GFP, Ran, Importin α1, β1, and β2, Nup205, and β-actin in control, arsenite- or sorbitol-treated cells, and arsenite-treated cells pre-treated with 5 μM GSK or 2 μM ISRIB, quantified on the right. (B) Control (row 1 and 3) or G3BP1/2 double knockout (G3BP KO, row 2 and 4) U-2 OS cells treated with arsenite (top two rows) or sorbitol (bottom two rows) were stained with TIA-1 (left column), Ataxin-2 (middle column), and G3BP (right column). (C) Non-stressed or arsenite-treated control or G3BP KO cells were immunoblotted for phospho-eIF2α or eIF2α, quantified on the right. (D) Non-stressed or arsenite-treated control or G3BP KO cells were treated with 10 μM puromycin for 15 minutes and immunoblotted for puromycin or subjected to Coomassie staining, quantified on the right. (E) Immunoblots of S-tdTomato, Ran, Importin α1 and β2, Nup205, and β-actin in non-stressed or arsenite-treated control or G3BP KO cells, quantified on the right. (F and G): Quantification of Figure 4E and F, respectively. Ctrl: control; KO: G3BP KO. Number of cells measured (n) for each condition indicated in graph. ns: not significant, *: p<0.05; **: p<0.01; ***: p<0.001, ****: p<0.0001. Data are represented as mean ± SEM.
Figure S5: Dipeptide repeat proteins and cytoplasmic TDP-43 cause nucleocytoplasmic transport factors to localize to stress granules (related to Figure 5). HEK293T Cells expressing GFP-tagged 50 repeats of poly-GR (top row) or poly-PR (middle row), or cytoplasmic TDP-43 (TDP(cyto)) (bottom row) were stained with nucleocytoplasmic transport factors (red), including Importin α1 (A), β1 (B), or β2 (C), Exportin-1 (D), or POM121 (E), together with Ataxin-2 (green) and DAPI (blue). White arrowheads indicate co-localization of Ataxin-2 with nucleocytoplasmic transport factors. Yellow arrowheads in (E) indicate co-localization of POM121 with nucleolar GR or PR.
Figure S6: Stress granules mediate the nucleocytoplasmic transport defects caused by poly-GR or PR or TDP(cyto) (related to Figure 6). (A) Additional cells to Figure 6A. (B) Control or G3BP1/2 double knockout (G3BP KO) U-2 OS cells expressing GFP-tagged 50 repeats of GR (column 1 and 2) or PR (column 3 and 4), or cytoplasmic TDP-43 (TDP(cyto)) (column 5 and 6) were stained with TIA-1 (top row), Ataxin-2 (mid row) and DAPI (blue, bottom row). GFP were shown in green (bottom row). (C and D) Dot blots of the levels of GFP-tagged 50 repeats of GR or PR or TDP(cyto) in HEK293T cells upon GSK or ISRIB treatment (C), or in control and G3BP KO cells (D), quantified below. Ctrl: control; KO: G3BP KO. Experiments repeated n=3 times; ns: not significant, **: p<0.01. Data are represented as mean ± SEM.
Figure S7: GSK, ISRIB, and Ataxin-2 ASO suppresses Ran defects in C9-ALS (related to Figure 7). (A) iPSN lines used in the study. (B) Nuclear Ran quantification of C9-ALS lines (#2-4) treated with DMSO, 5 μM GSK, or 2 μM ISRIB. (C) Ataxin-2 ASOs used in this study. (D) Nuclear Ran quantification of C9-ALS lines (#2-4) treated with scrambled (Scr) or Ataxin-2 ASO #1. N: nuclear; W: whole cell. (E) Immunoblot of phospho-eIF2α and eIF2α in control and C9-ALS iPSNs, quantified below (n is number of experiments; m is number of measurements). (F) Dot blot of GR in control and C9-ALS flies fed with DMSO, GSK, or ISRIB, quantified below. (G) Flight assays for flies expressing RanDN or Nup205 RNAi. Ctrl: control Number of cells measured (n) for each condition indicated in graphs. (H–M) Original Western blots. ns: not significant, *: p<0.05; **: p<0.01; ***: p<0.001, ****: p<0.0001. Data are represented as mean ± SEM.


