ABSTRACT
Mosaic interspecifically acquired alleles of the multiple transferable resistance (mtr) efflux pump operon correlate with increased resistance to azithromycin in Neisseria gonorrhoeae in epidemiological studies. However, whether and how these alleles cause resistance is unclear. Here, we use population genomics, transformations, and transcriptional analyses to dissect the relationship between variant mtr alleles and azithromycin resistance. We find that the locus encompassing the mtrR transcriptional repressor and the mtrCDE pump is a hot spot of interspecific recombination introducing alleles from Neisseria meningitidis and Neisseria lactamica into N. gonorrhoeae, with multiple rare haplotypes in linkage disequilibrium at mtrD and the mtr promoter region. Transformations demonstrate that resistance to azithromycin, as well as to other antimicrobial compounds such as polymyxin B and crystal violet, is mediated through epistasis between these two loci and that the full-length mosaic mtrD allele is required. Gene expression profiling reveals the mechanism of resistance in mosaics couples novel mtrD alleles with promoter mutations that increase expression of the pump. Overall, our results demonstrate that epistatic interactions at mtr gained from multiple neisserial species has contributed to increased gonococcal resistance to diverse antimicrobial agents.
KEYWORDS: Neisseria gonorrhoeae, antibiotic resistance, efflux pump, epistasis, gonorrhea, macrolide
IMPORTANCE
Neisseria gonorrhoeae is the sexually transmitted bacterial pathogen responsible for more than 100 million cases of gonorrhea worldwide each year. The incidence of resistance to the macrolide azithromycin has increased in the past decade; however, a large proportion of the genetic basis of resistance remains unexplained. This study is the first to conclusively demonstrate the acquisition of macrolide resistance through mtr alleles from other Neisseria species, demonstrating that commensal Neisseria bacteria are a reservoir for antibiotic resistance to macrolides, extending the role of interspecies mosaicism in resistance beyond what has been previously described for cephalosporins. Ultimately, our results emphasize that future fine-mapping of genome-wide interspecies mosaicism may be valuable in understanding the pathways to antimicrobial resistance. Our results also have implications for diagnostics and public health surveillance and control, as they can be used to inform the development of sequence-based tools to monitor and control the spread of antibiotic-resistant gonorrhea.
INTRODUCTION
The causal agent of gonorrhea, Neisseria gonorrhoeae, is a Gram-negative diplococcus and an exclusively human pathogen. The prevalence of N. gonorrhoeae with resistance to azithromycin has dramatically increased in recent years from just 0.6% in 2013 to 3.6% in 2016 in the United States (1), 0.8% in 2013 to 4.7% in 2016 in England and Wales (2), and 5.4% in 2013 to 7.1% in 2015 across Europe (3). Additionally, reports in 2015 from both China and Japan have documented resistance in as high as 30% of the gonococcal population in some regions (4). This spike in resistance is alarming, as azithromycin is one of the two first-line drugs, in conjunction with ceftriaxone, recommended as dual-antimicrobial therapy for uncomplicated cases of gonococcal infection by the Centers for Disease Control and Prevention (CDC) (1, 5). Azithromycin is a macrolide antibiotic that inhibits protein synthesis by binding to the 23S rRNA component of the 50S ribosome, and while the majority of resistance can be explained by mutations in the 23S rRNA azithromycin binding sites (C2611T and A2059G) (6–8), the genetic basis of a large fraction of resistance in the U.S. population is still unexplained (6), thus limiting the potential for development of molecular biology-based resistance diagnostics and restricting our understanding of the evolutionary paths to resistance.
Gonococci are adept at acquiring chromosomally encoded antimicrobial resistance determinants as a result of their natural competence for transformation, allowing for the spread of resistance and other adaptively advantageous alleles between lineages and even across species boundaries (9–12). Extensive intragenus gene exchange has led to the concept of Neisseria as a consortium of species interconnected by allele sharing, with “fuzzy” borders permitting rapid access to new adaptive solutions (12, 13). This produces genetic mosaicism within particular lineages, whereby some loci are the products of horizontal gene transfer and homologous recombination from other species. In gonococci, intragenus recombination is an important source of novel genetic variation with many observations of mosaic loci gained from other Neisseria species (14–18). However, aside from horizontal gene transfer facilitating the evolution of resistance to third-generation cephalosporins through acquisition of mosaic penA (8, 15), allelic exchange has not yet been demonstrated to be the basis for resistance to any other antibiotic class in this species.
Recent epidemiological studies from the United States, Canada, and Australia have reported an association between mosaic multiple transferable resistance (mtr) efflux pump alleles and increased resistance to azithromycin (8, 18–20). Mosaics at mtr appear to have originated through horizontal gene exchange from other Neisseria and have been identified by high sequence homology of the repressor of the pump (MtrR) to Neisseria meningitidis and Neisseria lactamica. Mosaic mtr alleles have previously been associated with an outbreak of azithromycin resistance in Kansas City, Missouri, from 1999 to 2000 (21, 22), and with the majority of azithromycin resistance reported in New South Wales, Australia (20). While correlation between mosaic mtr and azithromycin resistance suggests causality, there is little experimental evidence to confirm the association.
The Mtr efflux pump is comprised of the MtrC-MtrD-MtrE cell envelope proteins, which together export diverse hydrophobic antimicrobial agents such as antibiotics, nonionic detergents, antibacterial peptides, bile salts, and gonadal steroidal hormones from the cell (23–27). Overexpression of mtrCDE has been linked to Mtr-mediated resistance to diverse antimicrobial agents through increased drug export and also higher in vivo fitness in the murine genital tract (24, 28). Known mutations that alter expression of the pump include the mtrC120 substitution, an adenine-to-guanine transition located 120 bp upstream of the mtrC start codon which acts as an alternative promoter for mtrCDE (29); deletion of a single adenine (A deletion) in the mtrCDE promoter that has been shown to repress the transcription of mtrR while simultaneously enhancing transcription of mtrCDE (30); and mutations that abrogate the function of MtrR by inducing premature stop codons or radical amino acid substitutions in the DNA-binding motif (21, 31, 32). Finally, the presence or absence of a Correia element with an integration host factor (IHF) binding site inserted between mtrCDE and the mtrCDE promoter has also been shown to negatively impact mtrCDE transcription in N. meningitidis, perhaps through the formation of DNA secondary structure, which could act as a weak transcriptional terminator (33). However, it is unclear if resistance in mtr mosaics is derived from any of these mechanisms and alteration of mtrCDE transcription.
Here, we used a combination of population genomic and experimental approaches to dissect the mechanism of resistance in mosaics. We first assessed patterns of allelic diversity within the gonococcal population to define the boundaries of horizontal gene transfer at mtrRCDE and found that the entire mtr region is a hot spot of interspecies recombination which has introduced multiple rare and divergent mosaic alleles, likely from N. meningitidis and N. lactamica, into the gonococcal population. Strong linkage disequilibrium at mtrD and the mtr promoter region suggested the maintenance of epistatic allelic combinations; therefore, we tested for interaction effects within and between mtr loci via transformation. We discovered epistatic interactions across almost the entire mtrD gene, and also between mosaic mtrD and mosaic mtr promoter regions, that synergistically increased resistance to azithromycin and other substrates of the Mtr efflux pump. Furthermore, patterns of nucleotide diversity in this region coupled with experimental in vitro evidence suggest antibiotic-mediated selection may be acting to maintain these epistatic interactions at the population level in nature. Finally, we tested for regulatory evolution of pump components, as previous mechanisms of azithromycin resistance through the Mtr efflux pump have been demonstrated to be driven by expression. Our results support that inheritance of mosaic promoter regions increases the expression of mtrCDE while gaining mosaic mtrD alone does not. Thus, the likely mechanism of resistance in mosaics is a structural change to MtrD, which enhances the capacity of the protein to recognize or transport diverse antimicrobial agents, coupled with increased efflux through the amplified production of pump components.
RESULTS
Allelic diversity suggests increased interspecies admixture at mtrRCDE.
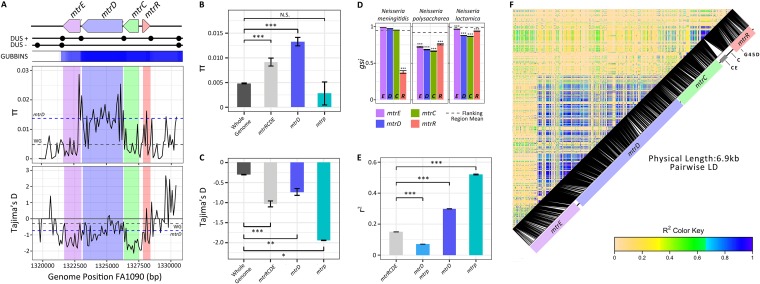
To gain insight into the evolutionary history of the mtrR transcriptional repressor and the mtrCDE pump, we analyzed patterns of diversity using the 1,102 Gonococcal Isolate Surveillance Project (GISP) isolates described by Grad et al. (8, 34). A significant increase in allelic diversity was observed across mtrRCDE compared to the entire genome, with the highest diversity at mtrD (Fig. 1A and B; see Table S1 in the supplemental material). We also detected a significant enrichment of rare alleles in the population across mtrRCDE (Fig. 1A and C; Table S1). Linkage disequilibrium was strongest at mtrD and the mtr promoter region in a comparison of all pairs of single-nucleotide polymorphisms (SNPs) within mtrRCDE, with the highest linkage observed at pairs of variant sites within each of these loci (Fig. 1E and F; Table S1).
FIG 1 .
Horizontal gene transfer (HGT) of mtr introduces novel adaptive genetic variation into Neisseria gonorrhoeae. (A) The FA1090 genomic position of the mtrRCDE locus is presented, with the locations of DNA uptake sequences (DUS) displayed (black circles). These DUS sequences are shared with closely related Neisseria species and increase the frequency of DNA uptake and recombination (58). The U.S. gonococcal population (n = 1,102 isolates) shows patterns of elevated allelic diversity as measured by π and elevated SNP densities as predicted by Gubbins (52) across mtrRCDE. Depressed values of Tajima’s D (Tajima’s D < 0) indicate an excess frequency of rare mutations across this region. The average values for these parameters are plotted for mtrD (blue dashed lines) and the genome-wide average (gray dashed lines). (B) The highest allelic diversity within mtrRCDE was found at mtrD and (C) an excess of rare alleles was detected across mtrRCDE (Tajima’s D < 0) compared to the rest of the genome. The values that are significantly different are indicated by a bar and asterisks as follows: *, P less than or equal to 0.05; **, P less than or equal to 0.001; ***, P less than or equal to 0.0001. Values that are not significantly different (N.S.) are indicated. mtrp, mtr promoter. (D) Depressed genealogical sorting index (gsi) values compared to those of a 25-kb mtr flanking region indicate extensive admixture of alleles between N. gonorrhoeae and other Neisseria across all mtr loci. Signatures of high allelic diversity, an excess of rare mutations, and interspecies admixture taken altogether suggest the recent introduction of novel genetic variation into this region. (E) Linkage disequilibrium was also observed, with the highest linkage measured by r2 occurring within mtrD and the mtr promoter regions. (F) r2 values plotted across mtrCDE in reference to their position in strain FA1090 are represented from 0 (yellow for low linkage) to 1 (blue for high linkage). Positions of the Correia element insertion (CE), A-to-C transversion in the mtrR promoter inverted repeat (C), and glycine to aspartic acid MtrR amino acid substitution (G45D) are indicated. Large tracts of high linkage suggest recent recombination of alleles from another source or maintenance of particular haplotypes due to selection. LD, linkage disequilibrium.
Population genetics statistics calculated for all mtr loci. Download TABLE S1, XLSX file, 0.04 MB (45.2KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To define interspecific admixture events within Neisseria, we characterized the genealogical sorting index (gsi [35]) to explore gene tree topology measures of species-specific phylogenetic exclusivity. gsi ranges from 0 (no exclusivity) to 1 (monophyletic) and serves as a metric to assess allele sharing that may arise through interspecific recombination or incomplete divergence from a recent split between species. We calculated gsi values for genes in a 25-kb window encompassing mtrRCDE with conserved microsynteny between N. gonorrhoeae and other Neisseria species (see Fig. S1 in the supplemental material). This region included 29 core genes excluding mtrRCDE. To define the region-specific gsi background, we calculated gsi values across 100 bootstrap replicates for each gene in the mtrRCDE flanking region by species. The mean gonococcal gsi values for flanking genes were 0.95 for N. meningitidis, 0.99 for N. lactamica, and 0.92 for Neisseria polysaccharea (Fig. 1D; Table S2). Significant reductions in gsi values were detected across mtrRCDE compared to the 29 loci within the surrounding 25-kb region (Fig. 1D). Significant allele sharing between N. gonorrhoeae and N. meningitidis was exclusively observed at mtrR (Fig. 1D; Table S2), while significant allele sharing between N. gonorrhoeae and both N. lactamica and N. polysaccharea occurred across all mtr loci (Fig. 1D; Table S2).
Fully sequenced, annotated genomes for N. gonorrhoeae (top), N. lactamica (middle), and N. meningitidis (bottom) were aligned with progressiveMauve (54). Syntenic blocks between these three species are illustrated by colored rectangles. For instance, the purple rectangles above the red arrows show a large (~100-kb) collinear genomic segment shared among the three Neisserial species. This syntenic block contains the mtr operon (positions 1,323,095 to 1,326,298). Download FIG S1, TIF file, 79.5 MB (81.4MB, tif) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genealogical sorting index (gsi) values as a measure of genealogical exclusivity and admixture. Download TABLE S2, XLSX file, 0.04 MB (44KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
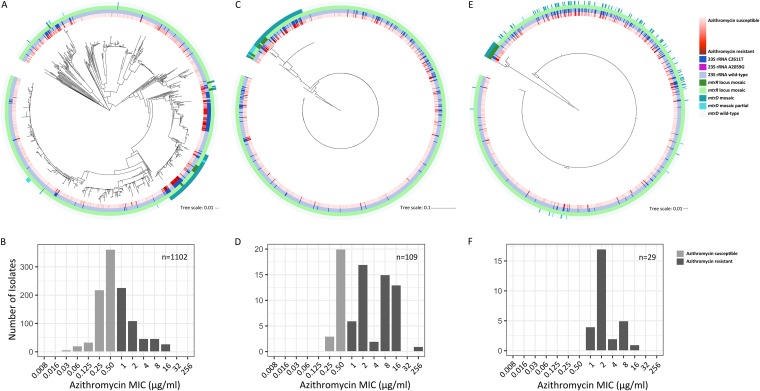
There were multiple recombined mosaic haplotypes present in the gonococcal population carrying the full-length mtrD (n = 80), mtrRCD (n = 9), and mtrRCDE (n = 20) genes and some isolates with partial mosaic mtrD with the majority of the gene homologous to native gonococcal sequence (n = 13) (Fig. 2). Of the 109 isolates with full-length mosaic mtrD, 4 isolates had mosaic mtrD genes that were 99% identical to the N. meningitidis gene, 5 isolates had mosaic mtrD alleles with 94 to 96% identity to the N. lactamica gene, and the remainder had alleles that aligned equally well to N. meningitidis and N. lactamica with identities ranging from 91 to 92%. Of the 29 isolates with mosaic promoter regions identified by Grad et al. (8), 24 isolates had mosaic promoter regions that were 96 to 98% identical to those in N. lactamica, 4 had regions that were 99% identical to the N. meningitidis region with the presence of a 153-bp Correia element insertion (21), and 1 isolate had a region that was 92% similar to the N. meningitidis region but lacked the Correia element that was present in the other 4 N. meningitidis-like isolates. While we find the highest sequence similarity of mosaic sequences to N. meningitidis and N. lactamica in the NCBI database, the paucity of sequences representing commensal Neisseria presents an issue with conclusive species annotation. However, it is clear that mosaic alleles are highly divergent from the native gonococcus-like alleles, which are relatively monomorphic. All isolates with full-length mosaic mtrD had azithromycin MICs of ≥0.25 µg/ml, while all isolates with full-length mosaic mtrD and a mosaic mtr promoter had azithromycin MICs of ≥1 µg/ml (Fig. 2).
FIG 2 .
Divergent mtrD and mtr promoter haplotypes are associated with increases in azithromycin MIC. (A) A maximum likelihood whole-genome-sequence phylogeny of 1,102 Neisseria gonorrhoeae isolates based on single-nucleotide polymorphisms generated from mapping to the FA1090 reference genome (8). The inner annotation ring shows azithromycin MICs on a continuous scale (red shading). The next annotation ring indicates isolates with at least two copies of the C2611T 23S rRNA mutation (dark blue) or isolates with four copies of the A2059G 23S rRNA mutation (magenta). The next annotation ring shows isolates that were identified as interspecies mosaics based on their sequence at mtrR by Grad et al. (8) (dark green). The outermost annotation ring shows isolates identified as mtrD mosaics in this study (teal). Tree scales or bars, 0.01 or 0.1 nucleotide substitutions per position. (B) The entire gonococcal population has a distribution of azithromycin MIC values which fall both above and below the defined resistance threshold (MIC ≥ 1 µg/ml). (C) A maximum likelihood phylogeny built on mtrD alignments shows 109 isolates with full-length mosaic alleles that are highly divergent from the wild-type gonococcal mtrD allele. (D) These 109 isolates are associated with elevated azithromycin MICs (MICs ≥ 0.25 µg/ml). (E) A maximum likelihood phylogeny built on the mtr promoter region shows that all 29 of the mtr promoter mosaics previously described by Grad et al. (8) also have inherited mosaic mtrD. (F) Isolates with both full-length mosaic mtrD and a mosaic mtr promoter region alleles have higher azithromycin MICs (MICs ≥ 1 µg/ml) than those isolates with mosaic mtrD alone.
Of the 29 mtr mosaics described by Grad et al. (8) and identified by a mosaic mtr promoter sequence, none had the mtrC120 substitution, A deletion, 23S rRNA mutation A2059G, mutations in rplD, rplV tandem duplications, or variants of the rRNA methylase genes ermC and ermB that have been associated with or experimentally confirmed to be involved in azithromycin resistance (7, 8, 19, 24, 29–32). However, four mosaic isolates had the premature stop codon mutations in mtrR, and five had the C2611T 23S rRNA mutation (6). While most of the isolates with full-length mosaic mtrD and a mosaic mtr promoter had azithromycin MICs ranging from 1 to 4 µg/ml, those with the mtrR premature stop codon or the C2611T 23S rRNA mutation had higher MICs ranging from 8 to 16 µg/ml, most likely due to the additive or epistatic effects of inheriting multiple mechanisms of resistance.
Epistasis between multiple mtr loci and within mtrD.
We exploited the natural competence of Neisseria to explore the potential for mosaic mtr alleles to produce an increase in resistance to azithromycin by transforming susceptible strains with either genomic DNA (gDNA) or PCR-amplified products from mosaic donors. Susceptible recipient strains for transformations included 28Bl (21, 36, 37), GCGS0353, and GCGS0465 (MIC ≤ 0.125 µg/ml; Table 1). Three strains with reported mosaic mtr alleles and azithromycin MICs of ≥1 µg/ml were selected as donors for DNA transfer (Table 1). These isolates included GCGS0276, GCGS0834, and GCGS0402. Of these mosaics, GCGS0276 had an mtrR sequence with the highest percent sequence identity to N. meningitidis isolates in the NCBI database, while GCGS0834 and GCGS0402 had mtrR genes most similar to N. lactamica isolates. Thus, we refer to these isolates as N. meningitidis-like and N. lactamica-like mtr mosaics, respectively. None of the selected donor strains had premature stop codons in mtrR or the C2611T mutation.
TABLE 1 .
Properties of strains used in the study
| Strain | Sourcea | MIC (µg/ml)b |
|||
|---|---|---|---|---|---|
| AZI | CV | PB | TX-100 | ||
| GCGS0276 | GISP isolate; Kansas City, MO | 1 | 8 | 1,000 | 16,000 |
| GCGS0402 | GISP isolate; Miami, FL | 2 | 8 | 500 | >16,000 |
| GCGS0834 | GISP isolate; Los Angeles, CA | 2 | 8 | 500 | >16,000 |
| GCGS0353 | GISP isolate; Dallas, TX | 0.03 | 1 | 60 | <250 |
| GCGS0465 | GISP isolate; Phoenix, AZ | 0.06 | 0.5 | 125 | <250 |
| 28Bl | Disseminated gonococcal infection isolate (CDC, 1974) |
0.125 | 4 | 60 | <250 |
GISP isolates are from the Gonococcal Isolate Surveillance Project. The geographical location where the specimen was collected is shown.
AZI, azithromycin; CV, crystal violet; PB, polymyxin B; TX-100, Triton X-100. MIC values are the modes from three independent replicate tests.
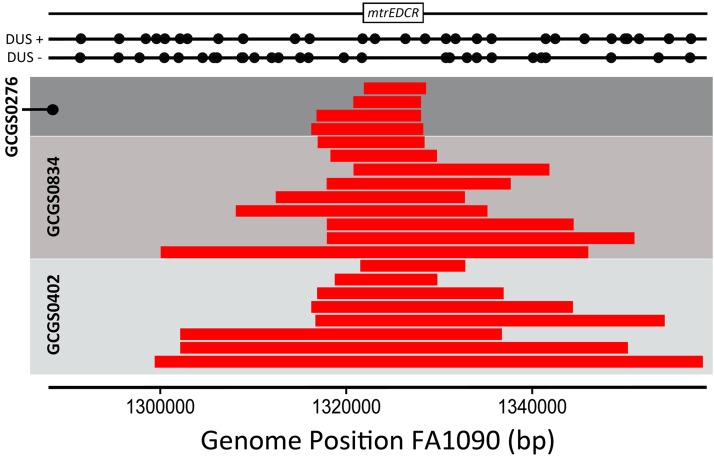
Genomic DNA from Neisseria GCGS0276, GCGS0402, and GCGS0834 donors was able to transform multiple susceptible recipient isolates to resistance on selective media containing azithromycin (Table 2, genomic DNA transformant strains). To identify the locus responsible, we sequenced the genomes of 28Bl cell lines transformed with gDNA from mosaic donors and characterized SNPs that had been inherited from donor strains that were not present in the 28Bl recipient. The only common region that had been inherited across all transformants contained the mtrRCDE locus (Fig. 3). Likewise, the susceptible GCGS0353 and GCGS0465 cell lines transformed with gDNA from mosaic donors and subsequently selected for resistance to azithromycin were also confirmed to have inherited mosaic mtrD and mtr promoter alleles (Table 2, genomic DNA transformant strains). Thus, we further characterized this region using both population genomic and experimental approaches to carefully detail the genetic basis of resistance at mtr in mosaic isolates.
TABLE 2 .
MIC values of transformant strains
| Strain | Recipient strain |
Transformation substrate | MIC (µg/ml)a |
Mosaic mtrDb |
Mosaic mtrpb |
|||
|---|---|---|---|---|---|---|---|---|
| AZI | CV | PB | TX-100 | |||||
| Genomic DNA (gDNA) transformant strains | ||||||||
| 28BlΔGCGS0276-gDNA | 28Bl | GCGS0276 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| GCGS0353ΔGCGS0276-gDNA | GCGS0353 | GCGS0276 gDNA | 1 | 16 | 1,000 | >16,000 | + | + |
| GCGS0465ΔGCGS0276-gDNA | GCGS0465 | GCGS0276 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| 28BlΔGCGS0402-gDNA | 28Bl | GCGS0402 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| GCGS0353ΔGCGS0402-gDNA | GCGS0353 | GCGS0402 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| GCGS0465ΔGCGS0402-gDNA | GCGS0465 | GCGS0402 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| 28BlΔGCGS0834-gDNA | 28Bl | GCGS0834 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| GCGS0353ΔGCGS0834-gDNA | GCGS0353 | GCGS0834 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| GCGS0465ΔGCGS0834-gDNA | GCGS0465 | GCGS0834 gDNA | 2 | 16 | 500 | >16,000 | + | + |
| PCR product transformant strains | ||||||||
| 28BlΔGCGS0276-mtrRpCDE | 28Bl | GCGS0276 mtrRpCDE | 1 | 16 | 500 | >16,000 | + | + |
| 28BlΔGCGS0276-mtrD | 28Bl | GCGS0276 partial mtrC (1185–1241 bp) and mtrD (1–3174 bp) |
0.5 | 8 | 250 | >16,000 | + | − |
| 28BlΔGCGS0276-mtrD/18-3174 | 28Bl | GCGS0276 partial mtrD (18–3174 bp) | 0.5 | 8 | 250 | 16,000 | + | − |
| 28BlΔGCGS0276-mtrD/+262-2724 | 28Bl | GCGS0276 partial mtrC (1185–1241 bp) and mtrD (1–2724 bp) |
0.5 | 8 | 250 | >16,000 | + | − |
| 28BlΔGCGS0276-mtrDCp | 28Bl | GCGS0276 mtrD, mtrC, mtr promoter | 1 | 16 | 500 | >16,000 | + | + |
| 28BlΔGCGS0402-mtrRpCDE | 28Bl | GCGS0402 mtrRpCDE | 2 | 16 | 500 | >16,000 | + | + |
| 28BlΔGCGS0402-mtrDCp | 28Bl | GCGS0402 mtrD, mtrC, mtr promoter | 2 | 16 | 500 | >16,000 | + | + |
AZI, azithromycin; CV, crystal violet; PB, polymyxin B; TX-100, Triton X-100. MIC values are the modes from three independent replicate tests.
The presence (+) of mosaic alleles was screened for by Sanger or whole-genome sequencing.
FIG 3 .
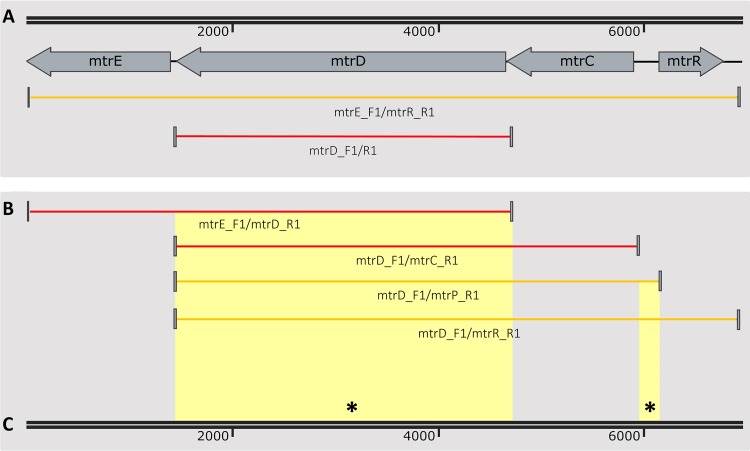
Unguided genomic DNA transformations demonstrate that the mosaic mtrRCDE region causes increased azithromycin resistance. The 28Bl azithromycin-susceptible recipient strain was transformed with gDNA from mosaic mtr donor strains and selected on plates containing 0.38 to 1 µg/ml azithromycin. Genomic sequencing revealed the boundaries of recombined DNA that azithromycin-resistant transformants had inherited from mosaic donor strains (red bars) in the 28Bl background, as identified by SNP homology using the FA1090 reference genome as a scaffold. The only genomic region that was consistently inherited from donor strains in the transformant cell lines encompassed mtrRCDE for both the N. meningitidis (GCGS0276) and N. lactamica-like (GCGS0402 and GCGS0834) mtr mosaics. Average in vitro recombination tract lengths were 9.5 ± 1.4 kb for strain GCGS0276 (n = 4), 24.3 ± 3.6 kb for GCGS0834 (n = 9), and 31.4 ± 6.0 kb for GCGS0402 (n = 8). The locations of all DNA uptake sequences (DUS) (black circles), as well as the mtr locus (white box), are mapped in reference to the FA1090 strain.
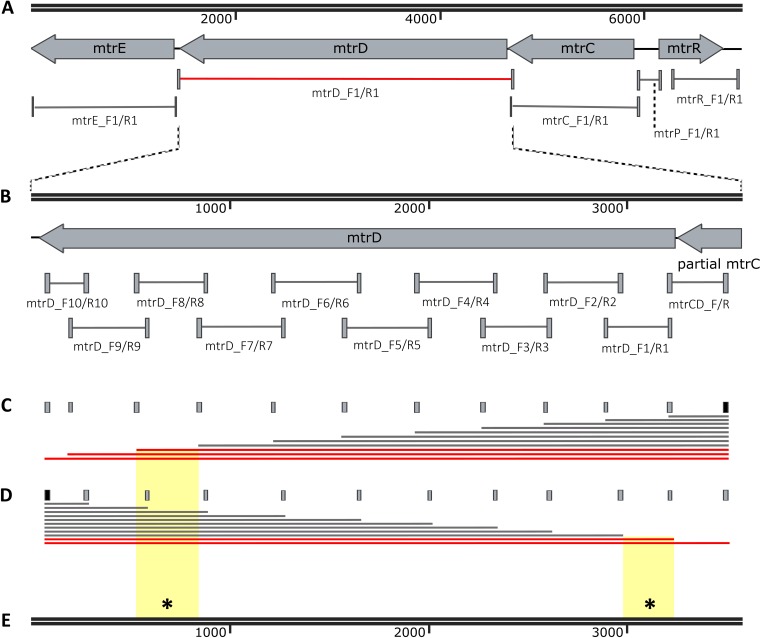
Primers were designed to amplify each locus in mtr for both a N. meningitidis-like mosaic (GCGS0276) and a N. lactamica-like mosaic (GCGS0402), and the resultant amplicons were then used to transform strain 28Bl (Fig. 4A; Table S3). For strain GCGS0276, the only locus within the mtrRCDE operon that was found to increase resistance to azithromycin independently was mtrD (Fig. 4A; Table 2, PCR product transformant strains). GCGS0276 mtrD in the 28Bl background raised the azithromycin MIC threefold from 0.125 to 0.5 µg/ml, yet no single region of mtrD was able to produce the 0.5 µg/ml phenotype independently (Fig. 4B). Population genomics results indicated the presence of high linkage disequilibrium across mtrD (Fig. 1E and F). Thus, to test for possible within-gene epistatic interaction effects, we designed a series of amplicons that incorporated increasingly larger fragments of the mtrD locus (Fig. 4C and D). Amplicons that contained both the 5′ end (18 to 356 bp) and 3′ end (2356 to 2724 bp) of GCGS0276 mtrD were together sufficient to increase the 28Bl azithromycin MIC to 0.5 µg/ml (Fig. 4E). There were four changes at the amino acid level between GCGS0276 and 28Bl in these regions, two in the PN1 domain of MtrD (I48T and G59D) and two in the PC2 domain (K823D and F854L) (Fig. 5), and in total, 20 amino acid changes between the GCGS0276 and 28Bl proteins (Fig. 5). Unlike the N. meningitidis-like mtrD allele from GCGS0276, the N. lactamica-like GCGS0402 mtrD allele was not able to produce resistance to azithromycin in the 28Bl background above the 0.38-µg/ml-concentration selection plates.
FIG 4 .
Epistatic interactions between multiple domains of mtrD contribute to elevated azithromycin MICs. (A) GCGS0276 mtrD in the 28Bl background elevated the azithromycin MIC from 0.125 µg/ml to 0.5 µg/ml (red). (B) Primer pairs designed to amplify ~300-bp fragments over the length of mtrD (see Table S4) resulted in no observed transformants on 0.38-µg/ml azithromycin selection plates, suggesting that multiple mutations across mtrD are needed for resistance. (C and D) To determine the regions that contributed to resistance, multiple fragment sizes were constructed by holding the rightmost forward primer (black) constant while adding different reverse primers (gray) (C) and by holding the leftmost reverse primer (black) constant while adding forward primers (gray) to separate reactions (D). The red lines in panels C and D indicate the PCR products that generated transformants on azithromycin selection plates. (E) At a minimum, SNPs at base pair positions 18 to 356 coupled with SNPs at positions 2356 to 2724 (indicated by yellow background and an asterisk) were required to raise the MIC from the 0.125 µg/ml of the recipient 28Bl strain to 0.5 µg/ml, though we are unable to exclude the possibility that additional SNPs between these two regions are also involved.
FIG 5 .
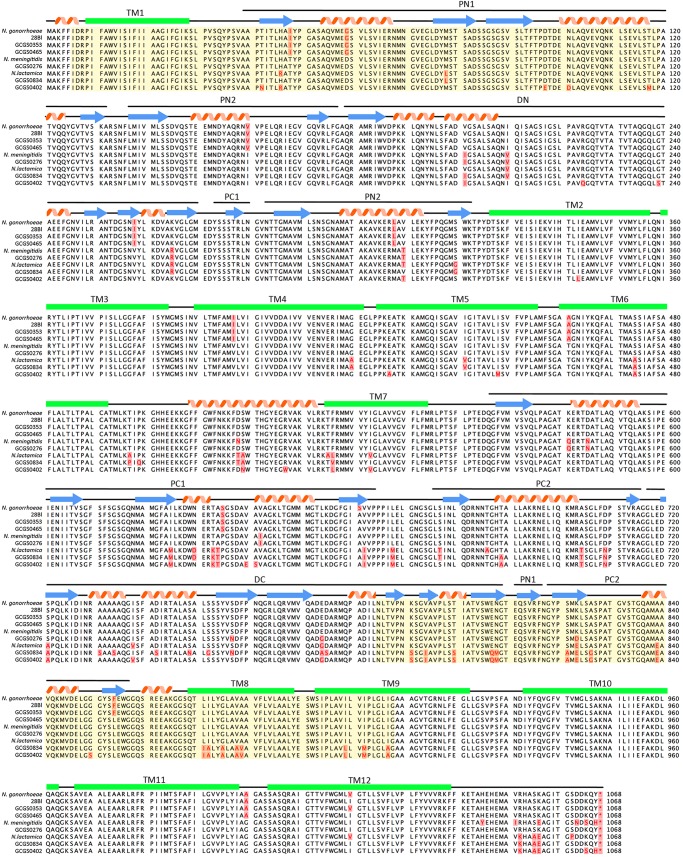
Alignment of wild-type and mosaic Neisseria gonorrhoeae MtrD with N. lactamica and N. meningitidis reference sequences. Amino acid alignment of MtrD sequences from N. gonorrhoeae FA1090 (AE004969.1), N. lactamica 020-06 (FN995097.1), and N. meningitidis NZ-05/33 (CP002424.1) with those of the gonococcal isolates used in this study are shown. The minor variants for each amino acid position are shown in red on a pink background. Though most of the amino acid changes in isolates with mosaic MtrDs are homologous to those found in either the chosen N. lactamica and N. meningitidis references, some amino acid changes are unique and thus have been either horizontally acquired from another source or represent de novo mutations. Overlaid on the alignment are the secondary structure elements of the protein: β pleated sheets (blue arrows); α-helices (red spirals); and transmembrane helices (TM) (green boxes) (40). Domains of MtrD are annotated and include the MtrE docking domain (DN and DC) and the MtrD pore domain (PN1, PN2, PC1, and PC2) (40). Epistatically interacting domains of MtrD that enhanced azithromycin MIC in mosaic isolates as deduced from transformation experiments are shown on yellow background.
Oligonucleotide primers and PCR conditions used for construction of the DNA templates for transformation and Sanger sequencing to confirm transformation. Download TABLE S3, XLSX file, 0.01 MB (10.8KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Though the mosaic GCGS0276 mtrD allele was clearly instrumental in enhancing resistance to azithromycin in strain 28Bl, it was not sufficient on its own to reproduce the level of resistance observed in the GCGS0276 donor strain. However, transformants that inherited the entire mtrRCDE operons of strains GCGS0276 and GCGS0402 had azithromycin MICs of 1 and 2 µg/ml, respectively, perfectly mirroring the donor strain resistance phenotypes (Table 2; Fig. 6A). Thus, in order to determine whether between-gene epistatic interactions resulted in donor strain resistance, amplicons were designed for each of these strains to amplify the mtrD locus in combination with other regions of the operon. Using this strategy, we found that for both strains GCGS0276 and GCGS0402, the donor strain azithromycin MICs of 1 and 2 µg/ml could be reproduced in strain 28Bl by transforming both mtrD and the mtr promoter region together (Fig. 6B; Table 2, PCR product transformant strains). Multiple mutations were present between isolates with mosaic and wild-type gonococcal mtr promoters; however, the only conserved mutation in all 29 mosaics was an A-to-C transversion in the mtrR promoter inverted repeat (Fig. 7).
FIG 6 .
Epistasis between mtrD and the mtr promoter region yields increased azithromycin resistance in mosaic isolates. (A) While transformation of the entire GCGS0276 mtrRCDE region perfectly reconstructed the donor strain MIC of 1 µg/ml in the 28Bl background (yellow line), GCGS0276 mtrD was the only region within mtrRCDE that could independently increase resistance to azithromycin (from 0.125 to 0.5 µg/ml [red line]). This suggested that epistasis across loci contributed to resistance in mtr mosaics. (B) To determine the additional locus needed to produce the high-level resistance seen in mtr mosaics, PCR products were designed to amplify mtrD in addition to other loci within mtrRCDE. Here, we found that the addition of the mtr promoter to constructs containing mtrD was sufficient to raise the MIC of 28Bl to the GCGS0276 phenotype of 1 µg/ml.
FIG 7 .
Alignment of wild-type and mosaic Neisseria gonorrhoeae mtr promoters with N. lactamica and N. meningitidis references. Nucleotide alignment of the mtr promoter region from N. gonorrhoeae FA1090 (AE004969.1), N. lactamica 020-06 (FN995097.1), and N. meningitidis NZ-05/33 (CP002424.1) with the gonococcal isolates used in this study. The minor allele variants for each position are shown in red on a pink background. Sequence features include the interleaved mtrCDE and mtrR promoters (yellow background), MtrR binding site (maroon bars), inverted repeat in the mtrR promoter (black bar), 153-bp Correia element insertion (red arrows), and integration host factor (IHF) binding site (blue bar) (21, 29, 30, 33). The C-to-T transition 120 bp upstream of the mtrC start codon (mtr120 [purple background]) has been demonstrated to increase expression of the efflux pump components and enhance resistance to various substrates of the pump (29); however, no T variants were detected at this site. Deletion of a single A (A deletion) in the mtrR promoter inverted repeated (blue background) has also been shown to increase expression of the efflux pump and enhance mtr-mediated resistance (30); no A-deletion variants were present. Interestingly, the N. lactamica reference, N. meningitidis reference, and all 29 isolates with mosaic mtr promoter regions had a C transversion in the second position within this repeat (blue background).
Mosaic mtr mutations contribute to resistance to diverse antimicrobial agents.
The Mtr efflux pump exports diverse antimicrobial agents from the cell (23–27). To determine whether mosaic mtr mutations were specific to enhancing azithromycin recognition or transport, or were capable of enhancing resistance to multiple substrates of the pump, we tested the susceptibilities of all strains and transformant lines to the dye crystal violet and the detergents Triton X-100 and polymyxin B.
All azithromycin-resistant mosaic mtr isolates had higher MICs for the additional antimicrobials tested than the azithromycin-susceptible GCGS0353, GCGS0465, and 28Bl isolates (Table 1). Genomic DNA transformants screened for the presence of both a mosaic mtrD and a mosaic mtr promoter region from either N. meningitidis-like or N. lactamica-like donors and transformants with mosaic mtrRCDE or mtrpCD from GCGS0276 or GCGS0402 had elevated MICs for these drugs compared to susceptible recipient strains (Table 2). GCGS0276 mtrD in the 28Bl genomic background increased MICs for crystal violet and polymyxin B by twofold (from 4 µg/ml to 8 µg/ml) and fourfold (from 60 µg/ml to 250 µg/ml), respectively, while addition of the mosaic mtr promoter increased MICs 1 dilution higher in both cases (Table 2).
Regulatory and structural mutations epistatically contribute to resistance.
We tested for the contribution of transcript regulatory variation to the mechanism of resistance by profiling gene expression via transcriptome sequencing (RNA-seq) of strains 28Bl, 28BlΔGCGS0276-mtrD, and 28BlΔGCGS0276-mtrRpCDE. As expression of the mtr efflux pump is inducible by exposure to antimicrobial agents (38, 39), we evaluated expression before azithromycin exposure and 120 min after the addition of a sub-MIC dose of azithromycin (0.125 µg/ml) to the culture media. Across 24 libraries, a total of 106 million 50-bp paired-end reads mapped to the FA1090 reference genome. Each library had on average 4.44 ± 3.49 million mappable reads.
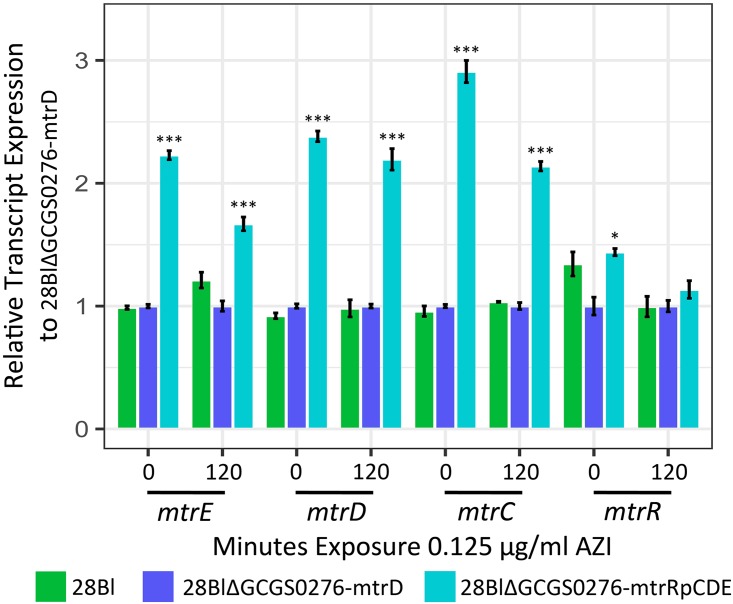
We assessed the impact of mosaic mtrD on mtrRCDE mRNA expression by comparing 28BlΔGCGS0276-mtrD transformants to 28Bl and found no significant differential regulation of transcripts encoding mtr efflux pump components (Fig. 8; Table S4). To determine the effect of the mosaic mtr promoter on pump expression, we compared 28BlΔGCGS0276-mtrD and 28BlΔGCGS0276-mtrRpCDE transformants (Fig. 8; Table S4). Here, the presence of a mosaic mtr promoter resulted in the significant upregulation of mtrC, mtrD, and mtrE across conditions (false-discovery rate [FDR] of <0.0001) and upregulation of mtrR in the absence of azithromycin (FDR of 0.003).
FIG 8 .
The N. meningitidis-like GCGS0276 mosaic mtr promoter sequence upregulates expression of mtr efflux pump component mRNAs. The 28Bl recipient strain (green), 28Bl transformants with mosaic GCGS0276 mtrD (blue), and 28Bl transformants with mosaic GCGS0276 mtrRCDE (teal) were exposed to sub-MIC (0.125 µg/ml) concentrations of azithromycin (AZI) for 120 min and subjected to whole-transcriptome sequencing before and after the addition of the antimicrobial. The 28BlΔGCGS0276-mtrD strain was then used as a reference to calculate the relative expression of transcripts for other strains by condition by gene. In both the presence and absence of drug, transformants with the presence of a mosaic mtr promoter region yielded significantly upregulated pump component mRNAs (false-discovery rate [FDR] of <0.0001), while transformants that inherited mosaic mtrD alleles did not significantly alter mtrCDE regulation from the 28Bl recipient strain levels. Values that are significantly different are indicated by asterisks as follows: *, P less than or equal to 0.05; *** P less than or equal to 0.0001.
Number of significantly differentially expressed (DE) genomic features between strains at a FDR of <0.01. Download TABLE S4, XLSX file, 0.01 MB (9.4KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Genomic and surveillance efforts from across the globe have shown an association between mosaic mtr alleles and azithromycin resistance (8, 18–20), but the causal role of these alleles in generating resistance has been unclear. Mtr-mediated resistance to macrolides has been attributed exclusively to increased mtrCDE expression (28–32). However, a majority of mtr mosaic isolates in the population genomic data set have none of the variants that alter regulation of mtrCDE, such as the mtrC120 substitution, deletion of a single adenine (A deletion) in the mtrR promoter inverted repeat, radical mtrR amino acid changes, or premature stop codon mutations in mtrR (28–32). Thus, using a combination of experimental and population genomic approaches, we set out to better characterize the mechanism of resistance in isolates with mosaic mtr sequences.
Population genomics and phylogenetic reconstruction results support mtrRCDE as a hot spot of interspecies allele sharing. Expectations for genomic signatures of recent horizontal gene transfer events include the following: (i) local elevation of allelic diversity and an excess of rare mutations prior to the fixation, rise to intermediate frequencies, or loss of new alleles from the population; (ii) a departure from species-specific phylogenetic tree topology, indicating the admixture of alleles for particular loci; and (iii) high linkage within the recombination tract as the combined effects of recombination and mutation do not have sufficient evolutionary time to break down local associations between sites. Our results demonstrate that each of these parameters departed from the genomic background at mtrRCDE, suggesting recent horizontal gene transfer as opposed to historical admixture in introducing novel haplotypes at this locus (Fig. 1). While historical admixture can also produce patterns of polyphyly between species, the rarity of mosaic alleles and their high linkage suggests a more recent origin in the gonococcal population.
At least 12 independent acquisitions of mosaic mtr alleles are supported by a whole-genome-based phylogeny of the U.S. gonococcal population samples (Fig. 2A), which have introduced multiple rare and divergent mtr haplotypes from N. meningitidis and N. lactamica into the gonococcal population. Multiple mosaic haplotypes were present in the population, some spanning exclusively over mtrD, while others extended over both mtrD and the mtr promoter region (Fig. 2C and E). Isolates with mosaic sequences were associated with increased azithromycin MIC, where isolates with full-length mosaic mtrD sequences had MICs of ≥0.25 µg/ml (Fig. 2D) and isolates with both mosaic mtrD and mtr promoters had MICs of ≥1 µg/ml (Fig. 2F). These results mirror reports of isolates with mosaic mtrR or mtr promoter regions having azithromycin MICs of ≥1 µg/ml; however, prior studies did not screen for mosaic mtrD (8, 18–20). Interestingly, while admixture between N. gonorrhoeae and N. polysaccharea is also supported by depressed gsi values across mtrRCDE, we find no evidence of N. polysaccharea-like mtr alleles in the gonococcal population sample. This may suggest unidirectional transfer of N. gonorrhoeae mtr alleles into the N. polysaccharea population via horizontal gene transfer, or one ancestrally shared mtr allele in both species has risen to high frequency in the N. gonorrhoeae population.
Though mosaic mtr alleles have previously been associated with resistance (8, 18–20), our results conclusively demonstrate that they are responsible for resistance to diverse antimicrobial agents and show that the mechanism of resistance involves multiple synergistically acting epistatic interactions, including epistasis between multiple sites within mtrD and epistasis between MtrD and the mtr promoter region. Surprisingly, despite the sequence divergence (8%) between N. meningitidis- and N. lactamica-like mtr mosaics, mtr sequences from both generated azithromycin resistance through the same epistatic mechanism. For the N. meningitidis-like GCGS0276 mosaic, mtrD alone was capable of raising the azithromycin MIC of strain 28Bl to 0.5 µg/ml, independent of transcriptional changes to the pump’s regulation, with the donor azithromycin resistance phenotype of 1 µg/ml reproduced only by adding the N. meningitidis mtr promoter region, which increased expression of mtrCDE (Table 2; Fig. 4, 6, and 8). Similarly, although acquisition of the N. lactamica-like GCGS0402 mtrD was not sufficient on its own to enhance azithromycin resistance, transformation of both the mtrD and the mtr promoter region yielded the donor’s azithromycin MIC of 2 µg/ml (Table 2, PCR product transformant strains). All transformants that inherited mosaic mtr alleles in addition to becoming resistant to azithromycin also became resistant to diverse antimicrobials (Table 2). Thus, the mechanism of resistance in mosaics is likely derived from both structural changes to MtrD, which nonspecifically export antimicrobials more efficiently, coupled with promoter mutations that alter regulation of mtrCDE, enhancing drug export as a result of the increased availability of Mtr pump complexes.
Addition of the GCGS0276 mosaic mtr promoter region in the 28Bl background increased expression of mtrCDE (Fig. 8); however, this isolate did not have the mtrC120 substitution, A deletion, or mutations in mtrR that have been previously demonstrated to increase mtrCDE transcription (28–32). While we were unable to pinpoint the precise mutation(s) responsible for increased mtrCDE expression in this mosaic, two polymorphisms present in strain GCGS0276 have previously been implicated in altered mtr regulation. The GCGS0276 mosaic has a 153-bp Correia element inserted between mtrC and the mtrCDE promoter with an IHF binding site, which has been demonstrated in meningococci to decrease mtrCDE transcription, possibly by acting as a weak transcriptional terminator (33). As we show that the addition of the mosaic promoter sequence increases mtrCDE expression in N. gonorrhoeae, the Correia element is either (i) not having the same phenotypic effect in N. gonorrhoeae and N. meningitidis or (ii) decreases expression of mtrCDE as in N. meningitidis while another mutation increases the transcription of mtrCDE to an extent that the level of transcriptional termination caused by the Correia element has no significant impact. The second polymorphism, a conserved A-to-C transversion in the mtrR promoter inverted repeat, was present in all 29 isolates with mosaic mtr promoters (Fig. 7). Interestingly, this mutation is located in the same inverted repeat as an A deletion that has been shown to both increase transcription of mtrCDE and increase resistance to diverse antimicrobials (30). Additionally, a previously described isolate with an azithromycin MIC of 2 µg/ml was noted to have an A-to-C transversion of another site within this repeat (29).
Nearly the full-length mtrD was required for azithromycin resistance in GCGS0276 transformants, indicating the role of within-gene epistasis, rather than a single point mutation within mtrD, in generating resistance. Transformants that inherited two regions at the 5′ and 3′ ends of mtrD from strain GCGS0276 increased the azithromycin MIC of strain 28Bl to 0.5 µg/ml (Fig. 4). These two regions are part of the central pore (PN1) that stabilizes the trimeric organization of the protein and the outer periplasmic region (PC2) of the protein that may interact with MtrC to form a functional pump complex (40). Though we show that the 5′ and 3′ regions were both required to increase resistance in the 28Bl background, we are currently unable to exclude the possibility that additional SNPs between these two regions are involved in the interaction. Twenty additional amino acid changes were present between the GCGS0276 and 28Bl MtrD proteins (Fig. 5). Of note, none of the mutations observed in strains 28Bl and GCGS0276 have been shown to contribute to macrolide resistance in the orthologous proteins AcrB and MexB in other species, nor are they located in the direct contact site (residue 616) for macrolide recognition, the multidrug binding site of the pump (residues 608 to 619), or the sites involved in forming the proton relay network of the pump (40–44). Additional studies will be needed to determine the combination of mutations in mtrD responsible for enhanced resistance, though it is clear that whatever the causal SNPs are, they are acting to enhance resistance nonspecifically to multiple substrates of the pump (Table 2).
The selective maintenance of epistasis between domains of MtrD in natural populations of gonococci is supported by our experimental and genomics results. Across mtrD, we observed local increases in linkage disequilibrium coupled with an increased frequency of rare mutations (Fig. 1). These signatures could be explained by the recent acquisition of neutral diversity from closely related species, with too little time for the effects of recombination and mutation to break down linkage of sites across imported DNA tracts or the spread of these mutations to higher frequencies (45, 46). However, our experimental results demonstrate strong purifying selection on plates containing azithromycin after inheritance of partial mtrD mosaic haplotypes (Fig. 4), suggesting that some of the linkage within mtrD observed in natural gonococcal populations may be driven by selection maintaining allelic combinations within the gene that increase resistance to antimicrobials. We speculate that this is due to the effects of selection against partial gene recombination events between highly divergent mtrD alleles, which could produce defective lethal or inefficient protein products when under antimicrobial selection.
Overall, our results defining the role of mosaic mtr in azithromycin resistance affirm the importance of other Neisseria species as an antibiotic resistance reservoir for N. gonorrhoeae. Moreover, whereas mosaic penA genes arise from interspecies recombinations within a single gene and confer cephalosporin resistance through novel structural forms (15, 47), our findings of horizontally acquired, epistatically interacting structural and regulatory variants in mtr point to the potential complexity by which antibiotic resistance can arise through the interactions of multiple loci. Interspecies mosaicism will be an important consideration for future development of sequence-based molecular resistance diagnostics, as markers designed to amplify gonococcus-specific sequence will overlook or incorrectly diagnose resistance phenotype. Thus, as the number of commensal neisserial genome sequences increase, analyses that map the patterns and extent of interspecies recombination may be a valuable guide in understanding pathways to resistance and in designing appropriate diagnostic tools.
MATERIALS AND METHODS
Genome sequencing and population genomics.
Sequencing libraries were prepared using a modification of Illumina’s Nextera XP protocol (48). Samples were dual-indexed and pooled (n = 15 per pool). Paired-end 150-bp sequencing was conducted on an Illumina MiSeq (Illumina Corp., San Diego, CA) platform located at the Harvard T. H. Chan School of Public Health to an average depth of 40×. Previously sequenced read libraries were obtained from the NCBI’s Short Read Archive (project PRJEB2090) and the European Nucleotide Archive (projects PRJEB2999 and PRJEB7904) (8, 34).
To determine the impact of interspecific recombination at mtrRCDE, we assessed patterns of allelic diversity across the mtrR transcriptional repressor and the mtrCDE pump compared to the rest of the genome for the 1,102 GISP gonococcal isolates (8, 34). Reads were aligned to the FA1090 reference using Bowtie2 v.2.2.4 (49), and variants were called using pilon v.1.16 (50). Vcftools v.0.1.12 (51) was used to merge resultant vcf files and calculate genome-wide values of π and Tajima’s D over 100-bp sliding windows, and r2 linkage by site. Gubbins v.2.2.0 (52) was used to predict regions of elevated single-nucleotide polymorphism (SNP) densities. For each isolate, BLASTn was used to identify the top hit and highest percent sequence identity for mtrD and the mtr promoter to all Neisseria within the NCBI database (E value of <10−40).
The extent of exclusive ancestry between Neisseria gonorrhoeae, N. meningitidis, N. lactamica, and N. polysaccharea was assessed using the genealogical sorting index (gsi) (35) for each gene across a 25-kb window surrounding mtrRCDE. In brief, we downloaded de novo assemblies and raw sequencing reads from NCBI for N. meningitidis (n = 431), N. lactamica (n = 326), N. polysaccharea (n = 37), and N. gonorrhoeae (n = 1102) (8, 34). Raw reads were assembled with SPAdes v.3.7.0 (53), and assemblies were aligned to the N. gonorrhoeae FA1090 reference genome (AE004969.1) using progressiveMauve (54) (snapshot 2015-02-13 for linux-x64), since a multigenome alignment for all genomes was not computationally tractable. The sequences that were aligned to each gene within 25 kb of mtrRCDE in strain FA1090 were then extracted with custom Perl scripts (available on request) and realigned with MAFFT v.7.309 (55). This method of pairwise alignments of de novo assemblies to the FA1090 reference identifies orthologs between N. gonorrhoeae and the other Neisseria species using both sequence identity and microsynteny, which is conserved in the genomic region surrounding mtrRCDE (see Fig. S1 in the supplemental material). We then used RAxML v.8.1.4 (56) to reconstruct the phylogeny for each gene, using 50 bootstrap replicates and the GTRCAT substitution model. With these multispecies phylogenies, we calculated gsi with the genealogicalSorting R package (35, 57).
Bacterial culture conditions.
N. gonorrhoeae isolates were provided by the CDC (Table 1). Isolates were cultured on GCB agar medium supplemented with 1% IsoVitaleX (Becton Dickinson Co., Franklin Lakes, NJ). After inoculation, the plates were incubated at 37°C in a 5% CO2 atmosphere incubator for 16 to 18 h. Antimicrobial susceptibility testing was conducted using the agar dilution method at a range of concentrations of azithromycin from 0 µg/ml to 16 µg/ml, crystal violet from 0.25 µg/ml to 32 µg/ml, Triton X-100 from 250 µg/ml to 16,000 µg/ml, and polymyxin B from 30 µg/ml to 1,000 µg/ml (1, 5). MICs were recorded after 24 h of growth. All isolate stocks were stored at −80°C in Trypticase soy broth containing 20% glycerol.
Transformation of mosaic mtr alleles.
Genomic DNA was extracted from isolates by lysing growth from overnight plates in TE buffer (10 mM Tris [pH 8.0], 10 mM EDTA) with 0.5 mg/ml lysozyme and 3 mg/ml proteinase K (Sigma-Aldrich Corp., St. Louis, MO). DNA was purified using the PureLink Genomic DNA minikit, treated with RNase A (Thermo Fisher Corp., Waltham, MA), and stored in water. Primers were designed to amplify regions of mtrRCDE. For primer pairs that did not amplify over a region containing a DNA uptake sequence, to enhance transformation efficiency, the 12-bp AT-DNA uptake sequence (AT-DUS) was added to the forward primer (5′-ATGCCGTCTGAA-3′) (58). PCR was conducted in 50-µl volumes using Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA) using the conditions listed in Table S3 in the supplemental material. Amplified products were run on a 0.8% agarose gel, excised, and purified with the QIAEX II gel extraction kit (Qiagen Inc., Valencia, CA) to remove genomic DNA (gDNA) contamination.
Transformations were conducted in liquid culture as described previously (21, 36, 37) at 37°C in a 5% CO2 atmosphere incubator. In brief, we inoculated GCP broth (7.5 g protease peptone 3, 0.5 g soluble starch, 2 g dibasic K2HPO4, 0.5 g monobasic KH2PO4, 2.5 g NaCl, and double-distilled water [ddH2O] to 500 ml; Becton Dickinson Co., Franklin Lakes, NJ) supplemented with 1% IsoVitaleX and 10 µM MgSO4 (Sigma-Aldrich Corp., St. Louis, MO) with naturally competent piliated cells to an optical density (OD) of ~0.5. Suspensions were then incubated with approximately 1 µg of gDNA or purified PCR product for 10 min to allow DNA uptake and homologous recombination and subsequently spread on GCB with 1% IsoVitaleX and incubated for 4 h to allow for expression of novel alleles. Transformants resistant to azithromycin were selected by plating cells on GCB supplemented with 1% IsoVitaleX and 0.38 to 1 µg/ml azithromycin and picking single colonies after 18 h.
All transformants were screened for successful transformation by testing the levels of resistance to known substrates of the Mtr efflux pump system as well as sequencing to couple the acquisition of novel mosaic alleles with gain of resistance. Transformants generated with targeted PCR products from mosaic donors were screened for mosaic alleles by sequencing mtrD and the mtr promoter region via Sanger sequencing using the GeneWiz sequencing service (GeneWiz Inc., Cambridge, MA) (Table 2). Undirected gDNA-transformed cell lines were sequenced via Illumina whole-genome sequencing as described above. To detect the alleles gained from mosaic donors, we searched for SNP homology between the transformants and mosaic donors that was not present in the 28Bl recipient strain (Fig. 3). Genomes were de novo assembled using SPAdes v.3.7.0 (53) and annotated with prokka v.1.11 (59). Alignment of transformant libraries to the 28Bl and mosaic references was conducted in Bowtie2 v.2.2.4 (49) using the “end-to-end” and “very-sensitive” options, which resulted in a higher percentage of uniquely mapped reads than any other combination of preset options. SNPs and indels between read libraries and the reference genomes were called with pilon v.1.16 (50), removing variant calls with qualities of <15.
Transcriptome construction.
Cells harvested from overnight plates were suspended in GCP supplemented with 1% IsoVitaleX and 0.042% sodium bicarbonate. Cultures were incubated at 37°C for 2 h to mid-log phase and then exposed to a sublethal dose of azithromycin (AZI) (0.125 µg/ml). RNA was extracted at 0 min (pre-AZI) and 120 min (post-AZI) exposure using the Direct-Zol kit (Zymo Research, Irvine, CA). Transcriptome libraries were prepared at the Broad Institute at the Microbial ‘Omics Core using a modified version of the RNAtag-seq protocol (60). Five hundred nanograms of total RNA was fragmented, depleted of genomic DNA, dephosphorylated, and ligated to DNA adapters carrying 5′-AN8-3′ barcodes of known sequence with a 5′ phosphate and a 3′ blocking group. Barcoded RNAs were pooled and depleted of rRNA using the RiboZero rRNA depletion kit (Epicentre, Madison, WI). Pools of barcoded RNAs were converted to Illumina cDNA libraries in two main steps: (i) reverse transcription of the RNA using a primer designed to the constant region of the barcoded adapter with addition of an adapter to the 3′ end of the cDNA by template switching using SMARTScribe (Clontech, Mountain View, CA) as described previously (61); (ii) PCR amplification using primers whose 5′ ends target the constant regions of the 3′ or 5′ adapter and whose 3′ ends contain the full Illumina P5 or P7 sequences. cDNA libraries were sequenced on the Illumina Nextseq 500 platform to generate 50-bp paired-end reads.
Barcode sequences were removed, and reads were aligned to the FA1090 reference genome. Read counts were assigned to genes and other genomic features using custom scripts. For the FA1090 reference genome, we mapped reads to either the sense or antisense strand for coding domain sequences (CDSs) (n = 1,894), tRNAs (n = 55), and rRNAs (n = 12). For intergenic regions (IGRs) (n = 1,722), we mapped to each antiparallel strand. Differential expression analysis was conducted in DESeq2 v.1.10.1 (62).
Data availability.
All genomic and transcriptomic read libraries generated in this study are available from the NCBI SRA database (accession numbers PRJNA475134 and PRJNA475139); all Sanger sequencing files are available in the NCBI database (accession numbers MH576918 to MH576947).
ACKNOWLEDGMENTS
This work was supported by the Richard and Susan Smith Family Foundation (http://www.smithfamilyfoundation.net/) and National Institutes of Health grant R01 AI132606 (https://www.nih.gov/).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank David Trees and Steve Johnson for training on gonococcal transformation design and Jonathan Livny for his technical assistance with transcriptome library preparation.
Footnotes
Citation Wadsworth CB, Arnold BJ, Sater MRA, Grad YH. 2018. Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. mBio 9:e01419-18. https://doi.org/10.1128/mBio.01419-18.
Contributor Information
William Shafer, Emory University School of Medicine.
Michael S. Gilmore, Harvard Medical School.
REFERENCES
- 1.Centers for Disease Control and Prevention 2016. Gonococcal Isolate Surveillance Project (GISP). Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/std/gisp/. Accessed 24 April 2018. [Google Scholar]
- 2.Public Health England 2017. Surveillance of antimicrobial resistance in Neisseria gonorrhoeae in England and Wales. Key findings from the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP). Antimicrobial resistance in Neisseria gonorrhoeae: data to June 2017, p 1–24. Public Health England, London, United Kingdom: http://www.gov.uk/government/publications/gonococcal-resistance-to-antimicrobials-surveillance-programme-grasp-report. [Google Scholar]
- 3.European Centre for Disease Prevention and Control 2017. Gonococcal antimicrobial susceptibility surveillance in Europe, 2015. ECDC Surveillance Report, p 1–44. European Centre for Disease Prevention and Control, Stockholm, Sweden: http://www.ecdc.europa.eu. [Google Scholar]
- 4.World Health Organization 2017. Gonococcal antimicrobial resistance in the Western Pacific Region, p 1–4. WHO fact sheet. WHO Regional Office for the Western Pacific, Manila, Philippines. [Google Scholar]
- 5.Workowski KA, Bolan GA, Centers for Disease Control and Prevention . 2015. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep 64:1–137. [PMC free article] [PubMed] [Google Scholar]
- 6.Ng LK, Martin I, Liu G, Bryden L. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 46:3020–3025. doi: 10.1128/AAC.46.9.3020-3025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chisholm SA, Dave J, Ison CA. 2010. High-level azithromycin resistance occurs in Neisseria gonorrhoeae as a result of a single point mutation in the 23S rRNA genes. Antimicrob Agents Chemother 54:3812–3816. doi: 10.1128/AAC.00309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grad YH, Harris SR, Kirkcaldy RD, Green AG, Marks DS, Bentley SD, Trees D, Lipsitch M. 2016. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis 214:1579–1587. doi: 10.1093/infdis/jiw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JM, Smith NH, O’Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc Natl Acad Sci U S A 90:4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Rourke M, Stevens E. 1993. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J Gen Microbiol 139:2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 11.O’Rourke M, Spratt BG. 1994. Further evidence for the non-clonal population structure of Neisseria gonorrhoeae: extensive genetic diversity within isolates of the same electrophoretic type. Microbiology 140:1285–1290. doi: 10.1099/00221287-140-6-1285. [DOI] [PubMed] [Google Scholar]
- 12.Corander J, Connor TR, O’Dwyer CA, Kroll JS, Hanage WP. 2012. Population structure in the Neisseria, and the biological significance of fuzzy species. J R Soc Interface 9:1208–1215. doi: 10.1098/rsif.2011.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer TF. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125–134. doi: 10.1016/S0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 14.Halter R, Pohlner J, Meyer TF. 1989. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J 8:2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spratt BG, Bowler LD, Zhang QY, Zhou J, Smith JM. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol 34:115–125. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 16.Feavers IM, Heath AB, Bygraves JA, Maiden MC. 1992. Role of horizontal genetic exchange in the antigenic variation of the class 1 outer membrane protein of Neisseria meningitidis. Mol Microbiol 6:489–495. doi: 10.1111/j.1365-2958.1992.tb01493.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J, Spratt BG. 1992. Sequence diversity within the argF, fbp and recA genes of natural isolates of Neisseria meningitidis: interspecies recombination within the argF gene. Mol Microbiol 6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]
- 18.Trembizki E, Doyle C, Jennison A, Smith H, Bates J, Lahra M, Whiley D, GRAND Study Investigators . 2014. A Neisseria gonorrhoeae strain with a meningococcal mtrR sequence. J Med Microbiol 63:1113–1115. doi: 10.1099/jmm.0.074286-0. [DOI] [PubMed] [Google Scholar]
- 19.Demczuk W, Martin I, Peterson S, Bharat A, Van Domselaar G, Graham M, Lefebvre B, Allen V, Hoang L, Tyrrell G, Horsman G, Wylie J, Haldane D, Archibald C, Wong T, Unemo M, Mulvey MR. 2016. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 54:1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiley DM, Kundu RL, Jennison AV, Buckley C, Limnios A, Hogan T, Enriquez R, El Nasser J, George CR, Lahra MM. 2018. Azithromycin-resistant Neisseria gonorrhoeae spreading amongst men who have sex with men (MSM) and heterosexuals in New South Wales, Australia, 2017. J Antimicrob Chemother 73:1242–1246. doi: 10.1093/jac/dky017. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SR, Sandul AL, Parekh M, Wang SA, Knapp JS, Trees DL. 2003. Mutations causing in vitro resistance to azithromycin in Neisseria gonorrhoeae. Int J Antimicrob Agents 21:414–419. doi: 10.1016/S0924-8579(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 22.McLean CA, Wang SA, Hoff GL, Dennis LY, Trees DL, Knapp JS, Markowitz LE, Levine WC. 2004. The emergence of Neisseria gonorrhoeae with decreased susceptibility to azithromycin in Kansas City, Missouri, 1999 to 2000. Sex Transm Dis 31:73–78. doi: 10.1097/01.OLQ.0000109514.91508.FC. [DOI] [PubMed] [Google Scholar]
- 23.Pan W, Spratt BG. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol 11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 24.Hagman KE, Shafer WM. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 177:4162–4165. doi: 10.1128/jb.177.14.4162-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delahay RM, Robertson BD, Balthazar JT, Shafer WM, Ison CA. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127–2133. doi: 10.1099/00221287-143-7-2127. [DOI] [PubMed] [Google Scholar]
- 26.Lucas CE, Balthazar JT, Hagman KE, Shafer WM. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol 179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagman KE, Lucas CE, Balthazar JT, Snyder L, Nilles M, Judd RC, Shafer WM. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117–2125. doi: 10.1099/00221287-143-7-2117. [DOI] [PubMed] [Google Scholar]
- 28.Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohneck EA, Zalucki YM, Johnson PJT, Dhulipala V, Golparian D, Unemo M, Jerse AE, Shafer WM. 2011. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2:e00187-11. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 31.Shafer WM, Balthazar JT, Hagman KE, Morse SA. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:907–911. doi: 10.1099/13500872-141-4-907. [DOI] [PubMed] [Google Scholar]
- 32.Zarantonelli L, Borthagaray G, Lee EH, Veal W, Shafer WM. 2001. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother 47:651–654. doi: 10.1093/jac/47.5.651. [DOI] [PubMed] [Google Scholar]
- 33.Rouquette-Loughlin CE, Balthazar JT, Hill SA, Shafer WM. 2004. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol Microbiol 54:731–741. doi: 10.1111/j.1365-2958.2004.04299.x. [DOI] [PubMed] [Google Scholar]
- 34.Grad YH, Kirkcaldy RD, Trees D, Dordel JD, Harris SR, Goldstein E, Weinstock H, Parkhill J, Hanage WP, Bentley S, Lipsitch M. 2014. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummings MP, Neel MC, Shaw KL. 2008. A genealogical approach to quantifying lineage divergence. Evolution 62:2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 36.Morse SA, Johnson SR, Biddle JW, Roberts MC. 1986. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob Agents Chemother 30:664–670. doi: 10.1128/AAC.30.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson SR, Grad Y, Ganakammal SR, Burroughs M, Frace M, Lipsitch M, Weil R, Trees D. 2014. In vitro selection of Neisseria gonorrhoeae mutants with elevated MIC values and increased resistance to cephalosporins. Antimicrob Agents Chemother 58:6986–6989. doi: 10.1128/AAC.03082-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouquette C, Harmon JB, Shafer WM. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol 33:651–658. doi: 10.1046/j.1365-2958.1999.01517.x. [DOI] [PubMed] [Google Scholar]
- 39.Zalucki YM, Dhulipala V, Shafer WM. 2012. Dueling regulatory properties of a transcriptional activator (MtrA) and repressor (MtrR) that control efflux pump gene expression in Neisseria gonorrhoeae. mBio 3:e00446-12. doi: 10.1128/mBio.00446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, Chou TH, Delmar JA, Lei HT, Rajashankar KR, Shafer WM, Yu EW. 2014. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One 9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wehmeier C, Schuster S, Fähnrich E, Kern WV, Bohnert JA. 2009. Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob Agents Chemother 53:329–330. doi: 10.1128/AAC.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eicher T, Cha H-J, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci 109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha HJ, Müller RT, Pos KM. 2014. Switch-loop flexibility affects transport of large drugs by the promiscuous AcrB multidrug efflux transporter. Antimicrob Agents Chemother 58:4767–4772. doi: 10.1128/AAC.02733-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ababou A, Koronakis V. 2016. Structures of gate loop variants of the AcrB drug efflux pump bound by erythromycin substrate. PLoS One 11:e0159154. doi: 10.1371/journal.pone.0159154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol 3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 46.Theunert C, Slatkin M. 2017. Distinguishing recent admixture from ancestral population structure. Genome Biol Evol 9:427–437. doi: 10.1093/gbe/evx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomberg J, Unemo M, Davies C, Nicholas RA. 2010. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 49:8062–8070. doi: 10.1021/bi101167x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group . 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Misawa K, Kuma K-I, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R Core Team 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 58.Ambur OH, Frye SA, Tønjum T. 2007. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol 189:2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 60.Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka C, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M, Livny J. 2015. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892–897. [DOI] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Population genetics statistics calculated for all mtr loci. Download TABLE S1, XLSX file, 0.04 MB (45.2KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fully sequenced, annotated genomes for N. gonorrhoeae (top), N. lactamica (middle), and N. meningitidis (bottom) were aligned with progressiveMauve (54). Syntenic blocks between these three species are illustrated by colored rectangles. For instance, the purple rectangles above the red arrows show a large (~100-kb) collinear genomic segment shared among the three Neisserial species. This syntenic block contains the mtr operon (positions 1,323,095 to 1,326,298). Download FIG S1, TIF file, 79.5 MB (81.4MB, tif) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genealogical sorting index (gsi) values as a measure of genealogical exclusivity and admixture. Download TABLE S2, XLSX file, 0.04 MB (44KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotide primers and PCR conditions used for construction of the DNA templates for transformation and Sanger sequencing to confirm transformation. Download TABLE S3, XLSX file, 0.01 MB (10.8KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of significantly differentially expressed (DE) genomic features between strains at a FDR of <0.01. Download TABLE S4, XLSX file, 0.01 MB (9.4KB, xlsx) .
Copyright © 2018 Wadsworth et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All genomic and transcriptomic read libraries generated in this study are available from the NCBI SRA database (accession numbers PRJNA475134 and PRJNA475139); all Sanger sequencing files are available in the NCBI database (accession numbers MH576918 to MH576947).