Abstract
Cryptochromes are blue light receptors regulated by light-dependent ubiquitination and degradation in both plant and animal lineages. The Arabidopsis genome encodes two cryptochromes, CRY1 and CRY2, of which CRY2 undergoes blue light-dependent ubiquitination and 26S proteasome-dependent degradation. The molecular mechanism regulating blue light-dependent proteolysis of CRY2 is still not fully understood. We found that the F-box proteins ZEITLUPE (ZTL) and Lov Kelch Protein2 (LKP2), which mediate blue light suppression of degradation of the CRY2 signaling partner CIB1, are not required for the blue light-dependent CRY2 degradation. We further showed that the previously reported function of the COP1–SPA1 protein complex in blue light-dependent CRY2 degradation is more likely to be attributable to its cullin 4 (CUL4)-based E3 ubiquitin ligase activity than its activity as the cryptochrome signaling partner. However, the blue light-dependent CRY2 degradation is only partially impaired in the cul4 mutant, the cop1-5 null mutant and the spa1234 quadruple mutant, suggesting a possible involvement of additional E3 ubiquitin ligases in the regulation of CRY2. Consistent with this hypothesis, we demonstrated that the blue light-dependent CRY2 degradation is significantly impaired in the temperature-sensitive cul1 mutant allele (axr6-3), especially under the non-permissive temperature. Based on these and other results presented, we propose that photoexcited CRY2 undergoes Lys48-linked polyubiquitination catalyzed by the CUL4- and CUL1-based E3 ubiquitin ligases.
Keywords: Arabidopsis thaliana, CRY2, Degradation, Ubiquitin E3 ligase
Introduction
Arabidopsis cryptochrome2 (CRY2) is a blue light receptor mediating light-dependent de-etiolation in young seedlings and photoperiod-dependent floral initiation in adult plants (Guo et al. 1998, Lin et al. 1998). CRY2 is a nuclear photoreceptor that mediates light regulation of plant growth and development by interacting with its signaling partners, including the CIB (CRY-interacting basic helix–loop–helix) transcription factors, PIFs (phytochrome-interacting factors) and the COP1 (CONSTITITIVE PHOTOMORPHOGENSIS 1)–SPA1 (SUPPRESSOR OF PHYA-105) complex (Liu et al. 2008, Keller et al. 2011, Zuo et al. 2011, Huang et al. 2014, Ma et al. 2016). Of the two Arabidopsis cryptochromes, CRY2 undergoes rapid ubiquitination and proteolysis in response to blue light (Ahmad et al. 1998, Lin et al. 1998). CRY2 is a stable protein in etiolated seedlings with a half-life of >24 h. However, CRY2 is rapidly degraded in etiolated seedlings exposed to blue light, with a markedly shortened half-life of approximately 25 min under a modest intensity of blue light (16 μmol m-2 s-1) (Yu et al. 2007a). It has been shown that CRY2 undergoes blue light-dependent polyubiquitination and 26S proteasome-dependent degradation that serves as a negative feedback regulatory mechanism of the CRY2 photoreceptor (Yu et al. 2007a, Liu et al. 2011). However, the molecular mechanism underlying blue light-dependent CRY2 polyubiquitination and degradation is still not fully understood.
For example, it has been hypothesized that CRY2 acts as its own photoreceptor mediating its blue light-dependent ubiquitination and degradation, because CRY2 degradation is dependent on blue light-induced phosphorylation, nuclear compartmentation and the light-induced conformational changes of CRY2. A more detailed analysis showed recently that the blue light-induced CRY2 degradation was reduced modestly in the phyA mutant (Weidler et al. 2012). The effect of phyA on CRY2 degradation is probably due to the regulatory role of phyA of SPA1, which is part of the COP1 E3 ligase complex known to be partly required for CRY2 degradation (Shalitin et al. 2002). However, it remains unclear whether the other blue light receptors, such as phototropins, ZEITLUPE (ZTL), Lov Kelch Protein2 (LKP2) or Flavin-binding Kelch Repeat F-box1 (FKF1), may be involved in the blue light-dependent CRY2 ubiquitination and degradation. The F-box LOV-domain proteins ZTL, LKP2 and FKF1 are FMN-containing blue light sensors known to regulate the nuclear proteins associated with light signaling and the circadian oscillator in plants (Ito et al. 2012). ZTL is the substrate receptor of the CUL1-based E3 ligase that targets TOC1 and PRR proteins for ubiquitination and degradation (Mas et al. 2003, Kiba et al. 2007, Harmon et al. 2008). Furthermore, ZTL and LKP2 also play an important role in CRY2 signaling. We have previously shown that the CRY2 signaling proteins CIB1, CIB2, CIB4 and CIB5 are ubiquitinated and degraded in the dark, and that ZTL/LKP2 mediate blue light inhibition of the degradation of CIB proteins (Liu et al. 2008, Liu et al. 2013). Given that CRY2 interacts with CIBs in response to blue light, that ZTL and LKP2 mediate blue light suppression of degradation of the CIB proteins and that ZTL is a known blue light receptor and the substrate receptor of the E3 ubiquitin ligase, it is of particular interest to determine whether ZTL and its related F-box proteins might play roles in the blue light-dependent ubiquitination and degradation of CRY2.
Protein polyubiquitination is accomplished by ubiquitin–ubiquitin linkages at any of the seven lysine residues of ubiquitin, including K63, K48, K33, K29, K11 and K6, whereby the isopeptide bonds form between the C-terminal glycine of the incoming ubiquitin monomer and the ϵ-amino group of a lysine residue of the ubiquitin attached to the substrate (Vierstra 2009, Kim et al. 2013, Walsh and Sadanandom 2014). Different types of ubiquitin–ubiquitin linkages have different consequences for the ubiquitinated substrates. K48- and K11-linked ubiquitin chains often target proteins for degradation by the 26S proteasome, whereas other types of linkages may serve other functions, including regulation of substrate compartmentation and activity (Vierstra 2009, Kim et al. 2013, Walsh and Sadanandom 2014). The Arabidopsis ubiquitylome contains at least 950 proteins, with the detectable abundance of K48 and K11 linkage changed most significantly in response to the treatment with the proteasome inhibitor MG132, suggesting their involvement in the 26S proteasome-dependent proteolysis (Kim et al. 2013). However, most of the well-established light signaling proteins, including CRY2, are not among the presently detectable ubiquitylome, presumably because of the relatively low abundance of the light signaling proteins. Therefore, the nature of the ubiquitin–ubiquitin linkage of the polyubiquitination of CRY2 remains unclear.
The cullin-based E3 ubiquitin ligases are the major groups of multimeric E3 ligases (Choi et al. 2014). The Arabidopsis genome encodes at least five cullins, CUL1, CUL2, CUL3a, CUL3b and CUL4 (Choi et al. 2014). It is not clear how many E3 ubiquitin ligases are involved in the blue light-dependent ubiquitination and degradation of CRY2 in Arabidopsis. Animal cryptochromes are often ubiquitinated by multiple E3 ligases. For example, at least two CUL1-based E3 ligases, SCFfbxl3 and SCFfbxl21, are known to catalyze the clock-dependent ubiquitination and proteolysis of mammalian cryptochromes in the nucleus and cytosol, respectively (Busino et al. 2007, Hirano et al. 2013, Yoo et al. 2013). Moreover, both CUL4-based and CUL1-based E3 ubiquitin ligases may be required for light-dependent ubiquitination and degradation of the Drosophila cryptochrome (Peschel et al. 2009, Ozturk et al. 2011, Ozturk et al. 2013). It has been shown that the COP1–SPA1 complex is required for the blue light-dependent CRY2 degradation (Shalitin et al. 2002, Weidler et al. 2012, Huang et al. 2014, Chen et al. 2015,). However, the COP1–SPA1 complex also plays significant roles in the cryptochrome-dependent signal transduction (Liu et al. 2008, Zuo et al. 2011, Huang et al. 2014). Therefore, it remains unclear whether the function of the COP1–SPA1 complex in CRY2 degradation is directly associated with its CUL4-based E3 ligase activity or indirectly associated with its function as the CRY2 signaling partner. For example, a recent discovery of a modest reduction of CRY2 degradation in the phyA mutant would be consistent with the role of phyA in regulating the activity of the SPA1–COP1 complex (Lau and Deng 2012, Weidler et al. 2012). This possibility further argues for the need to clarify the exact role of the COP1–SPA1 complex in blue light-dependent CRY2 degradation.
We report here a genetic study of blue light-dependent CRY2 ubiquitination and degradation. We found that the blue light receptors phototropins, ZTL, LKP2 and FKF1, are not required for the blue light-dependent CRY2 degradation. We established that CUL4 is required for the blue light-dependent CRY2 degradation, suggesting that the CUL4-based SPA1–COP1 E3 ligase is involved the blue light-induced CRY2 ubiquitination. However, we showed that the blue light-dependent CRY2 degradation is only partially impaired in both the spa1234 quadruple mutant and the cop1-5 null mutant, indicating the involvement of additional E3 ligase(s). Consistent with this hypothesis, we demonstrated that the blue light-dependent CRY2 proteolysis is impaired in the conditional axr6-3 allele of the cul1 mutation, although CRY2 degradation is not affected by the cul2 or cul3 mutations. These results argue that in addition to the CUL4-based COP1–SPA1 E3 ubiquitin ligases, a CUL1-based E3 ligase may also be involved in blue light-dependent CRY2 ubiquitination and degradation in Arabidopsis.
Results
The F-box LOV-domain proteins ZTL, LKP2 and FKF1 are not required for the blue light-dependent CRY2 ubiquitination
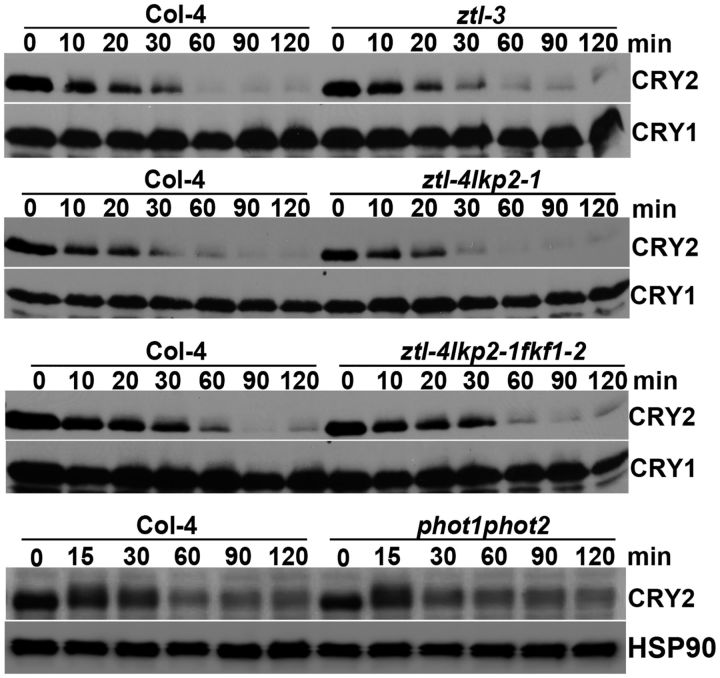
The F-box LOV-domain proteins ZTL, LPK2 and FKF1 have been shown to act as blue light receptors in plants (Kim et al. 2007, Ito et al. 2012). In addition, given that F-box proteins act as the substrate receptors of E3 ligases catalyzing cryptochrome ubiquitination in metazoa (Busino et al. 2007, Hirano et al. 2013, Yoo et al. 2013), that ZTL is a known E3 ligase targeting light-regulated proteins for ubiquitination and degradation (Mas et al. 2003), and that ZTL and LKP2 mediate blue light regulation of 26S-dependent degradation of the CRY2 signaling proteins CIBs (Liu et al. 2013), it is of particular interest to determine whether ZTL and the related F-box proteins might be involved in the blue light-dependent CRY2 degradation. We tested this possibility by comparing the CRY2 stability in the wild-type plant, the ztl-3 null mutant, the ztl/lkp2 double mutant and the ztl/lkp2/fkf1 triple mutant (Fig. 1). As expected, CRY2 is degraded in 7-day-old etiolated wild-type seedlings exposed to blue light (5 μmol m-2 s-1) (Fig. 1). However, CRY2 appears to degrade normally in response to blue light in the ztl-3 null mutant, the ztl/lkp2 double mutant and the ztl/lkp2/fkf1 triple mutant, suggesting the lack of direct involvement of these F-box blue light receptors in CRY2 ubiquitination. The phot1phot2 double mutant impaired in the other type of blue light receptors, PHOT1 and PHOT2, also seems not to affect the blue light-dependent CRY2 degradation. These results are consistent with the hypothesis that CRY2 mediates blue light regulation of its own ubiquitination and degradation.
Fig. 1.
The blue light receptors phototropins, ZTL, LKP2 and FKF1 are not required for blue light-dependent CRY2 degradation. Immunoblot showing blue light-dependent CRY2 degradation in the genotypes indicated. Seven-day-old etiolated seedlings of the wild type, and phot1-5phot2-1, ztl-3, ztl-4/lkp2-1 and ztl-4/lkp2-1/fkf1-2 mutants were exposed to blue light (5 μmol m-2 s-1) for the durations indicated. Samples were fractionated in 10% SDS–polyacrylamide gels. Immunoblots were probed with anti-CRY2 antibody (CRY2), stripped and re-probed with anti-HSP90 antibody (HSP90) or anti-CRY1 antibody (CRY1) as the loading control. Due to uncontrolled exposure time of ECL (enhanced chemiluminescence) between different immunoblots, the relative level of CRY2 is comparable only within the same immunoblot.
Fig. 2.
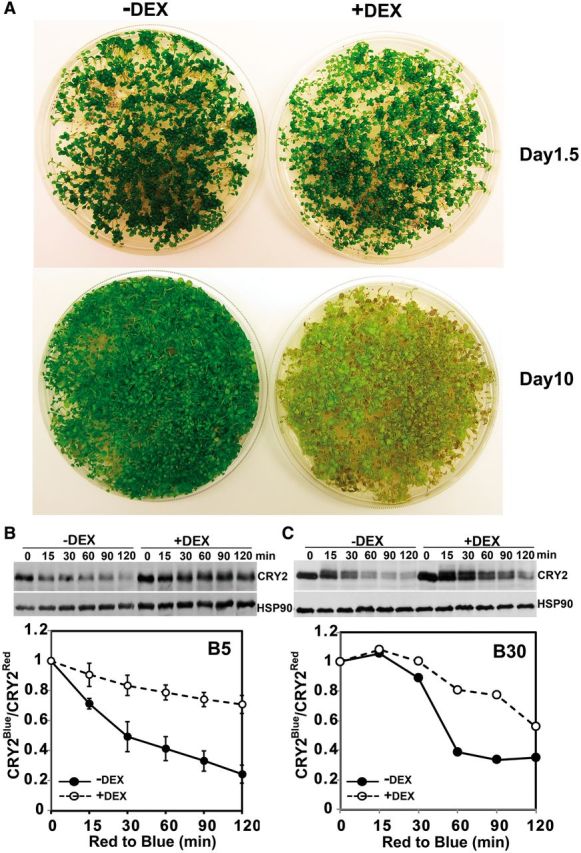
K48-linked polyubiquitination is required for blue light-dependent CRY2 degradation. (A) Morphological phenotypes of the transgenic plants expressing the dexamethasone (DEX)-induced ubR48 ubiquitin mutant. Five-day-old seedlings of the indicated genotypes grown in long-day (LD) photoperiods were treated with the mock solution (–DEX) or 0.3 mM DEX to induce the expression of ubR48 (+DEX). After DEX treatment, plants were left in continuous red light, and images were taken after 1.5 d (Day 1.5) or 10 d (Day 10). (B, C) Immunoblot comparison of the blue light-dependent CRY2 degradation in the ubR48-expressing seedlings treated with DEX (+DEX) or the mock solution (–DEX). Seedlings were treated with 0.3 mM DEX for 1.5 d in red light, exposed to blue light of different fluence rates (B5, 5 μmol m-2 s-1; B30, 30 μmol m-2 s-1) or left in red light for the times indicated. Immunoblots were probed with anti-CRY2, stripped and reprobed with anti-HSP90. The relative band intensities of CRY2 from the immunoblot were digitized and quantified. CRY2Blue, normalized (against HSP90) band intensity of CRY2 of blue light-treated samples; CRY2Red, normalized band intensity of CRY2 of red light-treated samples. For the results shown in (B), three experiments were performed; one representative immunoblot (top) and SDs (bottom, n = 3) are shown.
We next examined CRY2 degradation in mutants impaired in genes encoding important regulators of the circadian clock (CCA1, LHY, TOC1, ELF3 and GI), formation of the nuclear body (photobody) of phytochromes (HEMERA) and flowering time control (CO, FT, FD and FLC). Those genes are either regulated by the blue light receptors ZTL (such as TOC1) or CRY2 (such as FT) or they are involved in the cellular activities associated with the functions of ZTL (such as TOC1) or CRY2 (such as CO and HEMERA). Therefore, it is interesting to examine whether those genes may impose possible cross-talk or feedback regulation of CRY2 ubiquitination and degradation. Our results showed that none of the 11 mutants tested affects blue light-dependent CRY2 degradation (Supplementary Figs. S1, S2). For example, HEMERA is required for the formation of phytochrome photobodies, which interacts with phytochromes and PIF proteins to regulate phytochrome function and regulation (Chen et al. 2010b, Galvão et al. 2012, Qiu et al. 2015). Photoexcited CRY2 also forms a photobody, and it has been reported that formation of the CRY2 photobody is associated with CRY2 degradation in response to blue light (Mas et al. 2000, Yu et al. 2009). However, Supplementary Fig. S1 shows that the blue light-dependent CRY2 degradation is not affected in the hmr mutant, suggesting that the HEMERA-dependent photobody formation process is not required for the blue light-dependent CRY2 ubiquitination and degradation (Yu et al. 2009, Ozkan-Dagliyan et al. 2013). Similarly, no apparent defect in the blue light-dependent CRY2 degradation was detected in the circadian clock mutants toc1, cca1lhy, prr1579 and elf3 (Supplementary Fig. S1), or in the flowering-time mutants co, ft, tsf, gi, fd and flc (Supplementary Fig. S2). These results suggest that the phytochrome-dependent light signaling, the ZTL-dependent light signaling, the circadian clock and the flowering time regulators do not play significant roles in the blue light-dependent CRY2 degradation. Based on these results, we hypothesize that the blue light-dependent ubiquitination of CRY2 most probably results from conformational changes of the photoexcited CRY2.
CRY2 polyubiquitination involves K48 linkage of ubiquitins
CRY2 is known to be polyubiquitinated (Yu et al. 2007a), but the nature of the ubiquitin linkages of the blue light-dependent CRY2 polyubiquitination remains unclear. Polyubiquitination results from formation of the isopeptide bonds connecting the C-terminal glycine of the incoming ubiquitin monomer and the ϵ-amino group of one of the seven lysine residues (K63, K48, K33, K29, K11 and K6) of ubiquitin attached to the substrate protein (Vierstra 2009, Kim et al. 2013, Walsh and Sadanandom 2014). Because the K48 linkage is best known for targeting ubiquitinated proteins for degradation (Vierstra 2009, Kim et al. 2013, Walsh and Sadanandom 2014), we investigated the possible involvement of the K48 linkage in the blue light-dependent CRY2 degradation by a previously reported genetics approach, the ubR48 method (Schlögelhofer et al. 2006). In this experiment, the possible effect of expression of the ubR48 ubiquitin mutant, which contains arginine instead of lysine at position 48 (K48), on the blue light-dependent CRY2 degradation was examined in the transgenic plants expressing the dexamethasone (DEX)-inducible ubiquitin mutant ubR48 (Schlögelhofer et al. 2006). Because the mutant ubiquitin ubR48 cannot form the normal K48-dependent ubiquitin linkage and because ubR48 competes with the endogenous ubK48 ubiquitin, DEX-induced expression of ubR48 is expected to inhibit polyubiquitination of proteins polyubiquitinated via the K48-dependent ubiquitin linkage. As reported previously, the ubR48 transgenic seedlings grew normally in the absence of DEX (Fig. 2A, –DEX), or shortly after DEX treatment (Fig. 2A, +DEX, Day 1.5). In contrast, seedlings became yellowish upon treatment with DEX for a prolonged time (Fig. 2A, +DEX, Day 10), which has been previously reported to result from the ubR48-dependent impairment of protein ubiquitination and consequently programmed cell death (Schlögelhofer et al. 2006). To test the effects of induced expression of the ubR48 mutant on the blue light-dependent CRY2 degradation, seedlings were grown in long days (LDs) for 5 d, and treated with DEX (0.3 mM). After DEX treatment, seedlings were transferred to red light for 1.5 d, and exposed to blue light for 15–120 min before sample collection (Fig. 2B, C). As shown in Fig. 2, CRY2 was rapidly degraded in response to blue light in the ubR48 seedlings without DEX treatment. In contrast, the blue light-dependent CRY2 degradation is suppressed in the ubR48 seedlings treated with DEX undergoing the same blue light treatment as the control (Fig. 2B, C). Fig. 2 shows that CRY2 degradation is significantly reduced (P < 0.01) in the ubR48 plants treated with 0.3 mM DEX and exposed to blue light (5 μmol m-2 s-1) (Fig. 2C). CRY2 also exhibited markedly increased stability under a higher fluence rate of blue light in the DEX-treated ubR48-expressing seedlings (Fig. 2C). Our result is best explained by CRY2 being primarily polyubiquitinated via the K48 ubiquitin linkage in response to blue light.
The CUL4-based COP1–SPA1 E3 ubiquitin ligase is partially responsible for the blue light-dependent CRY2 degradation
The COP1–SPA1 complex plays an important role in CRY2 signal transduction, it has CUL4-based E3 ligase activity and it is required for blue light-dependent CRY2 degradation (Shalitin et al. 2002, Chen et al. 2010a, Lau and Deng 2012, Weidler et al. 2012). It is not known whether the E3 ligase activity or the cryptochrome signaling activity of the COP1–SPA1 complex is involved in the COP1–SPA1-dependent regulation of CRY2 degradation. We reasoned that if the involvement of the COP1–SPA1 complex in blue light-dependent CRY2 degradation is directly attributable to its CUL4-based E3 ligase activity, the stability of CRY2 should be affected by mutations of the CUL4 gene. Because a null cul4 mutant has not been reported, we analyzed CRY2 degradation in the cul4-1 hypomorphic allele, which has a T-DNA insertion in the 12th intron. The cul4-1 mutant is a weak allele that exhibited reduced CUL4 expression and a weak constitutive photomorphogenic phenotype. The constitutive photomorphogenic phenotype of the cul4-1 mutant was partially explained by impeding the function of the CUL4-based COP1–SPA1 E3 ubiquitin ligase activity (Bernhardt et al. 2006). We examined the blue light-dependent CRY2 degradation in the cul4-1 mutant using dark-adapted samples exposed to blue light (Fig. 3), which showed generally slower CRY2 degradation than that of the etiolated seedlings exposed to blue light (Fig. 1). As shown in Fig. 3, the blue light-dependent CRY2 degradation is indeed impaired in the cul4-1 mutant under the different fluence rates of blue light tested. In comparison with that of the wild-type plants, the protein stability of CRY2 is clearly elevated in cul4-1 mutant seedlings exposed to blue light of a modest fluence rate (15 μmol m-2 s-1) (Fig. 3A). Similarly, CRY2 degradation is significantly reduced (P < 0.01) in the cul4-1 mutant exposed to blue light with a higher fluence rate (30 μmol m-2 s-1) (Fig. 3B). These results argue strongly that the requirement for the COP1–SPA1 complex in blue light-dependent CRY2 degradation is most probably attributed to its CUL4-based E3 ubiquitin ligase activity.
Fig. 3.
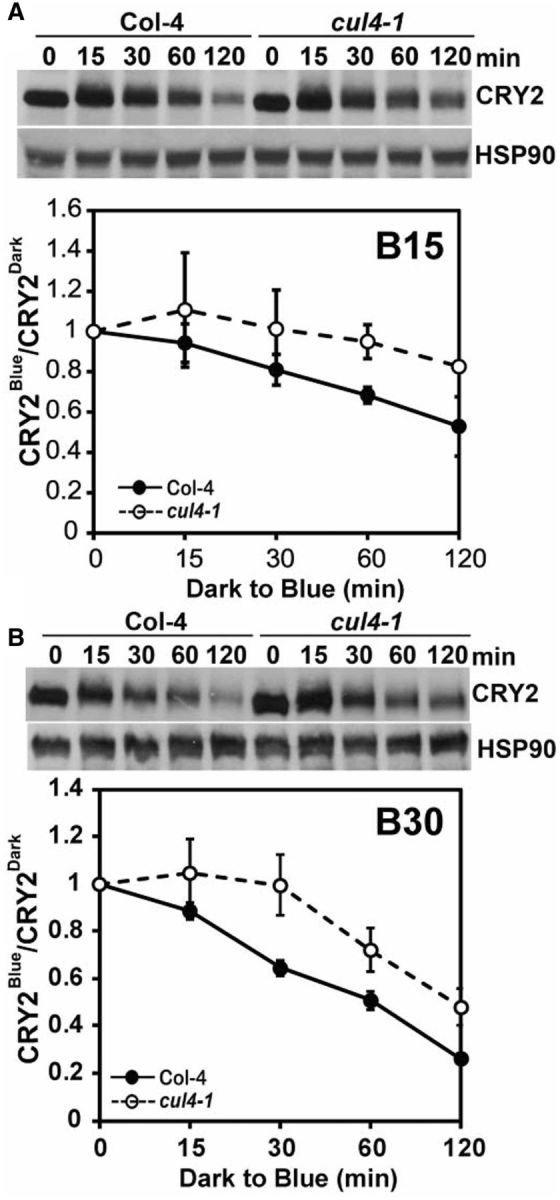
Cullin 4 is involved in blue light-dependent CRY2 degradation. (A) Immunoblot showing blue light-dependent CRY2 degradation in the cul4-1 mutant. Three-week-old wild-type (Col-4) and cul4-1 mutant plants grown in LD were adapted to dark for 24 h and treated with blue light (15 μmol m-2 s-1) for the times indicated before sample collection. The immunoblots were probed with anti-CRY2 antibody (CRY2) and reprobed with anti-HSP90 as the loading control. The relative band intensities of CRY2 were quantified and are shown with the SDs (n = 3). CRY2Blue, normalized (against HSP90) band intensity of CRY2 of blue light-treated samples, CRY2Dark, normalized band intensity of CRY2 of dark-treated samples. (B) Same as (A), except that plants were treated with blue light of 30 μμmol m-2 s-1. Three experiments were performed; one representative immunoblot and the relative levels of CRY2 (CRY2Blue/CRY2Dark) are shown with the SDs (n = 3).
The important role of COP1 in CRY2 degradation was previously revealed by a study of the cop1 hypomorphic alleles (cop1-4 and cop1-6) (Shalitin et al. 2002), but it remains unclear whether COP1 is the only E3 ligase catalyzing blue light-dependent CRY2 ubiquitination. To investigate this question, we examined CRY2 degradation in the cop1-5 null mutant allele (Ang and Deng 1994). Because the homozygous individuals of the cop1-5 allele are lethal at the seedling stage, this mutant is maintained as seeds harvested from heterozygous plants (Ang and Deng 1994). Because seeds homozygous for the cop1-5 mutation exhibit a dark-purple color (Ang and Deng 1994), we isolated the dark-purple cop1-5 seeds from populations of seeds harvested from the heterozygous cop1-5 parent. We germinated those cop1-5 homozygous mutant seeds in red light for 5 d, exposed them to blue light and analyzed CRY2 degradation (Fig. 4). Fig. 4 shows that, as reported previously in the cop1 weak alleles (Shalitin et al. 2002), the CRY2 protein is clearly more stable in the cop1-5 mutant plants than in the wild-type plants (Fig. 4). On the other hand, CRY2 still exhibited blue light-dependent degradation in the cop1-5 null mutant, especially when seedlings were exposed to higher fluence rates of blue light (Fig. 4B). This result indicates that the complete loss of function of COP1 in the cop1-5 mutant failed to abolish blue light-dependent CRY2 degradation completely. We next examined the effects of SPA proteins, which are components of the COP1–SPA ligases, in blue light-dependent CRY2 degradation. Arabidopsis encodes four SPA1-related proteins, referred to as SPA1–SPA4 (Laubinger et al. 2004), and we examined whether elimination of multiple SPA1-related genes might completely abolish the blue light-dependent CRY2 degradation. Under our experimental conditions, there seems to be no significant change in the stability of CRY2 in the spa123, spa134 and spa234 triple mutants examined (Fig. 5A). In contrast, the blue light-dependent CRY2 degradation is markedly impaired in the spa1234 quadruple mutants, suggesting that the four SPA proteins act redundantly in mediating COP1-dependent degradation. The spa triple mutants show much less CRY2 degradation than the wild-type seedlings under low fluence rates of blue light (Weidler et al. 2012), and in the present study higher fluence rates (at least 5 μmol m-2 s-1) were applied. Importantly, as observed in the cop1-5 null mutant (Fig. 4), the blue light-dependent CRY2 degradation was severely impaired but not completely abolished in the spa1234 quadruple mutants (Fig. 5B). This observation is consistent with the notion that the CUL4-based COP1–SPA ligases are not the only E3 ligase catalyzing blue light-dependent CRY2 ubiquitination and degradation. In addition to the COP1–SPA1 substrate receptor complex, a CUL4 E3 ligase also contains the DDB1 (DNA DAMAGED BINDING PROTEIN 1) bridge protein (Chen et al. 2010a, Lau and Deng 2012, Choi et al. 2014). However, similar to the SPA proteins, the DDB subunits of the CUL4-based ligase may also function redundantly, because CRY2 exhibited normal blue light-dependent degradation in the ddb1a, ddb1b and ddb2 monogenic mutants (Supplementary Fig. S3). Taking these results together, we propose that, in addition to the CUL4-based COP1–SPA E3 ubiquitin ligases, additional E3 ligase(s) are involved in the blue light-dependent CRY2 ubiquitination and degradation.
Fig. 4.
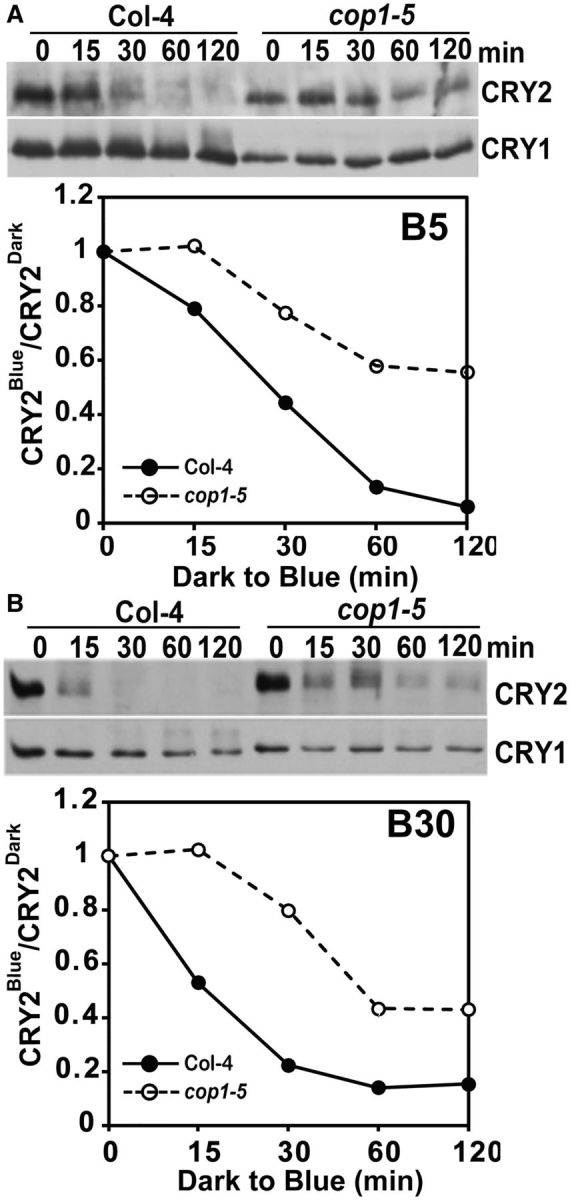
Blue light-dependent CRY2 degradation in the cop1-5 null mutant. Immunoblots showing reduced CRY2 degradation in response to blue light in the cop1-5 mutant. The wild-type (Col-4) or the cop1-5 mutant seedlings were grown in red light for 5 d and exposed to blue light of 5 μmol m-2 s-1 (A, B5) or 30 μmol m-2 s-1 (B, B30) for the durations indicated. The immunoblots were probed with anti-CRY2 antibody (CRY2), stripped and reprobed with anti-CRY1 (CRY1). The relative band intensities of CRY2 (normalized against CRY1) were quantified and presented as CRY2Blue/CRY2Red.
Fig. 5.
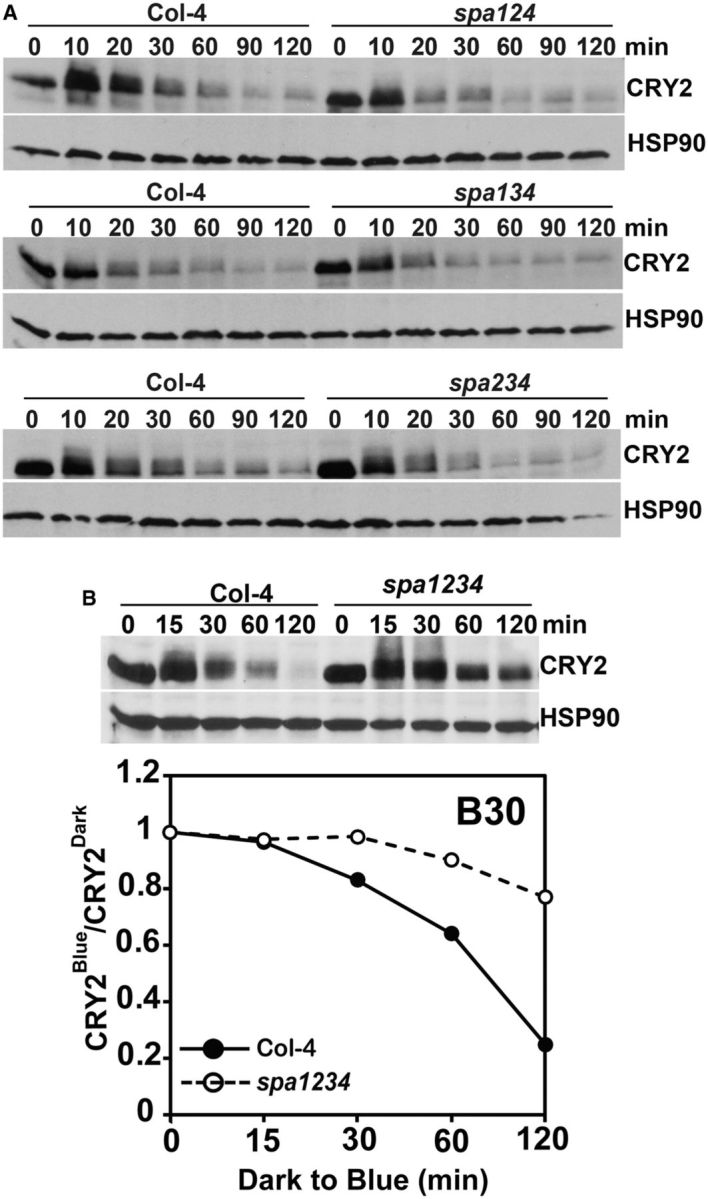
SPA proteins act redundantly to regulate blue light-dependent CRY2 degradation. (A) Immunoblot showing blue light-dependent CRY2 degradation in the genotypes indicated. Seven-day-old etiolated seedlings of wild-type (Col-4) or SPA triple mutants (spa124, spa134 or spa234) were exposed to blue light (5 μmol m-2 s-1) for the time indicated, samples were fractioned in 10% SDS–polyacrylamide gels and immunoblots, blotted, probed with anti-CRY2 antibody (CRY2), stripped and re-probed with anti-HSP90 antibody (HSP90) as the loading control. (B) Three-week-old wild-type (Col-4) and SPA quadruple mutant (spa1234) plants were grown in an LD photoperiod, adapted to the dark for 24 h, treated with blue light (30 μmol m-2 s-1) for the times indicated before sample collection. Samples were fractioned in 10% SDS–polyacrylamide gels and blotted. The immunoblots were probed with anti-CRY2 antibody (CRY2), stripped and re-probed with anti-HSP90 antibody (HSP90) as the loading control. The relative band intensities of CRY2 in the seedlings of Col-4 or spa1234 treated with blue light for the times indicated were normalized (against HSP90), quantified and aree presented as CRY2Blue/CRY2Dark.
A CUL1-based E3 ligase is involved in the blue light-dependent CRY2 ubiquitination and degradation
To test the hypothesis that E3 ligases in addition to the CUL4–COP1–SPA1 complex is involved in the blue light-dependent CRY2 ubiquitination and degradation, we analyzed CRY2 degradation in mutants impaired in genes encoding other cullin isoforms. Supplementary Fig. S4 shows that the blue light-dependent CRY2 degradation is not impaired in the cul2 mutant, suggesting that the CUL2-based E3 ubiquitin ligases may not be directly involved in CRY2 ubiquitination. We also examined the cul3a and cul3b monogenic mutants and the cul3acul3aCUL3bcul3b double mutant allele (Supplementary Fig. S4). Because the homozygous cul3acul3b double mutant is embryo lethal (Thomann et al. 2005), we examined cul3acul3aCUL3bcul3b that is homozygous for cul3a but heterozygous for cul3b (Supplementary Fig. S4). Again, no apparent defect of the blue light-dependent CRY2 degradation was detected in these mutants. Therefore, CUL3-based E3 ligase may also not participate directly in the blue light regulation of CRY2 stability.
We next examined whether CUL1-based ligases may be involved in the blue light-dependent CRY2 degradation in the temperature-sensitive cul1 (axr6-3) allele (Quint et al. 2005). The null alleles of the CUL1 gene are embryo lethal, whereas the axr6-3 allele is viable (Quint et al. 2005). The axr6-3 allele is a recessive missense mutation (E159K) of the CUL1 gene (Quint et al. 2005). The activity of CUL1 in the axr6-3 mutant declines progressively with increasing ambient temperature, resulting in more severe defects in CUL1-based E3 ligases at elevated (or non-permissive) temperatures (Quint et al. 2005). Among many different phenotypic defects, the axr6-3 mutant exhibited increased sensitivity to far-red light-dependent hypocotyl inhibition and reduced sensitivity of the far-red light-dependent PHYA degradation at 23 °C, indicating that the CUL1-based E3 ubiquitin ligase(s) plays important roles in photomorphogenesis even at normal temperatures (Quint et al. 2005).
We first compared the morphological phenotypes of the axr6-3 mutant and the wild-type plants grown at lower (20 °C) and higher (28 °C) ambient temperature under different light conditions. The adult plants of the axr6-3 mutant grown under a LD photoperiod illuminated by white light exhibited a dwarf phenotype at the normal growth temperature of 20 °C (Supplementary Fig. S5A). In the next experiment, the wild-type and axr6-3 seedlings were germinated and grown in the dark for 5 d at the normal temperature (20°C), then treated with two different temperatures (20 or 28°C) in the dark or continuous blue light (10 μmol m-2 s-1) for a further 3 d before phenotype analyses. Fig. 6 shows that, at 20°C, the axr6-3 mutant seedlings exhibited a slightly shorter hypocotyl than that of the wild-type seeding in both the dark and blue light (Fig. 6A, B). However, at 28 °C, the axr6-3 mutant seedlings exhibited a more obvious constitutive photomorphogenic phenotype in the dark, because the axr6-3 mutant seedlings developed expanded cotyledons and a shorter hypocotyl in the absence of light (Fig. 6A, B, Dark, 28 °C). When grown under continuous blue light at the higher temperature, the axr6-3 mutant exhibited a strong hypersensitivity to blue light by developing markedly shorter hypocotyls than that of the wild-type seedling (Fig. 6A, B, Blue, 28 °C). For example, the average lengths of hypocotyls of the axr6-3 mutant seedlings grown in the dark at 28 °C is about 55% that of the wild-type seedlings, whereas the average lengths of hypocotyls of the axr6-3 mutant seedlings is only about 15% that of the wild-type seedlings grown in blue light (10 μmol m-2 s-1) at 28 °C (Fig. 6B). The more severe constitutive or hypersensitive photomorphogenic phenotypes of the axr6-3 mutant grown at 28 °C are consistent with the more severe defect of this mutation in the CUL1-based protein ubiquitination processes at higher temperatures (Quint et al. 2005).
Fig. 6.
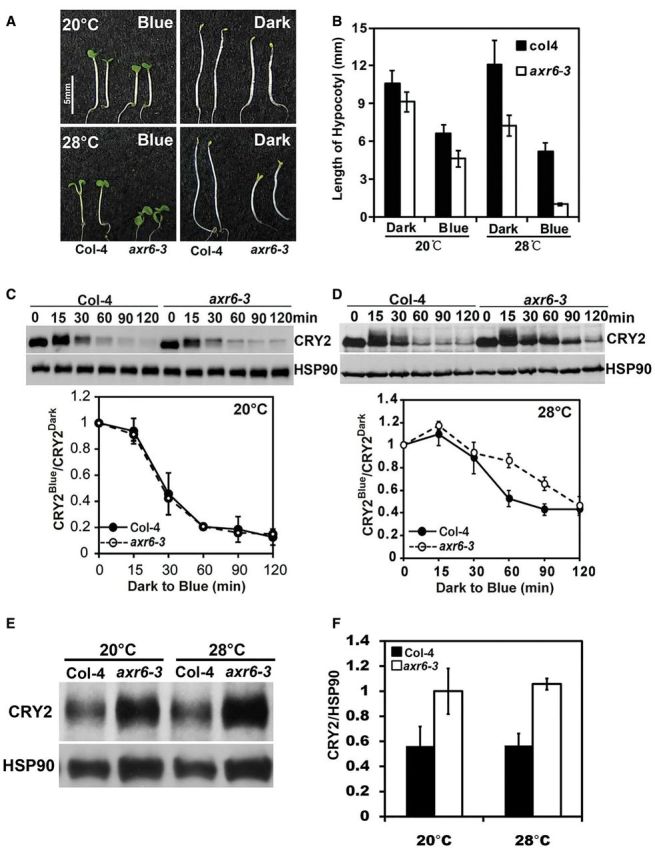
Cullin 1 is involved in blue light-dependent CRY2 degradation. (A) A comparison of the phenotype of axr6-3 and wild-type (Col-4) seedlings grown in the dark or continuous blue light (10 μmol m-2 s-1) at 20 °C for 5 d and then at 20 or 28 °C for an additional 3 d. The scale bar is 5 mm. (B) The hypocotyl lengths of the samples shown in (A). (C, D) Immunoblots showing blue light-dependent CRY2 degradation in the axr6-3 mutant. Five-day-old etiolated seedlings of the wild type (Col-4) or the axr6-3 mutant grown at 20 °C were moved to 28 °C or kept at 20 °C for 2 d, then transferred to blue light (5 μmol m-2 s-1) for the times indicated. Samples were fractioned in 10% SDS–polyacrylamide gels and the immunoblots were probed with anti-CRY2 antibody (CRY2), stripped and re-probed with anti-HSP90 antibody (HSP90) as the loading control. Three experiments were performed; one representative immunoblot (top) and the SD (bottom, n = 3) are shown. (E, F) Immunoblots showing the CRY2 protein level in Col-4 and axr6-3 mutants under continuous blue light (10 μmol m-2 s-1). Eight-day-old seedlings grown under different temperatures (20 or 28 °C) were collected. Samples were fractioned in 10% SDS–polyacrylamide gels and the immunoblots were probed with anti-CRY2 antibody (CRY2), stripped and re-probed with anti-HSP90 antibody (HSP90) as the loading control. Three independent experiments were carried out and the representative immunoblot (E) is shown. The relative band intensities of CRY2 were quantified, normalized against HSP90 and are presented as CRY2/HSP90, with the SD (n = 3) shown.
We next examined the blue light-dependent CRY2 degradation at different temperatures in the axr6-3 mutant. In this experiment, etiolated seedlings grown at 20 or 28°C were treated with blue light (5 μmol m-2 s-1) for 15–120 min, and CRY2 stability was analyzed by immunoblot (Fig. 6C, D). Fig. 6 shows that CRY2 was degraded normally in response to blue light in the axr6-3 mutant seedlings grown at 20 °C (Fig. 6C). In contrast, the blue light-dependent CRY2 degradation is modestly but significantly (P < 0.01) reduced in the axr6-3 seedlings grown at 28 °C (Fig. 6D). Given that the stability of CRY2 is determined by the blue light-dependent ubiquitination and 26S-dependent proteolysis (Shalitin et al. 2002, Yu et al. 2007b, Yu et al. 2009, Li et al. 2011, Wang et al. 2015), these results argue that a CUL1-based E3 ubiquitin ligase is probably also involved in the ubiquitination and degradation of CRY2 in response to blue light. We next compared the relative level of the CRY2 protein in the axr6-3 seedlings grown under continuous blue light (10 μmol m-2 s-1) (Fig. 6E, F). In contrast to that observed in etiolated seedlings exposed to blue light (Fig. 6C, D), the axr6-3 mutant seedlings grown under continuous blue light accumulated a significantly higher (P < 0.01) level of the CRY2 protein in the axr6-3 seedlings at both the permissive temperature of 20°C and the non-permissive temperature of 28°C (Fig. 6E, F). To examine whether this temperature-independent effect on CRY2 protein expression may result from changes of CRY2 mRNA expression, we compared levels of the CRY2 mRNA and CRY2 protein in the wild type and the axr6-3 mutant seedlings grown at 28°C under continuous blue light (10 μmol m-2 s-1) for 2, 3, 4, 5, 6 and 7 d after germination (Supplementary Fig. S5C, D). The results of this experiment show that the axr6-3 mutant seedlings grown under continuous blue light accumulated significantly higher levels of CRY2 mRNA in the axr6-3 seedlings. We also compared the CRY2 mRNA level in etiolated wild-type and the axr6-3 mutant seedlings grown at 28°C exposed to blue light (5 μmol m-2 s-1) for 30, 60 and 90 min (Supplementary Fig. S5E). No difference in the CRY2 mRNA level was observed between the wild type and the axr6-3 mutant. These results suggest that the temperature-independent increase of CRY2 protein accumulation in plants grown under continuous blue light in the axr6-3 mutant is caused by an increase of CRY2 mRNA accumulation. Why the axr6-3 mutant causes increased CRY2 mRNA expression under continuous blue light remains to be studied further. However, it is worth noticing that, although the temperature-sensitive dwarf phenotype of the axr6-3 mutant (Fig. 6A) hinted that the defective axr6-3 allele of the CUL1 gene may impair the activities of multiple E3 ligases to different extents at different temperatures, we failed to detect any significant change of CRY2 protein level in axr6-3 mutants under both permissive and non-permissive temperature conditions (Fig. 6E). One of the possible explanations for this observation might be that at the different temperatures, the impaired E3 ligases primarily affected the degradation speed in a sudden response rather than the steady-state level, which takes a long time to achieve, when the CRY2 concentration reached homeostasis in the cell.
Discussion
In the present study, we investigated genetic mechanisms underlying blue light-dependent ubiquitination and degradation of the blue light receptor CRY2. We showed that the blue light-dependent CRY2 degradation is suppressed by the conditional expression of the ubiquitin mutant ubR48, which is consistent with the hypothesis that CRY2 undergoes polyubiquitination primarily by the K48 ubiquitin–ubiquitin linkage. We also showed that neither the blue light receptor phototropins nor the blue light receptors ZTL, LKP2 and FKF1 is required for the blue light-dependent CRY2 degradation (Fig. 1). The lack of involvement of ZTL-type blue light receptors in the blue light regulation of CRY2 degradation is interesting because ZTL is a known CUL1-based E3 ubiquitin ligase that regulates ubiquitination and degradation of not only clock proteins but also the CRY2-interacting protein CIBs (Liu et al. 2013). This result, together with our finding that none of the genes closely associated with the function of the circadian clock, photobody formation or flowering time control is required for the blue light-dependent CRY2 degradation, supports the notion that CRY2 acts as the photoreceptor to mediate blue light regulation of its own ubiquitination and degradation (Yu et al. 2007a, Wang et al. 2015).
We further showed that the blue light-dependent degradation of CRY2 is impaired in the cul4-1 hypomorphic mutant, demonstrating that CUL4 is involved in CRY2 ubiquitination (Fig. 2). This observation provides strong evidence supporting the hypothesis that the involvement of COP1–SPA1 in CRY2 degradation is due to the CUL4-based E3 ubiquitin ligase activity of this protein complex. However, our results showing that the blue light-dependent CRY2 degradation was not completely abolished in the cop1-5 null mutant or in the spa1234 quadruple mutant argue for the involvement of an additional E3 ubiquitin ligase in the blue light-dependent CRY2 ubiquitination (Figs 4, 5). Consistent with this hypothesis, we showed that the blue light-dependent CRY2 degradation is also impaired in the temperature-sensitive cul1/axr6-3 mutant (Fig. 6), which suggests a possible involvement of the CUL1-based E3 ubiquitin ligase in the regulation of CRY2. The notion that multiple E3 ubiquitin ligases may be involved in the modification and regulation of Arabidopsis CRY2 is reminiscent of the cryptochrome ubiquitination systems in other organisms, which may suggest an evolutionary benefit for such systems. For example, both CUL1- and CUL4-based E3 ligases are likely to be involved in the blue light-dependent ubiquitination and degradation of Drosophila cryptochrome (Peschel et al. 2009, Ozturk et al. 2011, Ozturk et al. 2013). Two competing CUL1-based E3 ligases, SCFfbxl3 and SCFfbxl21, catalyze cryptochrome ubiquitination and degradation in different cellular compartments in mouse (Busino et al. 2007, Hirano et al. 2013, Yoo et al. 2013). It was proposed that SCFfbxl21 promotes degradation of mCRY in the cytosol, whereas it antagonizes the activity of the stronger SCFfbxl3 in the nucleus to balance the relative levels of nuclear mCRY that determines the circadian period of the clock in mammals (Yoo et al. 2013). We may expect similar utilities of the involvement of multiple E3 ubiquitin ligases in the blue light-dependent ubiquitination and degradation of plant cryptochromes. In Arabidopsis, the nuclear-localized CRY2 could form so-called photobodies in response to blue light, associated with its blue light-dependent degradation (Yu et al. 2009). Interestingly, CRY2 and SPA1 have been reported to co-localize in the nuclear bodies (Lian et al. 2011), which argues that SPA1–COP1-dependent degradation of CRY2 may occur in the nuclear bodies. Although there is no evidence to demonstrate the subcellular co-localization pattern of CUL1 and CRY2 proteins, it would be interesting to investigate the locations where CRY2 is degraded through the CUL1-dependent pathway. Finally, it is interesting that the axr6-3 mutant allele impairs both the far-red light-dependent degradation of phyA (Quint et al. 2005) and the blue light-dependent degradation of CRY2 (Fig. 6). These results suggest that the CUL1-based E3 ubiquitin ligase is involved in the light regulation of the stability of both phytochromes and cryptochromes. A direct test of this hypothesis would require identification of the possible CUL1-based E3 ligases involved in light-dependent degradation of phyA and CRY2.
Materials and Methods
Plant materials
The wild types used in this study are: Col-4 (Columbia-4), Ler (Landsberg erecta) and WS (Wassileskija). Mutants used here are: axr6-3 (Quint et al. 2005), cul2 (SALK_012144C), cul3a (SALK_050756C), cul3b (GABI_003D02), cul3acul3aCUL3bcul3b (Thomann et al. 2005), cul4-1 (Bernhardt et al. 2006), ddb-1a (SALK_038757C), ddb-1b (SALK_061944C), ddb2 (SALK_038892C), det1 (CS874643), spa123, spa124, spa134 and spa1234 (Laubinger et al. 2004), cop1-5 (CS6259), hmr-2 (Chen et al. 2010b), ubR48 (Schlögelhofer et al. 2006), toc1-2 (Ito et al. 2007), elf3-1 (Zagotta et al. 1992), cca1-1lhy1-1 (Mizoguchi et al. 2002), prr1579 (a gift from Takeshi Mizuno’s lab), fwa-1 (CS106), flc-3 (Redei 1962), ztl-3 (Somers et al. 2004), ztl-4lkp2-1 and ztl-4lkp2-1fkf1-2 (Baudry et al. 2010), phot1-5phot2-1 (Kagawa et al. 2001), co-9 (Balasubramanian et al. 2006), ft-10 (CS9869), gi-1 (CS3123), fd-1(CS53) and ft-10/tsf-1 (Jang et al. 2009).
Immunoblot analyses
To analyze levels of the CRY2 protein, seeds were sterilized for 15 min in 75% ethanol with 0.05% Triton X-100 (T9284, Sigma) and in 100% ethanol for 2 min, and dried in a vacuum machine for 30 min. After imbibition for 4 d in the dark at 4°C, seeds were exposed to white light for 24 h before being transferred to red light or darkness. After 2–7 d, seedlings were transferred to blue light of the indicated fluence rates and duration by LED (light-emitting diode) light. An LED was used to obtain monochromatic blue (peak 450 nm; half-bandwidth of 20 nm) and red light (peak 660 nm; half-bandwidth of 20 nm), and cool white fluorescent tubes were used for the white light in this study. After light treatment, seedling samples were collected for immunoblot analyses. The protein extracts were separated by 10% SDS–PAGE and transferred to an Immobilon NC Transfer Membrane (HATF00010, Millipore). The membrane was stained with Ponceau S red and blocked with 5% skimmed milk in PBST solution. After probing with primary and secondary antibodies, the membrane was incubated in the buffer supplied with the Super Signal West Pico Chemiluminescent substrate kit (Pierce) to detect signals derived from antibodies. The primary antibodies used in this study included: anti-CRY2 (1 : 3,000) (Guo et al. 1998), anti-CRY1 (1 : 3,000) (Guo et al. 1998) and anti-HSP90 (1 : 5,000, sc-33755, Santa Cruz).
Funding
This work is supported the National Institute of Health [GM56265 to C.L.; Fujian Agriculture and Forestry University research support to the Basic Forestry and Proteomics Research Center]; and Jilin University [research support to the Laboratory of Soil and Plant Molecular Genetics)].
Supplementary Material
Acknowledgments
The authors thank Drs. William Gray for the axr6-3 allele, Mark Esteller for the cul1-6 allele, Pascal Genschik for the cul3acul3aCUL3bcul3b, Takeshi Mizuno for the prr mutants, Ute Hoecker for all the spa mutants, Andreas Bachmair for the ubR48 transgenic line, Hanjo Hellmann for the cul4-1 allele and Xingwang Deng for the cop1-5 allele used in this study.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- CIB
CRY-interacting basic helix–loop–helix
- COP1
CONSTITITIVE PHOTOMORPHOGENSIS 1
- CRY2
cryptochrome2
- CUL
cullin
- DEX
dexamethasone
- FKF1
Flavin-binding Kelch Repeat F-box1
- LD
long days
- LKP2
Lov Kelch Protein2
- SPA1
(SUPPRESSOR OF PHYA-105
- ZTL
ZEITLUPE
References
- Ahmad M., Jarillo J.A., Cashmore A.R. (1998) Chimeric proteins between cry1 and cry2 arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Deng X.W. (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Ito S., Song Y.H., Strait A.A., Kiba T., Lu S., et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., Anders M., et al. (2006) CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47: 591–603. [DOI] [PubMed] [Google Scholar]
- Busino L., Bassermann F., Maiolica A., Lee C., Nolan P.M., Godinho S.I., et al. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316: 900–904. [DOI] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., et al. (2010a) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Galvão R.M., Li M., Burger B., Bugea J., Bolado J., et al. (2010b) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Lory N., Stauber J., Hoecker U. (2015) Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 11: e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.M., Gray W.M., Mooney S., Hellmann H. (2014) Composition, roles, and regulation of cullin-based ubiquitin E3 ligases. Arabidopsis Book 12: e0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão R.M., Li M., Kothadia S.M., Haskel J.D., Decker P.V., Van Buskirk E.K., et al. (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26: 1851–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Harmon F., Imaizumi T., Gray W.M. (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J. 55: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A., Yumimoto K., Tsunematsu R., Matsumoto M., Oyama M., Kozuka-Hata H., et al. (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152: 1106–1118. [DOI] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Deng X.W. (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 21: 96–103. [DOI] [PubMed] [Google Scholar]
- Ito S., Nakamichi N., Nakamura Y., Niwa Y., Kato T., Murakami M., et al. (2007) Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 48: 971–983. [DOI] [PubMed] [Google Scholar]
- Ito S., Song Y.H., Imaizumi T. (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Torti S., Coupland G. (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60: 614–625. [DOI] [PubMed] [Google Scholar]
- Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S., Kato T., et al. (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138–2141. [DOI] [PubMed] [Google Scholar]
- Keller M.M., Jaillais Y., Pedmale U.V., Moreno J.E., Chory J., Ballare C.L. (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Scalf M., Smith L.M., Vierstra R.D. (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., et al. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell 16: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang Q., Yu X., Liu H., Yang H., Zhao C., et al. (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc. Natl Acad. Sci. USA 108: 20844–20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., et al. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A.R. (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95: 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011) The action mechanisms of plant cryptochromes. Trends Plant Scie 16: 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Q., Liu Y., Zhao X., Imaizumi T., Somers D.E., et al. (2013) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. PProc. Natl. Acad. Sci. USA 110: 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., et al. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539. [DOI] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., et al. (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P., Devlin P.F., Panda S., Kay S.A. (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211. [DOI] [PubMed] [Google Scholar]
- Mas P., Kim W.Y., Somers D.E., Kay S.A. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.R., et al. (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Ozkan-Dagliyan I., Chiou Y.-Y., Ye R., Hassan B.H., Ozturk N., Sancar A. (2013) Formation of Arabidopsis cryptochrome 2 photobodies in mammalian nuclei: application as an optogenetic DNA damage checkpoint switch. J. Biol. Chem. 288: 23244–23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N., Selby C.P., Annayev Y., Zhong D., Sancar A. (2011) Reaction mechanism of Drosophila cryptochrome. Proc. Natl. Acad. Sci. USA 108: 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N., VanVickle-Chavez S.J., Akileswaran L., Van Gelder R.N., Sancar A. (2013) Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1–CUL4–ROC1 E3 ligase complex. Proc. Natl. Acad. Sci. USA 110: 4980–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N., Chen K.F., Szabo G., Stanewsky R. (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 19: 241–247. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li M., Pasoreck E.K., Long L., Shi Y., Galvão R.M., et al. (2015) HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27: 1409–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Ito H., Zhang W., Gray W.M. (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 43: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redei G. (1962) Supervital mutations of Arabidopsis. Genetics 47: 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlögelhofer P., Garzón M., Kerzendorfer C., Nizhynska V., Bachmair A. (2006) Expression of the ubiquitin variant ubR48 decreases proteolytic activity in Arabidopsis and induces cell death. Planta 223: 684–697. [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., et al. (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767. [DOI] [PubMed] [Google Scholar]
- Somers D.E., Kim W.Y., Geng R. (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A., Brukhin V., Dieterle M., Gheyeselinck J., Vantard M., Grossniklaus U., et al. (2005) Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 43: 437–448. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009) The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Walsh C.K., Sadanandom A. (2014) Ubiquitin chain topology in plant cell signaling: a new facet to an evergreen story. Front. Plant Sci. 5: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Barshop W.D., Bian M., Vashisht A.A., He R., Yu X., et al. (2015) The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol. Plant 8: 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidler G., zur Oven-Krockhaus S., Heunemann M., Orth C., Schleifenbaum F., Harter K., et al. (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. The Plant Cell 24: 2610–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-H., Mohawk J.A., Siepka S.M., Shan Y., Huh S.K., Hong H.-K., et al. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Klejnot J., Zhao X., Shalitin D., Maymon M., Yang H., et al. (2007a) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Sayegh R., Maymon M., Warpeha K., Klejnot J., Yang H., et al. (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Shalitin D., Liu X., Maymon M., Klejnot J., Yang H., et al. (2007b) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA104: 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Shannon S., Jacobs C., Meeks-Wagner D.R. (1992) Early-flowering mutants of Arabidopsis thaliana. Funct. Plant Biol. 19: 411–418. [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.