Abstract
Many current anti-cancer therapies rely on increasing the intracellular reactive oxygen and nitrogen species (RONS) contents with the aim to induce irreparable damage, which subsequently results in tumor cell death. A novel tool in cancer therapy is the use of cold atmospheric plasma (CAP), which has been found to be very effective in the treatment of many different cancer cell types in vitro as well as in vivo, mainly through the vast generation of RONS. One of the key determinants of the cell's fate will be the interaction of RONS, generated by CAP, with important proteins, i.e. redox-regulatory proteins. One such protein is cytoglobin (CYGB), a recently discovered globin proposed to be involved in the protection of the cell against oxidative stress. In this study, the effect of plasma-produced RONS on CYGB was investigated through the treatment of CYGB with CAP for different treatment times. Spectroscopic analysis of CYGB showed that although chemical modifications occur, its secondary structure remains intact. Mass spectrometry experiments identified these modifications as oxidations of mainly sulfur-containing and aromatic amino acids. With longer treatment time, the treatment was also found to induce nitration of the heme. Furthermore, the two surface-exposed cysteine residues of CYGB were oxidized upon treatment, leading to the formation of intermolecular disulfide bridges, and potentially also intramolecular disulfide bridges. In addition, molecular dynamics and docking simulations confirmed, and further show, that the formation of an intramolecular disulfide bond, due to oxidative conditions, affects the CYGB 3D structure, thereby opening the access to the heme group, through gate functioning of His117. Altogether, the results obtained in this study (1) show that plasma-produced RONS can extensively oxidize proteins and (2) that the oxidation status of two redox-active cysteines lead to different conformations of CYGB.
Abbreviations: CAP, Cold atmospheric plasma; CYGB, human cytoglobin; RONS, reactive oxygen and nitrogen species; ROS, reactive oxygen species; MS, mass spectrometry; CD, circular dichroism; MD, molecular dynamics; LC-MS/MS, liquid chromatography – tandem mass spectrometry; IM, ion mobility; ESI, nano-electrospray ionization; DTT, Dithiothreitol; TCEP, tris (2-carboxyethyl) phosphine; CID, collision-induced dissociation; RR, resonance raman; CYGBS-S, (monomeric cytoglobin with intramolecular disulfide bridge forming cysteines); CYGBSH…SH, (monomeric cytoglobin with free sulfhydryl groups on the cysteines); PSM, peptide spectrum matches
Keywords: Cold atmospheric plasma, Cytoglobin, Reactive oxygen and nitrogen species, Oxidative modifications, Disulfide bridges
Graphical abstract
1. Introduction
The use of cold atmospheric plasma (CAP) has attracted a lot of attention over the past few years and has already found many biomedical applications, including wound healing [1], sterilization [2], biofilm removal [3], blood coagulation [4], and more recently cancer therapy [5]. CAP is a plasma (i.e., ionized gas) in which the high-temperature electrons are in non-equilibrium with the low-temperature heavy particles, thereby keeping the gas itself near room temperature and making it suitable for treatment of living cells and tissues. Plasma is a mixture of various components, including charged particles (ions, electrons), reactive neutral species (reactive oxygen and nitrogen species; RONS), UV radiation, and electromagnetic fields [6]. Of those, RONS are hypothesized to mediate the effects observed in biological systems as they are known to be involved in rapid reactions with important biomolecules such as proteins and lipids [7], [8]. In the last few years, CAP was shown to be able to induce apoptotic cell death in numerous different cancer cell types [9], [10], [11], [12], [13], [14], [15]. Although not fully understood, a possible mechanism of action is through: (1) the production of reactive species by CAP, which then diffuse into the cells or create extra RONS inside the cells, e.g., due to lipid peroxidation; (2) an increase in the level of intracellular RONS that oxidize proteins, lipids, and DNA; (3) oxidation exceeding the anti-oxidant capacity of the cells leads to irreversibly damaged biomolecules, which in turn induces a physiological response through cell death. Therefore, to gain further insight into these processes, it is necessary to study the interaction between CAP and cells on a molecular level.
As proteins are the main facilitators of biological functions (and account for about 50% of the gross dry weight of cells), one could hypothesize that plasma-protein interactions will be key determinants of the cell's fate after plasma treatment. Recent investigations have shown that CAP can affect infectious prion protein [16], recombinant green fluorescent protein [17], lysozyme [18], and globins, such as hemoglobin and myoglobin [19] through modification of the higher-order structure and/or heme degradation. Proteins of a particular interest to investigate the plasma-protein interactions would be redox-sensitive proteins, as they are centrally involved in regulating the redox balance inside the cell, mainly through oxidative modification of redox-sensitive cysteine residues [20]. One such protein is cytoglobin (CYGB). CYGB is a recently discovered hexa-coordinated globin with many proposed functions, including oxygen transport, NO-dioxygenase activity [21], [22], reactive oxygen species (ROS)-scavenger [23], and tumor suppression [24]. Although some of these proposed functions are still under debate, there is a general consensus that CYGB can protect the cell from oxidative stress. This implies that CYGB is a redox-sensitive protein, of which the sensitivity has been suggested to occur through heme-coordination and/or cysteine residues (Cys38 and Cys83) on its surface, which can form intra- or intermolecular disulfide bridges [25], [26], [27]. Treating CYGB with plasma would allow for the verification and further clarification of the proposed models on how plasma-induced RONS affect biomolecules. Furthermore, it can shed new light on recent findings [28], [29], [30] concerning how CYGB is involved in cell protection against oxidative stress.
In this study, recombinant CYGB was treated with a cold plasma jet (kINPen IND) for different treatment times. Possible structural modifications were investigated by using electronic circular dichroism (CD), UV–Vis spectroscopy, and resonance Raman spectroscopy. Additionally, mass spectrometry (MS) experiments were performed on the native form and under denaturing conditions. Finally, molecular dynamics (MD) and docking simulations were performed to assess the effect of an intramolecular disulfide bond on the structure and function of monomeric CYGB. These combined experiments and simulations allow us to gain an insight into the effect of plasma treatment, and also oxidative stress, on structural features of proteins, which should contribute to a better understanding of the above-mentioned postulated functions of CYGB.
2. Experimental
2.1. Expression and purification of recombinant CYGB
The expression of recombinant wild-type CYGB was performed as previously described [31]. Briefly, human cDNA of CYGB was cloned in the pET3a expression vector. Inclusion bodies were solubilized in 6 M guanidinium hydrochloride and insoluble material was eliminated. Afterwards, CYGB was reconstructed by adding free hemin and dialyzed overnight. The samples were then purified with an Akta purifier system on a Hitrap DEAE-Sepharose column (both GE Healthcare, Life Sciences). The concentrated material was loaded onto a Sephacryl S-200 High resolution column (GE Healthcare). The final purity of the pooled CYGB was checked by absorbance spectra and SDS-PAGE.
2.2. CAP treatment of recombinant CYGB
A recombinant CYGB stock solution (19.3 mg/mL; 901 µM) was diluted either in deionized water or 50 mM tris-buffer (pH 8) to a final concentration of 20 µM. Plasma treatment was performed on 500 µL aliquots in a 24-well plate (on ice) for different time points (30 s, 1 min, 3 min, 5 min, 10 min) using the kINPen IND plasma jet (neoplas GmbH). The applied gas flow rate and gap distance were set at 3 L/min, and 10 mm, respectively, for all treatments. Argon was used as feed gas. After treatment, the evaporated volume of the aliquots was calculated and replaced by either water or tris-buffer. The treated samples were immediately put on ice before being handled for further analysis.
The pH of both used solvents was measured before and after treatment with CAP at different lengths of time to assess the effect of CAP treatment on the pH of the samples. The pH was measured with the pHenomenal pH 1100H pH-meter (VWR).
2.3. Mass spectrometry
2.3.1. Native and denatured MS
Before native MS analysis, 50 µL of each sample was buffer exchanged using a Micro Bio-Spin P-6 Gel column (Bio-Rad) equilibrated with 100 mM aqueous ammonium acetate (pH 6.8), according to the manufacturer's protocol. Afterwards, the samples were further diluted to a concentration of 10 µM.
3 µL of sample solution were loaded into an in-house produced, gold coated nano-electrospray ionization (ESI) glass capillaries and mounted in the needle holder of the instrument. MS spectra were acquired on a Synapt G2 HDMS instrument (Waters, Wilmslow, UK) using the following instrument parameters: 1.6 kV spray capillary voltage; 100 V sampling cone; 3 V extractor cone; 6.3 mbar backing pressure; 15 V and 5 V collision energy in the trap (2.4 × 10−2 mbar) and transfer (2.5 × 10−2 mbar) cell, respectively; 45 V trap DC bias; 180 mL/min and 90 mL/min He and IMS gas flow, respectively. The ions were separated in the ion mobility (IM) cell, using nitrogen gas and a wave velocity of 750 m/s and a wave height of 40 V.
The samples were also measured under denaturing conditions by diluting the sample 1:1 with acetonitrile containing 1% formic acid. The MS parameters for measuring the denatured samples were: 1.4 kV spray capillary voltage; 25 V sampling cone; 1 V extractor cone; 2.7 mbar backing pressure; and 5 V collision energy in trap (2.00 × 10−2 mbar) and transfer (2.2 × 10−2 mbar) cell. Settings for IM separation were the same as under native conditions. Additional native and denaturing measurements on the tris-diluted CYGB samples were also performed in 10 mM dithiothreitol (DTT).
2.3.2. In solution protein digestion and MS
To detect oxidative modifications at amino acid level, a tryptic digest was performed on each treated sample following our own protocol. Briefly, 40 µg of plasma-treated sample was used. 1% RapiGest SF Surfactant (Waters) was added to a final concentration of 0.1% before incubation for 5 min at 100 °C. 200 mM TCEP (tris (2-carboxyethyl) phosphine) was added to reduce the sample. After 1 h incubation at 55 °C, 375 mM 2-iodocetamide was added and further incubated for 30 min in the dark to alkylate the sample. After alkylation, the sample was precipitated with pre-chilled acetone and incubated overnight at − 20 °C. The acetone-precipitated pellet was resuspended in 100 mM triethylammonium bicarbonate to a final concentration of 1 µg/µL before MS grade trypsin protease (Thermo Scientific) was added to a final protease-protein ratio of 1:40 (w/w). Tryptic digest was performed overnight at 37 °C. The enzymatic reaction was stopped by freezing the samples at − 20 °C. The digested samples were measured using liquid chromatography – ion trap tandem mass spectrometry (LC-MS/MS).
The peptide mixture was separated by reversed phase C18 (RP-C18) liquid chromatography on a nanoAcquity UPLC system (Waters) using a Symmetry C18 trap column (5 µm particle size; 280 µm × 20 mm) connected to a BEH C18 analytical column (1.7 µm particle size; 100 µm × 100 mm) (Waters). Before loading, each sample was dissolved in 10 µL mobile phase A (0.1% formic acid in 100% water) and in total 1 µg of protein digest was loaded onto the column. A linear gradient of mobile phase B (0.1% formic acid in 100% acetonitrile) from 5% to 55% in 50 min followed by a steep increase to 100% mobile phase B in 3 min was used at a flow rate of 400 nL/min. The nano-UPLC system was coupled online with the mass spectrometer using a PicoTip Emitter (New objective) and a nanospray ion source (Thermo Scientific). The LTQ ion trap mass spectrometer (Thermo Scientific) was set up in MS mode, where a full MS1 scan (300–2000 m/z) was followed by collision-induced dissociation (CID) and a subsequent MS2 scan (100–2000 m/z). All scans were acquired in the linear ion trap with an automatic gain control (AGC) set at 3 × 104 ions for MS1 and 1 × 104 ions for MS2 scans. The normalized collision energy was 31% in CID, and a dynamic exclusion list of 30 s for data dependent acquisition was used. Between every sample a blank run was performed to exclude any possible carry-over effect.
Proteome Discoverer (2.1 SP1) software (Thermo Scientific) was used to perform database searching against the UniProt reference human database using both the Sequest and Mascot algorithms. Following settings were applied: precursor mass tolerance of 500 ppm, fragment mass tolerance of 0.50 Da. Trypsin was specified as digesting enzyme and 2 missed cleavages were allowed. Cysteine carbamidomethylation was defined as fixed modification and oxidations (on residues M/Y/D/F/H/W/N and P) were set as variable modifications. The results were filtered using the following settings: only medium and high confident peptides with a global FDR < 5% based on a target-decoy approach [16] and first ranked peptides were included in the results.
2.4. UV–vis spectroscopy
UV–vis circular dichroism (CD) and absorption spectra were collected using a Chirascan Plus qCD (LAAPD solid-state detector) spectrometer (Applied Photophysics Ltd.) operating with a bandwidth of 1 nm, a step size of 1 nm, a scanning speed of 0.5 s/nm, a sample temperature of 20 °C and a cuvette with a path length of 0.5 mm or 2.0 mm. The spectral region sensitive to secondary structure 180–280 nm was measured using a 0.5 mm path length and the spectral region 225–600 nm comprising the Soret band and Q-bands was measured with a path length of 2.0 mm.
2.5. Resonance Raman spectroscopy
Resonance Raman (RR) spectra were acquired using a Dilor XY-800 spectrometer in low-dispersion mode using a liquid N2-cooled CCD detector. The excitation source was a Spectra Physics (Mountain View) BeamLok 2060 Kr+ laser operating at 413.1 nm. The spectra were recorded at room temperature and the protein solutions were magnetically stirred at 500 rpm in order to avoid local heating and photochemical decomposition. The slit width used during the experiments was 200 µm. In general, 10–12 spectra were acquired with an integration time of 240 s each. Spikes due to cosmic rays were removed by omitting the highest and lowest data points for each frequency and by averaging the remaining values. Typical sample concentrations were in the order of 20 μM.
2.6. Computer simulations (molecular dynamics and docking)
Molecular dynamics (MD) and docking simulations were performed in order to elucidate the functionality of monomeric CYGB with and without an intramolecular disulfide bridge, at the molecular level. The simulations were carried out using the GROMACS [32] program package (version 5.1), applying the GROMOS 45a3 force field [33]. The coordinate file (with the initial configuration) of monomeric CYGB was obtained from the Protein Data Bank (PDB ID: 1V5H) [34]. The structure of CYGB without an initial disulfide bond, i.e. CYGBSH…SH, was placed in a dodecahedron box, with size chosen in such a way that there initially was 1.0 nm buffer distance between the protein and the boundary of the simulation box. This was done in order to avoid long distance interactions (i.e. Coulomb and van der Waals interactions) between CYGBSH…SH and its periodic images. The box was then filled with water molecules surrounding the CYGBSH…SH molecule, employing the SPC water model [35], and sodium counter ions were added to neutralize the system. Periodic boundary conditions were applied in all directions.
In order to study the effect of the abovementioned disulfide bond on the conformation of CYGB, this disulfide linkage was manually created using Avogadro software [36]. Subsequently, energy minimization was applied to optimize the modified structure, using the steepest descent algorithm. The measured length of the disulfide bond after the energy minimization was 2.05 Å, which is exactly the same as the disulfide bond length given in literature [37], [38]. The abovementioned MD procedure was then repeated (i.e., the creation of the box and addition of water molecules, as well as counter ions) to create the simulation system with CYGB containing a disulfide bridge, i.e. CYGBS-S.
Thus, two model systems were used (i.e., CYGBSH…SH and CYGBS-S) covered with water as a starting structure for the calculations. The systems were minimized in three steps, by applying three optimization algorithms, i.e., steepest descent, conjugate gradient and low-memory Broyden-Fletcher-Goldfarb-Shanno (L-BFGS) [39]. Subsequently, a 500 ps equilibration run was performed employing the NVT ensemble (i.e., a system with constant number of particles N, volume V and temperature T). Next, a 150 ns production run was performed using NPT dynamics (i.e., a system with constant number of particles N, pressure P and temperature T). The NPT simulations were carried out for 300 K and 1 bar pressure, employing the velocity-rescaling thermostat with a coupling constant of 0.1 ps and the isotropic Parrinello-Rahman barostat [40], [41], with a compressibility and coupling constant of 4.5 × 10−5 bar−1 and 2 ps, respectively. A 1.4 nm cut-off distance was applied for the van der Waals interactions. The long range electrostatic interactions are described by the particle mesh Ewald (PME) method [42], again using 1.4 nm cut-off for the real-space interactions in combination with a 0.15 nm spaced-grid for the reciprocal-space interactions.
In the docking simulations, binding hot spots of equilibrated CYGBSH…SH and CYGBS-S were predicted using the FTMap family of the web server [43]. FTMap samples the positions of 16 small probing organic molecules and identifies potential ligand binding regions of the protein based on the lowest binding free energy. These individual probing molecules may bind at various locations of the protein, and thus their clusters specify binding hot spots (so-called binding pockets) of the protein. The probing molecules used in the FTMap server are ethane, ethanol, isopropanol, isobutanol, acetonitrile, methanamine, N,N-dimethylformamide dimethyl ether, benzaldehyde, benzene, cyclohexane, phenol, acetamide, acetone, acetaldehyde and urea. They vary in shape, size and polarity, and can be considered as drug-like molecules [43]. The hot spot of the protein was identified by means of a mapping algorithm that consisted of the following four steps: (1) each probe molecule samples on a grid around the fixed protein. The energy function considers van der Waals energy with attractive and repulsive contributions and an electrostatic as well as solvation term characterized by the Poisson–Boltzmann continuum model. The algorithm choses the 2000 best positions of each probing molecule and proceeds to the next stage. (2) These best positions of the probe molecules with the protein complex are energy minimized using the CHARMM potential [44] with an analytical continuum electrostatics model [45]. During the energy minimization run, the protein atoms are fixed while the probing molecules are free to move. (3) The minimized structures are clustered within a 4 Å radius, considering the lowest energy structure. For each probe molecule, six clusters are retained with the lowest energy, which are ranked by their Boltzmann averaged energies. (4) The clusters of the 16 different probe molecules are then clustered into consensus clusters. This clustering stage defines the center of mass for each cluster probe. The highest ranked consensus cluster is selected, using the distance between the centers of mass of the cluster centers as measure for the distance and 4 Å as the clustering radius. VMD [46], and Pymol [47] visualizing tools were used for preparation of the images.
3. Results
3.1. Spectroscopic changes upon plasma treatment of CYGB
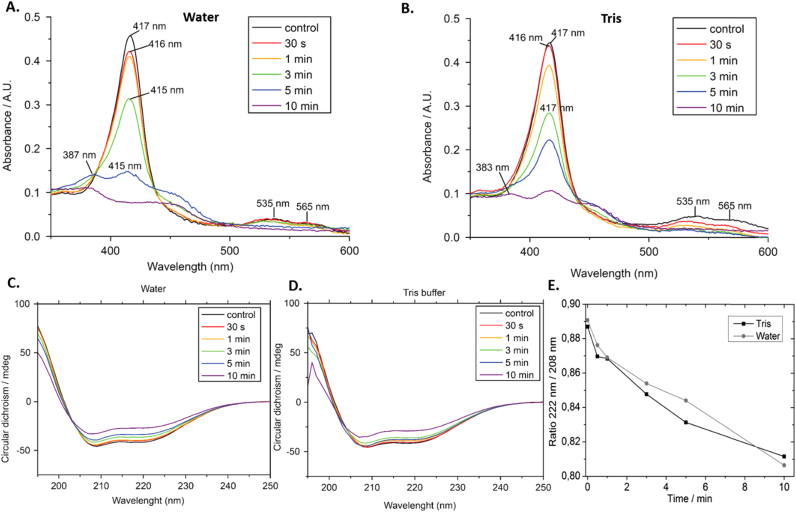
As an initial study of the structural effects of plasma treatment on CYGB, UV–vis absorption and CD spectra were recorded and are presented in Fig. 1. The UV–vis absorption spectrum of the untreated sample contains a Soret (γ) band at 417 nm, and β and α bands in the visible region of the spectrum at 535 and 565 nm, respectively, indicating that the heme iron is in a ferric low-spin HisF8-Fe(III)-HisE7 hexa-coordination state [48]. The treatment of CYGB with plasma induces significant spectroscopic changes. Most noteworthy is the time-dependent disappearance of the Soret band, but also the steady decrease of the intensity of the β and α bands with longer treatment times. When comparing the spectra, acquired in the two solvents that were employed, both similarities and differences can be observed. For both, a time-dependent decrease in intensity of the Soret band can be observed. However, the Soret band of the samples diluted in tris-buffer is still clearly visible after 5 min exposure to plasma. Even after 10 min, there is still positive intensity discernible at 417 nm in tris-buffer, while this is not the case in water. Even more so, in water, a new peak appears at 387 nm after 5 min of treatment, indicating that free heme is present in the solution [49]. In tris-buffer, the same peak (383 nm) only starts to appear after 10 min treatment, again indicating the presence of free heme.
Fig. 1.
Spectroscopic changes of CYGB after plasma treatment. UV–vis spectra of untreated and plasma-treated CYGB in (A.) water, and (B.) tris-buffer, in the 350–600 nm region with the Soret band (417 nm) and β (535 nm) and α (565 nm) bands clearly visible. (C) and (D): CD spectra of untreated and plasma-treated CYGB samples in the 190–250 nm region, depicting the double minima at 208 nm and 222 nm typical for alpha helical structures. (E): Change of the 208/222 nm ratio in function of the treatment time with both used solvents.
The CD spectra of CYGB before and after treatment are shown in Fig. 1C and D. All spectra display the typical double minima at 208 and 222 nm, characteristic for protein structures that are predominantly α-helical [50]. Plasma treatment, besides altering the intensity of the double minima, does not seem to influence the spectra significantly, suggesting that the secondary structure remains largely unaltered by the plasma treatment. As the ϴ222/ϴ208 ratio is sensitive to structural changes, the ratio was plotted over treatment time (Fig. 1E) [50]. The plot shows that, with both solvents, the ratio drops depending on the treatment time. The change in the ratio, although very minor, indicates a slight unfolding of the protein.
pH measurements indicated that the pH of the samples dissolved in water significantly drops after plasma treatment, whereas with tris-buffer, no change in pH could be observed (Fig. S3). As the change in pH of water could have a profound effect on CYGB, and thus on the results obtained with this solvent, we decided to focus further only on the results obtained with tris-buffer.
3.2. Mass spectrometry experiments
Following the UV–vis and CD measurements, mass spectrometry (under native and denaturing conditions) was used to assess changes in the tertiary structure of CYGB samples as well as chemical modifications after treatment with plasma.
3.2.1. Native MS
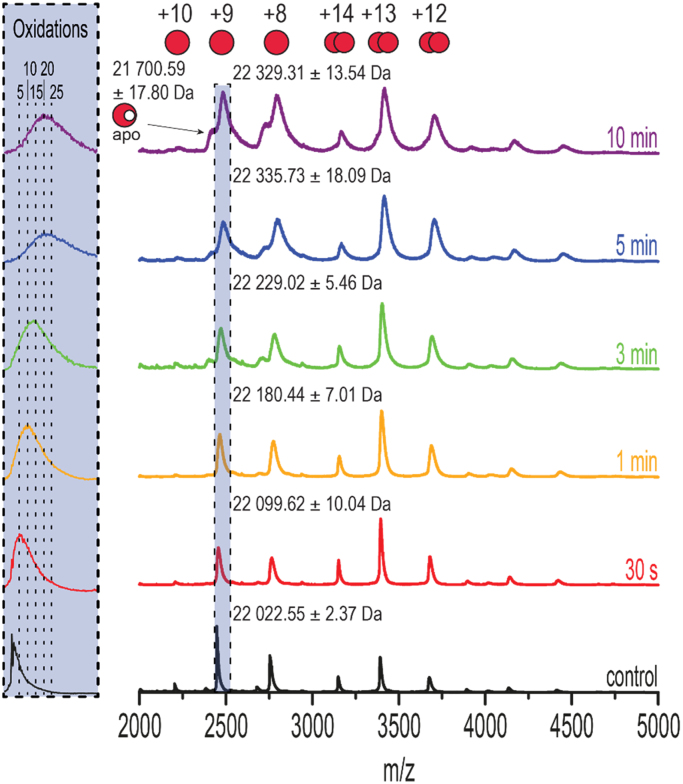
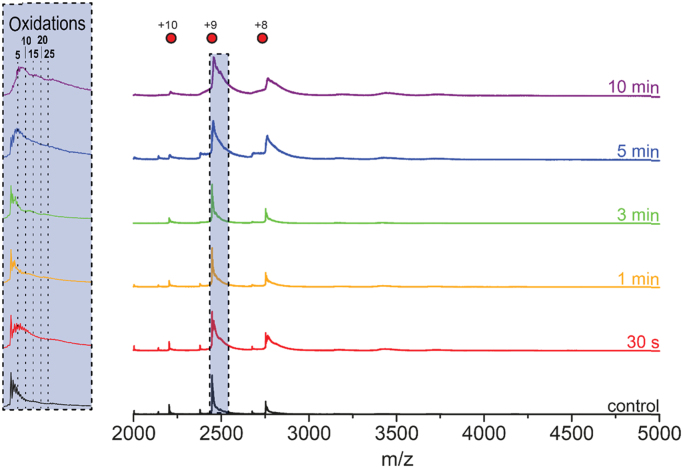
Fig. 2 shows the native mass spectra of untreated and treated CYGB. The spectrum of the untreated sample (“control”) clearly exhibits a charge state distribution for monomeric and dimeric species, with the monomeric species accounting for the majority of the signal (see also Fig. S1). As can be seen, plasma treatment not only leads to peak broadening and a shift to higher m/z values, but also to a small increase of dimeric species. The peak shift and broadening is highlighted (in blue) for the monomeric + 9 species in the m/z region of 2400–2600 (Fig. 2, inset). Here, it can be seen that plasma treatment of only 30 s already causes a mass shift to higher m/z values and peak broadening, which indicates an unresolved distribution of oxidized species. Not much change is observed up to 3 min treatment of CYGB. However, a major peak shift is visible after 5 and 10 min of treatment, which is probably caused by binding of an additional ligand to the heme (see further, Fig. 3). In addition, a small fraction of apo-CYGB is also present, in agreement with UV–vis data that showed heme loss after 10 min as well.
Fig. 2.
Native MS. Native MS spectra of 10 µM holo CYGB in 100 mM ammonium acetate after different duration of plasma treatment (20 µM CYGB in 50 mM tris-buffer). Monomeric (+10, +9, +8) and dimeric (+14, +13, +12) species are identified. The monomeric +9 species are highlighted in blue. Inset: zoomed in section of the +9 monomers, highlighting mass shifts and peak broadening caused by plasma-induced oxidations. The theoretical mass of holo CYGB is 22,021.11 Da.
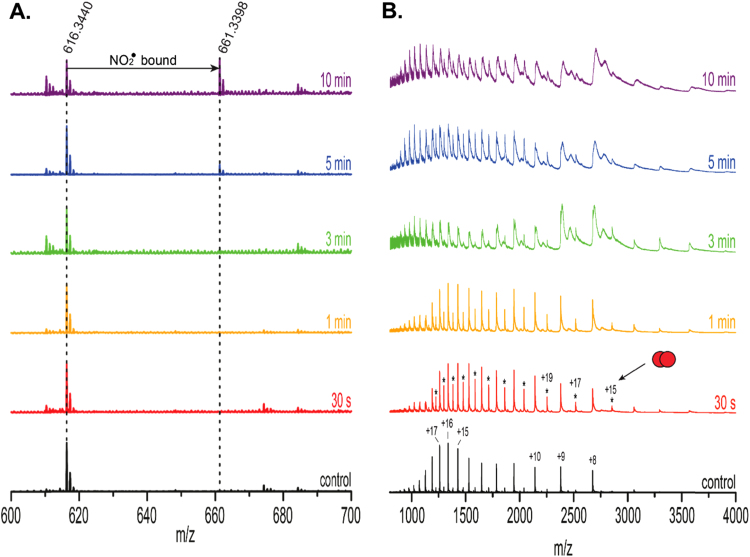
Fig. 3.
Denaturing MS of CYGB. ESI spectra of 10 µM CYGB (diluted 1:1 in acetonitrile containing 1% formic acid) after different duration of plasma treatment (20 µM CYGB in 50 mM tris-buffer). (A.): Zoomed in m/z region from 600 to 700. The peak at 616.3440 m/z represents the free heme group. 5–10 min of treatment induces binding of a molecule, i.e. •NO2 to the heme group (661.3398 m/z). (B.): Full spectrum ranging from 800 to 4000 m/z showing the apo-form of the protein. The appearance of smaller peaks (asterisks) in between the monomers highlights the presence of covalent dimers in the sample. The theoretical mass of CYGB in the apo-form (without heme) is 21 404.62 Da compared to the experimental mass of 21 407.29 ± 4.16 Da in the control.
3.2.2. Denatured MS
Plasma-treated samples were also measured under denaturing conditions, in order to determine if the observed dimers were noncovalent complexes, or covalently linked, e.g. via disulfide bridges (Fig. 3). As can be seen in Fig. 3(B) (bottom panel, “control”) and Fig. S2, unfolding leads to a much broader charge state distribution for the monomer that no longer contains the heme group. The spectra also show minor peaks in between the monomers (Fig. 3B), highlighting the presence of dimeric species that still remain after denaturation. This indicates that the dimerization occurs via covalent bonds. In Fig. 3(A), the m/z region between 600 and 700 is shown with the peak corresponding to the free heme group (616m/z). A plasma treatment time of up to 3 min does not show much effect; however after 5 min a new peak appears at 661m/z, indicating the binding of a molecule to the heme group (while also replacing a hydrogen atom) which we interpret as •NO2 (+45 Da).
We note that dimers are already present in the native spectra (Fig. 2) without plasma treatment, but almost entirely absent in the denatured spectrum (Fig. 3A). In order to investigate whether pre-existing noncovalent CYGB dimers are converted to covalent, disulfide-bound dimers under the influence of plasma oxidation, measurements in tris-buffer, under native conditions, were repeated after adding 10 mM of the reducing DTT to the samples. Fig. 4 shows the full spectra and zoomed in section of the monomeric +9 peak (inset, highlighted in blue). As can be observed, addition of DTT leads to a loss of the dimeric species, indicating that dimerization is stabilized by disulfide bonds. Furthermore, the addition of DTT also reduces the plasma-induced oxidations. This is clearly visible when comparing the monomeric +9 peaks in Fig. 4 with the ones depicted in Fig. 2, under otherwise identical conditions. The peak shifts after 5 and 10 min of plasma treatment again indicate binding of an additional ligand to the heme group.
Fig. 4.
Native MS after reduction with DTT. Native MS spectra of 10 μM CYGB in 100 mM ammonium acetate after adding 10 mM DTT. Monomeric +9 is highlighted. Inset: the zoomed in section of the monomeric +9 peak. Addition of DTT leads to a loss of the dimers via reduction of disulfide bridges. DTT also seems to reduce the number of oxidations (narrowing of the peaks). See also Fig. 2 for comparison.
3.3. Amino acid modification sites of CYGB
Next, we aimed to map plasma-induced oxidative modification sites using LC-orbitrap MS/MS of trypsin-digested samples. Table 1 gives an overview of different types of amino acids that were found to be modified after plasma treatment of a specific duration. CAP was able to oxidize all different amino acids that were selected for the analysis (Table 1), with some seeming to be more readily oxidized than others. Of the investigated amino acids, methionine, tryptophan, and tyrosine were the three found to be preferably oxidized, as reported previously [51]. Surprisingly, no clear treatment-time-dependent increase in the number of oxidized amino acids could be observed, indicating that oxidation may occur rapidly. In the suppl. Table S2, the complete amino acid sequence of CYGB is given together with the specific location of each identified modification, for each exposure time. Table S2 shows that some of the amino acids, at certain locations, were never oxidized. For example, two of the six methionine and two of the five tyrosine (Met66, Met72, Tyr59, and Tyr123) were not found to be oxidized, under any condition.
Table 1.
Oxidative modifications of CYGB. A list of the number of different amino acids in CYGB (# AA in sequence) and the number of oxidized amino acids of each type before (control) and after different treatment times. Eight amino acids (M, Y, D, F, H, W, N, and P) were chosen for analysis. Samples were measured with Ion trap LC-MS/MS and spectra were analyzed with Mascot and Sequest algorithms.
|
Tris-buffer | |||||||
|---|---|---|---|---|---|---|---|
| Amino Acid | # AA in sequence | Control | 30 s | 1 min | 3 min | 5 min | 10 min |
| Methionine (M) | 6 | 2 | 3 | 4 | 4 | 3 | 4 |
| Tyrosine (Y) | 5 | 2 | 3 | 3 | 2 | 3 | 2 |
| Tryptophan (W) | 3 | 1 | 2 | 2 | 2 | 2 | |
| Phenylalanine (F) | 8 | 1 | 1 | 1 | 1 | 1 | 1 |
| Aspartic acid (D) | 5 | 1 | 2 | 2 | 1 | ||
| Asparagine (N) | 5 | 3 | 3 | 1 | 2 | ||
| Proline (P) | 13 | 1 | 3 | ||||
| Histidine (H) | 6 | 1 | 1 | 1 | 1 | ||
| TOTAL | 51 | 5 | 12 | 16 | 13 | 13 | 14 |
Table 2 gives the total number of identified peptide fragments (so-called ‘Peptide Spectrum Matches’, PSM), together with the number of oxidized PSMs. Here, a clear treatment-time-dependent increase in oxidized peptides can be observed, illustrating the increasing oxidation potential with increasing treatment time. It is also noticeable (Table 1, Table 2 & S2) that in the control samples some amino acids (M,Y and F) are also oxidized. This oxidation is likely due to the exposure of the sample to the surrounding atmosphere, or may occur during electrospray.
Table 2.
Oxidative modifications of CYGB. A list of the number of oxidized peptide fragments before and after plasma treatment. The number of Peptide Spectrum Matches (PSM) is the total number of identified peptide spectra matched for the protein. Samples were measured with Ion trap LC-MS/MS and spectra were analyzed with Mascot and Sequest algorithms.
| Sample | # PSM in total | #Oxidation PSM | % Oxidation | |
|---|---|---|---|---|
| Tris-buffer | Control | 90 | 15 | 17% |
| 30 s | 121 | 23 | 19% | |
| 1 min | 223 | 44 | 20% | |
| 3 min | 117 | 34 | 29% | |
| 5 min | 109 | 38 | 35% | |
| 10 min | 138 | 54 | 39% |
3.4. MD and docking simulations
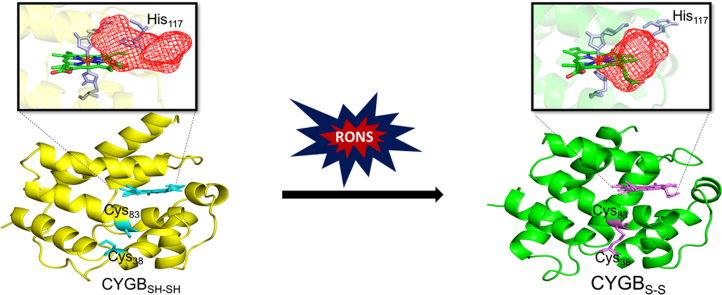
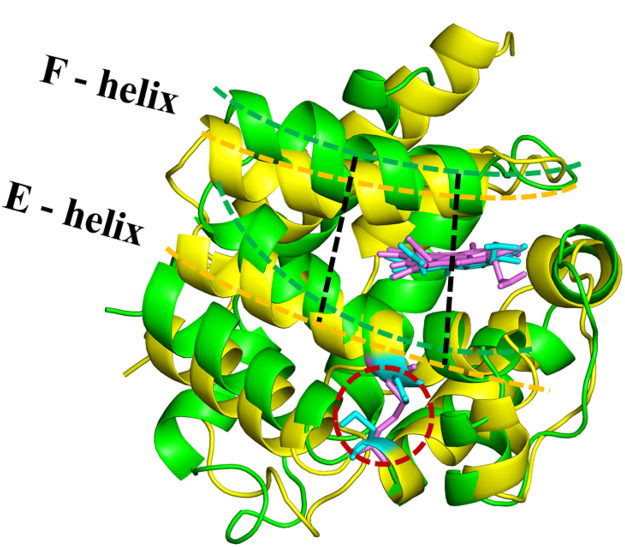
CYGB possesses two cysteine residues (Cys38 and Cys83), which are known to be able to form disulfide bridges, either inter- or intramolecularly [25], [26]. As CYGB is proposed to be a monomer in vivo [52], and the formation of an intramolecular disulfide bridge can influence the activity of CYGB, we chose to investigate the effect of the formation of an intramolecular disulfide bridge in between the cysteine amino acids of monomeric CYGB (i.e., CYGBS-S) with MD simulations. We have shown experimentally that an intermolecular disulfide bridge is present in the dimeric fraction. However, we can also speculate that the monomeric fraction may contain an intramolecular disulfide bridge. Fig. 5 illustrates the last snapshots of the equilibrated CYGBSH…SH (i.e. CYGB monomer without an initial disulfide bond) and CYGBS-S molecules (i.e. CYGB monomer with an intramolecular disulfide bond) depicted in yellow and green, respectively. The F-helix of CYGB is located at the proximal side of the heme group, while the E-helix is positioned at the distal side. The disulfide linkage in CYGBS-S clearly influences its conformation and hence, the positions of the E- and F-helices are shifted relative to CYGBSH…SH (cf. the yellow and green dashed lines). In order to calculate the distances between the helices, residues Ala82 and Ala88 in the F-helix and Ala107 and Ala112 in the E-helix were chosen. These amino acids are positioned at the end of the black dashed lines but they are not shown in the figure for the sake of clarity. The calculated distances between these amino acids located in the E- and F-helices are also given in Table S1, indicating the minor change in the modified CYGBS-S. It is also obvious from Fig. 5 that there is a negligible shift of the heme group in the modified CYGBS-S molecule compared to native CYGBSH…SH. Hence, the formation of the disulfide bridge between the cysteine residues does not alter the local orientation of the heme group and imidazole planes of the histidines (i.e., His81, His113) bound to the heme: the imidazole planes do not rotate versus each other. This was also reported in literature by experimental investigations [53].
Fig. 5.
Aligned structures of CYGBSH…SH (yellow) and CYGBS-S (green). The distances between Ala88-Ala107 and Ala82-Ala112 are depicted by black dashed lines. The yellow and green dashed lines are plotted to indicate shifted positions of the E-and F-helices. The heme group and cysteine residues of both CYGBSH…SH and CYGBS-S are presented in licorice view in blue and pink, respectively. The disulfide (S-S) bridge is shown within the red dashed circle.
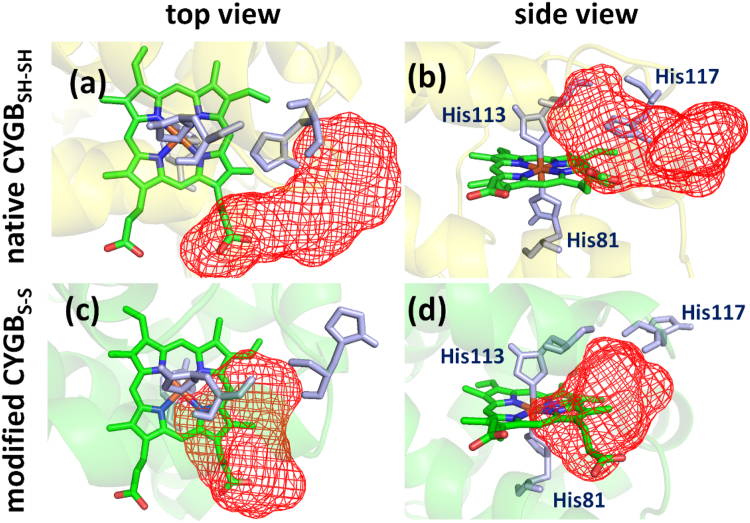
Comparing the consecutive MD frames of both monomeric CYGBSH…SH and CYGBS-S, indicates that the His117 residue located nearby the heme group (see Fig. 6) substantially changes its orientation in CYGBS-S. Indeed, the calculated distance between the heme iron and one of the nitrogen atoms of His117 (located in the imidazole ring) almost doubled in CYGBS-S (see Table. S1), which is due to the conformational change caused by the intramolecular disulfide bond. Besides the change in orientation of His117, it is noticeable that the propionate group of the heme closest to His117 flips its position upon disulfide bond formation (facing downward in CYGBS-S; see Fig. 6(b) and (d)). To get more insight into the structural changes caused by the intramolecular disulfide bond, the binding hot spots of both CYGBSH…SH and CYGBS-S were investigated as described in Section 2.
Fig. 6.
Binding hot spots. Binding hot spot of CYGBSH…SH (a-b) and (c-d) CYGBS-S presented as red meshed regions. The helices in the structures are represented in pale colors, for the sake of clarity. The heme group and the histidine residues (81, 113, 117) are shown in green and light purple colors and licorice views, respectively. It is clear that in CYGBSH…SH the access to the heme group is more limited than in CYGBS-S.
10 CYGB structures with 1 ns time interval were extracted using the last 10 ns trajectory of the equilibrated (so-called production) run. These structures were employed to examine the structural dependencies of the binding hot spots of CYGB. Here, only one of the results out of ten are presented, as the other nine results were very similar. Three different pockets (with lowest binding energies) were obtained in both CYGBSH…SH and CYGBS-S, which are depicted as meshed regions in Fig. 6. The other two meshed regions are not shown as we are focusing on the catalytic site of CYGB (i.e., the heme pocket). As mentioned above, the formation of a disulfide bond between the cysteine residues leads to a change of the CYGB conformation, which results in a doubling of the distance between the highly localized His117 residue and the heme group in CYGBS-S (see Table S1 and cf. Fig. 6(a) and (c) or Fig. 6(b) and (d)). Accordingly, this results in the heme group to become more accessible to the probing molecules (see overlap between heme group and meshed regions in Fig. 6(c) and (d)). In addition, the flipping of the position of the heme propionate upon disulfide formation further contributes to the accessibility of the heme pocket (Fig. 6(b) and (d)). Our binding hot spot analysis indeed shows that in the case of CYGBS-S the binding pocket is located around the heme group, whereas in CYGBSH…SH it is found with limited access to the heme group, i.e., around the helices (cf. Fig. 6(a) and (c), or Fig. 6(b) and (d)). This again confirms that in CYGBSH…SH the access to the heme group is more limited than in CYGBS-S.
4. Discussion
In this study, it was shown that the treatment of recombinant CYGB with CAP resulted in chemical modifications of the protein. The spectral characteristics described for the untreated recombinant CYGB are identical to the ones described previously, showing that the protein is in the hexacoordinated ferric form [48]. Upon plasma treatment, spectral changes occur, which appear to be dependent on the duration of the treatment. These changes were more prominent when water was used as a solvent than in tris-buffer. This suggests that in water, the effect of plasma, which is mainly through reactive species production, is greater than in tris-buffer. However, there is one main difference between the two solvents that were employed: deionized water has no buffering capacity, whereas tris-buffer does. As plasma treatment induces many different reactive species (i.e. •OH, H2O2, •NO, O2−, ONOOH, •NO2, NO2-, NO3-) [54], these species can contribute to the acidification of liquids. Indeed, pH measurements highlighted that plasma treatment induces a significant drop in the pH of deionized water, whereas no such effect could be observed with tris-buffer (Fig. S3). Thus, with water, there is a secondary effect of plasma treatment, through the acidification of the sample. Low pH can, depending on the protein, lead to protein denaturation and aggregation. In Fig. 1(A), after 5 min treatment, the Soret peak at 415 nm has almost completely disappeared, and a new peak has emerged at 387 nm, indicating free heme in the solution [49]. Additionally, the ultracentrifugation of the 5 min treated sample showed a clear brown precipitate, indicating the aggregation and denaturation of the sample upon plasma treatment. In tris-buffer, on the other hand, no clear precipitate after ultracentrifugation of the 5 min treated sample could be observed, yet still, clear spectral changes were observed. Altogether, this confirms that in water, plasma treatment acidifies the solution, leading to structural changes in the protein, and the eventual (partial) denaturation of the protein sample. This has some implications for the further interpretation of the results from this study. As the main aim of this study was to investigate the effects of plasma-generated RONS on CYGB, the results obtained from treating CYGB with plasma in tris-buffer are of higher relevance and will be discussed further in more detail (see Suppl. Figs. S6 & S7 for MS spectra in water).
From the absorption spectra of the samples treated in tris-buffer, it can be concluded that plasma induces a decrease in intensity of the Soret band at 417 nm. This could indicate two things: the first option would be that the protein is continuously degrading more with longer treatment times, with the eventual, almost complete denaturation of the protein at 10 min treatment time. Although the appearance of a peak at 383 nm (after 10 min treatment) indicates the leaking of the heme out of the protein (and thus denaturing of the protein), CD and MS data (Figs. 1E and 2) however shows that the largest fraction of the protein is still folded and contains the heme group. The slight unfolding we see in the CD-spectra may be explained by the MD data where it is shown that the formation of an intramolecular disulfide bridge in CYGBS-S induces a minor shift of the E- and F-helices, relative to CYGBSH…SH (see Table S1 & Fig. 5). Furthermore, measurement of the drift time of the monomeric (+9 peak) species showed that there was no significant alteration of the collision cross section (CCS) of the treated protein (Fig. S4), indicating that the three-dimensional shape, i.e. the tertiary structure, is not significantly affected upon plasma treatment, and thus excluding this option.
The second option would be that plasma treatment induces specific chemical modifications in the protein that lead to the observed spectral changes. The Soret band is emblematic for heme-containing proteins such as globins and various cytochromes, and arises primarily due to π-π * transitions in the heme group [55]. Binding of molecules to the heme structure or close environment could affect the (electronic) structure and lead to a decrease and/or shift of the Soret peak. From the MS data, it is clear that there is indeed covalent modification of CYGB upon plasma treatment. In Fig. 2, it can be seen that there is a peak shift and broadening, indicating that plasma treatment induces numerous oxidations, either at specific amino acids, or at the heme group. Moreover, the MS spectra of the denatured sample (Fig. 3) clearly show that, with 5 or 10 min treatment, there is binding of a molecule, most likely •NO2, to the protein bound heme group. Binding of •NO2 has already been shown to occur in vitro in other globins, i.e. myoglobin [56], hemoglobin [57], and more recently leghemoglobin [58], by exposing the proteins to excess NaNO2. In these studies, it was shown that heme nitration originated from the substitution of a hydrogen atom by •NO2 from one of the vinyl groups. The subsequent formation of a nitrovinyl group results in a modified visible spectrum (a Soret band with lower intensity) and a heme group with m/z 661, similar to the results that were obtained in this study. Addition of a 1000-fold excess of NaNO2 to CYGB (data not shown) led to the same modified spectra observed after plasma treatment of 5 and 10 min, confirming the nitration of the heme. Furthermore, resonance Raman (RR) experiments further confirmed the binding of •NO2 to the heme (Fig. S5). The high-frequency region of the RR spectra revealed the appearance of a strong band at ~ 1325 cm−1, which is specific for a nitroaromatic group [59], [60], [61], again indicating the binding of •NO2 to the vinyl groups of the heme. Plasma treatment of liquids does indeed yield the formation of •NO2 [62], [63], [64]. Thus, upon plasma treatment for 5 min and longer, •NO2, and most probably other related nitrogen species, are sufficiently produced in order to induce the binding of •NO2 to the heme. Furthermore, the fact that •NO2 can bind to the CYGB heme could implicate that it also performs a similar biological role, i.e. the involvement of CYGB in NO homeostasis. Previous reports already proposed a role for CYGB in NO regulation through its NO dioxygenase activity [26]. However, under conditions of oxygen deprivation, CYGB could act as a source of NO by switching to a nitrite reductase function. The switching between these two functions is regulated through the oxidation of the two surface-exposed cysteine residues [65].
Besides nitration of the heme, plasma induces the oxidation of specific amino acid residues of CYGB. Here, the three most oxidized amino acids were methionine, tryptophan, and tyrosine. In Takai et al. [66] reported that plasma was able to chemically modify amino acids through the oxidation of their side chains, with sulfur-containing (i.e. Met and Cys) and aromatic amino acids (i.e. Trp, Tyr, and Phe) preferentially modified. Furthermore, they found that the relative reactivity towards plasma treatment was Met > Cys > Trp > Phe > Tyr. This is not entirely in agreement with our findings (Table 1). However, in this study, a complete protein was treated with plasma, whereas in the study of Takai et al., a mixture of amino acids was treated with plasma. Therefore, it is highly likely that in CYGB, due to its 3D-structure, some amino acids, e.g. Phe, are less solvent accessible compared to when individual amino acids are studied, leading to different results. This is further highlighted in Table S2, where it can be seen that certain locations of preferably oxidized amino acids, such as Met and Trp were never oxidized, indicating that the access of certain amino acids is hindered due to the tertiary structure of CYGB. This observed reactivity is in agreement with an increasing body of evidence from other experimental approaches which utilize ROS to cause oxidation of the solvent-accessible surface area of proteins, such as hydroxyl radical footprinting by synchrotron radiation [51] as well as fast photochemical oxidation of proteins [67] and other oxidative labelling techniques [68].
As for cysteine, during the digestion of the protein sample prior to measuring with LC-MS/MS, the cysteines are reduced by TCEP and carbamidomethylated. This modification is introduced on purpose to prevent disulfide bridges from forming again, and was therefore not selected for the analysis of oxidative modifications [69]. Yet, from the MS data (Fig. 2, Fig. 4) it is clear that cysteines do become oxidized (i.e., formation of disulfide bridge). CYGB possesses two cysteine residues (Cys38 and Cys83), which are known to be able to form disulfide bridges, either inter- or intramolecularly [25], [26]. Such a formation of CYGB dimers upon plasma treatment can indeed be observed in native mass spectra (Fig. 2), and even more clearly under denaturing conditions in Fig. 3. Moreover, the addition of the reducing agent DTT to both the untreated and treated samples led to the disappearance of the dimeric peaks in the spectrum (Fig. 4). However, there is still a significant portion of CYGB that stays in the monomeric form (Fig. 2). It is highly likely that the monomers that are still present after treatment have formed intramolecular disulfide bridges. Although the dimeric form of CYGB has been suggested to be less active than the monomeric form (see paragraph below), this observed dimerization after treatment could be an artefact of the experimental setup. In vivo, the concentration of CYGB lies somewhere between 0.1 and 5 µM, at which CYGB is proposed to be a monomer [70]. Higher concentrations, like the one that was used here, could bring CYGB molecules closer together and subsequently stimulate the formation of dimers. Nonetheless, from the abovementioned, we can conclude that plasma treatment leads to the formation of intermolecular disulfide bridges, thereby stabilizing the CYGB dimers, but presumably also intramolecular disulfide bridges in the remaining monomers. This would confirm the two cysteine residues as redox-active cysteines.
The state of the two surface-exposed cysteine residues, either free, intra- or intermolecularly linked, has been proposed to be critical in determining the biochemical and redox properties of CYGB [71]. A recent study demonstrated a clear difference in the ability of CYGB to bind lipids between the monomeric and dimeric form of the protein, with the monomer, containing an intramolecular disulfide bridge, more easily able to bind lipids [25]. This difference is suggested to occur because the presence of an intramolecular disulfide bond leads to the formation of a more open heme pocket structure, thereby facilitating access for external ligands, such as lipids, whereas an intermolecular disulfide bond does not alter the environment of the heme pocket [25]. This is perfectly in line with the MD and docking simulations performed in this study on monomeric CYGB. Here, it is seen that in CYGBSH…SH, the access to the heme group is more limited than in CYGBS-S. We further found that the His117 residue plays a vital role in either limiting or increasing the access to the heme group, thereby acting like a gate function for CYGBSH…SH and CYGBS-S. Thus, our investigations confirm, and further show, that the formation of an intramolecular disulfide bond affects the CYGB structure, thereby opening the access to the heme group, through gate functioning of His117, which in turn could lead to a higher binding affinity. In addition the redox state of the Cys38 and Cys83 residues appears to act as a ‘molecular switch’, activating the protein only under the correct conditions, i.e. oxidative stress. This would support CYGB's proposed cytoprotective role in oxidative stress conditions, which is probably linked to the lipid peroxidase activity of the protein [25], [49].
5. Conclusion
In conclusion, the treatment of CYGB with CAP leads to chemical modifications of the protein, yet the overall structure of the protein remains largely intact.
Short treatment times were already sufficient to induce the oxidative modification of mainly sulfur-containing and aromatic amino acids. Longer treatment times also lead to the nitration of the heme, through the binding of •NO2 to the vinyl groups, which proposes a role for CYGB in NO homeostasis. Upon plasma treatment the two cysteine residues of CYGB were oxidized, which stabilizes the dimeric CYGB, through intermolecular disulfide bridges, but most likely also monomeric CYGB containing an intramolecular disulfide bond. The simulations performed in this study also confirm the importance of the oxidation of Cys38 and Cys83 residues in regulating the ligand-binding properties, and thus also the activity, of CYGB. Furthermore, His177 was found to play a vital role in regulating the access to the heme group.
As mentioned in the introduction, the aim of this study was to elucidate the effects of plasma-generated (and in general) RONS on the structure and, to some extent, the function(s) of CYGB. Although some protein denaturing occurred, our results nonetheless confirm that (plasma-generated) RONS can indeed induce chemical modifications at specific sites, which lead to different conformations of CYGB. These different conformations are mediated by the oxidation status of two redox-active cysteines. As globins (more particularly hexacoordinated globins) are increasingly hypothesized to play a role in redox biology, findings here seem to further support the idea of CYGB functioning as a redox-active protein.
This study is of great interest because it provides further support and new insights that CYGB, on a molecular basis, has the potential to play a role in the cellular response against oxidative stress. However, further research on a cellular level is needed to validate these findings and to unravel the cellular pathways in which CYGB is involved. Such research should focus on elucidating the effects of (plasma-generated) RONS on cells and the role of CYGB in the response to these RONS.
Acknowledgements
M.Y. and N.K. gratefully acknowledge financial support from the Research Foundation – Flanders (FWO), Grant nos. 1200216N and 12J5617N. The computational work was carried out using the Turing HPC infrastructure at the CalcUA core facility of the Universiteit Antwerpen (UA), a division of the Flemish Supercomputer Center VSC, funded by the Hercules Foundation, the Flemish Government (department EWI). C.M acknowledges the financial support provided by the Flemish Community and the University of Antwerp (BOF-NOI) for the pre-doctoral scholarship is under grant number/project ID: 28465. S.V.D., S. D. and Z.H. acknowledge the FWO (Grant G.0687.13) and the GOA-BOF UA 2013–2016 (project-ID 28312) for funding. The computational resources and services used in this work were provided by the HPC core facility CalcUA of the Universiteit Antwerpen, and VSC (Flemish Supercomputer Center), funded by the Research Foundation - Flanders (FWO) and the Flemish Government – department EWI.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.07.019.
Contributor Information
Joey De Backer, Email: joey.debacker@uantwerpen.be.
Sylvia Dewilde, Email: sylvia.dewilde@uantwerpen.be.
Appendix A. Supplementary material
Supplementary material
References
- 1.Kubinova S. Non-thermal air plasma promotes the healing of acute skin wounds in rats. Sci. Rep. 2017;7:45183. doi: 10.1038/srep45183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J. Inactivation of Staphylococcus aureus in water by pulsed spark discharge. Sci. Rep. 2017;7(1):10311. doi: 10.1038/s41598-017-10784-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patange A. Controlling Brochothrix thermosphacta as a spoilage risk using in-package atmospheric cold plasma. Food Microbiol. 2017;66:48–54. doi: 10.1016/j.fm.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Fridman G. Blood coagulation and living tissue sterilization by floating-electrode dielectric barrier discharge in air. Plasma Chem. Plasma Process. 2006;26(4):425–442. [Google Scholar]
- 5.Yan D., Sherman J.H., Keidar M. Cold atmospheric plasma, a novel promising anti-cancer treatment modality. Oncotarget. 2017;8(9):15977–15995. doi: 10.18632/oncotarget.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauvin J. Analysis of reactive oxygen and nitrogen species generated in three liquid media by low temperature helium plasma jet. Sci. Rep. 2017;7(1):4562. doi: 10.1038/s41598-017-04650-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandamme M. ROS implication in a new antitumor strategy based on non-thermal plasma. Int. J. Cancer. 2012;130(9):2185–2194. doi: 10.1002/ijc.26252. [DOI] [PubMed] [Google Scholar]
- 8.Graves D.B. Reactive species from cold atmospheric plasma: implications for cancer therapy. Plasma Process. Polym. 2014;11(12):1120–1127. [Google Scholar]
- 9.Kalghatgi S. Effects of non-thermal plasma on mammalian cells. PLoS One. 2011;6(1):e16270. doi: 10.1371/journal.pone.0016270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeylen S. Cold atmospheric plasma treatment of melanoma and glioblastoma cancer cells. Plasma Process. Polym. 2016;13(12):1195–1205. [Google Scholar]
- 11.Hattori N. Effectiveness of plasma treatment on pancreatic cancer cells. Int. J. Oncol. 2015;47(5):1655–1662. doi: 10.3892/ijo.2015.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim C.-H. Effects of atmospheric nonthermal plasma on invasion of colorectal cancer cells. Appl. Phys. Lett. 2010;96(24):243701. doi: 10.1063/1.3449575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S.J. Induction of apoptosis in human breast cancer cells by a pulsed atmospheric pressure plasma jet. Appl. Phys. Lett. 2010;97(2):023702. [Google Scholar]
- 14.Lee S. Cold atmospheric plasma restores tamoxifen sensitivity in resistant MCF-7 breast cancer cell. Free Radic. Biol. Med. 2017;110:280–290. doi: 10.1016/j.freeradbiomed.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Canal C. Plasma-induced selectivity in bone cancer cells death. Free Radic. Biol. Med. 2017;110:72–80. doi: 10.1016/j.freeradbiomed.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Julák J. Inactivation of prions using electrical DC discharges at atmospheric pressure and ambient temperature. Plasma Process. Polym. 2011;8(4):316–323. [Google Scholar]
- 17.Yasuda H. States of biological components in bacteria and bacteriophages during inactivation by atmospheric dielectric barrier discharges. Plasma Process. Polym. 2008;5(6):615–621. [Google Scholar]
- 18.Takai E. Protein inactivation by low-temperature atmospheric pressure plasma in aqueous solution. Plasma Process. Polym. 2012;9(1):77–82. [Google Scholar]
- 19.Attri P. Influence of reactive species on the modification of biomolecules generated from the soft plasma. Sci. Rep. 2015;5:8221. doi: 10.1038/srep08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr. Opin. Struct. Biol. 2004;14(6):679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Gardner A.M., Cook M.R., Gardner P.R. Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in Rat Hepatocytes. J. Biol. Chem. 2010;285(31):23850–23857. doi: 10.1074/jbc.M110.132340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X. Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration. Biochemistry. 2012;51(25):5072–5082. doi: 10.1021/bi300291h. [DOI] [PubMed] [Google Scholar]
- 23.Fordel E. Neuroglobin and cytoglobin expression in mice. Evidence for a correlation with reactive oxygen species scavenging. FEBS J. 2007;274(5):1312–1317. doi: 10.1111/j.1742-4658.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 24.Shivapurkar N. Cytoglobin, the newest member of the globin family, functions as a tumor suppressor gene. Cancer Res. 2008;68(18):7448–7456. doi: 10.1158/0008-5472.CAN-08-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckerson P. Cytoglobin ligand binding regulated by changing haem-co-ordination in response to intramolecular disulfide bond formation and lipid interaction. Biochem. J. 2015;465(1):127–137. doi: 10.1042/BJ20140827. [DOI] [PubMed] [Google Scholar]
- 26.Tsujino H. Disulfide bonds regulate binding of exogenous ligand to human cytoglobin. J. Inorg. Biochem. 2014;135:20–27. doi: 10.1016/j.jinorgbio.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Zhou D. Oxygen binding and nitric oxide dioxygenase activity of cytoglobin are altered to different extents by cysteine modification. FEBS Open Biol. 2017;7(6):845–853. doi: 10.1002/2211-5463.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latina A. DeltaNp63 targets cytoglobin to inhibit oxidative stress-induced apoptosis in keratinocytes and lung cancer. Oncogene. 2016;35(12):1493–1503. doi: 10.1038/onc.2015.222. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S. Cytoglobin promotes cardiac progenitor cell survival against oxidative stress via the upregulation of the NFkappaB/iNOS signal pathway and nitric oxide production. Sci. Rep. 2017;7(1):10754. doi: 10.1038/s41598-017-11342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou L. Recombinant human cytoglobin prevents atherosclerosis by regulating lipid metabolism and oxidative stress. J. Cardiovasc Pharmacol. Ther. 2018;23(2):162–173. doi: 10.1177/1074248417724870. [DOI] [PubMed] [Google Scholar]
- 31.Dewilde S. Expression, purification, and crystallization of neuro- and cytoglobin. Methods Enzymol. 2008;436:341–357. doi: 10.1016/S0076-6879(08)36019-4. [DOI] [PubMed] [Google Scholar]
- 32.Van Der Spoel D. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 33.Schuler L.D., Daura X., Gunsteren W.F. Van. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 2001;22(11):1205–1218. [Google Scholar]
- 34.Sugimoto H. Structural basis of human cytoglobin for ligand binding. J. Mol. Biol. 2004;339(4):873–885. doi: 10.1016/j.jmb.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 35.H.J. Berendsen, et al., Interaction models for water in relation to protein hydration, in: Intermolecular Forces, Springer, 1981, pp. 331–342.
- 36.Hanwell M.D. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Chemin. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katti S.K., LeMaster D.M., Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J. Mol. Biol. 1990;212(1):167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 38.A.P. Wiita, et al., Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques, Proc. Natl. Acad. Sci. USA, vol. 103(19), pp. 7222–7227. [DOI] [PMC free article] [PubMed]
- 39.Shanno D.F. Conditioning of quasi-Newton methods for function minimization. Math. Comput. 1970;24(111):647–656. [Google Scholar]
- 40.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. [Google Scholar]
- 42.Essmann U. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. [Google Scholar]
- 43.Kozakov D. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015;10(5):733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks B.R. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4(2):187–217. [Google Scholar]
- 45.Schaefer M., Karplus M. A comprehensive analytical treatment of continuum electrostatics. J. Phys. Chem. 1996;100(5):1578–1599. [Google Scholar]
- 46.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.W.L. DeLano, The PyMOL Molecular Graphics System. 〈http://pymol.org〉.
- 48.Sawai H. Characterization of the heme environmental structure of cytoglobin, a fourth globin in humans. Biochemistry. 2003;42(17):5133–5142. doi: 10.1021/bi027067e. [DOI] [PubMed] [Google Scholar]
- 49.Reeder B.J., Svistunenko D.A., Wilson M.T. Lipid binding to cytoglobin leads to a change in haem co-ordination: a role for cytoglobin in lipid signalling of oxidative stress. Biochem. J. 2011;434(3):483–492. doi: 10.1042/BJ20101136. [DOI] [PubMed] [Google Scholar]
- 50.Kelly S.M., Jess T.J., Price N.C. How to study proteins by circular dichroism. Biochim Biophys. Acta. 2005;1751(2):119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Xu G., Chance M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007;107(8):3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 52.Lechauve C. Cytoglobin conformations and disulfide bond formation. FEBS J. 2010;277(12):2696–2704. doi: 10.1111/j.1742-464X.2010.07686.x. [DOI] [PubMed] [Google Scholar]
- 53.Vinck E. Structural change of the heme pocket due to disulfide bridge formation is significantly larger for neuroglobin than for cytoglobin. J. Am. Chem. Soc. 2004;126(14):4516–4517. doi: 10.1021/ja0383322. [DOI] [PubMed] [Google Scholar]
- 54.Lu X. Reactive species in non-equilibrium atmospheric-pressure plasmas: generation, transport, and biological effects. Phys. Rep. 2016;630(Suppl. C):S1–S84. [Google Scholar]
- 55.Dayer M.R., Moosavi-Movahedi A.A., Dayer M.S. Band assignment in hemoglobin porphyrin ring spectrum: using four-orbital model of Gouterman. Protein Pept. Lett. 2010;17(4):473–479. doi: 10.2174/092986610790963645. [DOI] [PubMed] [Google Scholar]
- 56.Bondoc L.L., Timkovich R. Structural characterization of nitrimyoglobin. J. Biol. Chem. 1989;264(11):6134–6145. [PubMed] [Google Scholar]
- 57.Otsuka M. Covalent modifications of hemoglobin by nitrite anion: formation kinetics and properties of nitrihemoglobin. Chem. Res Toxicol. 2010;23(11):1786–1795. doi: 10.1021/tx100242w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navascués J. Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proc. Natl. Acad. Sci. USA. 2012;109:2660–2665. doi: 10.1073/pnas.1116559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. Lin-Vien, et al., Chapter 11 - The Nitro Group, in: The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules, Academic Press, San Diego, 1991, pp. 179–189.
- 60.Lambrou A., Pinakoulaki E. Resonance Raman detection of the myoglobin nitrito heme Fe-O-N˭O/2-nitrovinyl species: implications for helix E-helix F interactions. Phys. Chem. Chem. Phys. 2015;17(5):3841–3849. doi: 10.1039/c4cp04352a. [DOI] [PubMed] [Google Scholar]
- 61.Ioannou A. Nitrite coordination in myoglobin. J. Inorg. Biochem. 2017;166:49–54. doi: 10.1016/j.jinorgbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 62.Van Boxem W. Anti-cancer capacity of plasma-treated PBS: effect of chemical composition on cancer cell cytotoxicity. Sci. Rep. 2017;7(1):16478. doi: 10.1038/s41598-017-16758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Girard P.M. Synergistic effect of H2O2 and NO2 in cell death induced by cold atmospheric He plasma. Sci. Rep. 2016;6:29098. doi: 10.1038/srep29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurake N. Cell survival of glioblastoma grown in medium containing hydrogen peroxide and/or nitrite, or in plasma-activated medium. Arch. Biochem Biophys. 2016;605:102–108. doi: 10.1016/j.abb.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 65.Reeder B.J., Ukeri J. Strong modulation of nitrite reductase activity of cytoglobin by disulfide bond oxidation: implications for nitric oxide homeostasis. Nitric Oxide. 2018;72(Suppl. C):S16–S23. doi: 10.1016/j.niox.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Takai E. Chemical modification of amino acids by atmospheric-pressure cold plasma in aqueous solution. J. Phys. D: Appl. Phys. 2014;47(28):285403. [Google Scholar]
- 67.Li K.S., Shi L., Gross M.L. Mass spectrometry-based Fast Photochemical Oxidation of Proteins (FPOP) for higher order structure characterization. Acc. Chem. Res. 2018;51(3):736–744. doi: 10.1021/acs.accounts.7b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roeser J. Oxidative protein labeling in mass-spectrometry-based proteomics. Anal. Bioanal. Chem. 2010;397(8):3441–3455. doi: 10.1007/s00216-010-3471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lippincott J., Apostol I. Carbamylation of cysteine: a potential artifact in peptide mapping of hemoglobins in the presence of urea. Anal. Biochem. 1999;267(1):57–64. doi: 10.1006/abio.1998.2970. [DOI] [PubMed] [Google Scholar]
- 70.K. Yoshizato, et al., Discovery of cytoglobin and its roles in physiology and pathology of hepatic stellate cells, Proc. Japan Acad. Ser. B Phys. Biol. Sci., vol. 92(3), pp. 77–97. [DOI] [PMC free article] [PubMed]
- 71.Tsujino H. Disulfide bonds regulate binding of exogenous ligand to human cytoglobin. J. Inorg. Biochem. 2014;135(Suppl. C):S20–S27. doi: 10.1016/j.jinorgbio.2014.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material