 A concise synthetic protocol enables rapid receptor screening of hydroxy fatty acid.
A concise synthetic protocol enables rapid receptor screening of hydroxy fatty acid.
Abstract
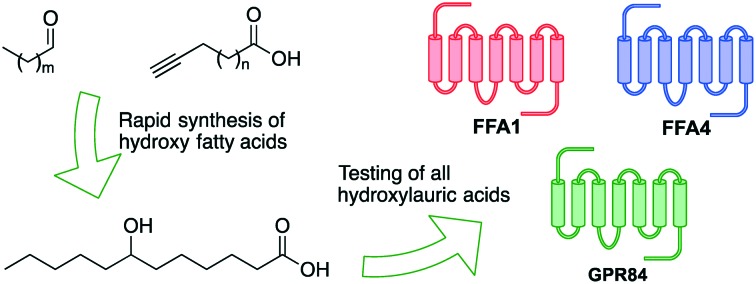
Saturated hydroxy fatty acids make up a class of underexplored lipids with potentially interesting biological activities. We report a succinct and general synthetic route to saturated hydroxy fatty acids hydroxylated at position 6 or higher, and exemplify this with the synthesis of hydroxylauric acids. All regioisomers of hydroxylauric acids were tested on free fatty acid receptors FFA1, FFA4 and GPR84. The results show that the introduction of a hydroxy group and its position have a high impact on receptor activity.
Introduction
Fatty acids are fundamental components of structure and energy storage, precursors of a range of signalling molecules, and have in recent years also turned out to have a role in the direct regulation of metabolism and inflammation through activation of free fatty acid receptors.1 Polyunsaturated hydroxy fatty acids include potent chemical mediators such as hydroxyoctadecadienoic acids, lipoxins, hydroxyeicosatetraenoic acids, hydroxyeicosapentaenoic acids, hydroxydocosapentaenoic acids, resolvins, protectins and marseins, derived by oxidation of linoleic acid, arachidonic acid, and omega-3 fatty acids.2–5 Saturated hydroxy fatty acids have been less studied as potential signalling molecules but are relatively common. For example, 2-hydroxyfatty acids are abundant in sphingolipids,6 and 3-hydroxyfatty acids are ubiquitous intermediates in fatty acid synthesis and β-oxidation and are constituents of inflammatory lipopolysaccharides.7 4-Hydroxyfatty acids are formed by oxidation of saturated fatty acids in mammary glands,8 4-hydroxylauric acid activates immune natural killer cells,9 and 9-hydroxystearic acid is an endogenously produced histone deacetylase inhibitor that has potent anti-proliferative effects in cancer cells.10 Recently, fatty acid esters of hydroxy fatty acids (FAHFAs), mainly composed of saturated fatty acids, have appeared as a novel lipid class reported to have anti-inflammatory and anti-diabetic properties.11
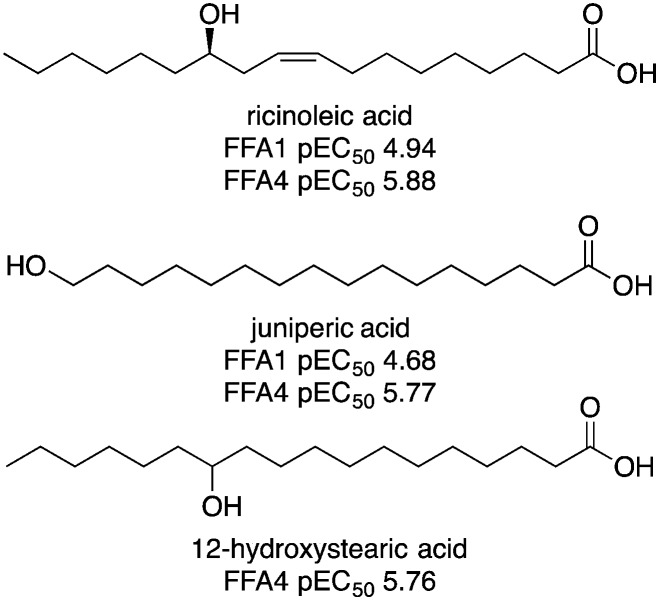
Free fatty acid receptors, in particular, the medium-to-long-chain fatty acid receptors FFA1 (GPR40) and FFA4 (GPR120), have become established as interesting potential therapeutic targets for treatment of metabolic diseases.1 The recognition that dietary fatty acids activate therapeutic targets suggests the likely identification of therapeutic or ‘nutraceutical’ food constituents.12 In our work towards characterizing food constituents and metabolites on free fatty acid receptors, we observed that some hydroxy fatty acids, such as ricinoleic acid, juniperic acid and 12-hydroxystearic acid, tended to have higher potency than the parent fatty acids on FFA4 and FFA1 (Fig. 1).13 Similarly, Suzuki et al. have reported that certain hydroxy fatty acids have higher activity than the corresponding parent fatty acids on the medium-chain fatty acid receptor GPR84.14 These observations triggered our interest in exploring the activity of saturated hydroxy fatty acids on free fatty acid receptors more systematically. Hydroxylauric acids (HLAs) were selected for the initial studies since lauric acid has previously been shown to be active on FFA1, FFA4 and GPR84.13,15
Fig. 1. Hydroxy fatty acids with activity on FFA1 and FFA4.13.
Only ω-hydroxylauric acid (12-HLA) and the lactones of 4-HLA and 5-HLA were commercially available, thus, the remaining regioisomers could only be accessed by synthesis. A wide range of methods are available for the synthesis of 2-hydroxyfatty acids and 3-hydroxyfatty acids.16–19 For 6-hydroxyfatty acids and beyond, however, there is a surprising paucity of efficient and systematic synthetic methods. There are several reports on biocatalytic hydroxylation of the end positions of fatty acids but, apart from hydroxylation at the ω-position, these methods generally suffer from a lack of selectivity.20–23 The only systematic approach involves reaction of an acyl chloride in the presence of the ester-protected carboxylate with a Grignard reagent or alkyl cadmium followed by reduction of the resulting ketone and hydrolysis.24,25 With the number of possible saturated hydroxy fatty acids in mind, we wished to develop a simpler and more direct approach, preferably avoiding protecting groups. Thus, we here report a concise synthetic route to racemic saturated fatty acids hydroxylated at position 6 or further up, exemplified with the synthesis of HLAs. We furthermore report the activity of all these HLAs on medium- and long-chain fatty acid receptors FFA1, FFA4 and GPR84. This confirmed that the introduction of a hydroxyl group into a saturated fatty acid can have strong effects on its agonist activity with the fatty acid receptors, depending on the position of the hydroxyl group and the identity of the receptor.
Results and discussion
Synthesis
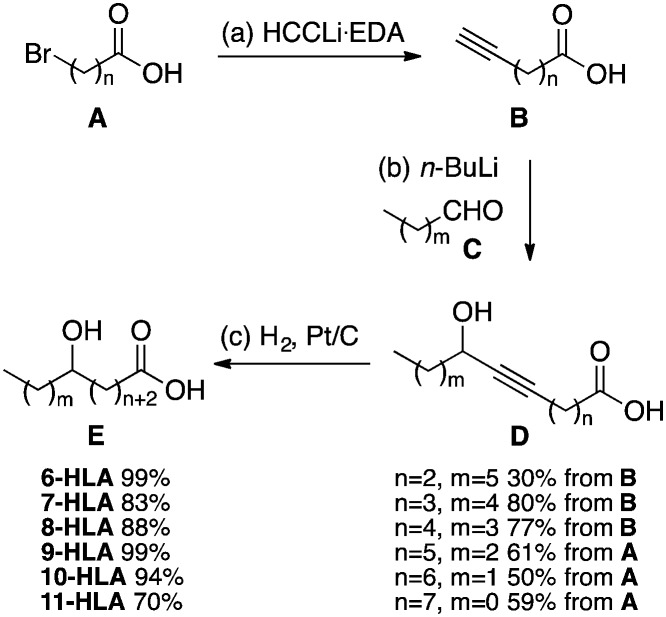
With the goal to identify a general and concise route that avoided protecting group steps, we decided to explore nucleophilic addition of a carbon nucleophile to an aldehyde. It was envisioned that doubly deprotonated terminal alkynylcarboxylic acids could be reacted with aldehydes without interference from the carboxylate. Thus, double deprotonation of terminal alkynoic acids B (n = 2–4) and reaction with aldehydes C (m = 4–6) gave propargylic alcohols D that were hydrogenated over platinum to provide the desired HLAs E (Scheme 1). Propargylic alcohol precursors (D) of 6-HLA, 7-HLA and 8-HLA were obtained in 30–80% yield from commercial terminal alkynoic acids (B). Propargylic alcohol precursors of 9-HLA, 10-HLA and 11-HLA were synthesized from ω-bromofatty acids A by reaction with lithium acetylide–ethylenediamine (EDA) followed by deprotonation of the crude product and reaction with aldehydes C to provide D in a 50–61% overall yield (Scheme 1). Hydrogenation of propargylic alcohols D over platinum gave the hydroxyfatty acids E in 70–99% yield.
Scheme 1. Reagents and conditions: (a) HCCLi–EDA, DMSO, rt; (b) n-BuLi (2 equiv.), THF, –78 °C to rt; then aldehyde C, 1 h, rt (30–80% from B, 50–61% from A over 2 steps); (c) H2, Pt/C, NaOH (1.1 equiv.), MeOH, rt (70–99%).
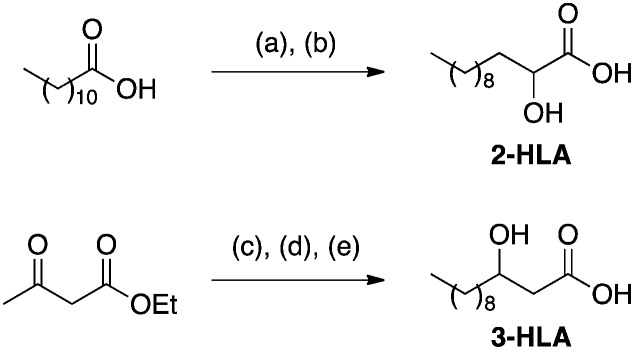
The remaining 2-HLA was obtained by a Hell–Volhard–Zelinsky-type α-bromination followed by hydrolysis with sodium hydroxide in 53% overall yield and 3-HLA was synthesized by γ-alkylation of doubly deprotonated ethyl acetoacetate followed by reduction of the β-ketone and ester hydrolysis (Scheme 2). 4-HLA and 5-HLA were obtained from the commercially available corresponding lactones and 12-HLA was purchased.
Scheme 2. Synthesis of 2-OH and 3-OH fatty acids. Reagents and conditions: (a) SOCl2, Br2, 50 °C. (b) NaOH (2 M, aq.), 85 °C, 53% over two steps. (c) NaH, n-BuLi, n-C9H19Br, THF, 0 °C to rt, 95%. (d) NaBH4, THF, EtOH. (e) LiOH, H2O : THF (1 : 2), 36% over two steps.
Biological results
The activity of the HLAs on human FFA1 and FFA4 was evaluated in β-arrestin-2 recruitment assays, as described previously,13 and on human GPR84 in a [35S]GTPγS assay. The parent compound lauric acid has EC50 of approximately 10 μM on FFA1 and FFA4. Interestingly, 2-HLA and 6-HLA displayed partial agonist activity on FFA1 with potency similar to lauric acid, whereas the remaining HLAs were essentially inactive (Table 1 and Fig. S1†). On FFA4, none of the compounds showed significant activity compared with lauric acid.
Table 1. Activity of HLAs on FFA1, FFA4 and GPR84.
| FFA1 a pEC50 | FFA4 a pEC50 | GPR84 b pEC50 | |
| Lauric acid | 4.94 ± 0.13 c 13 | 5.12 ± 0.07 (ref. 13) | — d |
| 2-HLA | 5.17 ± 0.38 | <4.0 | 4.89 ± 0.10 |
| 3-HLA | <4.0 e | <4.0 | 5.28 ± 0.07 |
| 4-HLA | <4.0 | <4.0 | 4.42 ± 0.12 |
| 5-HLA | <4.0 | <4.0 | <4.0 |
| 6-HLA | 4.73 ± 0.56 | <4.0 | <4.0 |
| 7-HLA | <4.0 | <4.0 | <4.0 |
| 8-HLA | <4.0 | <4.0 | <4.0 |
| 9-HLA | <4.0 | <4.0 | <4.0 |
| 10-HLA | <4.0 | <4.0 | <4.0 |
| 11-HLA | <4.0 | <4.0 | <4.0 |
| 12-HLA | <4.0 | <4.0 | 4.68 ± 0.09 |
Suzuki et al. have previously found 2-HLA and 3-HLA to act as agonists on GPR84.14 We confirmed the agonistic activity of these compounds and in addition found 4-HLA and 12-HLA to act as GPR84 agonists with similar efficacy, the latter being in contrast to the observations by Suzuki et al. The remaining HLAs were inactive (Table 1 and Fig. S2†).
Wang et al. reported lauric acid to be a GPR84 agonist with pEC50 of 4.98 in a [S35]GTPγS assay and 5.06 in a cAMP assay.15 Suzuki et al. subsequently found that lauric acid had some activity, albeit with significantly lower potency and efficacy than 2-HLA and 3-HLA, in a phosphoinositide accumulation assay but was inactive up to 100 μM in the [S35]GTPγS assay.14 We found lauric acid to be inactive in the cAMP assay up to 100 μM. Combined, these results indicate the introduction of hydroxy groups at specific positions has significant effect upon the activity of fatty acids at FFA1 and, especially, at GPR84.
None of the HLAs displayed activity on any of the receptors that warranted further exploration. It was therefore decided not to proceed with acquisition and testing of pure enantiomers. The results did however confirm that the introduction of a hydroxyl group into a saturated fatty acid can have a strong effect on its potency on free fatty acid receptors, and that the position of the hydroxyl group is critical for this effect. It is expected that, if necessary, the synthetic method can be adapted to access pure enantiomers, e.g. by implementation of Carreira's asymmetric propargylic alcohol synthesis,26 although straight-chain aldehydes tend to be more challenging substrates for this protocol, or by resolution at the propargylic alcohol stage.27
Experimental
Chemistry
Commercial starting materials and solvents were used without further purification unless otherwise stated. Anhydrous THF was freshly distilled from sodium/benzophenone. DMSO was dried over 3 Å molecular sieves and stored under argon. TLC was performed on TLC silica gel 60 F254 plates and visualized by staining in basic KMnO4-solution. Column chromatography was performed using silica gel 60 (0.040–0.063 mm, Merck). 1H and 13C NMR spectra were recorded at 400 and 101 MHz, respectively, on a Bruker Avance 400 at 300 K. All spectra were calibrated relative to the solvent residual peak. High-resolution mass spectrometry was performed on a Bruker micrOTOF II. The purity of all test compounds were >95% as assessed by 1H and 13C NMR spectroscopy.
General procedures for synthesis of hydroxyfatty acids
Step A: substitution of ω-bromofatty acid by lithium acetylide–ethylenediamine
A lithium acetylide–ethylenediamine (EDA) complex (3 equiv.) was dissolved in DMSO (0.4 mL mmol–1). To this was added ω-bromofatty acid (1 equiv.) in DMSO (0.8 mL mmol–1). The reaction was stirred at room temperature for 20–24 h. The reaction was then acidified with 2 M aq. HCl to approximately pH 1, brine was added and the mixture was extracted with ethyl acetate. The organic phase was washed with brine/1 M aq. HCl (1 : 1), dried over Na2SO4 and evaporated in vacuo. The crude material was carried on directly to step B.
Step B: synthesis of propargylic alcohols
The ω-alkynoic acid (1.1 equiv.) was dissolved in THF (10 mL mmol–1) and cooled to –78 °C. n-Butyl lithium (2.5 M in hexanes, 2.2 equiv.) was added dropwise. After 10 minutes the suspension was allowed to reach room temperature. After an additional 20 minutes, the aldehyde (1 equiv.) was added in one portion. After stirring for 30–60 minutes the reaction mixture was acidified with 2 M aq. HCl to pH 1 and brine was added. The aqueous phase was extracted with ethyl acetate. The combined organic phase was dried over Na2SO4 and evaporated in vacuo. The crude product was purified by column chromatography (1 : 1 : 100 AcOH : MeOH : DCM).
Step C: hydrogenation of propargylic alcohols
To the hydroxydodecynoic acid in methanol (0.8 mL mmol–1) was added NaOH (1.1 eq.) and Pt/C (0.05 eq., 5% w/w). The reaction mixture was placed under an atmosphere of H2 and stirred for 20 hours before acidification with 1 M HCl to pH 1 and addition of Na2SO4. The mixture was filtered through a pad of Celite, dried over Na2SO4 and evaporated in vacuo. If necessary, the product was purified by column chromatography (1 : 1 : 100 AcOH : MeOH : DCM).
6-Hydroxy-4-dodecynoic acid
6-Hydroxy-4-dodecynoic acid was prepared according to the general procedure (GP) step B using 4-pentynoic acid (97 mg, 0.85 mmol), butyl lithium (0.77 mL, 1.9 mmol) and heptanal (120 μL, 0.86 mmol), with the modification of 1,3-dimethyltetrahydropyrimidin-2(1H)-one (DMPU, 0.23 mL, 1.9 mmol) added as a deaggregator during the deprotonation and the reaction was only allowed to reach 0 °C, to give 55 mg (32%) of the title compound as a white amorphous solid: 1H NMR (400 MHz, CD3OD) δ 4.24 (t, J = 6.7 Hz, 1H), 2.49 (s, 4H), 1.68–1.51 (m, 2H), 1.48–1.26 (m, 8H), 0.91 (dd, J = 8.7, 4.9 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 175.7, 83.8, 83.2, 63.0, 39.3, 34.5, 33.0, 30.1, 26.4, 23.7, 15.4, 14.4; ESI-HRMS calcd for C12H20NaO3 (M + Na+) 235.1305, found 239.1300.
6-Hydroxylauric acid (6-HLA)28
6-Hydroxylauric acid was prepared according to GP step C using 6-hydroxy-4-dodecynoic acid (30 mg, 0.14 mmol), NaOH (7 mg, 0.2 mmol) and Pt/C (27 mg, 6.9 μmol Pt) to give 33 mg (99%) of the title compound as an amorphous white solid: 1H NMR (400 MHz, CD3OD) δ 3.55–3.47 (m, 1H), 2.30 (t, J = 7.4 Hz, 2H), 1.68–1.55 (m, 2H), 1.49–1.28 (m, 14H), 0.91 (t, J = 6.8 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.6, 72.3, 38.5, 38.1, 35.0, 33.1, 30.6, 26.8, 26.4, 26.2, 23.7, 14.42; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found 239.1609.
7-Hydroxy-5-dodecynoic acid
7-Hydroxy-5-dodecynoic acid was prepared according to GP step B using 5-hexynoic acid (125 μL, 1.13 mmol), butyl lithium (0.88 mL, 2.2 mmol) and hexanal (120 μL 1.0 mmol) to give 170 mg (80%) of the title compound as a colorless oil: 1H NMR (400 MHz, CDCl3) δ 4.35 (tt, J = 6.6, 1.9 Hz, 1H), 2.49 (t, J = 7.3 Hz, 2H), 2.31 (td, J = 6.9, 1.9 Hz, 2H), 1.84 (p, J = 7.1 Hz, 2H), 1.73–1.58 (m, 2H), 1.49–1.23 (m, 6H), 0.90 (t, J = 6.9 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ 178.6, 83.9, 82.5, 62.7, 38.0, 32.7, 31.5, 24.9, 23.5, 22.6, 18.1, 14.0; ESI-HRMS calcd for C12H20NaO3 (M + Na+) 235.1305, found 235.1311.
7-Hydroxylauric acid (7-HLA)
7-Hydroxylauric acid was prepared according to GP step C using 7-hydroxy-5-dodecynoic acid (51 mg, 0.24 mmol), NaOH (11 mg, 0.28 mmol) and Pt/C (45 mg, 12 μmol Pt) to give 43 mg (83%) of the title compound as a white wax: 1H NMR (400 MHz, CD3OD) δ 3.58–3.44 (m, 1H), 2.29 (t, J = 7.4 Hz, 2H), 1.67–1.56 (m, 2H), 1.51–1.23 (m, 14H), 0.91 (t, J = 6.9 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.7, 72.4, 38.4, 38.3, 35.0, 33.2, 30.3, 26.5, 26.5, 26.1, 23.7, 14.4; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found: 239.1609.
8-Hydroxy-6-dodecynoic acid
8-Hydroxy-6-dodecynoic acid was prepared according to GP step B using 6-heptynoic acid (160 μL, 1.26 mmol), butyl lithium (1.0 mL, 2.5 mmol) and pentanal (125 μL, 1.18 mmol) to give 192 mg (77%) of the title compound as a colorless oil: 1H NMR (400 MHz, CDCl3) δ 4.35 (tt, J = 6.6, 1.9 Hz, 1H), 2.39 (t, J = 7.4 Hz, 2H), 2.25 (td, J = 7.0, 1.9 Hz, 2H), 1.80–1.51 (m, 6H), 1.46–1.28 (m, 4H), 0.91 (t, J = 7.2 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ 179.1, 84.6, 81.9, 62.7, 37.8, 33.4, 27.9, 27.4, 23.8, 22.4, 18.4, 14.0; ESI-HRMS calcd for C12H20NaO3 (M + Na+) 235.1305, found 235.1311.
8-Hydroxylauric acid (8-HLA)
8-Hydroxylauric acid was prepared according to GP step C using 8-hydroxy-6-dodecynoic acid (49 mg, 0.23 mmol), NaOH (18 mg, 0.45 mmol) and Pt/C (45 mg, 1.2 μmol Pt) to give 44 mg (88%) of the title compound as a white amorphous solid over two steps: 1H NMR (400 MHz, CD3OD) δ 3.55–3.46 (m, 1H), 2.28 (t, J = 7.4 Hz, 2H), 1.67–1.56 (m, 2H), 1.50–1.24 (m, 14H), 0.92 (t, J = 7.1 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.7, 72.4, 38.4, 38.2, 35.1, 30.5, 30.3, 29.1, 26.7, 26.1, 23.9, 14.4; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found 239.1615.
9-Hydroxy-7-dodecynoic acid (9)
9-Hydroxy-7-dodecynoic acid was prepared according to GP steps A and B using lithium acetylide–EDA (378 mg, 4.11 mmol), 6-bromohexanoic acid (256 mg, 1.31 mmol), butyl lithium (0.82 mL, 2.1 mmol) and butanal (83 μL, 0.92 mmol) and purified by column chromatography (1 : 1 : 100, AcOH : MeOH : DCM) to give 122 mg (61%) of the title compound as a thick oil: 1H NMR (400 MHz, CDCl3) δ 4.36 (tt, J = 6.6, 1.9 Hz, 1H), 2.37 (t, J = 7.4 Hz, 2H), 2.22 (td, J = 6.8, 1.9 Hz, 2H), 1.73–1.40 (m, 10H), 0.94 (t, J = 7.4 Hz, 3H); 13C NMR (101 MHz, CDCl3) δ 179.1, 85.0, 81.6, 62.5, 40.3, 33.8, 28.1, 28.1, 24.1, 18.5, 18.5, 13.7; ESI-HRMS calcd for C12H20NaO3 235.1305 (M + Na+), found 235.1299.
9-Hydroxylauric acid (9-HLA)29
9-Hydroxylauric acid was prepared according to GP step C using 9-hydroxy-7-dodecynoic acid (29 mg, 0.14 mmol), NaOH (7 mg, 0.16 mmol) and Pt/C (28 mg, 7 μmol Pt) to give 30 mg (99%) of the title compounds a white amorphous solid: 1H NMR (400 MHz, CD3OD) δ 3.56–3.47 (m, 1H), 2.28 (t, J = 7.4 Hz, 2H), 1.66–1.56 (m, 2H), 1.47–1.29 (m, 14H), 0.93 (t, J = 7.0 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.8, 72.2, 40.7, 38.4, 35.0, 30.7, 30.4, 30.2, 26.7, 26.1, 19.9, 14.5; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found 239.1606.
10-Hydroxy-8-dodecynoic acid
10-Hydroxy-8-dodecynoic acid was prepared according to GP steps A and B using lithium acetylide–EDA (325 mg, 3.53 mmol), 7-bromoheptanoic acid (248 mg, 1.19 mmol), n-BuLi (0.68 mL, 1.7 mmol) and propanal (110 μL, 1.52 mmol) and purified by column chromatography (1 : 1 : 100, AcOH : MeOH : DCM) to give 82 mg (50%) of the title compound as a colorless oil: 1H NMR (400 MHz, CD3OD) δ 4.20 (tt, J = 6.6, 1.9 Hz, 1H), 2.29 (t, J = 7.4 Hz, 2H), 2.21 (td, J = 6.8, 1.9 Hz, 2H), 1.72–1.28 (m, 10H), 0.98 (t, J = 7.4 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.7, 85.3, 82.5, 64.4, 34.9, 32.4, 30.1, 29.7, 29.5, 26.0, 19.3, 10.0; ESI-HRMS calcd for C12H20NaO3 (M + Na+) 235.1305, found 235.1307.
10-Hydroxylauric acid (10-HLA)
10-Hydroxylauric acid was prepared according to GP step C using 10-hydroxy-8-dodecynoic acid (51 mg, 0.24 mmol), NaOH (12 mg, 0.30 mmol) and Pt/C (47 mg, 12 μmol Pt) to give 48 mg (94%) of the title compound as a white amorphous solid: 1H NMR (400 MHz, CD3OD) δ 3.37–3.29 (m, 1H), 2.18 (t, J = 7.4 Hz, 2H), 1.53–1.46 (m, 2H), 1.42–1.20 (m, 14H), 0.83 (t, J = 7.4 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.8, 73.9, 37.9, 35.0, 31.0, 30.8, 30.6, 30.4, 30.2, 26.8, 26.1, 10.3; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found 239.1620.
11-Hydroxy-9-dodecynoic acid
11-Hydroxy-9-dodecynoic acid was prepared according to GP step A and B using lithium acetylide–EDA (310 mg, 3.37 mmol), 8-bromooctanoic acid (252 mg, 1.13 mmol) and butyl lithium (0.70 mL, 1.75 mmol) and ethanal (0.15 mL, 2.7 mmol) and purified by column chromatography (1 : 1 : 100, AcOH : MeOH : DCM) to give 99 mg (59%) of the title compound as a white amorphous solid: 1H NMR (400 MHz, CDCl3) δ 4.52 (qt, J = 6.5, 1.9 Hz, 1H), 2.35 (t, J = 7.5 Hz, 2H), 2.19 (td, J = 7.0, 1.9 Hz, 2H), 1.69–1.58 (m, 2H), 1.54–1.45 (m, 2H), 1.44–1.32 (m, 9H); 13C NMR (101 MHz, CDCl3) δ 179.3, 84.6, 82.3, 58.6, 33.9, 28.8, 28.6, 28.5, 28.5, 24.7, 24.6, 18.6; ESI-HRMS calcd for C12H20NaO3 (M + Na+) 235.1305, found 235.1311.
11-Hydroxylauric acid (11-HLA)30
11-Hydroxylauric acid was prepared according to GP step C using 11-hydroxy-9-dodecynoic acid (49 mg, 0.24 mmol), NaOH (12 mg, 0.30 mmol) and Pt/C (46 mg, 12 mmol Pt) to give 36 mg (70%) of the title compounds as a white amorphous solid: 1H NMR (400 MHz, CD3OD) δ 3.76–3.65 (m, 1H), 2.28 (t, J = 7.4 Hz, 2H), 1.59 (dd, J = 14.4, 7.2 Hz, 2H), 1.43–1.29 (m, 14H), 1.14 (d, J = 6.2 Hz, 3H); 13C NMR (101 MHz, CD3OD) δ 177.8, 68.6, 40.2, 35.0, 30.8, 30.7, 30.6, 30.4, 30.2, 26.9, 26.1, 23.5; ESI-HRMS calcd for C12H24NaO3 (M + Na+) 239.1618, found 239.1620.
Biology
[35S]GTPγS binding assay
Cells expressing FLAG-GPR84-eYFP were homogenized in 10 mM Tris-HCl (pH 7.4) and 0.1 mM EDTA followed by centrifugation at 1000 × g for 5 min at 4 °C to remove nuclei and cellular debris. Membrane fractions were collected by spinning the supernatant at 38 000 × g for 45 min and resuspending the pellet in 20 mM HEPES (pH 7.5) with 5 mM MgCl2. 10 μg of membrane were incubated at 30 °C for 1 h in assay buffer (20 mM HEPES, 5 mM MgCl2, 160 mM NaCl, 0.05% fatty acid free bovine serum albumin, pH 7.5) containing 1 μM GDP and 0.1 nM [35S]GTPγS (PerkinElmer Life Sciences) in the absence or presence of compounds. Reactions were terminated by vacuum filtration through GF/C filters, and the retained radioactivity was quantified on a liquid scintillation counter.
Conclusions
We have developed a general and concise protocol for synthesis of racemic saturated hydroxy fatty acids with the hydroxyl group at position 6 or higher and exemplified this with the synthesis of HLAs. The protocol will be useful in further exploration of saturated fatty acids and the FAHFA lipid class. HLAs were tested on the fatty acid receptors FFA1, FFA4 and GPR84. The results showed that a hydroxy group in specific positions preserved the activity of the parent fatty acid, including the confirmed activity of compounds previously reported as GPR84 agonists, whereas the remaining HLAs displayed low or no activity. Apart from 2-HLA, which was active on both FFA1 and GPR84, the active HLAs differed with the identity of the receptor, consistent with the low sequence similarity between the receptors.
Supplementary Material
Acknowledgments
We thank Professor John Nielsen (Univ. of Copenhagen) for valuable discussions. The study was supported by the Danish Council for Strategic Research (grant 11-116196) and the University of Southern Denmark.
Footnotes
†The authors declare no competing interests.
‡Electronic supplementary information (ESI) available: Synthetic procedures and compound characterization and NMR spectra. See DOI: 10.1039/c7md00130d
References
- Milligan G., Shimpukade B., Ulven T., Hudson B. D. Chem. Rev. 2017;117:67. doi: 10.1021/acs.chemrev.6b00056. [DOI] [PubMed] [Google Scholar]
- Masoodi M., Mir A. A., Petasis N. A., Serhan C. N., Nicolaou A. Rapid Commun. Mass Spectrom. 2008;22:75. doi: 10.1002/rcm.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primdahl K. G., Stenstrøm Y., Hansen T. V., Vik A. Chem. Phys. Lipids. 2016;196:1. doi: 10.1016/j.chemphyslip.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Primdahl K. G., Aursnes M., Walker M. E., Colas R. A., Serhan C. N., Dalli J., Hansen T. V., Vik A. J. Nat. Prod. 2016;79:2693. doi: 10.1021/acs.jnatprod.6b00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C. N. Nature. 2014;510:92. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2010;1801:405. [Google Scholar]
- Raetz C. R., Whitfield C. Annu. Rev. Biochem. 2002;71:635. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimick P. S., Walker N. J., Patton S. Biochem. J. 1969;111:395. doi: 10.1042/bj1110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-J., Vijaya Krishna R., Tsai C.-C., Wu W.-H., Chao L. K., Hwang K.-H., Chien C. M., Chang H.-Y., Chen S.-T. Bioorg. Med. Chem. 2010;18:6896. doi: 10.1016/j.bmc.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Parolin C., Calonghi N., Presta E., Boga C., Caruana P., Naldi M., Andrisano V., Masotti L., Sartor G. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2012;1821:1334. doi: 10.1016/j.bbalip.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Yore M. M., Syed I., Moraes-Vieira P. M., Zhang T., Herman M. A., Homan E. A., Patel R. T., Lee J., Chen S., Peroni O. D., Dhaneshwar A. S., Hammarstedt A., Smith U., McGraw T. E., Saghatelian A., Kahn B. B. Cell. 2014;159:318. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulven T., Christiansen E. Annu. Rev. Nutr. 2015;35:239. doi: 10.1146/annurev-nutr-071714-034410. [DOI] [PubMed] [Google Scholar]
- Christiansen E., Watterson K. R., Stocker C. J., Sokol E., Jenkins L., Simon K., Grundmann M., Petersen R. K., Wargent E. T., Hudson B. D., Kostenis E., Ejsing C. S., Cawthorne M. A., Milligan G., Ulven T. Br. J. Nutr. 2015;113:1677. doi: 10.1017/S000711451500118X. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Takaishi S., Nagasaki M., Onozawa Y., Iino I., Maeda H., Komai T., Oda T. J. Biolumin. Chemilumin. 2013;288:10684. doi: 10.1074/jbc.M112.420042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Wu X. S., Simonavicius N., Tian H., Ling L. J. Biolumin. Chemilumin. 2006;281:34457. doi: 10.1074/jbc.M608019200. [DOI] [PubMed] [Google Scholar]
- Vasilakaki S., Barbayianni E., Leonis G., Papadopoulos M. G., Mavromoustakos T., Gelb M. H., Kokotos G. Bioorg. Med. Chem. 2016;24:1683. doi: 10.1016/j.bmc.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y., Mori K. Eur. J. Org. Chem. 2005;2005:4789. [Google Scholar]
- Galleano I., Schiedel M., Jung M., Madsen A. S., Olsen C. A. J. Med. Chem. 2016;59:1021. doi: 10.1021/acs.jmedchem.5b01532. [DOI] [PubMed] [Google Scholar]
- De Vleeschouwer M., Sinnaeve D., Van den Begin J., Coenye T., Martins J. C., Madder A. Chem. – Eur. J. 2014;20:7766. doi: 10.1002/chem.201402066. [DOI] [PubMed] [Google Scholar]
- Chiang C.-H., Ramu R., Tu Y.-J., Yang C.-L., Ng K. Y., Luo W.-I., Chen C. H., Lu Y.-Y., Liu C.-L., Yu S. S. F. Chem. – Eur. J. 2013;19:13680. doi: 10.1002/chem.201302402. [DOI] [PubMed] [Google Scholar]
- Kühnel K., Maurer S. C., Galeyeva Y., Frey W., Laschat S., Urlacher V. B. Adv. Synth. Catal. 2007;349:1451. [Google Scholar]
- Durairaj P., Malla S., Nadarajan S. P., Lee P.-G., Jung E., Park H. H., Kim B.-G., Yun H. Microb. Cell Fact. 2015;14:45. doi: 10.1186/s12934-015-0228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri Y., Hannemann F., Girhard M., Kappl R., Hutter M., Urlacher V. B., Bernhardt R. FEBS J. 2015;282:74. doi: 10.1111/febs.13104. [DOI] [PubMed] [Google Scholar]
- Ebert C., Felluga F., Forzato C., Foscato M., Gardossi L., Nitti P., Pitacco G., Boga C., Caruana P., Micheletti G., Calonghi N., Masotti L. J. Mol. Catal. B: Enzym. 2012;83:38. [Google Scholar]
- Abraham S., Lan Y., Lam R. S. H., Grahame D. A. S., Kim J. J. H., Weiss R. G., Rogers M. A. Langmuir. 2012;28:4955. doi: 10.1021/la204412t. [DOI] [PubMed] [Google Scholar]
- Frantz D. E., Fässler R., Carreira E. M. J. Am. Chem. Soc. 2000;122:1806. [Google Scholar]
- Birman V. B., Guo L. Org. Lett. 2006;8:4859. doi: 10.1021/ol061906y. [DOI] [PubMed] [Google Scholar]
- Shimotori Y., Aoyama M., Tsukasa H., Miyakoshi T. Heterocycles. 2012;85:1061. [Google Scholar]
- Ahmad I., Sherwani M. R. K., Hasan S. Q., Ahmad, Jr. M. S., Osman S. M. Phytochemistry. 1983;22:493. [Google Scholar]
- Voss G., Gerlach H. Helv. Chim. Acta. 1983;66:2294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.