Main Text
Fluorescence correlation spectroscopy (FCS) is a well-established technique based on the analysis of fluorescence intensity fluctuations. FCS has become a popular biophysical method for the investigation of molecular properties in solutions and in cells (1). In FCS, the fluctuations in the recorded fluorescence intensity F(t) are characterized by an autocorrelation function that contains average information about the typical width and relative magnitude of the fluctuations:
| (1) |
where the brackets indicate averaging over the entire measurement. The strength of correlation spectroscopy is that the averaging process of many noisy fluctuations yields a robust estimation of the characteristic parameters describing the origin of the fluctuations. Examples of biophysical parameters measurable by FCS are diffusion coefficients, molecular concentrations, aggregation rates, molecular interactions, fluorescence brightness, and binding constants (1).
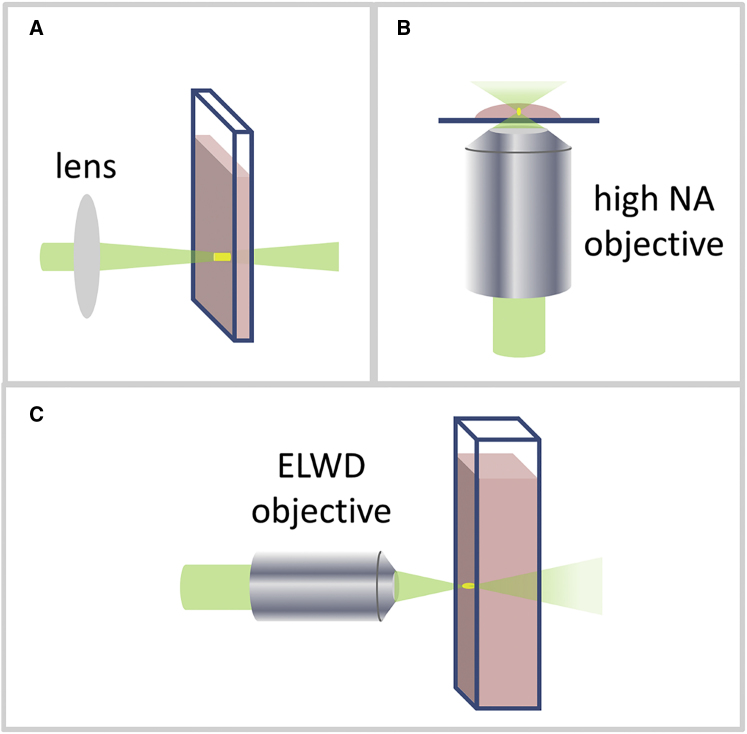
The relative amplitude of the fluctuations generated by the fluorescent molecules scales as ∼1/N, where N is the number of molecules in the observation volume. In real experiments, if this number is too high, the fluctuations generated by the fluorescent molecules may be overshadowed by the presence of other systematic fluctuations. For this reason, FCS data are typically recorded at small values of N, i.e., in femtoliter (1 fL = 10−15 L) observation volumes and at low concentrations (<1 μM). Notably, in the very first experimental implementation of FCS dating back to 1974, an observation volume of ∼103 fL was generated by laser illumination of a flat, thin-walled cuvette containing a ∼1 nM sample solution (2) (Fig. 1 A). However, after this first experiment in cuvette, it was soon realized that the confocal microscope was the ideal instrument for FCS because of its high photon detection efficiency and good rejection of background fluorescence (3). As a matter of fact, most FCS measurements are currently performed on confocal microscopes equipped with a high NA objective (Fig. 1 B). Nevertheless, the microscope-based configuration may represent a limitation for certain types of experiments, for instance those requiring high temperatures, stirring of the sample, or use of nonaqueous solvents (4).
Figure 1.
(A) Schematic of the configuration used by Magde et al. (2) in their first experimental demonstration of FCS. A lens was used to produce a focal spot size of 5.7 μm, and the optical path of the flat cuvette was either 25 or 150 μm. (B) A schematic of the FCS configuration used in confocal microscopes is shown, equipped with a high NA objective. (C) A schematic of the configuration used by Sahoo et al. in their cuvette-FCS setup is shown. The use of an extra-long working distance objective makes FCS feasible with standard cuvettes. To see this figure in color, go online.
In their article appearing in this issue of Biophysical Journal, Sahoo et al. (5) describe a novel experimental setup for performing FCS directly from solutions placed inside cuvettes. They use an extra-long working distance (>1.8 mm) objective in horizontal geometry to get enough sensitivity to perform FCS inside conventional cuvettes (Fig. 1 C). With an effective observation volume of ∼1.8 fL and a molecular brightness of up to ∼50 KHz, the performance of this cuvette-FCS setup is comparable to those of most microscope-based FCS setups. The use of a cuvette seems advantageous for measurements in controlled temperature ranges or in combination with automatic titrators, as demonstrated by Sahoo et al. In fact, they study urea-dependent unfolding of a tetramethylrhodamine-labeled Nt-apoE4 protein by coupling the cuvette with automatic titration and stirring, suggesting that this setup could find extensive application in single-molecule studies of denaturant-dependent folding and unfolding of proteins (6, 7). The novel fluorescence correlation spectrometer could certainly become a convenient alternative to whoever wants to perform single-point FCS in solutions.
Compared to other fluorescence spectroscopy techniques that were first established on cuvette and only later combined with microscopy (for instance, fluorescence lifetime spectroscopy, which evolved into fluorescence lifetime imaging), FCS has become a mature technique in the confocal microscope. This has naturally led to many important technical developments, such as, for instance, scanning FCS (8), image correlation spectroscopy (9), and a huge number of applications in live cells. Now, with FCS moving back from microscope to cuvette, single-molecule fluctuation measurements could be performed as easily as with a spectrofluorometer. Still, it remains to be seen if some of the concepts developed for microscope-based FCS will find application in the new cuvette-based configuration. For instance, a potential issue of the cuvette-FCS setup is represented by the huge difference between the small observation volume required to detect single-molecule fluctuations (∼1.8 fL) and the total cuvette volume (∼3.5 mL). In this respect, mechanical scanning of the cuvette could be useful to explore a larger portion of the cuvette, especially in the case of very dilute samples (10, 11). This idea has been demonstrated only with large, bright particles such as fluorescently labeled bacteria and yeast cells (10, 11). The introduction of a cuvette-FCS setup sensitive to single-molecule fluctuations opens interesting perspectives toward detection of very low concentrations of toxic agents.
Editor: Jochen Mueller.
References
- 1.Elson E.L. Fluorescence correlation spectroscopy: past, present, future. Biophys. J. 2011;101:2855–2870. doi: 10.1016/j.bpj.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 3.Koppel D.E., Axelrod D., Webb W.W. Dynamics of fluorescence marker concentration as a probe of mobility. Biophys. J. 1976;16:1315–1329. doi: 10.1016/S0006-3495(76)85776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zettl H., Häfner W., Krausch G. Fluorescence correlation spectroscopy of single dye-labeled polymers in organic solvents. Macromolecules. 2004;37:1917–1920. [Google Scholar]
- 5.Sahoo B., Sil T.B., Karmakar B., Garai K. A fluorescence correlation spectrometer for measurements in cuvettes. Biophys. J. 2018;115:455–466. doi: 10.1016/j.bpj.2018.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay K., Saffarian S., Frieden C. Measuring unfolding of proteins in the presence of denaturant using fluorescence correlation spectroscopy. Biophys. J. 2005;88:1413–1422. doi: 10.1529/biophysj.104.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman E., Itkin A., Haran G. Using fluorescence correlation spectroscopy to study conformational changes in denatured proteins. Biophys. J. 2008;94:4819–4827. doi: 10.1529/biophysj.107.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruan Q., Cheng M.A., Mantulin W.W. Spatial-temporal studies of membrane dynamics: scanning fluorescence correlation spectroscopy (SFCS) Biophys. J. 2004;87:1260–1267. doi: 10.1529/biophysj.103.036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen N.O., Höddelius P.L., Magnusson K.E. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys. J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altamore I., Lanzano L., Gratton E. Dual channel detection of ultra low concentration of bacteria in real time by scanning FCS. Meas. Sci. Technol. 2013;24:65702. doi: 10.1088/0957-0233/24/6/065702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skinner J.P., Swift K.M., Tetin S.Y. Simplified confocal microscope for counting particles at low concentrations. Rev. Sci. Instrum. 2013;84:074301. doi: 10.1063/1.4812782. [DOI] [PMC free article] [PubMed] [Google Scholar]