Abstract
The spatial distribution of the human epidermal growth factor 2 (HER2) receptor in the plasma membrane of SKBR3 and HCC1954 breast cancer cells was studied. The receptor was labeled with quantum dot nanoparticles, and fixed whole cells were imaged in their native liquid state with environmental scanning electron microscopy using scanning transmission electron microscopy detection. The locations of individual HER2 positions were determined in a total plasma membrane area of 991 μm2 for several SKBR3 cells and 1062 μm2 for HCC1954 cells. Some of the HER2 receptors were arranged in a linear chain with interlabel distances of 40 ± 7 and 32 ± 10 nm in SKBR3 and HCC1954 cells, respectively. The finding was tested against randomly occurring linear chains of six or more positions, from which it was concluded that the experimental finding is significant and did not arise from random label distributions. Because the measured interlabel distance in the HER2 chains is similar to the 36-nm helix-repetition distance of actin filaments, it is proposed that a linking mechanism between HER2 and actin filaments leads to linearly aligned oligomers.
Introduction
HER2 is a member of the HER (ErbB) family of receptors, a subfamily of four closely related receptor tyrosine kinases: HER1 (EGFR, ErbB-1), HER2 (ErbB-2), HER3 (ErbB-3), and HER4 (ErbB-4). This family of membrane proteins regulates cell proliferation, survival, and various other processes. Under physiological circumstances, most HER family members are activated by ligands and dimerize with another activated HER protein. The activated dimer protein complex then triggers intracellular processes. This situation is different for the family member HER2, which is overexpressed in a particularly aggressive subtype of breast cancer (1). It is an orphan receptor that does not have a known ligand, and it resides in the plasma membrane in an open conformation, ready to form homodimers or heterodimers with other members of the HER receptor family. As a consequence, intracellular signaling and cell growth is frequently dysregulated in HER2-overexpressing cells (2). The signaling-active homodimerized form of HER2 preferentially resides in membrane ruffles in the plasma membrane (3), which serve as junctions for cellular signaling and are known to drive motility, invasiveness, and metastasis of cancer cells (4). We hypothesize that in HER2-overexpressing cells, distinct spatial arrangements, aside from the known dimerization of the signaling-active receptor and its preference to form larger clusters (5), might contribute to its functionality in overexpressing cancer cells.
In this study, we examined the spatial distribution of HER2 in large areas (∼1000 μm2) of the intact plasma membrane for two different cell lines. Our aim was to study the receptor distribution beyond its mere presence as homodimers found in membrane-ruffled areas versus monomers in flat areas of the plasma membrane (3); the distribution was found to alter upon incubation with the antibody-based drug trastuzumab (6). For the detection of membrane-bound HER2, we applied specific labeling with affibodies and quantum dots (QDs) suitable for correlative fluorescence and electron microscopy (7). Using environmental scanning electron microscopy (ESEM)-scanning transmission electron microscopy (STEM), it is possible to detect the labels with nanometer precision while the cells are kept in a liquid state so that the plasma membrane remains intact (8). Of key importance for the study of receptor distributions is to avoid relying on selected regions of single cells, given that large differences occur between cells due to cancer cell heterogeneity (9). Membrane areas of a representative size of multiple whole cells must be examined, whereas at the same time single-molecule detection is required. Two human breast cancer cell lines were used, SKBR3 and HCC1954. Both are known to overexpress HER2 and serve as a model system of HER2+ breast cancer in numerous studies (10). The HCC1954 cell line is a trastuzumab-resistant cell line. Among many thousands of labels, it was observed that HER2 sometimes aligned in linear chains with regular interlabel distances, and the observation of these chains was highly significant with respect to randomly occurring patterns.
Materials and Methods
Materials
SKBR3 and HCC1954cells were obtained from ATCC (Manassas, VA), and authenticated by genotyping (Multiplexion, Heidelberg, Germany). Biotin-conjugated anti-HER2 affibody molecule (HER2-AFF-B, type: (ZHER2:477)2) was purchased from Affibody AB, Bromma, Sweden. Dulbecco’s phosphate-buffered saline was from Lonza Cologne, Cologne, Germany. Dulbecco’s modified Eagle’s medium GlutaMAX with high glucose and pyruvate, RPMI Medium 1640 GlutaMAX, minimum essential medium nonessential amino acids 100× solution, QD Qdot 655 streptavidin conjugate (strep-QD), and fetal bovine serum were from Life Technologies, Carlsbad, CA. Normal goat serum (GS) was purchased from Rockland Immunochemicals, Gilbertsville, PA. High-performance liquid chromatography (HPLC)-grade acetone and ethanol, phosphate-buffered saline (PBS) 10× solution, electron-microscopy-grade glutaraldehyde 25% solution, D-glucose, D-saccharose, glycine, biotin-free and molecular-biology-grade albumin Fraktion V (BSA), and sodium cacodylate trihydrate were from Carl Roth KG, Karlsruhe, Germany. Electron-microscopy-grade formaldehyde 16% solution was from Electron Microscopy Sciences, Hatfield, PA. HPLC-grade deionized water, poly-L-lysine (hydrobromide (mol wt 70,000–150,000), sodium tetraborate, boric acid, and gelatin from cold-water fish skin (GEL) were from Sigma-Aldrich Chemie, Munich, Germany. Custom-designed silicon microchips were purchased from DENSsolution, Delft, the Netherlands. The microchips had outer dimensions of 2.0 × 2.6 × 0.4 mm3, and each contained a silicon nitride (SiN) membrane window with dimensions of 50 × 400 μm2 and a membrane with a thickness of 50 nm.
Preparation of microchips with adhered cells
Microchips with thin SiN windows were used as a support for the SKBR3 cells for all experiments and were designed for ESEM-STEM under wet conditions. The cells were seeded onto these microchips, and the samples were prepared using a protocol described elsewhere in detail (11, 12). In brief, new microchips contained a protective photoresistant coating that was removed with subsequent 2 min washes in acetone and ethanol. The microchips were then plasma cleaned for 5 min to render the SiN surfaces hydrophilic and placed in 0.01% poly-L-lysine for 5 min, rinsed two times in water, and placed in an individual well of a 96-well plate filled with cell media. The cells were seeded on the microchips by adding a droplet of ∼20 μL cell suspension to each well. They settled on the window within several minutes with a density of 5–15 per window area, as was verified from inspection using an upright microscope. Finally, the microchips were incubated for several hours and inspected using a phase contrast light microscope (Leica, Mannheim, Germany). If the density of cells had reached the required number of 5–15 per SiN window, and if the cells had flattened out, indicating cell adherence, the chips were ready for protein labeling.
Labeling of HER2 on SKBR3 or HCC1954 cells
A two-step labeling method was used (3, 12). First, the cells were incubated with a biotinylated anti-HER2-affibody that binds to a single epitope at the HER2-receptor (13), and second, the cells were fixed and incubated with strep-QDs that bind in a one/one ratio to the biotin moiety of the affibody (13). Before labeling, the cells were incubated in cell media without fetal bovine serum for 24 hr in a CO2 incubator. Serum starvation was performed to improve labeling efficiency and ensure reproducibility between the different experiments. 20 μM anti-HER2-affibody-biotin stock solution was added to GS-GEL-BSA-PBS (1% GS, 0.5% BSA, 0.1% gelatin) to obtain a 200-nM HER2-affibody-biotin labeling solution. The microchips with adherent cells were rinsed twice in serum-free media, rinsed and incubated for 5 min in GS-GEL-BSA-PBS, and subsequently incubated in the 200-nM HER2-affibody-biotin solution for ∼10 min at room temperature. Excess labeling agent was removed by rinsing four times with PBS and once with 0.1 M cacodylate buffer, 0.1 M sucrose, pH 7.4 (CB), followed by fixation with 3% formaldehyde in CB. The microchips were then rinsed once with CB and three times with PBS and placed in GLY-PBS (0.1% glycine in PBS) for 2 min. 1 μM strep-QD655 stock solution was diluted in 40 mM Borate buffer and added to 1% bovine serum albumin (hBSA)-PBS (1% BSA) to obtain a 5 nM solution. The fluorescent QD label was allowed to bind to the biotin moiety by incubating the samples in the strep-QD solution for 12 min. Note that without fixation before the incubation with strep-QD655, label-induced clustering might occur (14). Finally, all microchips were rinsed four times with hBSA-PBS. After fluorescence microscopy (see next paragraph), the cells were fixed with 2% glutaraldehyde in CB to stabilize the samples for electron microscopy, and then rinsed once with CB and three times with hBSA-PBS and stored in hBSA-PBS at 4°C until ESEM imaging. A control experiment was performed using the same process and quantities described above, except that the cells were not incubated in the anti-HER2-affibody-biotin solution before QD labeling, verifying that no unspecific binding of QDs occurred.
Fluorescence microscopy
The microchips with cells were imaged in an inverted fluorescence microscope (DMI6000B Leica; Leica), in which the microchips were placed upside down in a cell culture glass-bottom dish in hBSA-PBS after the formaldehyde fixation. Cells were imaged with 20× and 40× objectives. Direct interference contrast (DIC) images were acquired to study the cell topography, whereas the fluorescence signal revealed the localization of regions with a bound label.
ESEM
Intact cells in a liquid state were studied in an ESEM (Quanta 400 FEG; FEI, Hillsboro, OR), using STEM in dark-field mode (8, 15), to observe the individual QD-HER2 positions. The cells were maintained in a liquid state under a thin layer of water by cooling the sample stage to 3°C and adjusting to a pressure of 720–760 Pa. To maintain the cells in this wet state (12), the microchip was rinsed three times with HPLC-grade water and then placed in the sample stage of the microscope. Before the vacuum chamber was closed, 3 μL of HPLC-grade water was dropped on the sample, and three droplets of 3 μL HPLC-grade water were placed on the stage near the sample to provide moisture for the pumping and purging cycles. The vacuum chamber was then evacuated to 900 Pa and cycled four to five times between 900 and 1500 Pa. After the purging cycle, the chamber pressure was set to 810 Pa, and a closed water layer was visible in the image. The pressure was then adjusted to obtain optimal imaging conditions by regulating it down until the water layer was thin enough for the electrons to pass through to the STEM detector at the outer edges of the cells, providing sufficient contrast to observe the QDs and maintaining a liquid layer covering the sample (8). The pressure was adjusted to a value between 720 and 760 Pa depending on the sample.
An electron energy of 30 kV, spot size of ∼2 nm, probe current 0.6 nA, and working distances between 6 and 7 mm were used for ESEM. First, overview ESEM-STEM images were acquired, including an overview image of the entire SiN window, which were compared to the previously recorded fluorescence and DIC images. This correlative method allowed for quick relocation of the cells during electron microscopy as well as choosing representative cells for the high-resolution STEM imaging. Images of cells showing individual labels were acquired at a magnification of 60,000×, a pixel size of 2.2 nm, and a pixel dwell-time of 50 μs. The image size was 2048 × 1768 pixels, yielding a scanning area of 17.5 μm2 per image. The electron dose was 4 × 104 e−/nm2, within the known limit of radiation damage (8). Images of each analyzed cell were acquired before and after zooming in to 60,000×.
Particle detection
The QD positions were automatically determined using a locally designed plugin in ImageJ (National Institutes of Health), as discussed elsewhere (3). Briefly, the image was first noise filtered using a Gaussian filter with a radius of one pixel. The background variations in the image were then removed by applying a Fourier filter adjusted for the passage of spatial frequencies between a factor of three smaller and a factor of three higher than those of the approximate QD size of 8 nm. The image was binarized using an automated threshold with a maximal entropy setting. Finally, the particles were automatically detected using the Analyze Particles tool, which selects particles with an area within a factor of 5 of the expected area. The coordinates of detected particles of one image were stored in a data set. The precision of the particle detection amounted to a pixel size of 2.2 nm. The pair correlation function was calculated using a plugin implemented in ImageJ based on an algorithm described elsewhere (3).
Detection of linear particle chains
The obtained data sets with particle coordinates were analyzed for the presence of linear particle chains. The following criteria were used to identify linear sequences of particles. 1) The sequence should consist of at least six particles to reduce the likelihood of random line detection. The value was chosen experimentally from visually analyzing the images for rarely occurring lines; it was initially estimated that chains of six particles would not occur by random chance, and this was later verified in a statistical analysis. 2) The distance between every two consecutive particles in the sequence should be 36 ± 15 nm, which centers around the actin-filament-repetition distance and accounts for error in labeled positions and particle detection. 3) The particles in the sequence should be positioned on a line, with the angle of connecting lines between three adjacent particles ranging between 150 and 210°. The ranges of variation in the interparticle distance and the angles between particles were chosen from the approximate variability of the distances and angles in lines as determined from visually analyzing the images. To perform this analysis, a program was developed in MATLAB (MATLAB and Statistics Toolbox Release 2015b, The MathWorks, Natick, MA) based on the concept of an adjacency matrix constructed as follows. Firstly, for a data set with N particles labeled p1…pN, the distance matrix D was defined as a matrix with dimensions N × N. Every entry Di,j contained the distance from particle pi to particle pj. Next, the adjacency matrix A was constructed, also with dimensions N × N. The entries of A were defined as
The matrix A thus contained a value of one for every pair of particles that could be reached with a step of size 36 ± 15 nm and zero for every other pair of particles. The matrix A was considered as a graph in which every row corresponded to a vertex and every nonzero entry corresponded to an edge. Every path of length k except for cyclic paths corresponded to a sequence of k particles in which each two consecutive particles were separated by a distance of 36 ± 15 nm. Two other distance ranges were tested as well, 36 ± 15 and 75 ± 15 nm. The goal was then to find all such paths in the graph. This was achieved by exploiting a specific property of the adjacency matrix. Considering Ak (i.e., A × A × … k times), it holds that Aki,j is equal to the number of paths of length k from particles pi to pj (16). By computing Ak, the number of paths of length k from pi to pj can be determined for every pair of particles pi and pj. Particularly for Aki,j > 0, it is known that at least one path of length k exists from pi to pj. In these cases, Yen’s algorithm (17), which computes k shortest loopless paths for a graph with a non-negative edge cost, was used to compute each path.
The remaining criterion was that particles were positioned approximately linearly, and this was enforced by filtering the resulting matrix. For every identified path, the Pearson product-moment correlation coefficient (18) was employed as a measure of linearity, and paths with an absolute linearity <0.9 were discarded. As an additional local criterion, the angle between every consecutive line segment of the path was computed. Paths for which a line segment formed an angle of >30° with neighboring line segments were also discarded, which helped to remove outliers from the data.
Simulation of random data
To safeguard against random detection events and statistically evaluate our results, synthetic random data were generated. The data set was designed to preserve the large features of the specimen but randomize the positions of particles. This analysis allowed for the observation of the localization and arrangement of HER2 receptors separately from the probability that particles at random positions will also form linear structures. The synthetic data were generated as follows. Each experimental image was divided into a grid of 8 × 8 segments. The particles in each segment were randomized so that the segments contained the same number of particles as the actual image but at random positions; this approach preserved the large-scale structure of the image, such as local protein densities, but discarded the exact positioning of the particles. The statistical analysis described above was executed on both experimental and randomized data. The approach allowed us to compare the number of linear structures detected in the experimental data to the number of random occurrences in the synthetic data to exclude random effects as the source of our observations.
Results
Detecting HER2 positions in SKBR3 cells
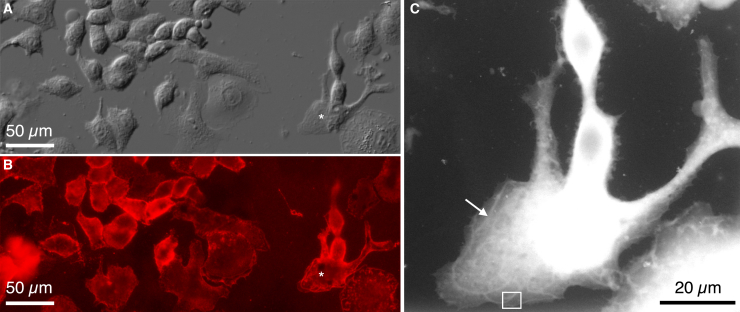
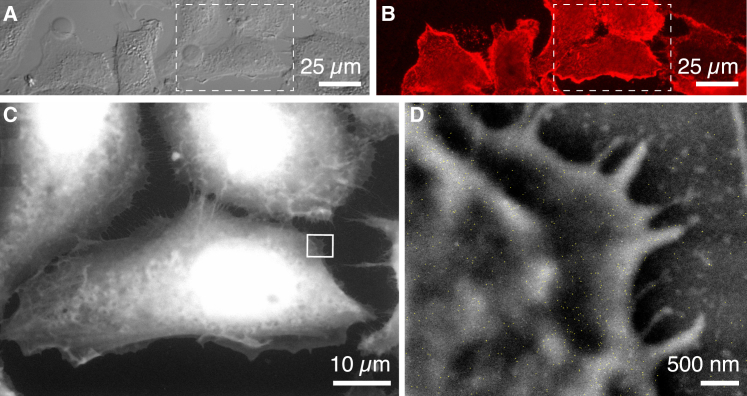
ESEM operated in wet STEM mode (15) was used to study the localization of individual HER2 proteins in the intact plasma membranes of SKBR3 breast cancer cells (3, 8). QD protein labels were used to indicate the presence of HER2, because their cores consist of an electron-dense material that provides atomic-number contrast in STEM (19, 20). QDs also generate a fluorescence signal, allowing for the testing of successful labeling using light microscopy. SKBR3 cells grown on microchips suitable for ESEM-STEM were incubated with a specific label consisting of biotinylated HER2-affibody and strep-QDs. Using a two-step labeling method (3, 12), it was possible to label each HER2 receptor with only one QD (3). The aim of the experiment was to study correlations in the localization of the HER2 receptors. Fig. 1, A and B show the DIC image and fluorescence signal, respectively, from an area of a SiN window coated with several flattened SKBR3 cells. The cell confluency was ∼50%, such that most cells touched each other at several locations, but the individual cells had enough space to fully extend on the SiN substrate, providing thin peripheral regions suitable for examination using STEM. HER2 in the plasma membrane, represented by the red fluorescent signal in Fig. 1 B, exhibited a predominantly heterogeneous distribution. The overall abundance of HER2 differed not only between cellular regions but also between the individual cells. The largest density of HER2 occurred on membrane ruffles and outer edges of cells. Membrane ruffles, or invadopodia, are highly dynamic structures involved in migration and metastasis in cancer cells (21), with typical widths of 0.5–1.5 μm and lengths of up to several tens of micrometers. These are visible, for example, as thin elongated shapes in the cell marked with an asterisk in both the DIC and fluorescence images in Fig. 1, A and B. These areas consist of denser cellular material resulting in stronger electron scattering and thus appear brighter in the image.
Figure 1.
Micrographs showing SKBR3 cells grown on a silicon nitride (SiN) membrane. (A) A direct interference contrast (DIC) image of several cells flattened out on the SiN supporting membrane is shown. (B) The fluorescence signal originating from quantum-dot (QD)-labeled HER2 proteins in the plasma membranes of the cells is shown. (C) An image of the same sample acquired with environmental scanning electron microscopy (ESEM) using scanning transmission electron microscopy (STEM) detection in a wet environment is shown. The cells were in a liquid state and covered by a thin water layer. The large cell is the same as the cell marked with an asterisk in (A) and (B). The bright elongated shape marked by an arrow is an example of a membrane ruffle. To see this figure in color, go online.
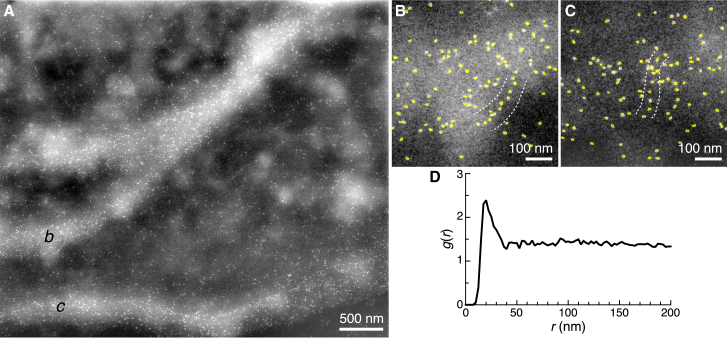
Cellular regions with high HER2 concentrations in the membrane were selected for ESEM-STEM. The cell marked with an asterisk in Fig. 1, A and B is shown at higher magnification in the ESEM image of Fig. 1 C. This cell exhibits membrane ruffles close to its edge, visible as bright lines in the cell topography and indicated by an arrow. The thicker region around the cell nucleus appears white under dark-field STEM contrast and was too thick for high-resolution electron microscopy. Fig. 2 A depicts a high-resolution image of the region outlined by a rectangle in Fig. 1 C. The bright shapes spanning the image from the top right and splitting up to the bottom left reflect a membrane ruffle, whereas another ruffle extends from the left to the right side in the lower part of the image. Individual QD positions are visible as bright dots. HER2 was highly abundant in the membrane ruffles, confirming previously published results (3). The QD-label positions were automatically detected for further examination. Fig. 2 B shows a selected region (indicated with b in Fig. 2 A) with QDs outlined by yellow shapes resulting from automated position detection, each reflecting an individual HER2 protein. The QD positions were detected with a precision of 2 nm. HER2 appeared to be distributed into many individual labels and pairs as well as some larger clusters. Because the labeling was not 100% efficient, a higher abundance of HER2 proteins may have been present than indicated by the label positions.
Figure 2.
ESEM-STEM micrograph of a region of a SKBR3 cell with QD-labeled HER2. (A) An image of the plasma membrane region at the edge of the cell shown within the rectangle in Fig. 1C is given. The small white spots represent individual QDs. The brighter shade in the background reflects the membrane ruffle. The image was filtered to improve the visibility of the QDs (Gauss filter with a size of 0.5 pixel and fast Fourier transform band-pass filter with a range of 1–400 pixels). (B) The highlighted outlines of automatically detected QDs at location b in (A) are shown. Several QDs are positioned in a linear chain between the dashed line. (C) QDs at location c in (A) with a linear chain of QDs between the dashed line are shown. (D) The pair correlation function g(r) as function of the radial distance between labels r calculated for all detected label positions in (A) is shown. To see this figure in color, go online.
A statistical analysis of the distances between pairs of labels was conducted to detect possible preferred arrangements. For this purpose, the pair correlation function g(r) (22) was calculated for all detected labels in Fig. 2 A using a method described elsewhere (3). The value of g(r) equals 1 for a random label distribution. The g(r) shown in Fig. 2 D exhibits a peak at r = 20 nm, indicating that a label pair distance of 20 nm occurs more frequently than in a random distribution. This peak indicates the presence of signaling-active HER2 homodimers, given that the center-to-center distance between two QD labels attached to such dimers equals 20 nm, with some variation due to linker flexibility (3). The finding that this section of the plasma membrane with membrane ruffles contains HER2 homodimers is consistent with previously published results (3).
Data were acquired of five cells on two different microchips, with a total of 48 images collected, representing a membrane area of 840 μm2 and an average QD label density of 86/μm2. The experiment was then repeated on two cells and six images for a total analyzed area of 151 μm2, yielding an average QD density of 121/μm2. These experiments are summarized in Table 1.
Table 1.
Summarization of Data for SKBR3 and HCC1954 Cells for Both Initial and Repeated Experiments
| Number of Cells | Number of Images | Image Area μm2 | Number of QD Labels | Label Density | Number of Chains Found | Chain Density | |
|---|---|---|---|---|---|---|---|
| SKBR3, Experiment 1 | 5 | 48 | 840 | 72,240 | 86/μm2 | 85 | 0.10/μm2 |
| SKBR3, Experiment 2 | 2 | 6 | 151 | 18,271 | 121/μm2 | 34 | 0.23/μm2 |
| HCC1954, Experiment 1 | 3 | 51 | 860 | 38,640 | 46/μm2 | 2 | 0.0023/μm2 |
| HCC1954, Experiment 2 | 3 | 8 | 202 | 21,614 | 107/μm2 | 15 | 0.084/μm2 |
The linear chains of six or more labels were spaced with an interval of 36 ± 15 nm. Overlapping lines were not counted.
Arrangement of HER2 in linear chains
An intriguing HER2 spatial distribution was observed in several images. The dashed lines in Fig. 2, B and C enclose seemingly linear chains of QD labels. It is highly unlikely that linear chains of proteins formed by chance, indicating that an underlying mechanism drives this specific arrangement. It is known from previous work that there is a correlation between epidermal-growth-factor-receptor (EGFR) localization and the actin-filament-helix-repetition distance of 36 nm (23). It is also known that the structures of EGFR and HER2, both members of the EGFR-family, are very similar. It was therefore hypothesized that the repetition length of the actin filament structure could correlate to the measured interlabel distance, with variation due to the structural flexibility of both the receptor protein and the QD linker.
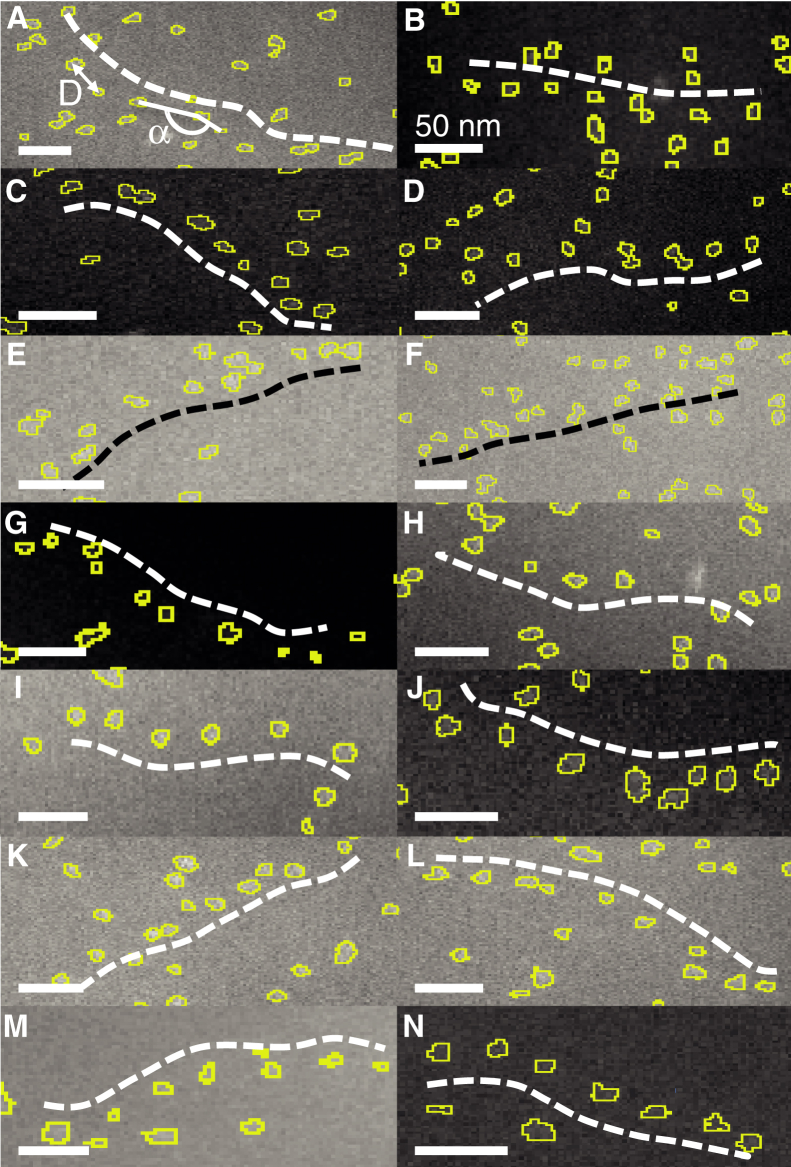
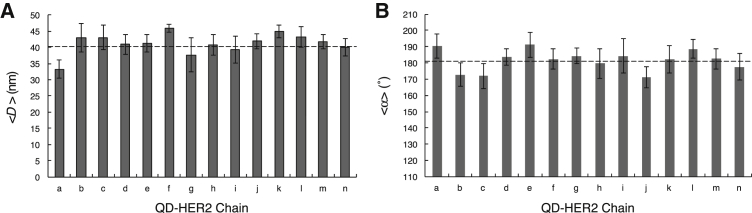
Fourteen examples of linear chains are shown in Fig. 3. Using this labeling and imaging method, it was possible to investigate the localization and interlabel distances between HER2 proteins oriented in linear chains (indicated by D in Fig. 3 A). The angle between the two line segments connecting three adjacent HER2 proteins is indicated by α in Fig. 3 A. The presence of linear chains of HER2 proteins was accordingly analyzed from a series of images; the criteria for a linear chain were the detection of six or more QDs with an interlabel distance of 36 ± 15 nm and a linear correlation factor of at least 0.9 (see Materials and Methods). The average interlabel spacing for the lines shown in Fig. 3 was measured to be 41 ± 6 nm (Fig. 4 A), with the error margin describing the SD. The linearity of the chains was analyzed using the angle between the two line segments connecting three particles (Fig. 4 B). For the 14 plotted examples, these angles are 182 ± 14° on average, indicating that there is no preferred curvature. The analysis of all images from both experiments (see Table 1) revealed the presence of 119 linear HER2 chains in a total of 54 images, with a total area of analyzed SKBR3 cells of 991 μm2. The linear chains thus had a density of 0.12/μm2, much lower than the overall density of QD labels. The average label distance in all lines amounted to 40 ± 7 nm, and the average angle was 181 ± 16°.
Figure 3.
Selected regions of ESEM-STEM images showing QD-HER2 labels arranged in linear chains in the plasma membranes of different SKBR3 cells. (A) A chain of labels is shown along the dashed white line. (D) reflects the distance between two QDs. The angle of the line connecting three adjacent QDs is indicated by α. (B)–(N) Further examples of linear chains of QD-HER2 positions are shown. All scale bars, 50 nm. To see this figure in color, go online.
Figure 4.
Analysis of QD-HER2 chain positions for the images shown in Fig. 3. (A) Average interlabel distance <D> measured from all QD positions in a single chain. The chain indicator (a–n) is the same as the panel indicator in Fig. 3. The average value of all measurements is included as a dashed line. (B) The average angle <α> between all lines connecting three QDs for each chain is given. The average for all measurements is indicated by the dashed line.
Safeguarding against false positives
A fundamental problem with all sufficiently sensitive pattern-detection algorithms is that similar patterns may also be found with some probability in random data, an effect generally known as false-positive identification. Thus, the presence of particle chains found in the experimental data does not necessarily imply successful labeling of linear chains of proteins but could possibly be a statistical effect of the detection algorithm. So, one must ask if the number of particle chains exceeded the expected value for random particle positions. To verify this, we conducted control experiments on randomized data. For each image, 100 randomized data sets were generated with a similar local density of the labels. The same particle chain detection as was used for the experiments was then performed for the randomized data sets. The data for SKBR3 cells are included in Table 2.
Table 2.
Comparison of Linear Particle Chains with an Interval of 36 ± 15 nm Found in Experimental Data with Linear Chains Found in Simulated Random Data for SKBR3 Cells
| Data Set |
L = 6 |
L ≥ 7 |
||||||
|---|---|---|---|---|---|---|---|---|
| Number of Chains | Number of Chains in Random Data | N | p-Value | Number of Chains | Number of Chains in Random Data | N | p-Value | |
| 1 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 2 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 3 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 4 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 5 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 6 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 7 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 8 | 6 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 9 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 10 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 11 | 3 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 12 | 2 | 0 | 100× | <0.01 | 1 | 0 | 100× | <0.01 |
| 13 | 2 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 14 | 4 | 0 | 100× | <0.01 | 3 | 0 | 100× | <0.01 |
| 15 | 8 | 0 | 96× | <0.01 | 5 | 0 | 100× | <0.01 |
| – | 1 | 3× | – | – | 0 | 100× | – | |
| – | 2 | 1× | – | – | 0 | 100× | – | |
| 16 | 7 | 0 | 97× | <0.01 | 1 | 0 | 100× | <0.01 |
| 2 | 3× | 0 | 100× | |||||
| 17 | 1 | 0 | 99× | <0.02 | 0 | 0 | 100× | <0.01 |
| – | 3 | 1× | – | – | 0 | 100× | – | |
| 18 | 7 | 0 | 99× | <0.01 | 4 | 0 | 100× | <0.01 |
| – | 1 | 1× | – | – | 0 | 100× | – | |
| 19 | 3 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 20 | 16 | 0 | 89× | <0.01 | 2 | 0 | 98× | <0.02 |
| – | 1 | 7× | – | – | 1 | 1× | – | |
| – | 2 | 1× | – | – | 2 | 1× | – | |
| – | 4 | 2× | – | – | – | – | – | |
| 21 | 1 | 0 | 99× | <0.02 | 31 | 0 | 10× | <0.1 |
| – | 1 | 1× | – | – | – | – | – | |
| 22 | 1 | 0 | 99× | <0.02 | 0 | 0 | 100× | <0.01 |
| – | 1 | 1× | – | – | – | – | – | |
| 23 | 28 | 0 | 75× | <0.01 | 6 | 0 | 97× | <0.01 |
| – | 1 | 12× | – | – | 1 | 1× | – | |
| – | 2 | 8× | – | – | 2 | 2× | – | |
| – | 3 | 5× | – | – | – | – | – | |
Particle chains of a length L = 6 and L ≥ 7 were considered. For each experimental data set, 100 random data sets were simulated and analyzed, preserving the local density of labels. For each data set, we list the number of chains found in the original image (column Number of chains). Each random data set might contain a different number of particle chains, so we list the number of chains found in the random data sets (column Number of lines in random data) and in how many random data sets this number occurred (column N). Additionally, we give the probability of finding at least the number of particle chains found in the original image in a randomized data set, i.e., by chance (column p-value).
Of 23 images in which lines occurred in the original data and in the repeated experiment, the expected number of linear chains of six particles was 4.2 per image; in the corresponding randomly generated data with 100 data sets for each of the 23 images, the expected number of linear chains was only 0.033 per image. A similar analysis on six images with linear chains of seven or more particles yielded 3.1 linear chains per image in the original data and 0.01 linear chains per image in the randomly generated data. The density of linear chains in the random data was 9.2 × 10−4/μm2, over two orders of magnitude lower than the linear chains found in the original data. The p-values did not exceed 0.02 for any of the data sets, indicating that the number of linear chains in the original data is statistically significant and that linear chains formation in SKBR3 cells did not occur because of random chance.
Test for different intervals in linear chains
In addition to the proposed interval between HER2 labels of 36 ± 15 nm, we also tested particle distances of 15 ± 15 and 75 ± 15 nm to ensure that the proposed interval is meaningful. The test was performed for the data set of the first experiment involving 48 images of five cells in total. In 23 of those images, a total of 85 linear chains were found (Table 1). These 23 images were analyzed for the different intervals. Because a change in the interval also changes the probability of finding random chains, the tests were repeated for randomized data sets with the new intervals.
In the interval 15 ± 15 nm, we found a single linear chain with six particles in four different images. The corresponding tests of the randomized data sets (100 for each image) resulted in a single linear chain in one image and no linear chains in all other experiments. It can thus be concluded that these linear chains found in the experimental images are significant. The number of linear chains was much smaller than the 85 linear chains found for the interval of 36 ± 15 nm.
In the interval 75 ± 15 nm, we found 37 linear chains in the images (Table 3). However, the larger interval also resulted in a larger number of linear chains found in the randomized data. The p-values were smaller than 0.05 for only four of the images, each of which included one linear chain. The other images exhibited more than two linear chains in the experimental image. However, several linear chains were found in the randomized images with similar label density. The lines found in those images were thus not statistically significant. In effect, this interval showed only four significant linear chains. It can therefore be concluded that the interval of 36 ± 15 nm is meaningful.
Table 3.
Comparison of Linear Particle Chains Found for an Interval of 75 ± 15 nm in Experimental Images of SKBR3 Cells Compared with Simulated Random Data for SKBR3 Cells
| Data Set | Number of Chains | Number of Chains in Random Data | N | p-Value |
|---|---|---|---|---|
| 8 | 1 | 0 | 100× | <0.01 |
| 11 | 1 | 0 | 100× | <0.01 |
| 3 | 1 | 0 | 98× | <0.03 |
| – | 1 | 2× | – | |
| 9 | 1 | 0 | 98× | <0.03 |
| 1 | 2× | |||
| 1 | 2 | 0 | 93× | >0.05 |
| 1 | 7× | |||
| 2 | 3 | 0 | 95× | >0.05 |
| 1 | 5× | |||
| 17 | 6 | 0 | 63× | >0.05 |
| 1 | 7× | |||
| 15 | 7 | 0 | 72× | >0.05 |
| 1 | 22× | |||
| 2 | 3× | |||
| 3 | 3× | |||
| 16 | 15 | 0 | 50× | >0.05 |
| 1 | 12× | |||
| 2 | 7× | |||
| 3 | 1× |
Particle chains of a length L = 6 were considered. For each experimental data set, N random data sets were simulated and analyzed, preserving the local density of labels. The data set numbers correspond with those in Table 2.
HER2 arrangement in HCC1954 cells
To evaluate whether the HER2 linear chains would also appear in a different cell line, the same two-step labeling method was used to investigate the localization of HER2 in HCC1954 breast cancer cells (Fig. 5 A). Like for SKBR3 cells, fluorescence microscopy indicated that quantum dots primarily concentrated at the edges of cells and in membrane ruffles (Fig. 5 B); these areas were the focus of ESEM imaging. Fig. 5 C shows several cells in an overview ESEM-STEM image; Fig. 5 D is an example of a high-resolution image in which the QD labels are visible, with a slightly higher density in brighter regions at the cell edge. In the first experiment, a total of 51 images and a membrane area of 861 μm2 were examined from three different cells, yielding a density of QD labels of 46/μm2, a factor of two lower than that of the SKBR3 cells. Note that the edges of a few images were discarded because they contained clipped signals. The QD density of unspecific labeling from the control experiment, which excluded the HER2-affibody-biotin labeling step, was measured to be 0.3 QDs/μm2, two orders of magnitude lower than for the HER2 labeling experiments. The analysis was repeated on eight images obtained from three additional cells for a total analyzed area of 202 μm2 and a QD density of 107/μm2. A total of 17 linear chains were found in this data, as summarized in Table 1.
Figure 5.
Analysis of HCC1954 cells with QD-labeled HER2. (A) A DIC overview image showing several adhered cells with flat edges is presented. (B) A fluorescence image revealing the locations of QD-labeled HER2 is shown. (C) An ESEM-STEM micrograph of regions of HCC1954 cells with QD-labeled HER2 at the location of the dashed rectangle in (A) and (B) is shown. (D) A high-resolution image of the region indicated with the rectangle in (C) is shown. Automatically detected label positions are indicated as colored spots, and the image was filtered as described in Fig. 2. To see this figure in color, go online.
The linear chains in HCC1954 cells exhibited a somewhat smaller interlabel distance of 32 ± 10 nm. The relative SD was higher than for the SKBR3, consistent with a lower number of linear chains observed for the HCC1594 cells. The density of linear chains was 0.016/μm2 for this cell type, much lower than the density of linear chains in SKBR3 cells (Table 1). It is not surprising that the density of linear chains is lower than in SKBR3 cells due to the lower density of QD labels; perhaps the labeling efficiency was insufficient in this experiment. Additionally, HCC1594 tends to have fewer membrane ruffles and different phenotypic features than SKBR3, which likely leads to a different HER2 distribution and localization.
The same analysis on random chains was performed for this cell line (Table 4). 17 linear chains of six particles were found in six images in the original and repeated experiment, leading to an expected value of three per image in which linear chains were found; the random data sets, again from 100 sets per image, yielded 0.021 chains per image when linear chains occurred in the original image. The random analysis yielded a density of 1.5 × 10−4 chains/μm2, a factor of 100 lower than in the original images. The simulated random control data contained significantly (all p-values < 0.03) fewer chains compared to the measured data for HCC1954 cells, as shown in Table 4, indicating that the experimentally observed linear arrangement is statistically significant. We can thus exclude the possibility that the detected particle chains are a statistical effect of the detection algorithm. This analysis of HCC1954 cells suggests that although the density HER2 line likely differs from the SKBR3 cell line due to different phenotypic behavior, the linear particle chains were present in a significant quantity in both cell lines.
Table 4.
Comparison of Linear Particle Chains Found in Experimental Data with Linear Chains with an Interval of 36 ± 15 nm Found in Simulated Random Data for HCC1954 Cells
| Data set |
L = 6 |
L ≥ 7 |
||||||
|---|---|---|---|---|---|---|---|---|
| Number of Chains | Number of Chains in Random Data | N | p-Value | Number of Chains | Number of Chains in Random Data | N | p-Value | |
| 1 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 2 | 1 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 3 | 1 | 0 | 98× | <0.03 | 0 | 0 | 100× | <0.01 |
| 2 | 2× | |||||||
| 4 | 8 | 0 | 96× | <0.01 | 0 | 0 | 100× | <0.01 |
| 2 | 3× | |||||||
| 5 | 1× | |||||||
| 5 | 4 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
| 6 | 2 | 0 | 100× | <0.01 | 0 | 0 | 100× | <0.01 |
Discussion
to date, only a very few reports exist of membrane receptor proteins organized in chain-like arrangements, among them rhodopsin (24), TGR5, a bile acid-sensing G-protein coupled receptor (25), and EGFR (26); interestingly, all of these receptors function as dimers, which also applies to HER2. We report here on the linear spatial arrangement of HER2 detected in HER2-overexpressing breast cancer cell lines. Chain-like arrangements of HER2 with interlabel distances of 40 ± 7 and 32 ± 10 nm were found in SKBR3 and HCC1954 cells, respectively. These linear structures were found on the same membrane structures earlier identified as membrane ruffles, in which HER2 resided preferentially in homodimeric form (3).
Liquid-phase electron microscopy was utilized to directly image specifically labeled endogenous, single HER2 molecules in the plasma membrane of intact cells in a wet environment. HER2 was labeled with a small probe consisting of an affibody and a QD (3), resulting in a ≤1:1 ratio of label to molecule. Liquid-phase STEM provides a planar view of the membrane, thereby allowing the detection of many thousands of labeled HER2 proteins in single cells. The study of the spatial arrangement of membrane proteins at the nanoscale was possible because of the 2-nm precision of the ESEM-STEM method in combination with the capability to analyze large areas approaching 1000 μm2 of intact plasma membrane. Several types of liquid-phase electron microscopy systems exist for studying membrane proteins in cells (3, 27, 28, 29, 30). Most other methods for analyzing membrane proteins in cells do not achieve sufficient spatial resolution to resolve individual protein positions for endogenous membrane proteins, as is the case for super-resolution fluorescence microscopy (31, 32), or are incapable of handling a series of whole cells, such as in cryo transmission electron microscopy, which typically focuses on analyzing a subsection of a cell (33). Indirect optical methods have been used extensively to study receptors of the HER family, but various open questions about their spatiotemporal organization remain (34) because membrane proteins are notoriously difficult to study in their native environment in the plasma membrane.
The observed densities of linear chains of six successively labeled proteins (0.12/μm2 and 0.018/μm2 for SKBR3 and HCC1954 cells, respectively) were rather low compared to the density of the individual labels of ∼100/μm2. The actual number of linear chains was potentially much larger, but presumably not all HER2 receptors were observed due to a limited labeling efficiency η; supposing that η = 0.5, the chance to observe a chain of six would amount to η6 = 0.016. In other words, if all HER2 proteins were assembled in linear chains of 6, only 1.6% would be observed. Longer chains have an even lower probability of detection. Because the labeling efficiency never reaches 100%, a large plasma membrane area must be analyzed to observe the linear chains. This would also be the case for genetically engineered fluorescent tags, for which bleaching and other factors lead to a reduction in detected particles. The detection of linear chains in the experimental data was proven to be highly significant compared to random simulated data with similar density patterns (Tables 2 and 3). Note that our analysis does not preclude nor exclude the arrangement of HER homodimers in linear chains.
A possible explanation for the linear organization of HER2 is a linkage between HER2 and actin filaments. Actin, the main component of the cytoskeleton, is known to colocalize with HER2 in membrane ruffles of HER2-overexpressing cells, as was shown by fluorescence microscopy (35, 36) and indirectly by immunoprecipitation. The latter biochemical method does not provide spatial information, but the results indicate a direct interaction between HER2 and actin (37). However, direct interaction seems to occur exclusively in cells with HER2 overexpression such as the cancer cell lines used in this study. Besides direct binding, several membrane-bound and cytosolic proteins can serve as linking molecules between HER2 and actin, for instance, through integrins (38), which are transmembrane receptors essential for mediating cell-extracellular matrix and cell-cell interactions. The linking mechanism may involve more than one protein, as found for instance in reporting by Wang et al. on the formation of a HER2/actin complex in conjunction with guanine nucleotide exchange factor (Vav2), Ras-related C3 botulinum toxin substrate 1 (Rac1), serine/threonine-protein kinase (Pak1), and actinin (37). Jeong et al. found indications of the formation of a HER2/actin complex involving two ion channels, namely sodium-hydrogen exchanger regulatory factor 1 (NHERF1) and plasma membrane calcium ATPase2 (PMCA2), together with chaperone HSP90 (39).
The average interlabel distance found in the HER2 chains (23) agrees, within error, with the 36-nm helix-repetition distance of actin filaments, which is reflected in the interval length observed for myosin (40, 41) and for talin proteins that bind to cross-bridging actin filaments (42). The organization into linear chains was also proposed for the closely related EGFR, a membrane receptor belonging to the same family of tryrosine kinases as HER2 (43).
The chain-line arrangement with regular protein distances may point to the assembly of HER2 in a larger oligomer. For the case of EGFR, such a larger oligomeric structure enhances the potential for autophosphorylation, receptor kinase activation, and phosphorylation, thereby increasing the signaling potency of the receptor (44). The suggested critical limit of at least four participating EGFRs would result in a length of 32 nm (26), matching the average HER2 repeat value found in our images for the HCC1954 cell line. Using double-immunofluorescence labeling, it has been demonstrated that the EGFR colocalizes with actin filaments, both in control and EGF-stimulated cells (45). Residues 984–996 in the C-terminus of EGFR have been identified as a binding site for actin and may well be involved in the formation of higher-order receptor oligomers and/or receptor clustering after ligand activation of the kinase domain (46). The arrangement of EGFR and HER2 along actin filaments could have a functional role for connecting the cytoskeleton to the extracellular matrix via integrin (47).
The organization of HER2 into linear chains attached to actin filaments points toward a fundamental principle behind the spatial-functional organization of growth factor receptors in membrane ruffles in the plasma membrane via assembly into larger oligomeric structures.
Author Contributions
All authors designed research and wrote the manuscript. K.P., V.T., and D.P. performed research. P.T. and T.D. contributed analytic tools. K.P., P.T., and T.D. analyzed data.
Acknowledgments
We thank Eduard Arzt for his support through INM.
Research was supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB1027, the Leibniz Competition 2014, and by the Krupp Internship Program for Stanford Students in Germany.
Editor: Joseph Zasadzinski.
Footnotes
Kelly Parker and Patrick Trampert contributed equally to this work.
References
- 1.Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology. 2001;61(Suppl 2):1–13. doi: 10.1159/000055396. [DOI] [PubMed] [Google Scholar]
- 2.Brennan P.J., Kumagai T., Greene M.I. HER2/neu: mechanisms of dimerization/oligomerization. Oncogene. 2000;19:6093–6101. doi: 10.1038/sj.onc.1203967. [DOI] [PubMed] [Google Scholar]
- 3.Peckys D.B., Korf U., de Jonge N. Local variations of HER2 dimerization in breast cancer cells discovered by correlative fluorescence and liquid electron microscopy. Sci. Adv. 2015;1:e1500165. doi: 10.1126/sciadv.1500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brix D.M., Clemmensen K.K., Kallunki T. When good turns bad: regulation of invasion and metastasis by ErbB2 receptor tyrosine kinase. Cells. 2014;3:53–78. doi: 10.3390/cells3010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy P., Jenei A., Jovin T.M. Activation-dependent clustering of the erbB2 receptor tyrosine kinase detected by scanning near-field optical microscopy. J. Cell Sci. 1999;112:1733–1741. doi: 10.1242/jcs.112.11.1733. [DOI] [PubMed] [Google Scholar]
- 6.Peckys D.B., Korf U., de Jonge N. Liquid-phase electron microscopy of molecular drug response in breast cancer cells reveals irresponsive cell subpopulations related to lack of HER2 homodimers. Mol. Biol. Cell. 2017;28:3193–3202. doi: 10.1091/mbc.E17-06-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer P., Hoogenboom J.P., Giepmans B.N. Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods. 2015;12:503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- 8.Peckys D.B., Baudoin J.P., de Jonge N. Epidermal growth factor receptor subunit locations determined in hydrated cells with environmental scanning electron microscopy. Sci. Rep. 2013;3:2626. doi: 10.1038/srep02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedard P.L., Hansen A.R., Siu L.L. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes N.E., Dey J.H. PI3K inhibition overcomes trastuzumab resistance: blockade of ErbB2/ErbB3 is not always enough. Cancer Cell. 2009;15:353–355. doi: 10.1016/j.ccr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Ring E.A., Peckys D.B., de Jonge N. Silicon nitride windows for electron microscopy of whole cells. J. Microsc. 2011;243:273–283. doi: 10.1111/j.1365-2818.2011.03501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peckys D.B., de Jonge N. Studying the stoichiometry of epidermal growth factor receptor in intact cells using correlative microscopy. J. Vis. Exp. 2015;103:e53186. doi: 10.3791/53186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eigenbrot C., Ultsch M., Härd T. Structural basis for high-affinity HER2 receptor binding by an engineered protein. Proc. Natl. Acad. Sci. USA. 2010;107:15039–15044. doi: 10.1073/pnas.1005025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown E., Verkade P. The use of markers for correlative light electron microscopy. Protoplasma. 2010;244:91–97. doi: 10.1007/s00709-010-0165-1. [DOI] [PubMed] [Google Scholar]
- 15.Bogner A., Thollet G., Gauthier C. Wet STEM: a new development in environmental SEM for imaging nano-objects included in a liquid phase. Ultramicroscopy. 2005;104:290–301. doi: 10.1016/j.ultramic.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Diestel R. Springer; New York: 2000. Graph Theory. [Google Scholar]
- 17.Yen J.Y. An algorithm for finding shortest routes from all source nodes to a given destination in general networks. Q. Appl. Math. 1970;27:526–530. [Google Scholar]
- 18.Fisher R.A. On the ‘probable error’ of a coefficient of correlation deduced from a small sample. Metron. 1921;1:3–32. [Google Scholar]
- 19.Dukes M.J., Peckys D.B., de Jonge N. Correlative fluorescence microscopy and scanning transmission electron microscopy of quantum-dot-labeled proteins in whole cells in liquid. ACS Nano. 2010;4:4110–4116. doi: 10.1021/nn1010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giepmans B.N., Deerinck T.J., Ellisman M.H. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 21.Weaver A.M. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 2006;23:97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 22.Stoyan D., Stoyan H. Estimating pair correlation functions of planar cluster processes. Biom. J. 1996;38:259–271. [Google Scholar]
- 23.Needham S.R., Zanetti-Domingues L.C., Clarke D.T. Structure-function relationships and supramolecular organization of the EGFR (epidermal growth factor receptor) on the cell surface. Biochem. Soc. Trans. 2014;42:114–119. doi: 10.1042/BST20130236. [DOI] [PubMed] [Google Scholar]
- 24.Fotiadis D., Liang Y., Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 25.Greife A., Felekyan S., Seidel C.A. Structural assemblies of the di- and oligomeric G-protein coupled receptor TGR5 in live cells: an MFIS-FRET and integrative modelling study. Sci. Rep. 2016;6:36792. doi: 10.1038/srep36792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Needham S.R., Roberts S.K., Martin-Fernandez M.L. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat. Commun. 2016;7:13307. doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano K., Kinoshita T., Sato C. Electron microscopy of primary cell cultures in solution and correlative optical microscopy using ASEM. Ultramicroscopy. 2014;143:52–66. doi: 10.1016/j.ultramic.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Liv N., van Oosten Slingeland D.S., Hoogenboom J.P. Electron microscopy of living cells during in situ fluorescence microscopy. ACS Nano. 2016;10:265–273. doi: 10.1021/acsnano.5b03970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahmke I.N., Verch A., de Jonge N. Graphene liquid enclosure for single-molecule analysis of membrane proteins in whole cells using electron microscopy. ACS Nano. 2017;11:11108–11117. doi: 10.1021/acsnano.7b05258. [DOI] [PubMed] [Google Scholar]
- 30.DE Jonge N. Membrane protein stoichiometry studied in intact mammalian cells using liquid-phase electron microscopy. J. Microsc. 2018;269:134–142. doi: 10.1111/jmi.12570. [DOI] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz J., Manley S. Putting super-resolution fluorescence microscopy to work. Nat. Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shivanandan A., Deschout H., Radenovic A. Challenges in quantitative single molecule localization microscopy. FEBS Lett. 2014;588:3595–3602. doi: 10.1016/j.febslet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 33.Hoenger A., McIntosh J.R. Probing the macromolecular organization of cells by electron tomography. Curr. Opin. Cell Biol. 2009;21:89–96. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valley C.C., Lidke K.A., Lidke D.S. The spatiotemporal organization of ErbB receptors: insights from microscopy. Cold Spring Harb. Perspect. Biol. 2014;6:a020735. doi: 10.1101/cshperspect.a020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong J., Kim W., Wysolmerski J.J. HER2 signaling regulates HER2 localization and membrane retention. PLoS One. 2017;12:e0174849. doi: 10.1371/journal.pone.0174849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lal S., Kersch C., Neuwelt E.A. Interactions between αv-integrin and HER2 and their role in the invasive phenotype of breast cancer cells in vitro and in rat brain. PLoS One. 2015;10:e0131842. doi: 10.1371/journal.pone.0131842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S.E., Shin I., Arteaga C.L. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor β. Cancer Res. 2006;66:9591–9600. doi: 10.1158/0008-5472.CAN-06-2071. [DOI] [PubMed] [Google Scholar]
- 38.Soung Y.H., Clifford J.L., Chung J. Crosstalk between integrin and receptor tyrosine kinase signaling in breast carcinoma progression. BMB Rep. 2010;43:311–318. doi: 10.5483/bmbrep.2010.43.5.311. [DOI] [PubMed] [Google Scholar]
- 39.Jeong J., VanHouten J.N., Wysolmerski J.J. The scaffolding protein NHERF1 regulates the stability and activity of the tyrosine kinase HER2. J. Biol. Chem. 2017;292:6555–6568. doi: 10.1074/jbc.M116.770883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikawa S., Homma K., Ikebe M. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- 41.Steffen W., Smith D., Sleep J. Mapping the actin filament with myosin. Proc. Natl. Acad. Sci. USA. 2001;98:14949–14954. doi: 10.1073/pnas.261560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Robson R.M., Stromer M.H. Talin can crosslink actin filaments into both networks and bundles. Biochem. Biophys. Res. Commun. 1996;218:530–537. doi: 10.1006/bbrc.1996.0095. [DOI] [PubMed] [Google Scholar]
- 43.Needham S.R., Hirsch M., Martin-Fernandez M.L. Measuring EGFR separations on cells with ∼10 nm resolution via fluorophore localization imaging with photobleaching. PLoS One. 2013;8:e62331. doi: 10.1371/journal.pone.0062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozer N., Barua D., Clayton A.H. Exploring higher-order EGFR oligomerisation and phosphorylation--a combined experimental and theoretical approach. Mol. Biosyst. 2013;9:1849–1863. doi: 10.1039/c3mb70073a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.den Hartigh J.C., van Bergen en Henegouwen P.M., Boonstra J. The EGF receptor is an actin-binding protein. J. Cell Biol. 1992;119:349–355. doi: 10.1083/jcb.119.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jorissen R.N., Walker F., Burgess A.W. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 47.Hamidi H., Pietilä M., Ivaska J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br. J. Cancer. 2016;115:1017–1023. doi: 10.1038/bjc.2016.312. [DOI] [PMC free article] [PubMed] [Google Scholar]