Abstract
Background
Enterococcus faecalis and Enterococcus faecium are ubiquitous opportunistic pathogens found in the guts of humans and farmed animals. This study aimed to determine the occurrence, antimicrobial resistance, virulence, biofilm-forming ability and genotypes of E. faecalis and E. faecium from swine farms. Correlations between the genotypes, virulotypes, antibiotic resistance, and the environmental factors such as locality of farms and farm hygiene practice were explored.
Methods
E. faecalis and E. faecium strains were isolated from the oral, rectal and fecal samples of 140 pigs; nasal, urine and fecal samples of 34 farmers working in the farms and 42 environmental samples collected from seven swine farms located in Peninsular Malaysia. Antibiotic susceptibility test was performed using the disk diffusion method, and the antibiotic resistance and virulence genes were detected by Polymerase Chain Reaction. Repetitive Extragenic Palindromic-Polymerase Chain Reaction and Pulsed-Field Gel Electrophoresis were performed to determine the clonality of the strains. Crosstab/Chi-square test and DistLM statistical analyses methods were used to determine the correlations between the genotypes, virulence factors, antibiotic resistance, and the environmental factors.
Results
A total of 211 E. faecalis and 42 E. faecium were recovered from 140 pigs, 34 farmers and 42 environmental samples collected from seven swine farms in Peninsular Malaysia. Ninety-eight percent of the strains were multidrug-resistant (resistant to chloramphenicol, tetracycline, ciprofloxacin and erythromycin). Fifty-two percent of the strains formed biofilms. Virulence genes efa, asaI, gelE, esp, cyl and ace genes were detected. Virulence genes efa and asaI were most prevalent in E. faecalis (90%) and E. faecium (43%), respectively. Cluster analyses based on REP-PCR and PFGE showed the strains were genetically diverse. Overall, the strains isolated from pigs and farmers were distinct, except for three highly similar strains found in pigs and farmers. The strains were regional- and host-specific.
Discussion
This study revealed alarming high frequencies of multidrug-resistant enterococci in pigs and swine farmers. The presence of resistance and virulence genes and the ability to form biofilm further enhance the persistence and pathogenicity of the strains. Although the overall clonality of the strains were regionals and host-specific, strains with high similarity were found in different hosts. This study reiterates a need of a more stringent regulation to ensure the proper use of antibiotics in swine husbandry to reduce the wide spread of multidrug-resistant strains.
Keywords: Enterococcus faecium, Enterococcus faecalis, MDR, Farmers, Virulence genes, Pigs
Introduction
Enterococci are Gram-positive facultative anaerobes and are part of microbiota of humans and animals (Lebreton, Willems & Gilmore, 2014; Kristich, Rice & Arias, 2014). Antibiotics such as macrolides, trimethroprim, fluoroquinolones and tetracyclines are commonly used for animal husbandry and human medicine in Southeast Asia, and resistances towards these antibiotics have been reported for Enterococcus spp. (Daniel et al., 2015; Health Action International Asia Pacific, 2013).
Livestock industry plays an important role in the transmission of multidrug-resistant (MDR) Enterococci strains due to the close interaction between farmers, livestock and the farm environment (Novais et al., 2013; Woolhouse et al., 2015). Antimicrobial agents are used as prophylactics in feeds and the pervasive selection of resistant bacteria in the swine gut facilitates the persistence and dissemination of MDR strains to other animals and humans (Woolhouse et al., 2015; Hwang et al., 2009). The emergence of MDR bacteria in the livestock industry is of public health concern as transmission of these bacteria to humans has been reported (Compassion in World Farming, 2011). The spread of such MDR strains can occur through direct (consumption of contaminated food, direct contact of farmers and veterinarians) or indirect (animal waste handling, contaminated ground water or surfaces) routes (Daniel et al., 2015). The resistance determinants carried by the MDR strains could also be transmitted to the other commensal strains in the host and cause further complications (Price et al., 2018). In addition, infections caused by MDR strains have been associated with long hospital stays, high morbidity and mortality rate (Beganovic et al., 2018).
The two most common enterococcal species, Enterococcus faecalis and Enterococcus faecium, present diverse virulence factors such as aggregation substance, gelatinase, cytolysin, enterococcal surface protein and collagen binding cell wall protein which are encoded by their respective genes asa1, gelE, cylA, esp and ace (in plasmids) or hyl (in chromosome) (Vidana et al., 2015). Enterococcal surface protein, encoded by esp gene, is one of the factors associated with biofilm formation (Toledo-arana et al., 2001). The biofilm formation ability can enhance the persistence and endurance of the strains by providing extra protection to the bacteria cells and confers higher tolerance to the antibiotics (Holmberg & Rasmussen, 2016).
The objectives of the study were to determine the occurrence of MDR E. faecalis and E. faecium isolated from pigs, swine farmers and swine farm environment in Peninsular Malaysia. The virulotypes, biofilm-forming ability, and the genotypes (based on PFGE and REP-PCR) were determined.
Materials and Methods
Sample collection
Samplings were carried between August to December 2013 in seven selected swine farms in the northern (PF1, PF2, PF3, PF4 and PF5) and central (SF1 and SF2) regions of Peninsular Malaysia (Fig. 1). The selected regions are high density pig farming areas. Ten piglets and 10 weaning pigs were randomly selected from each farm and 140 oral swabs, 140 rectal swabs and 91 fecal samples were collected from 140 pigs from seven swine farms. All samples were collected under veterinary supervision. Thirty-four nasal swabs, 34 urine and 13 fecal samples were collected from 34 swine farmers who worked in the seven participating farms. The human subjects were advised to collect midstream catch urine samples. Environmental samples included 14 drinking water samples, 14 feed samples and 14 pen swabs were also collected from each farm. Background information of the samples were collected based on questionnaire and observation of the same attending veterinarian. The information such as farm location, hygiene practice, gender of the pigs, body temperature and health condition of the pigs (healthy/unhealthy) were collected and used for statistical analysis. The farm hygiene practices were arbitrarily divided into three categories: HP1 refers to application of fundamental hygiene practice (clean the pens with detergents only). HP2 refers to a better level of hygiene practice, includes cleaning the pens and disinfecting equipment. HP3 refers to the best hygiene practice, includes personal hygiene practice of farmers, foot-dip practices and all above mentioned hygiene practices. Physical examination was performed by the field veterinarian to determine the health status of pigs. Pigs that presented with abnormal clinical signs, behavior and elevated body temperature were categorized as unhealthy. This study was approved by the Animal Care and Use Committee (ACUC), Faculty of Veterinary Medicine, UPM (UPM/IACUC/FYP-AUP-T006/). The human samples collection was approved by Medical Ethics Committee, University Malaya Medical Centre (Ethic committee/IRB reference number: 1010.41), with written informed consent of the human subjects. All swabs were collected using sterile Transwab® Amies Charcoal (Medical Wire & Equipment Co, Corsham, UK). All the samples were transported on ice and processed within 24 h upon collection.
Figure 1. The map of Peninsular Malaysia showing the locations of the seven swine farms.
Number of strains in each farm: PF1 (nE. faecalis = 40; nE. faecium = 3); PF2 (nE. faecalis = 31; nE. faecium = 7); PF3 (nE. faecalis = 13; nE. faecium = 0); PF4 (nE. faecalis = 24; nE. faecium = 4); PF5 (nE. faecalis = 12; nE. faecium = 1); SF1 (nE. faecalis = 43; nE. faecium = 23); SF2 (nE. faecalis = 48; nE. faecium = 4).
Bacterial isolation and identification
All samples except fecal samples were enriched in Brain Heart Infusion (BHI) medium containing 6.5% NaCl followed by plating on the CHROMagar Orientation (CHROMagar Company, Paris, France). For fecal samples, one gram of feces was suspended in two ml of 0.85% saline before being streaked onto CHROMagar Orientation.
Approximately 3,450 presumptive colonies were obtained in the initial screening based on the manufacturer’s guidelines. Due to the large numbers of isolates, 10% of turquoise blue colored colonies (approximately 10–15 colonies per plate) were selected for further identification using Vitek-MS (Biomérieux, Marcy I’Etoile, France) according to the manufacturer’s protocol. The Escherichia coli (ATCC 8739) strain was used as the control. PCR targeting sodA genes was used to confirm the identity of the strains (Jackson, Fedorka-Cray & Barrett, 2004).
Antibiotic susceptibility test
Antibiotic susceptibility test for non-repeat strains was determined with commercial antimicrobial discs (Oxoid, Basingstoke, United Kingdom) by using the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2015). The bacterial strains were tested against a panel of antimicrobial agents which are commonly prescribed for enterococcal infections and frequently used in the swine farms including ampicillin (AMP, 10 µg), vancomycin (VAN, 30 µg), chloramphenicol (C, 30 µg), tetracycline (TET, 30 µg), ceftiofur (EFT, 30 µg), ciprofloxacin (CIP, 5 µg), erythromycin (E, 15 µg), low-level gentamicin (CN, 10 µg), high-level gentamicin (HLG, 120 µg), linezolid (LZD, 30 µg), penicillin (P, 10U) and teicoplanin (TEC, 30 µg). Strains with zones of inhibition under intermediate and resistant categories were defined as non-susceptible in this study. Staphylococcus aureus ATCC 25923 and E. faecalis ATCC 29212 were used as control strains. Multidrug resistance of the strain was determined by antibiotics which are not intrinsically resistant by E. faecalis, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
PCR detection of antibiotic resistance genes and virulence genes
Detection of antibiotic resistance genes and virulence genes were used to determine the resistance and virulence potential of the strains. Boiled cell lysates were used as DNA templates. Antibiotic resistance and virulence genes were identified by Polymerase Chain Reaction (PCR) using primers and condition as previously described (File S1).
Biofilm assay
To study the biofilm forming ability, crystal violet assay was performed using the protocol described by Baldassarri et al. (2001) with slight modifications. Biofilm was allowed to grow at 37 °C for 48 h and the optical density (OD) of eluted crystal violet stain was measured at OD570 nm. The true OD readings of each strain were acquired after deducting the negative control, which contained only the growth medium. The biofilm forming ability of the studied strains was scored as previously described (Stepanovic et al., 2000). A strong biofilm forming strain VREr5 from Lim, Teh & Thong (2017) was used as a positive control.
Repetitive extragenic palindromic-PCR (REP-PCR)
REP-PCR was performed using primer (GTG)5 as described by Versalovic et al. (1994). DNA extraction was performed using Genomic DNA Mini kit (Yeastern Biotech Co., Taiwan) according to the manufacturer’s guidelines. PCR was carried out in a total volume of 25 µl master mix containing 1X buffer, 2.5 mM of MgCl2, 0.8 mg/ml of bovine serum albumin (BSA), 0.1 mM of dNTPs, 1.6 µM of primers and 1.0 U of Taq polymerase (Promega, Madison, USA). The PCR protocol involved an initial denaturation of 7 min at 95 °C, followed by 30 cycles of 90 °C for 30 s, 40 °C for 1 min, 65 °C for 8 min and a final elongation of 16 min at 65 °C. The amplicons generated were electrophoresed on a 1.5% agarose gel at 100 V for 6 h. A 1 kb DNA marker (Promega, Madison, WI, USA) was used as the molecular size standard. The gels were stained with Gel-Red and visualized with Gel Doc XR imaging system (Bio-Rad, Hercules, CA, USA). The banding patterns generated were analyzed with BioNumerics 6.0 (Applied Maths, Kortrijk, Belgium). All the PCR fingerprints profiles were assigned an arbitrary designation. The quantitative difference among the profiles was defined by the Dice coefficient, Hierarchical clustering analysis was carried out based on unweighted pair group with arithmetric averages (UPGMA) using 1.5% tolerance. REP-PCR was repeated twice to ensure the reproducibility.
Pulsed-Field Gel Electrophoresis (PFGE)
PFGE for SmaI-digested genomic DNA was performed in a CHEF Mapper (Bio-Rad, Hercules, CA, USA) as described by Turabelidze et al. (2000) with some modification in the DNA preparation. In brief, the bacteria cells were lysed with a combination of lysozyme (2.5 mg/ml) and mutanolysin (1,250 U/ml) for 4 h at 37 °C. Bacterial cells were lysed overnight in cell lysis buffer containing 0.5M EDTA, 1% sarcosyl and 50 µg/ml of proteinase K. The plugs were washed thoroughly with TE buffer. Slices of DNA plugs were digested overnight with SmaI at 25 °C and XbaI at 37 °C. PFGE was run using pulse times 3.5 s to 25 s for 12 h (block 1, 6V, 120°) and 1 s to 5 s for 10 h (block 2, 6V, 120°). XbaI-digested Salmonella enterica serovar Braenderup H9812 was used as the size marker. PFGE was repeated once to ensure reproducibility. The PFGE profiles were analyzed by BioNumerics version 6.0 software (Applied Maths, Kortrijk, Belgium). A dendrogram was constructed based on Dice coefficient of similarity (F) and unweighted pair group method with arithmetic mean (UPGMA) algorithm with 1.5% position tolerance.
Statistical analysis
The environmental factors used for correlation study included sample subjects, sample matrix, sampling region and farm hygiene practice. The association of genotypes with resistotypes, virulotypes, biofilm-forming ability and the environmental factors were determined using distance-based linear model (DistLM) by PRIMER 6 with PERMANOVA add on package (PRIMER-E, UK). The correlation between the resistotypes and virulotypes with other factors were tested with crosstabs/chi-square analysis using SPSS version 21 (IBM SPSS Statistics; IBM corporation, Armonk, NY, USA).
Results
Occurrence of E. faecalis and E. faecium
A total of 289 Enterococcus strains (232 from pigs, 54 from farmers and three from farm environments) were isolated. These strains were affiliated with five species, which included E. faecalis (n = 211), E. faecium (n = 42), E. hirae (n = 15), E. gallinarium (n = 11) and E. casseliflavus (n = 10). All E. faecalis and E. faecium strains were confirmed with PCR targeting the sodA genes (Jackson, Fedorka-Cray & Barrett, 2004).
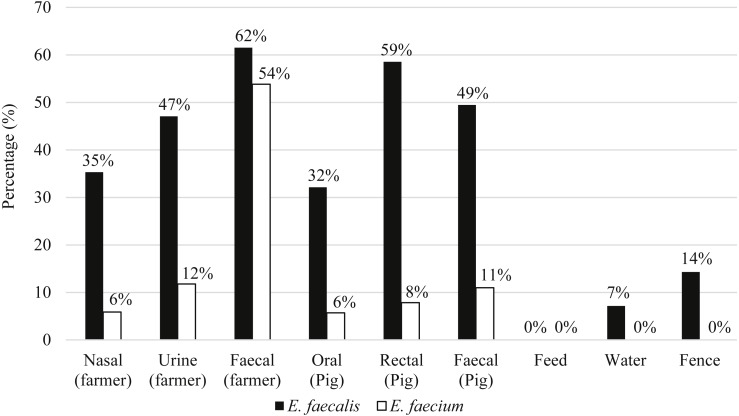
Eighty-seven pigs and 21 farmers harbored either E. faecalis, E. faecium or both species of Enterococcus. Figure 2 shows the distribution of E. faecalis and E. faecium in each type of sample matrix. Higher occurrence of enterococci was observed in humans compared to pigs and only three E. faecalis strains were isolated from the environmental samples (Fig. 2). E. faecalis was isolated from all participating farms but E. faecium was restricted to the swine samples collected from farms PF2, PF4, PF5, SF1 and SF2 as well as the human samples from PF1, PF4 and SF1.
Figure 2. Percentage of distribution of E. faecalis and E. faecium in each sample matrix.
Antimicrobial susceptibility of E. faecalis and E. faecium
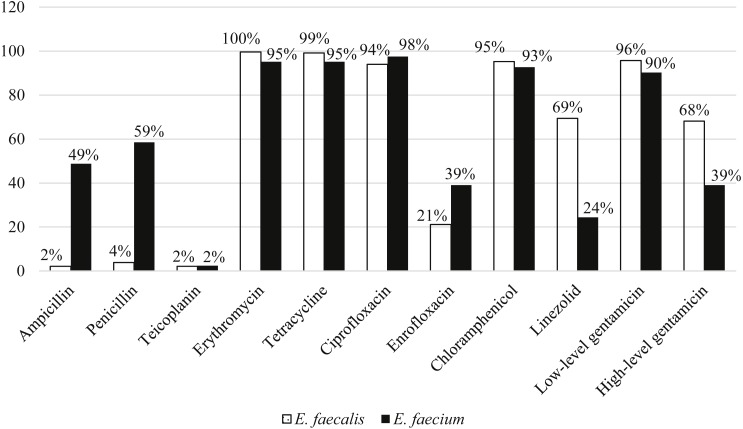
Among the 211 E. faecalis strains, 98% and 96% of them were susceptible to ampicillin and penicillin, respectively. However, E. faecium were more resistant to ampicillin (49%) and penicillin (59%) compared to E. faecalis. All the strains were susceptible to vancomycin and only two percent of strains from each species were resistant to teicoplanin. E. faecalis (68%) strains were relatively more resistant to high-level gentamicin (120 µg) compared to E. faecium (39%). Overall, more than 90% of both species were resistant to erythromycin, tetracycline, ciprofloxacin, chloramphenicol and low-level gentamicin (Fig. 3).
Figure 3. Percentage of antibiotic resistance of E. faecalis and E. faecium in this study.
Ninety-eight percent of the strains were MDR (non-susceptible to at least 4 classes of antibiotics) and were resistant to chloramphenicol, tetracycline, ciprofloxacin, gentamicin and erythromycin. There were 47 resistotypes observed in this study. Thirty-four and 23 resistotypes were observed in E. faecalis (Table 1) and E. faecium (Table 2), respectively. Ten resistotypes were shared by both species of bacteria. The resistance patterns observed in swine strains were more diverse than human strains. Resistotypes R20 and R26 were most prevalent among the E. faecalis strains and were resistant to erythromycin, tetracycline, ciprofloxacin, chloramphenicol and gentamicin. Resistance genes detection by PCR showed that all the strains tested in study harbored at least one of the antibiotic resistance genes tested. For the genes encoding for erythromycin resistance, ermB was the most prevalent compared to ermA and msr. A total of 96% and 95% of E. faecalis and E. faecium strains, respectively, harbored ermB. Besides, 90% of both E. faecalis and E. faecium strains possessed tetL and 98% of E. faecalis and 95% of E. faecium contained tetM gene encoding tetracycline resistance. At least one aminoglucoside resistance gene was present in 89% of the E. faecalis and 83% of the E. faecium strains. Among the six aminoglycoside resistance genes, aac(6′)-Ie-aph(2″)-Ia and aph(3′)-IIIa were detected in most of the strains (64% with aac(6′)-Ie-aph(2″)-Ia and 87% with aph(3′)-IIIa). Notably, four strains harbored vancomycin resistance genes (van B and van C1) but none were resistant to vancomycin based on disk diffusion method.
Table 1. Resistotypes of Enterococcus faecium strains based on the classes of antibiotics tested.
| Resistotypes (nP, nH, nE)a | Classes of antibiotic resistancesb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | |||||
| P | AMP | VAN | TEC | E | TET | CIP | ENR | C | LZD | CN | HLG | |
| R1 (nP = 1, nH = 0, nE = 0) | R | R | ||||||||||
| R2 (nP = 1, nH = 0, nE = 0) | R | R | R | |||||||||
| R3 (nP = 0, nH = 1, nE = 0) | R | R | R | |||||||||
| R4c(nP = 6, nH = 0, nE = 0) | R | R | R | R | ||||||||
| R5 (nP = 1, nH = 0, nE = 0) | R | R | R | R | ||||||||
| R6 (nP = 2, nH = 0, nE = 0) | R | R | R | R | R | |||||||
| R7 (nP = 8, nH = 0, nE = 1) | R | R | R | R | R | |||||||
| R8 (nP = 2, nH = 0, nE = 0) | R | R | R | R | R | |||||||
| R9 (nP = 1, nH = 1, nE = 0) | R | R | R | R | R | |||||||
| R10 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | |||||||
| R11 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | |||||||
| R12 (nP = 0, nH = 2, nE = 0) | R | R | R | R | R | R | ||||||
| R13c(nP = 24, nH = 1, nE = 0) | R | R | R | R | R | R | ||||||
| R14c(nP = 32, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R15 (nP = 3, nH = 2, nE = 1) | R | R | R | R | R | R | ||||||
| R16 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | ||||||
| R17c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R18c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R19c(nP = 7, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R20c(nP = 61, nH = 9, nE = 1) | R | R | R | R | R | R | R | |||||
| R21 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | |||||
| R22 (nP = 0, nH = 3, nE = 0) | R | R | R | R | R | R | R | |||||
| R23 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R24 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R25 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R26c(nP = 11, nH = 10, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R27 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R28 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R29c(nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R30 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R31 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R32 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | R | R | |||
| R33 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | R | |||
| R34c(nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | R | R | ||
Notes.
nP, number of swine isolates; nH, number of human isolates, nE: number of environmental isolates.
C1, penicillin; C2, glycopeptides; C3, macrolides; C4, tetracyclines; C5, fluoroquinolones; C6, phenicols; C7, oxazolidinones; C8, aminoglycosides; AMP, ampicillin; P, penicillin; TEC, teicoplanin; E, erythromycin; TET, tetracycline; CIP, ciprofloxacin; ENR, enrofloxacin; C, chloramphenicol; LZD, linezolid; CN, gentamicin; HLG, high-level gentamicin; R, resistant.
Resistotypes shared by both E. faecalis and E. faecium.
Table 2. Resistotypes of Enterococcus faecalis strains based on the classes of antibiotics tested.
| Resistotypesa | Classes of antibiotic resistancesb | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | |||||
| P | AMP | VAN | TEC | E | TET | CIP | ENR | C | LZD | CN | HLG | |
| R4c(nP = 0, nH = 1, nE = 0) | R | R | R | R | ||||||||
| R13c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R14c(nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R17c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R18c(nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | ||||||
| R19c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R20c(nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R26c(nP = 0, nH = 4, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R29c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R34c(nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | R | R | ||
| R35 (nP = 0, nH = 2, nE = 0) | R | R | R | |||||||||
| R36 (nP = 1, nH = 2, nE = 0) | R | R | R | R | ||||||||
| R37 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | ||||||
| R38 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R39 (nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | ||||||
| R40 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R41 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R42 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | |||||
| R43 (nP = 0, nH = 1, nE = 0) | R | R | R | R | R | R | R | |||||
| R44 (nP = 5, nH = 0, nE = 0) | R | R | R | R | R | R | R | |||||
| R45 (nP = 1, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R46 (nP = 3, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | ||||
| R47 (nP = 2, nH = 0, nE = 0) | R | R | R | R | R | R | R | R | R | |||
Notes.
nP, number of swine isolates; nH, number of human isolates; nE, number of environmental isolates.
C1, penicillin; C2, glycopeptides; C3, macrolides; C4, tetracyclines; C5, fluoroquinolones; C6, phenicols; C7, oxazolidinones; C8, aminoglycosides; AMP, ampicillin; P, penicillin; TEC, teicoplanin; E, erythromycin; TET, tetracycline; CIP, ciprofloxacin; ENR, enrofloxacin; C, chloramphenicol; LZD, linezolid; CN, gentamicin; HLG, high-level gentamicin; R, resistant.
Resistotypes shared by both E. faecalis and E. faecium.
Virulotyping
A total of 99% and 57% of the E. faecalis and E. faecium strains, respectively, harbored at least one of the five virulence genes screened in this study. efa was the most prevalent virulence gene detected in E. faecalis strains (90%), followed by asaI (68%), gelE (63%), esp (40%), cyl (40%) and ace (15%). In contrast, asaI was most prevalent in E. faecium (43%). Other virulence genes including esp, gelE, efa and cyl were also present in 21%, 19%, 17% and 14% of E. faecium, respectively. None of the strains harbored hyl gene.
Biofilm assay
Overall, about 62% of the strains from pigs and humans are capable of producing biofilms. Among the 142 biofilm-forming E. faecalis strains, 37%, 34% and 29% were weak, moderate and strong biofilm formers, respectively. Eight E. faecium strains formed weak biofilm and notably, half of them were isolated from human urine samples. All three environmental strains could form strong biofilm.
Genotypic characterization by REP-PCR and PFGE
REP-PCR subtyped the 211 non-repeat E. faecalis strains into 145 REP-profiles with 9 to 21 DNA fragments ranging in size from 500 to 6,000 bp. Reproducible patterns were observed. Based on 85% similarity, the E. faecalis strains were grouped into 24 clusters and 13 unique patterns (Files S2). From the dendrogram, the strains isolated from different samples matrix (oral swabs, nasal swabs, rectal swabs, urine and fecal samples) were grouped in the same cluster. However, distinct clustering was observed for strains originated from different regions (northern and central region of Peninsular Malaysia). Strains isolated from both regions were observed in Clusters C4, C5 and C9. Overall, REP-patterns of strains from central region were host-specific (Clusters C2, C19, C23 and C24 comprised of strains isolated from humans and Clusters C7, C13, C20, C21 and C22 comprised of strains isolated from pigs). Three environmental E. faecalis strains were clustered with the swine and human strains. For instance, Enfs99 recovered from a pen swab in PF1 was clustered with human nasal strain from the same farm in C15 (Files S2). Enfs103 isolated from a pen swab was 95% similar to the swine oral strains isolated from the same farm, PF1. Enfs98 from the water sample of PF3 formed cluster C10 with the swine fecal strains of the same farm.
E. faecium strains were grouped into 11 clusters and 11 unique patterns based on 85% similarity of the REP-profiles (Files S3). Similar to E. faecalis, the strains were regional specific. Strains in Clusters C1, C2, C3 and C5 were from the northern region and Clusters C4, C6 to C11 were made up of strains isolated from the central region only. The E. faecium strains from the central region were also host specific. Human strains were grouped in Clusters C4 and C9 while swine-related strains were grouped in Clusters C6, C7, C8, C10 and C11. In contrast, Cluster C2 consisted of both human- and swine-related strains isolated from the northern region.
Similar clustering and clonality of the strains were observed based on PFGE. PFGE generated 126 reproducible pulsotypes for E. faecalis with 12 to 26 bands and grouped into 27 clusters and 11 unique patterns based on 85% similarity (Files S4). Major clusters were represented by Clusters C15, C16 and C22, comprised of strains isolated from both humans and pigs. However, regional- and host-specific clusters were also observed in the dendrogram (Files S4). Meanwhile, 35 pulsotypes of E. faecium strains were grouped into 11 clusters and 6 unique patterns (Files S5). All the clusters were regional specific and Cluster C2 was made up of strains from both pigs and humans.
Antibiotic resistance profiles of E. faecalis and E. faecium strains
Correlations between the antibiotic resistance of E. faecalis and E. faecium strains with different factors were determined by chi-squared test (Table 3). Ampicillin resistance of E. faecalis was correlated to the sampling region and biofilm-forming ability. High-level gentamicin resistance of E. faecalis was correlated with farm locality, sample matrix, farm hygiene practice and the biofilm-forming ability of the strains. In contrast, ampicillin resistance of E. faecium was correlated with all the environmental factors. Penicillin and high-level gentamicin resistance were correlated with the biofilm-forming ability of the strains. Strains isolated from different regions possessed different ampicillin and erythromycin resistance. The farm hygiene practice was found to correlate with the strains’ resistance towards ampicillin, penicillin, tetracycline and enrofloxacin (Table 3).
Table 3. Significance of correlation between the antibiotic resistances of different samples classification based of Chi-squared test.
| P-values (P < 0.05 = rejectnull)a,b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Groupsc | AMP 10 | P 10 | TEC 30 | E 5 | TET 30 | CIP 5 | ENR 5 | C 30 | LZD 30 | CN 10 | CN 120 |
| E. faecalis: | |||||||||||
| Host | x | x | x | x | 0.007 | x | x | x | 0.014 | x | x |
| Biofilm | 0.002 | x | x | x | x | x | x | x | x | 0.031 | 0.023 |
| Matrix | x | x | x | 0.001 | 0.010 | 0.006 | x | x | 0.006 | x | 0.018 |
| Region | 0.006 | x | x | x | x | x | x | x | x | x | 0.001 |
| Hygiene | x | x | x | x | x | x | x | x | x | x | 0.010 |
| Virulotypes | x | 0.030 | x | x | x | x | x | 0.001 | 0.018 | x | x |
| E. faecium: | |||||||||||
| Host | 0.002 | 0.001 | x | x | x | x | x | x | x | x | x |
| Biofilm | x | 0.015 | x | x | x | x | x | x | x | x | 0.038 |
| Matrix | 0.033 | x | x | x | x | x | 0.010 | x | 0.007 | x | 0.006 |
| Region | 0.023 | x | x | 0.040 | x | x | x | x | x | x | x |
| Hygiene | x | 0.032 | x | x | 0.035 | x | 0.004 | x | x | x | x |
| Virulotypes | x | 0.030 | x | x | x | x | x | 0.001 | 0.018 | x | x |
Notes.
AMP, ampicillin; P, penicillin; TEC, teicoplanin; E, erythromycin; TET, tetracycline; ENR, enrofloxacin; CIP, ciprofloxacin; C, chloramphenicol; LZD, linezolid; CN, gentamicin.
x, No significant correlation.
Subject, human host and swine host; Biofilm refers to biofilm former and non-former; Matrix, oral, rectal, nasal, urine and fecal; Region, northern region and central region; hygiene, HP1, HP2, HP3.
The presence of ace gene in E. faecalis strains was dependent on the sample matrix and sampling region while the efa gene was correlated with sampling region, sample matrix, farm hygiene practice and resistotypes (Table 4). On the other hand, the presence of efa gene in E. faecium was correlated with the sampling region and virulotypes of the strains.
Table 4. Correlation between the presence of virulence genes with environmental factors, virulotypes and resistotypes.
| P-values (P < 0.05 = rejectnull)a,b | ||||||
|---|---|---|---|---|---|---|
| Groupsc | ace | efa | gelE | asa | esp | cyl |
| E. faecalis: | ||||||
| Host | x | x | x | x | x | x |
| Biofilm | x | x | x | x | x | x |
| Matrix | 0.034 | 0.019 | x | x | x | x |
| Region | 0.049 | 0.001 | 0.021 | x | 0.004 | x |
| Hygiene | x | 0.033 | x | x | 0.006 | x |
| Resistotypes | x | 0.037 | 0.022 | x | x | x |
| Virulotypes | x | x | x | x | x | x |
| E. faecium: | ||||||
| Host | x | x | x | x | x | x |
| Biofilm | x | x | x | x | x | x |
| Matrix | x | x | x | x | x | x |
| Region | x | 0.038 | x | x | x | x |
| Hygiene | x | x | x | x | x | x |
| Resistotypes | x | x | x | x | 0.009 | x |
| Virulotypes | x | 0.020 | x | x | x | x |
Notes.
ace, collagen binding cell wall protein; efa, endocarditis specific antigen; gelE, gelatinase; asa, aggregation substance; esp, enterococcal surface protein; cyl, cytolysin
x, No significant correlation.
Subject: human host and swine host; Biofilm refers to biofilm former and non-former; Matrix, oral, rectal, nasal, urine and fecal; Region, northern region and central region; Hygiene, HP1, HP2, HP3.
Distribution of the E. faecalis and E. faecium strains across locations, host and sample type, and its relationship to the biofilm-forming ability
To determine the association between the genotypes, resistotypes, virulotypes and different environmental factors, a composite similarity matrix of the two typing methods was used to perform statistical analysis on the correlation between the genotype of the strains and different factors. Based on the DistLM results, the DNA fingerprints of strains isolated from both pigs and humans were significantly different among each other (Pseudo-F = 2.5204, P = 0.020). We further evaluated the factors which might affect the genotypes of the strains such as sample matrix, sampling region, farm hygiene practice, biofilm-forming ability as well as the resistotypes and virulotypes of the strains (Table 5). For the E. faecalis isolated from pigs, a significant correlation was detected for all tested factors except biofilm-forming ability while for human strains, no association was found between the genotypic patterns in relation to farm hygiene practice, sample matrix and biofilm-forming ability. Conversely, E. faecium isolated from swine samples were significantly associated with the sampling region and hygiene practice. The strains also exhibited significant genotypic difference in association with resistotypes and virulotypes. E. faecium strains isolated from human samples was the only group of strains which was significantly correlated to biofilm-forming ability and similar to E. faecalis, they were unaffected by the farm hygiene practice. Overall, the statistical analysis supported the finding that the strains isolated in this study are host and regional specific. Strong correlations were also found between the genotypes and the resistance and virulence profiles of the strains.
Table 5. Correlation between environmental factors, virulotypes and resistotypes with composition phylogenetic relationship inferred using REP-PCR and PFGE.
| Species | Subjects | Factorsa | Composite of PFGE and REP-PCR | |
|---|---|---|---|---|
| Pseudo F | P | |||
| E. faecalis | Pigs | Region | 26.229 | 0.001 |
| Hygiene practice | 24.392 | 0.001 | ||
| Sample matrix | 5.6083 | 0.001 | ||
| Resistotypes | 2.8807 | 0.014 | ||
| Virulotypes | 4.7217 | 0.001 | ||
| Biofilm | 1.3114 | 0.244 | ||
| Humans | Region | 4.9953 | 0.001 | |
| Hygiene practice | 0.5268 | 0.767 | ||
| Sample matrix | 1.7178 | 0.126 | ||
| Resistotypes | 2.3653 | 0.048 | ||
| Virulotypes | 6.0838 | 0.001 | ||
| Biofilm | 1.1187 | 0.361 | ||
| E. faecium | Pigs | Region | 9.3722 | 0.001 |
| Hygiene practice | 7.7394 | 0.001 | ||
| Sample matrix | 0.2937 | 0.902 | ||
| Resistotypes | 1.6440 | 0.159 | ||
| Virulotypes | 4.5310 | 0.006 | ||
| Biofilm | 2.2920 | 0.055 | ||
| Humans | Region | 6.7071 | 0.001 | |
| Hygiene practice | 2.8370 | 0.050 | ||
| Sample matrix | 4.7811 | 0.010 | ||
| Resistotypes | 2.6958 | 0.046 | ||
| Virulotypes | 2.9966 | 0.037 | ||
| Biofilm | 4.4749 | 0.007 | ||
Notes.
Subject, human host and swine host; Biofilm refers to biofilm former and non-former; Matrix, oral, rectal, nasal, urine and fecal; Region: northern region and central region; Hygiene, HP1, HP2, HP3.
Discussion
More than 90% of the enterococci studied were MDR. Traditionally, the first line treatment for enterococcal infection is a combination of cell wall-active agents and aminoglycosides (Kristich, Rice & Arias, 2014). The occurrence of resistance to high-level gentamicin compromises the efficacy of this drug and the increasing MDR status further limits the antimicrobial therapy. The degree of antibiotic resistance in the herd is strongly correlated with the antibiotic usage as the routine application of antibiotics serves as a selective pressure for resistant bacteria (Woolhouse et al., 2015; Benjamin, Bruce & Lance, 2017). According to the National Pharmaceutical Control Bureau of Ministry of Health, Malaysia, there are 97 antimicrobials registered for the use in livestock. A majority of these antimicrobials are used in poultry and swine farms despite the fact that some of these drugs are classified as Critically Important Antimicrobials by WHO (Health Action International Asia Pacific, 2013). In Malaysia, the Department of Veterinary Services (DVS) is responsible for certification, inspection, accreditation and implementation of legislation to ensure the production of quality livestock. They also ensure that the use of antibiotics is in accordance with the Feed Act 2009 of Malaysia. Taking into consideration the effect of antimicrobial resistance in public health and livestock industry, the Malaysian government has embarked multiple strategies against the antimicrobial resistance threat, which included increase public awareness and education on appropriate use of antibiotics, expedite surveillance and research, and improve infection prevention and control (MOH Malaysia, 2017). The unregulated use of antibiotics in swine husbandry, the co-transfer of resistance genes between enterococcal strains from different origins might have contributed to the high prevalence of resistant strains in this study (Silveira et al., 2013). The strains isolated in this study were mainly resistant to chloramphenicol, tetracycline, ciprofloxacin, gentamicin and erythromycin. Similar results were also observed in study on swine meat chain (Rizzotti, Rossi & Torriani, 2016). E. faecium was more resistant to penicillin and ampicillin while E. faecalis was more resistant to aminoglycosides. Larsen et al. (2011) reported that porcine-origin E. faecalis strains were genetically related to the strains isolated from infective endocarditis patients, suggesting that pigs can be a reservoir of human pathogens. Farm workers, veterinarians and those who are in close contact with the animals are at a higher risk of being colonized or infected by resistant bacteria harbored by the animal hosts. The resistance and virulence genes can also be transferred to other pathogenic bacteria such as Staphylococcus aureus, causing severe bacteremia (Kristich, Rice & Arias, 2014; Health Action International Asia Pacific, 2013). Strains with a particular resistance phenotype harbored the corresponding antibiotic resistance genes. Notably, E. faecalis strains showing high-level gentamicin resistance harbored two resistance genes, which are the aac(6′)-Ie-aph(2″)-Ia and aph(3′)-IIIa. Strains with only one aminoglycoside resistance gene were either susceptible or resistant to low-level gentamicin only. However, this trend was only observed in the E. faecalis strains. Vancomycin-resistant enterococcus (VRE) have been previously reported from swine farms located in Selangor, Perak, Johor and Penang (Getachew et al., 2010; Getachew et al., 2012; Getachew et al., 2013). However, these studies mainly focused on vancomycin resistance phenotype as VRE strains are important nosocomial pathogens and molecular evidence have also suggested that animals are reservoir of VRE (Courvalin, 2005). Although VRE was not found in our study, our findings showed that enterococcal strains found in the swine farms were multidrug-resistant. This indicates the importance of screening other antibiotics apart from vancomycin.
Biofilms facilitate antibiotic resistance and host colonization of pathogens (Lebreton, Willems & Gilmore, 2014). Our results showed that E. faecalis has a higher potential to form biofilm than E. faecium. Biofilm-forming E. faecalis found in this study possessed higher rates of resistance towards ampicillin, penicillin, chloramphenicol and high-level gentamicin. A mature enterococcal biofilm contains more bacteria and is more tolerant to antibiotics (Holmberg & Rasmussen, 2016). This probably explains the higher infection rate of E. faecalis compared to E. faecium, because the biofilm-forming E. faecalis strains are more persistent than E. faecium. Toledo-arana et al. (2001) reported that esp gene is involved in the biofilm formation of enterococcal strains. However, such correlation was not found in our study. In contrast, efa and gelE were found to have significant correlation with the resistotypes of E. faecalis strains.
The statistical analysis of the composite genotypic data showed that E. faecalis and E. faecium were significantly grouped according to their hosts (humans vs. pig), suggesting a host specific distribution. However, there are exceptions. Certain E. faecalis strains isolated from different hosts with high percentage of similarity (>90%) were observed (Files S2 and S4). For instance, Enfs114 from nasal swab of a farmer and Enfs66 from a swine oral swab from PF4 possessed 100% and 91% similarity in REP and PFGE banding patterns, respectively. This result is consistent with the report by Freitas et al. (2011), which showed that certain E. faecalis and E. faecium strains in humans and pigs were indistinguishable by PFGE and MLST. It is also noteworthy that the strains isolated from swine oral and human nasal swabs were highly similar with the environmental strains, indicating a potential of inter-host dispersal of the enterococci strains, as strains could be transmitted from one host to another host through the environment. Detection of regional-specific strains could be explained by different swine farm management implemented in each farm as different disinfection and antibiotic exposure could act as a selective pressure.
Conclusion
This study showed a high occurrence of MDR E. faecalis and E. faecium in the studied swine farms. MDR Enterococci strains from livestock industry harboring various antibiotic resistance and virulence genes pose a significant health risk to the public. Ubiquitous strains shared by different hosts were identified although majority of the strains were regional- and host-specific. A more stringent regulation is needed to ensure the proper use of antibiotics in swine husbandry to reduce the wide spread of MDR strains.
Supplemental Information
Acknowledgments
We thank the University of Malaya for facilities support.
Funding Statement
This research was funded by a High Impact Research (HIR) grant [UM.C/625/1/HIR-MOHE/CHAN/11/02] and a UMRG grant RP022-2012B of the University of Malaya. The first author, SC Tan was supported by the University of Malaya Fellowship Scheme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Shiang Chiet Tan conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Chun Wie Chong, Cindy Shuan Ju Teh, Peck Toung Ooi and Kwai Lin Thong conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Human Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The human samples collection was approved by Medical Ethics Committee, University Malaya Medical Centre (Ethic committee/IRB reference number: 1010.41).
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
This study was approved by the Animal Care and Use Committee (ACUC), Faculty of Veterinary Medicine, UPM (UPM/IACUC/FYP-AUP-T006/2013).
Data Availability
The following information was supplied regarding data availability:
The raw data are provided in the Supplemental Files.
References
- Baldassarri et al. (2001).Baldassarri L, Cecchini R, Bertuccini L, Ammendolia MG, Iosi F, Arciola CR, Montanaro L, Di Rosa R, Gherardi G, Dicuonzo G, Orefici G, Creti R. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Medical Microbiology and Immunology. 2001;190:113–120. doi: 10.1007/s00430-001-0096-8. [DOI] [PubMed] [Google Scholar]
- Beganovic et al. (2018).Beganovic M, Luther MK, Rice LB, Arias CA, Rybak MJ, LaPlante KL. A review of combination antimicrobial therapy for Enterococcus faecalis bloodstream infections and infective endocarditis. Clinical Infectious Diseases. 2018;67(2):303–309. doi: 10.1093/cid/ciy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, Bruce & Lance (2017).Benjamin JK, Bruce AH, Lance BP. Food-animal production and the spread of antibiotic resistance: the role of ecology. Frontiers in Ecology and the Environment. 2017;15(6):309–318. doi: 10.1002/fee.1505. [DOI] [Google Scholar]
- CLSI (2015).Clinical Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; twenty third informational supplement M100-S23 CLSI. Clinical Laboratory Standards Institute; Wayne: 2015. [Google Scholar]
- Compassion in World Farming (2011).Compassion in World Farming (CIWF) Antibiotics in animal farming: public health and animal welfare. Surrey, CIWFhttps://www.ciwf.org.uk/media/3758863/Antibiotics-in-Animal-Farming-Public-Health-and-Animal-Welfare.pdf. [15 January 2016];2011
- Courvalin (2005).Courvalin P. Antimicrobial drug resistance: “prediction is very difficult, especially about the future”. Emerging Infectious Diseases. 2005;11(10):1503–1506. doi: 10.3201/eid1110.051014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel et al. (2015).Daniel DS, Lee SM, Dykes GA, Rahman S. Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Applied and Environmental Microbiology. 2015;81:6090–6097. doi: 10.1128/AEM.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas et al. (2011).Freitas AR, Coque TM, Novais C, Hammerum AM, Lester CH, Zervos MJ, Donabedian S, Jensen LB, Francia MV, Baquero F, Peixe L. Human and swine hosts share vancomycin-resistant Entercoccus faecium CC17 and CC5 and Enterococcus faecium CC2 clonal clusters harboring Tn1546 on indistinguishable plasmids. Journal of Clinical Microbiology. 2011;49(3):925–931. doi: 10.1128/JCM.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getachew et al. (2013).Getachew Y, Hassan L, Zakaria Z, Aziz SA. Genetic variability of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis isolates from humans, chickens, and pigs in Malaysia. Applied and Environmental Microbiology. 2013;79(15):4528–4533. doi: 10.1128/AEM.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getachew et al. (2010).Getachew Y, Hassan L, Zakaria Z, Lokman N. Species distribution and resistance phenotypes of vancomycin-resistant Enterococcus isolated from pigs in Pulau Pinang, Malaysia. Pertanika Journal of Tropical Agricultural Science. 2010;33(1):15–25. [Google Scholar]
- Getachew et al. (2012).Getachew Y, Hassan L, Zakaria Z, Zaid CZM, Yardi A, Shukor RA, Marawin LT, Embong F, Aziz SA. Characterization and risk factors of vancomycin-resistant Enterococci (VRE) among animal-affiliated workers in Malaysia. Journal of Applied Microbiology. 2012;113:1184–1195. doi: 10.1111/j.1365-2672.2012.05406.x. [DOI] [PubMed] [Google Scholar]
- Health Action International Asia Pacific (2013).Health Action International Asia Pacific (HAIAP) Antibiotic use and antibiotic resistance in food animals in Malaysia: a threat to human and animal health. Third World Network (TWN) Penang in association with Consumers’ Association of Penang, Malaysiahttp://www.haiasiapacific.org/wp-content/uploads/2014/06/Memo-on-Antibiotics-in-animal-feeds-the-case-for-Malaysia-21-Nov-2013-V1.pdf. [28 December 2016];2013
- MOH Malaysia (2017).Ministry of Health (MOH) Malaysia Malaysian action plan on antimicrobial resistance (MyAP-AMR) 2017–2021. http://www.moh.gov.my/index.php/file_manager/dl_item/554756755a584a6961585268626938794d4445334c306468636d6c7a49464268626d5231595734765457467359586c7a615746755830466a64476c76626c395162474675583039755830467564476c7461574e7962324a7059577866556d567a61584e305957356a5a56386f54586c425543314254564970587a49774d5463744d6a41794d5335775a47593d. [03 January 2018];2017
- Holmberg & Rasmussen (2016).Holmberg A, Rasmussen M. Mature biofilms of Enterococcus faecalis and Enterococcus faecium are highly resistant to antibiotics. Diagnostic Microbiology and Infectious Disease. 2016;84:19–21. doi: 10.1016/j.diagmicrobio.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Hwang et al. (2009).Hwang IY, Ku HO, Lim SK, Park CK, Jung GS, Jung SC, Nam HM. Species distribution and resistance patterns to growth-promoting antimicrobials of enterococci isolated from pigs and chickens in Korea. Journal of Veterinary Diagnostic Investigation. 2009;21:858–862. doi: 10.1177/104063870902100616. [DOI] [PubMed] [Google Scholar]
- Jackson, Fedorka-Cray & Barrett (2004).Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. Journal of Clinical Microbiology. 2004;42(8):3558–3565. doi: 10.1186/s12866-015-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich, Rice & Arias (2014).Kristich CJ, Rice LB, Arias CA. Enterococcal infection—treatment and antibiotic resistance. In: Gilmore MS, Clewell DB, Ike Y, Shanker N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. pp. 123–184. [PubMed] [Google Scholar]
- Larsen et al. (2011).Larsen J, Schonheyder HC, Singh KV, Lester CH, Olsen SS, Porsbo LJ, Garcia-Migura L, Jensen LB, Bisgaard M, Murray BE, Hammerum AM. Porcine and human community reservoirs of Enterococcus faecalis, Denmark. Emerging Infectious Diseases. 2011;17(12):2395–2397. doi: 10.3201/eid17120101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton, Willems & Gilmore (2014).Lebreton F, Willems R, Gilmore M. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shanker N, editors. Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. pp. 5–64. [PubMed] [Google Scholar]
- Lim, Teh & Thong (2017).Lim SY, Teh CSJ, Thong KL. Biofilm-related diseases and omics: global transcriptional profiling of Enterococcus faecium reveals different gene expression patterns in the biofilm and planktonic cells. OMICS A Journal of Integrative Biology. 2017;21(10):592–601. doi: 10.1089/omi.2017.0119. [DOI] [PubMed] [Google Scholar]
- Novais et al. (2013).Novais C, Freitas AR, Silveira E, Antunes P, Silva R, Coque TM, Peixe L. Spread of multidrug-resistant Enterococcus to animals and humans: an underestimated role for the pig farm environment. Journal of Antimicrobial Chemotherapy. 2013;68:2746–2754. doi: 10.1093/jac/dkt289. [DOI] [PubMed] [Google Scholar]
- Price et al. (2018).Price VJ, McBride SW, Duerkop B, Palmer KL. CRISPR-Cas blocks antibiotic resistance plasmid transfer between Enterococcus faecalis strains in the gastrointestinal tract. bioRxiv. 2018 doi: 10.1101/312751. [DOI]
- Rizzotti, Rossi & Torriani (2016).Rizzotti L, Rossi F, Torriani S. Biocide and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from the swine meat chain. Food Microbiology. 2016;60:160–164. doi: 10.1016/j.fm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Silveira et al. (2013).Silveira E, Freitas AR, Antunes P, Barros M, Campos J, Coque TM, Peixe L, Novais C. Co-transfer of resistance to high concentrations of copper and first-line antibiotics among Enterococcus from different origins (humans, animals, the environment and foods) and clonal lineages. Journal of Antimicrobial Chemotherapy. 2013;69(4):899–906. doi: 10.1093/jac/dkt479. [DOI] [PubMed] [Google Scholar]
- Stepanovic et al. (2000).Stepanovic S, Vucovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of Microbiological Methods. 2000;40(2):175–179. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- Toledo-arana et al. (2001).Toledo-arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I. The enterococcal surface protein, esp, is involved in Enterococcus faecalis biofilm formation. Applied and Environmental Microbiology. 2001;67(10):4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turabelidze et al. (2000).Turabelidze D, Kotetishvili M, Kreger A, Morris Jr JG, Sulakvelidze A. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. Journal of Clinical Microbiology. 2000;38(11):4242–4245. doi: 10.1128/jcm.38.11.4242-4245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic et al. (1994).Versalovic J, Schneider M, De Bruijn F, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in Molecular and Cellular Biology. 1994;5:25–40. [Google Scholar]
- Vidana et al. (2015).Vidana R, Rashid MU, Ozenci V, Weintraub A, Lund B. The origin of endodontic Enterococcus faecalis explored by comparison of virulence factor patterns and antibiotic resistance to that of isolates from stool samples, blood cultures and food. International Endodontic Journal. 2015;49(4):343–351. doi: 10.1111/iej.12464. [DOI] [PubMed] [Google Scholar]
- Woolhouse et al. (2015).Woolhouse M, Ward M, Van Bunnik B, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370(1670):20140083. doi: 10.1098/rstb.2014.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Aminov, Garrigues-JeanJean & Mackie (2001).Aminov RI, Garrigues-JeanJean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Applied and Environmental Microbiology. 2001;67(1):22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark et al. (1993).Clark NC, Cooksey RC, Hill BC, Swenson JM, Tenover FC. Characterization of glycopeptide-resistant enterococci from US hospitals. Antimicrobial Agents and Chemotherapy. 1993;37:2311–2317. doi: 10.1128/AAC.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka Malen, Evers & Courvalin (1995).Dutka Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. Journal of Clinical Microbiology. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gever et al. (2003).Gever D, Danielsen M, Huys G, Swings J. Molecular characterization of tet (M) genes in Lactobacillus isolates from different types of fermented dry sausages. Applied and Environmental Microbiology. 2003;69(2):1270–1275. doi: 10.1128/Aem.69.2.1270-1275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannu et al. (2003).Mannu L, Pab A, Daga E, Comunian R, Zanetti S, Dupre I, Sechi LA. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. International Journal of Food Microbiology. 2003;88:291–304. doi: 10.1016/S0168-1605(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Satake et al. (1997).Satake S, Clark N, Rimland D, Nolte FS, Tenover FC. Detection of vancomycin-resistant enterococci in fecal samples by PCR. Journal of Clinical Microbiology. 1997;35(9):2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar et al. (1999).Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infection and Immunity. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko et al. (2003).Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos MJ, Lerner SA, Chow JW. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrobial Agents and Chemotherapy. 2003;47(4):1423–1426. doi: 10.1128/AAC.47.4.1423-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven et al. (2004).Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asaI, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Entercoccus faecium. Journal of Clinical Microbiology. 2004;42(10):4473–4479. doi: 10.1128/JCM.42.40.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are provided in the Supplemental Files.