ABSTRACT
In recent years, coherent with growing biologics portfolios also the number of complex and thus difficult‐to‐express (DTE) therapeutic proteins has increased considerably. DTE proteins challenge bioprocess development and can include various therapeutic protein formats such as monoclonal antibodies (mAbs), multi‐specific affinity scaffolds (e.g., bispecific antibodies), cytokines, or fusion proteins. Hence, the availability of robust and versatile Chinese hamster ovary (CHO) host cell factories is fundamental for high‐yielding bioprocesses. MicroRNAs (miRNAs) have emerged as potent cell engineering tools to improve process performance of CHO manufacturing cell lines. However, there has not been any report demonstrating the impact of beneficial miRNAs on industrial cell line development (CLD) yet. To address this question, we established novel CHO host cells constitutively expressing a pro‐productive miRNA: miR‐557. Novel host cells were tested in two independent CLD campaigns using two different mAb candidates including a normal as well as a DTE antibody. Presence of miR‐557 significantly enhanced each process step during CLD in a product independent manner. Stable expression of miR‐557 increased the probability to identify high‐producing cell clones. Furthermore, production cell lines derived from miR‐557 expressing host cells exhibited significantly increased final product yields in fed‐batch cultivation processes without compromising product quality. Strikingly, cells co‐expressing miR‐557 and a DTE antibody achieved a twofold increase in product titer compared to clones co‐expressing a negative control miRNA. Thus, host cell engineering using miRNAs represents a promising tool to overcome limitations in industrial CLD especially with regard to DTE proteins. Biotechnol. Bioeng. 2017;114: 1495–1510. © 2017 The Authors. Biotechnology and Bioengineering Published by Wiley Periodicals, Inc.
Keywords: microRNA, miR‐557, Chinese hamster ovary (CHO) cells, difficult‐to‐express proteins, cell engineering, monoclonal antibody
Introduction
Monoclonal antibodies (mAbs) represent the largest group of biopharmaceuticals and most therapeutic proteins that have already gained approval account for mAbs. However, the number of complex and clinically potent biologicals such as multi‐specific antibody formats and fusion proteins has increased dramatically (Jost and Pluckthun, 2014; Spiess et al., 2015). Compared to classical mAbs, multi‐specific antibodies, and fusion proteins are artificially designed therapeutics which have never undergone an evolutionary optimization. Thus, it seems less surprising that these sophisticated molecules often turn out to be difficult‐to‐express (DTE) (Johari et al., 2015). Although state‐of‐the‐art industrial cell line development (CLD) workflows often generate cell clones surpassing volumetric productivities of 3 g/L, standard mAbs are also frequently found to be challenging in terms of expression using mammalian cell factories such as Chinese hamster ovary (CHO) cells (Bentley et al., 1998; Pybus et al., 2014). It is known that small variations in the amino acid sequence of the variable domain of an antibody can have substantial impact on its affinity to the target antigen. However, it is surprising that variations in the rather small variable domains also dramatically influence expression levels and thus manufacturability (Mason et al., 2012; Seeliger et al., 2015). Nonetheless, there is an urgent requirement to prepare current industrial CHO host cell lines for such future manufacturing challenges to provide effective therapeutics and to address unmet medical needs. In order to provide access to sufficient amounts of these innovative drugs, host cell engineering of CHO cells represents a promising approach to overcome limited production yields of complex biologics.
In recent years, microRNAs (miRNAs) have become an important pillar in CHO cell engineering due to substantial scientific efforts to unravel the function of these small but impactful regulators of gene expression in CHO cells (Barron et al., 2011b; Fischer et al., 2015b; Hackl et al., 2012a; Jadhav et al., 2013; Muller et al., 2008; Stiefel et al., 2015). miRNAs are small non‐protein coding RNAs that post‐transcriptionally regulate gene expression in most eukaryotic cells including CHO cells. From a biotechnological perspective the miRNA technology represents a very smart genetic tool as miRNAs are not translated into proteins and therefore miRNA overexpression does not add translational burden to the production cell line. Furthermore, individual miRNAs are capable of regulating entire cellular pathways (Fischer et al., 2015a; Hackl et al., 2012a), and thus might have a substantial impact on CHO cell phenotypes. Currently available literature on miRNA research in CHO cells mostly focusses on the identification of miRNAs to improve recombinant protein production (Emmerling et al., 2015; Fischer et al., 2014, 2015d; Jadhav et al., 2014; Kelly et al., 2015; Loh et al., 2014), or cell growth (Barron et al., 2011a; Druz et al., 2013; Sanchez et al., 2014), while other studies were dedicated to basic principles of miRNA biogenesis and function (Diendorfer et al., 2015; Fischer et al., 2015a,2015c; Hackl et al., 2012b, 2011; Hernandez Bort et al., 2012). We have previously identified miR‐557 to increase productivity of a recombinant easy‐to‐express mAb in CHO cells (Strotbek et al., 2013). In order to explore how miRNAs can enhance an industrial CHO cell line generation process, we generated novel CHO host cells stably expressing miR‐557 and compared their CLD performance to control host cells in two independent CLD campaigns. For this purpose, we selected two different antibody candidates, an easy‐to‐express mAb that can be produced in the multi‐gram per liter range as well as a very DTE mAb exhibiting productions yields less than 500 mg/L. CLD performance was substantially enhanced in the presence of miR‐557 resulting in increased numbers of high‐producing clones as well as enhanced final antibody productivities of the established cell clones in fed‐batch cultivations. We demonstrate that stable manipulation of miRNA expression in industrial CHO host cell lines represents a powerful opportunity to increase success in CHO CLD, especially regarding DTE proteins. The novel miRNA technology might enable access to sufficient amounts of sophisticated therapeutic proteins even if they may exhibit inherent difficulties in expression levels. Application of miRNAs in industrial manufacturing cell lines might therefore increase success in biopharmaceutical drug development in the future.
Materials and Methods
Cell Culture
Suspension adapted Boehringer Ingelheim (BI) proprietary glutamine synthetase deficient (GS−/−) CHO host and recombinant CHO production cell lines (derived from both GS and DHFR deficient CHO host cells) were cultivated in shake flasks (Corning, Corning, NY) unless otherwise stated. An overview on the recombinant CHO production cell lines used for transient miRNA transfection experiments are listed in Table I. Cells were cultivated at 37°C, 5% CO2 and with agitation at 125 rpm (50 mm orbit) in an orbital shaker incubator (Infors, Bottmingen, Switzerland). BI proprietary serum‐free, chemically‐defined, and animal component free cell culture media were used for cultivation. Seeding cell density of stock cultures was set to 0.3 × 106 viable cells per milliliter and cells were passaged in a 4/3 day rhythm. Cell concentration and viability during routine stock culture cultivation was assessed using a Cedex HiRes Analyzer™ (Roche Diagnostics, Mannheim, Germany) by means of trypan blue exclusion.
Table I.
Overview on recombinant CHO cell lines used for transient miRNA transfection studies
| Cell line ID | Selection system | Type of protein | mAb expression level |
|---|---|---|---|
| CHO‐mAb1 | GS | IgG antibody | High |
| CHO‐mAb2 | GS | Bispecific antibody | Medium |
| CHO‐mAb3 | DHFR | IgG antibody | High |
| CHO‐mAb4 | GS | Bispecific IgG‐scFv fusion | Medium |
| CHO‐mAb5 | GS | IgG antibody | Very low |
| CHO‐mAb6 | GS | IgG antibody | Low |
| CHO‐mAb7 | GS | Bispecific antibody | Low |
Transfection
Transfection of small RNAs such as miRNA mimics or siRNAs (Qiagen, Hilden, Germany) was performed as previously described using ScreenFect® A plus (InCella, Eggenstein‐Leopoldshafen, Germany) (Fischer et al., 2013). Briefly, ScreenFect® A plus was first diluted in ScreenFect® Dilution Buffer (InCella) before the respective amount of miRNA or siRNA (50 nM final RNA concentration) was added followed by a complexation for 20 min at room temperature. The resulting transfection complexes were transferred to a U‐bottom shaped 96‐deepwell plate (Enzyscreen, Haarlem, the Netherlands). Exponentially growing CHO cells were pelleted, resuspended in fresh culture medium, and added to each well containing transfection complexes at a cell concentration of 0.5 × 106 viable cells per milliliter. 96‐deepwell culture plates were placed onto a mini‐orbital digital shaker (Bellco, Vineland) located inside a cell culture incubator and cultured at 37°C, 5% CO2, 90% relative humidity and with agitation at 800 rpm (1.6 mm orbit). MiScript miRNA mimics, AllStars Negative Control siRNA (NT‐siRNA) as well as CHO‐specific cell death control siRNAs were purchased from Qiagen.
Transfection of plasmid DNA was performed using the Cell Line Nucleofector® Kit V (Lonza, Vervier, Belgium) on a Nucleofector® (Lonza) according to the instructions provided by the manufacturer. Briefly, 5.0 × 106 cells were pelleted and resuspended in transfection solution. After addition of 5 μg of endotoxin‐free plasmid DNA cells were transfected and seeded in pre‐warmed culture medium. Cell culture medium was exchanged for selection medium 24 h post transfection. Transfected cells were cultivated at 37°C and 5% CO2 in humidified atmosphere until single cell cloning (SSC). Each transfection was performed in biological triplicates (n = 3).
Quantitative Reverse‐Transcription Real‐Time PCR (qRT‐PCR)
CHO cells stably overexpressing hsa‐miR‐557 (pcDNA6.2‐GW/emGFP‐miR557‐miR557) or a negative control construct (pcDNA6.2‐GW/emGFP‐miR‐NT) were subcultivated every 2–3 days with seeding densities of 0.3 × 106 cells/mL. One day after subcultivation, total RNA was extracted from 1 × 106 cells using the RNeasy Plus Mini Kit (Qiagen) according to the manufactureŕs protocol. RNA concentration was measured on a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and 10 ng of RNA was reverse transcribed into cDNA using the TaqMan® MicroRNA Assay (#4427975, Thermo Fisher Scientific) and TaqMan microRNA Reverse Transcription Kit (#4366596, Thermo Fisher Scientific) according to the manufacturer's instructions. qRT‐PCR was performed with DyNAmo Color Flash Probe qPCR Kit (Biozym, Hessisch Oldendorf, Germany) using a Cfx96 device (Biorad, Hercules, CA). U6 snoRNA was used as reference. Calculation of ΔCq values was done with the single threshold method (Biorad CFX manager software 2.1).
Product Quantification
Concentration of standard mAbs, bispecific antibodies, and fusion proteins possessing an Fc part were determined via bio layer interferometry using Protein A coupled biosensors on an Octet® QK384 system (Pall Life Science, Dreieich, Germany). Cell culture supernatant was diluted in cultivation media to a total volume of 200 μL. The Protein A coupled biosensors were incubated in cultivation media for 15 min for equilibration. Recuperation of biosensors between each measurement was achieved in a regeneration solution containing 10 mM glycine at a pH of 1.5. A standard curve using the respective antibody was used to calculate product concentrations in culture supernatant.
Cell Line Development
An overview on the CLD process used in this study is shown in Figure 3A. Briefly, stable mAb producing cell pools were subjected to SSC using fluorescent activated cell sorting (FACS) on a FACSAria™ III (BD Biosciences). For each miRNA overexpressing host cell line, 1200 cell clones were deposited in 384‐well plates (Corning). Automated fluorescence microscopy was conducted on days 0, 7, and 14 post SSC using a Cellavista™ high‐end RS cell imager (Synentec bio Services GmbH, Muenster, Germany) to ensure monoclonality as well as to monitor cell growth after SSC. After analysis of mAb concentration in the culture supernatant best production clones were transferred to 96‐well plates and cultivated for several days until cultures were further expanded to 24‐well and 6‐well plates, respectively, thereby decreasing the number of clones down to 20 based on performance. Based on growth and mAb productivity, cell lines were further reduced.
Figure 3.
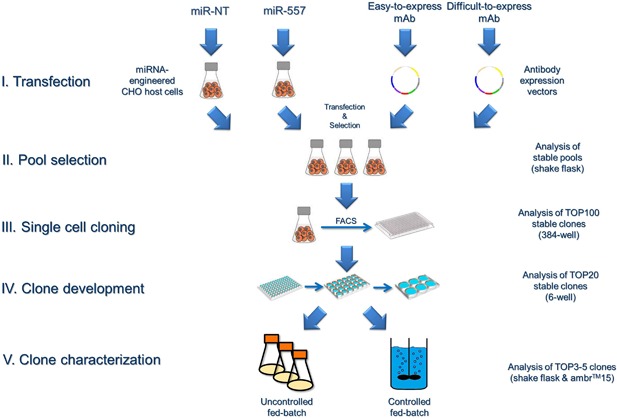
Overview on the cell line development process used in this study. Respective analyses performed during each step are indicated. Bioprocess performance of final production clones was examined both using uncontrolled (shake flask) and controlled (ambr®15) fed‐batch cultivation processes.
Fed‐Batch Cultivation
Analysis of bioprocess performance of the five best clones was performed in uncontrolled and of the three best clones in controlled 10‐day fed‐batch cultivations. Uncontrolled fed‐batch cultivations were performed in 250 mL shake flasks (Corning, Glendale, AZ) while controlled cultivations were conducted in an advanced micro bioreactor system (ambr®15) (TAP Biosystems, Hertfordshire, UK). Briefly, cells were seeded at 0.3 × 106 viable cells per mL in a proprietary production medium. Feeding was started at day three following inoculation using a proprietary feed medium and included 30 mL of feed medium per liter of cultivation volume per day. Glucose boli were added to the cultures if glucose concentrations dropped below 3 g/L. Process parameters (cell concentration, viability, pH, pO2, glucose, and lactate concentration) as well as mAb concentration were determined every 48 h (shake flasks) or daily (ambr®15). Cell concentration and viability was analyzed using a Cedex HiRes Analyzer™ (Roche Diagnostics). Glucose and lactate concentration was determined on a Biosen C‐Line System (EKF Diagnostic, Barleben, Germany), while pH and pO2 were analyzed using a RAPIDLab®248 system (Siemens Healthcare, Erlangen, Germany). Antibody concentration was determined as described above on an Octet® QK384 system (Pall Life Science).
Statistics
Unless otherwise stated, an unpaired two‐tailed t‐test was applied (*P < 0.05; **P < 0.01; ***P < 0.001) to determine statistical significance between two independent sets of samples. Statistical determination of the probability to identify a high‐producing cell clone was calculated based on volumetric mAb productivity of clones cultivated in 6‐well plates. Toward this end, mAb titer data of both antibody candidates from miR‐NT (n = 25) and miR‐557 (n = 25) expressing cell clones was analyzed using Dell Statistica data analysis software version 12 (Dell, Round Rock, TX) and fitted to a normal distribution function. The normal distribution function was determined by the following formula:
where, m is the mean, s is the standard deviation, e is the base of the natural logarithm and p is the constant Pi. After determining the mean productivity for CHO‐miR‐NT derived control cells a high‐producing cell clone was generally defined to exhibit an at least a 1.5‐fold increased volumetric productivity compared to the mean productivity of control cells.
Analysis of Product Quality
Antibodies were purified from cell‐free cell culture supernatant using 1 mL MediaScout® MiniColumns (Atoll, Weingarten, Germany) filled with MabSelect resin (GE Healthcare, Munich, Germany) run on an ÄKTAxpress chromatography system (GE Healthcare). The low pH used for elution was neutralized to pH 5.5 with Tris(hydroxymethyl)‐aminomethan (TRIS) to prevent for antibody denaturation. The final protein concentration was determined using a NanoDrop 2000c (Thermo Fisher Scientific). mAb aggregation and fragmentation was analyzed using ultra performance size exclusion chromatography (UP‐SEC). UP‐SEC was performed on an ACQUITY Bio H Class UPLC system (Waters, Eschborn, Germany) using a BEH200 SEC column (Waters) and run with an L‐arginine/ammonium sulfate/isopropanol buffer at pH 7.3. Resulting chromatograms were integrated using the EMPOWER 3 chromatography data software (Waters). Integrity of the products was analyzed by microchip‐based capillary electrophoresis (CE) with the Protein Express Assay run on a LabChip GXII instrument (PerkinElmer, Rodgau, Germany) according to the manufacturer's protocol. Purity of the samples was determined under reduced conditions by adding 33 mM DTT to the provided sample buffer. Electropherograms were analyzed using the LabChip GX software (PerkinElmer).
N‐linked glycosylation at the Fc region of the antibodies was analyzed after buffer exchange by ultrafiltration using 10 kDa MWCO PES Vivaspin 500 filter units (Sartorius, Goettingen, Germany) to pure water using CE with the ProfilerPro Glycan Profiling system on a LabChip GXII instrument (PerkinElmer) according to the manufacturer's protocol. Electropherograms were analyzed by the LabChip GX software (PerkinElmer) to identify and quantify known N‐glycan structures.
Results
Transient Transfection of miR‐557 Enhances Productivity in Seven Different Recombinant CHO Cell Lines
A previously performed large scale miRNA mimics screen in a mAb‐producing CHO cell line revealed miR‐557 to increase antibody productivity (Strotbek et al., 2013). To investigate whether miR‐557 is capable of enhancing productivity of recombinant proteins regardless of the molecule type we transiently transfected seven different recombinant CHO production cell lines with either miR‐557 mimics or non‐targeting control miRNAs (miR‐NT). The selected production cell lines included different CHO host cell types (CHO DG44, CHO‐K1), different selection systems (DHFR/MTX, GS/MSX), different molecule types (standard IgGs, bispecific IgGs, fusion proteins) as well as different expression levels (high, medium, low). An overview on cell lines used for transient miRNA mimic transfections is listed in Table I. Production cell lines transfected with miRNA mimics were analyzed for changes in recombinant protein production at 96 h post miRNA transfection. Transient introduction of miR‐557 mimics significantly increased productivity in all cell lines tested compared to control cells transfected with a non‐targeting miRNA (Fig. 1). The extent of productivity enhancement was highly comparable between different production cell lines and accounted for a relative increase of approximately 40% compared to control cultures. These results underscore that also stable expression of miR‐557 might enhance the production capacity for recombinant proteins in diverse CHO host cell lines. Furthermore, these data also imply that application of miR‐557 may likely be functional regardless of the CHO cell background and type of therapeutic protein.
Figure 1.
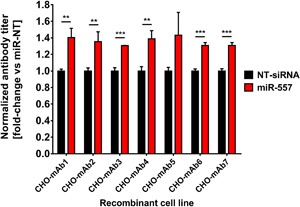
Transient transfection of miRNA mimics in seven different recombinant CHO production cell lines. Cells were transfected either with hsa‐miR‐557 mimics or non‐target siRNAs (NT‐siRNA) and cultured for 96 h in agitated 96‐well format. Product quantification was performed using Protein A coupled biosensors on an Octet® QK384. mAb productivity of miR‐557 transfected cells was normalized to NT‐siRNA transfected control cells. Data are presented as mean value and error bars represent the standard deviation (SD) of three independent transfections. Testing of statistical significance between control and miRNA transfected CHO production cell lines was performed by applying an unpaired two‐tailed t‐test (**P < 0.01; ***P < 0.001).
Generation of Stable miRNA Overexpressing CHO Host Cells
The successful application of the miRNA technology relies on establishment of stable miRNA overexpressing CHO host cell lines. In order to generate a miR‐557 overexpressing CHO cell line, we used the commercially available pcDNA6.2GW‐emGFP‐miR vector system for miRNA overexpression in mammalian cells which encodes a green fluorescent protein (GFP) reporter gene harboring a miRNA expression cassette in its 3′ untranslated region (3′UTR). Notably, miR‐557 is not endogenously expressed in CHO cells. In order to induce strong expression of miR‐557, we inserted a tandem miRNA expression construct comprising two pre‐miR‐557 loci in close proximity to each other (Fig. 2A). As negative control, we used a non‐targeting miRNA sequence instead of miR‐557. Following transfection of the miRNA expression plasmids stable cell pools were established by antibiotic selection. To generate a more homogenous miRNA overexpressing cell population, GFP‐positive cells were enriched by FACS sorting. Figure 2B shows representative flow cytometry histograms of stable transfected and sorted cell pools. Established miRNA overexpressing cells displayed high and homogenous expression of GFP regardless of the miRNA expressed compared to non‐transfected CHO host cells. In addition, high‐level expression of mature miR‐557 was confirmed by qRT‐PCR for CHO‐miR‐557 compared to negative control (CHO‐miR‐NT) cells (Fig. 2C). Taken together these data confirmed a successful establishment of miRNA overexpressing CHO host cells.
Figure 2.
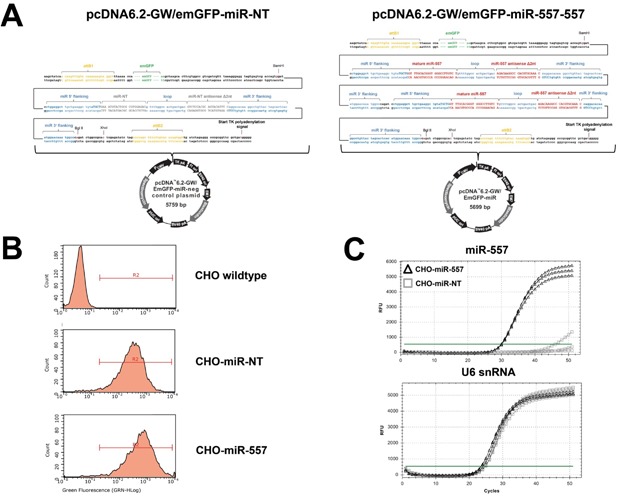
Generation of stable miRNA expressing CHO host cells. CHO host cells were stably transfected with miRNA expression plasmids followed by antibiotic selection in order to generate stable miRNA overexpressing cells. (A) Overview on the pcDNA6.2‐GW/emGFP‐miR expression vector system used for this study. For strong expression of mature miR‐557 a tandem expression construct comprising two pre‐miR‐557 expression cassettes was used. Respective miRNA sequences, flanking regions as well as restriction enzyme cleavage sites are indicated. (B) FACS analysis of emGFP expression of CHO host cells stably expressing pcDNA6.2‐GW/emGFP‐miR expression plasmids. Non‐transfected wildtype CHO host cells served as negative control. (C) Analysis of mature miRNA expression of stable engineered CHO host cells using qRT‐PCR. QRT‐PCR amplifications curves are shown for miR‐557 (black triangles) as well as miR‐NT expressing control cells (grey squares). Total RNA was isolated from exponentially growing host cells followed by reverse transcription. Expression of mature miR‐557 (upper graph) was normalized to the expression levels of U6 snoRNA (lower graph). Analysis was performed in technical triplicates which are indicated by separate amplification curves.
Stable Expression of miR‐557 Improves Cell Line Development Process
So far, there is no evidence that stable co‐expression of a beneficial miRNA and a therapeutic protein is capable of enhancing industrial CLD processes. For this reason, we made use of our newly established CHO host cells stably expressing the pro‐productive microRNA miR‐557 and tested them in two entire CLD campaigns (Fig. 3). To determine, if the functionality of the miRNA engineered host cells is product independent we used two different monoclonal antibody candidates exhibiting completely different expression behavior (easy‐to‐express vs. DTE). The CLD process started with transfection of an antibody construct harboring both heavy and light chain of the antibody on a single expression plasmid followed by selection of stable pools. Established stable pools were then compared for bioprocess performance in a 10‐day fed‐batch cultivation process to determine if stable pools derived from miR‐557 expressing host cells might already display a superior phenotype. Indeed, already stable mAb‐producing cell pools co‐expressing miR‐557 displayed significantly improved process performance and demonstrated increased volumetric mAb productivity (Fig. 4). Here, especially cell pools producing the DTE antibody were observed to exhibit higher peak VCD as well as better harvest viabilities. Similar to results obtained in transient miRNA transfection experiments using different stable mAb‐producing CHO cell lines (Fig. 1), positive effects of miR‐557 on productivity were observed to be independent of the product.
Figure 4.
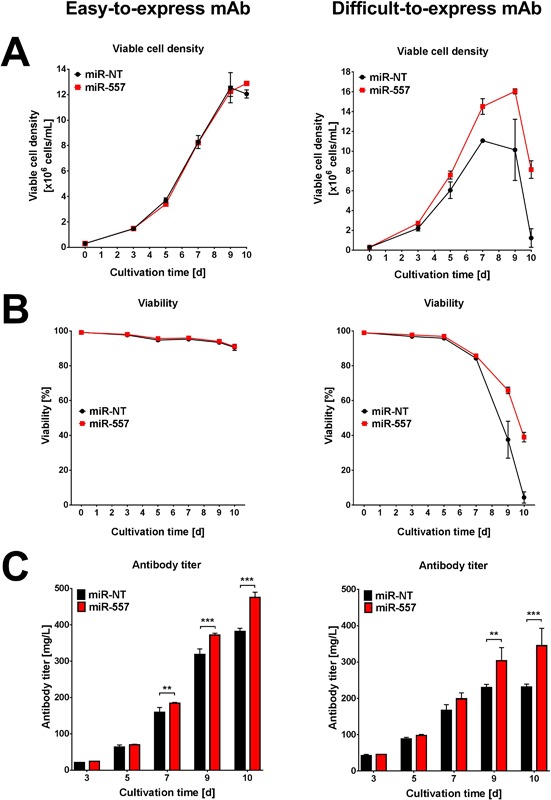
Analysis of fed‐batch performance of stable cell pools either producing an easy‐to‐express or difficult‐to‐express antibody candidate. (A) Viable cell density, (B) viability and (C) volumetric antibody titer of pools co‐expressing either miR‐557 (red) or miR‐NT (black) are shown for a 10‐day fed‐batch cultivation process in 250 mL shake flasks. The left panel represents results of an easy‐to‐express and the right panel for a difficult‐to‐express mAb. Data are presented as mean value and error bars represent the SD of three independent cultivations. For statistical analysis a two‐way ANOVA was applied comparing the mean values of the different miRNA overexpressing cell pools at each time point (**P < 0.01; ***P < 0.001).
Stable pools were subjected to SSC using an in‐house established FACS‐assisted single cell sorting procedure where individual cells were deposited into 384‐well plates and monitored for monoclonality immediately after sorting via automated high‐throughput microscopy. After approximately 2 weeks, mAb concentration was quantified in culture supernatant of wells containing growing cell populations. Figure 5A shows volumetric antibody titers at an early stage in the CLD process (384‐well plates) from the TOP100 cell clones derived from either miR‐557 or control miRNA expressing CHO host cells. Cell clones co‐expressing miR‐557 demonstrated significantly increased antibody titers in 384‐well plates regardless of the mAb candidate. Although mean productivity of the DTE mAb was about twofold lower than of the easy‐to‐express antibody, presence of miR‐557 partly led to volumetric productivities that were comparable to values of the easy‐to‐express mAb produced by negative control cells.
Figure 5.
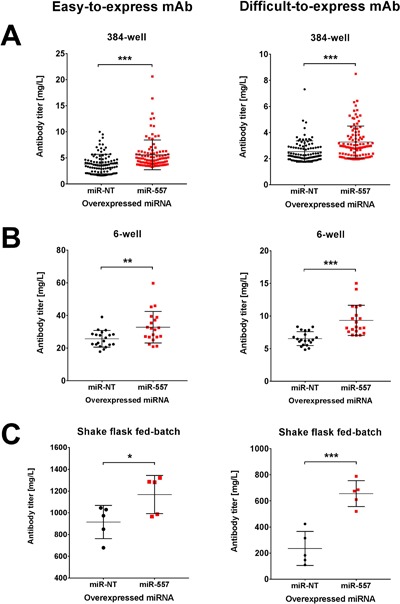
Cell line development performance of miRNA engineered CHO host cells. (A) mAb productivity of TOP 100 stable clones in 384‐well plates at 14 days following single cell cloning (SSC). (B) mAb productivity of TOP 20 stable clones cultivated in 6‐well plates 6 weeks post SSC. (C) Final mAb titer of the best five stable clones at day 10 of uncontrolled fed‐batch cultivations (shake flasks). Stable production cell clones derived from miR‐557 expressing host cells (red) were compared to clones derived from negative control host cells (black). Product quantification was performed using Protein A coupled biosensors on an Octet® QK384. For statistical analysis an unpaired two‐tailed t‐test was applied (*P < 0.05; **P < 0.01; ***P < 0.001).
For both mAb candidates we selected a subset of cell clones to be further expanded and compared the productivity of miR‐557 expressing versus negative control derived cell clones again at a later stage in the CLD process (6‐well format). Analysis of the TOP20 clones confirmed previous findings of 384‐well plates and presence of miR‐557 resulted in significantly enhanced mAb titers for both antibody candidates (Fig. 5B). According to their performance, we selected the TOP5 clones and compared the different cell lines in uncontrolled fed‐batch cultivation processes using shake flasks. Regarding the easy‐to‐express antibody, final production titers of negative control cells reached 915 ± 68 mg/L, while miR‐557 expressing cell clones exhibited significantly elevated mAb titers of 1168 ± 78 mg/L (Fig. 5C). Positive effects of miR‐557 on mAb productivity were much more pronounced in cells expressing the DTE antibody candidate. Here, constitutive expression of miR‐557 led to titers of 656 ± 44 mg/L compared to 235 ± 58 mg/L achieved with negative control cells (Fig. 5C).
Finally, using product titers of cells cultivated in 6‐well format during clone development, we calculated a theoretical probability to identify high‐producing cell clones. Cell clones exhibiting ≥1.5‐fold increased mAb titers compared to the mean productivity of control cultures expressing a non‐targeting miRNA were ascertained as high‐producers. We determined that regardless of the antibody candidate, presence of miR‐557 considerably increased the probability to identify hyper‐productive CHO cell clones (Fig. 6). Stable expression of miR‐557 in CHO cells increased the probability of generating a high‐producing cell clone by 27.3% for the easy‐to‐express mAb and by 40.3% for the DTE mAb. Taken together these results indicate that expression of miR‐557 in CHO cells may significantly enhance success in CHO CLD through overall improvement of recombinant protein production and increased probability in identifying high‐producing clones.
Figure 6.
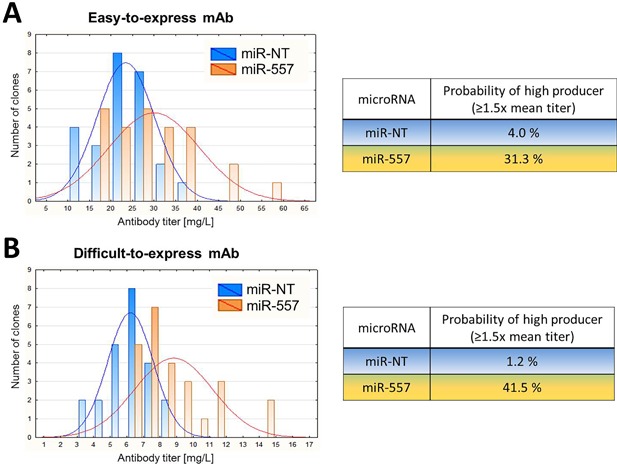
Calculated probability to identify a high‐producing cell clone. Determination of a theoretical probability to identify a high‐producing cell clone from cells expressing miR‐557 (orange) or control cells expressing a non‐targeting miRNA (blue). A high‐producing cell clone was defined to exhibit at least a 1.5‐fold increased volumetric mAb titer compared to the mean productivity of control cells. Histograms illustrate the distribution of volumetric antibody productivity for an (A) easy‐to‐express and (B) difficult‐to‐express antibody. Volumetric mAb productivity of cells present in the late phase of clone development (6‐well plate) was used for calculation and statistical analysis.
Fed‐Batch Performance of CHO Cell Lines Is Substantially Enhanced in Presence of miR‐557
Stable expression of miR‐557 could be demonstrated to significantly improve CLD processes using CHO cells. Consequently, we investigated process performance of the TOP3 selected clones of each CLD campaign (easy and DTE mAb) and compared established production clones expressing miR‐557 with control cells in a fully controlled fed‐batch cultivation process. Toward this end, we used an advanced micro bioreactor (ambr™15) system since this instrument allowed assessment of bioprocess performance of a large number of cell clones in parallel. Cell clones expressing miR‐557 and the easy‐to‐express mAb demonstrated a mean improvement in volumetric productivity of 19% in a 10‐day fed‐batch process compared to cell clones expressing a non‐targeting miRNA (Fig. 7A). Furthermore, detailed analysis of the best clones for each miRNA revealed that in presence of miR‐557 the TOP clone not only showed enhanced mAb titer (Fig. 7B), but also improved cell growth (Fig. 7C and D) and prolonged high viabilities compared to the best miR‐NT expressing control cell clone (Fig. 7E). Lactate formation was found to be highly comparable throughout the entire cultivation process indicating that miR‐557 expressing cells produced less lactate on a per cell basis compared to miR‐NT expressing cells (Fig. 7F). This suggests that miRNAs such as miR‐557 are even capable of further enhancing the production of an easy‐to‐express antibody. However, positive effects on cellular antibody productivity of miR‐557 on CHO cells were even more pronounced in cells expressing a DTE antibody candidate. Here, expression of miR‐557 resulted in final mAb titers of 962 ± 22 mg/L which corresponded to an increase in volumetric mAb productivity of approximately twofold compared to control cells which only reached 558 ± 5 mg/L (Fig. 8A). Notably, all of our previously performed clone development efforts for this particular DTE mAb candidate never surpassed volumetric productivities of 500 mg/L (data not shown). This suggests that miR‐NT expressing CHO host cells used in this study demonstrated highly comparable performances compared to unmodified CHO host cells. Similar to results obtained with the easy‐to‐express antibody comparison of the best cell clones illustrate that miR‐557 expressing clones exhibit improved overall process performance and demonstrated higher mAb titer, improved growth values, prolonged viability, and decreased cell‐specific lactate production (Fig. 8B–F).
Figure 7.
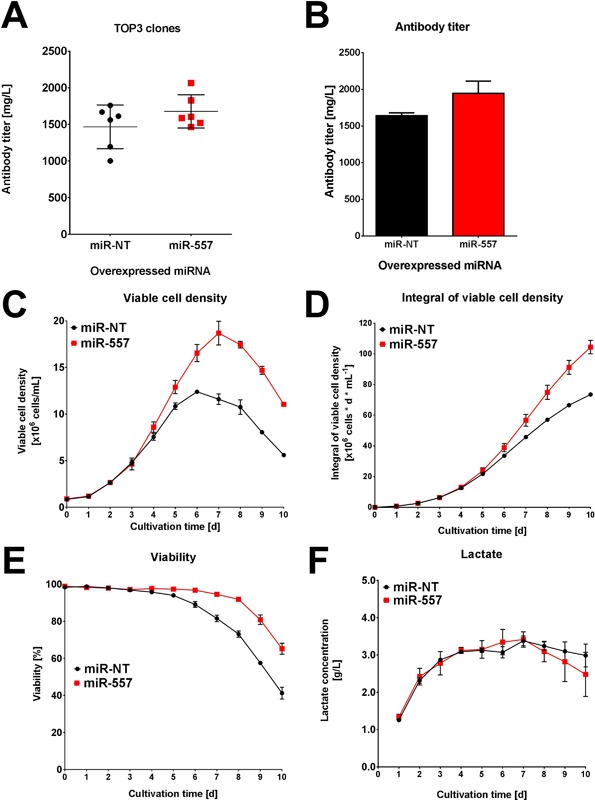
Fed‐batch performance of engineered CHO host cells expressing an easy‐to‐express mAb. The three TOP clones derived from either miR‐557 expressing (red) or negative control host cell lines (black) were cultivated for 10 days in fully controlled fed‐batch cultivation using an ambr™15 bioreactor system. Each clone was cultivated in two biological replicates. (A) Volumetric antibody productivity of the three TOP clones regarding an easy‐to‐express mAb. Each data point represents one bioreactor run. (B–F) Fed‐batch cultivation process data of the respective TOP clone derived from either miR‐557 (red) or miR‐NT (black) engineered host cells (n = 2). Process data shown include volumetric productivity, viable cell density, integral of viable cell density (biomass), viability, and lactate concentration.
Figure 8.
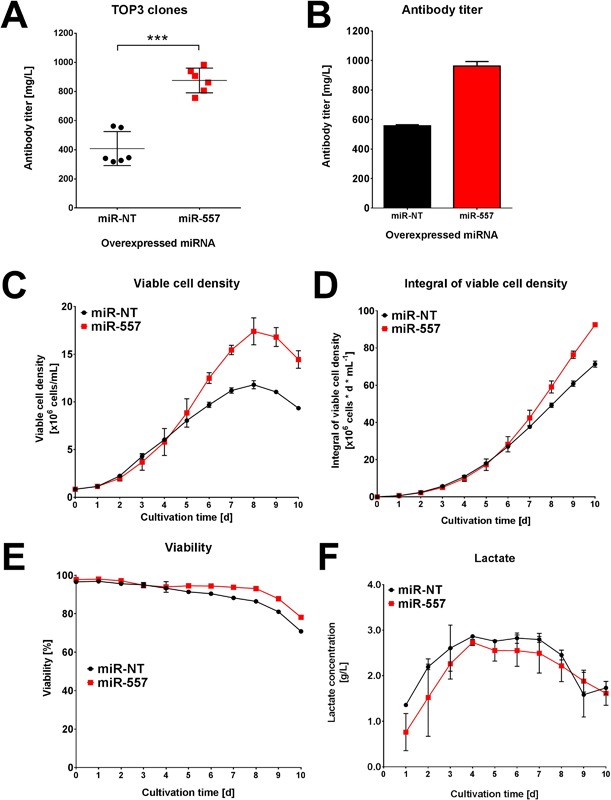
Fed‐batch performance of engineered CHO host cells expressing a difficult‐to‐express mAb. The three TOP clones derived from either miR‐557 expressing (red) or negative control host cell lines (black) were cultivated for 10 days in fully controlled fed‐batch cultivation using an ambr™15 bioreactor system. Each clone was cultivated in two biological replicates. (A) Volumetric antibody productivity of the three TOP clones regarding a difficult‐to‐express mAb. Each data point represents results of one bioreactor run. (B–F) Fed‐batch cultivation process data of the respective TOP clone derived from either miR‐557 (red) or miR‐NT (black) engineered host cells (n = 2). Process data shown include volumetric productivity, viable cell density, integral of viable cell density (biomass), viability, and lactate concentration.
Expression of miR‐557 in CHO Cells Has No Negative Impact on Product Quality Attributes
Changes in product quality can have substantial influences on safety and efficacy of a therapeutic protein. Since, miRNAs regulate the expression of hundreds of different genes concomitantly in a cell, it was crucial to investigate the potential impact of miR‐557 expression on product quality. Toward this end, we analyzed both recombinant antibody candidates with respect to N‐linked glycosylation, mAb aggregation/fragmentation as well as purity, and integrity of the antibody chains. We collected culture supernatant of controlled small‐scale fed‐batch cultivations at day of harvest and analyzed the purified antibodies for changes in product quality. Expression of miR‐557 did not negatively affect aggregation propensity of both mAb candidates compared to miR‐NT expressing control cell lines (Fig. 9A). In addition, we did not observe any impacts on mAb fragmentation if miR‐557 was present (Fig. 9A). In line with this, also the purity and integrity of the produced antibodies did not vary between cell clones expressing miR‐557 or control miRNA, respectively (Fig. 9B). Finally, also N‐glycosylation pattern were highly comparable between miR‐557 expressing and control cells (Fig. 9C). Furthermore, overall N‐glycan distribution was very similar between both antibody candidates. In order to validate these investigations on product quality, we additionally analyzed antibodies produced in previously performed uncontrolled fed‐batch cultivations (shake flasks) which could nicely confirm results obtained in ambr®15 cultivations for both antibody candidates (Supplementary Fig. S1). These data suggest that expression of miR‐557 in CHO cells does not negatively affect product quality but substantially increases success in cell line and process development due to improved growth and productivity.
Figure 9.
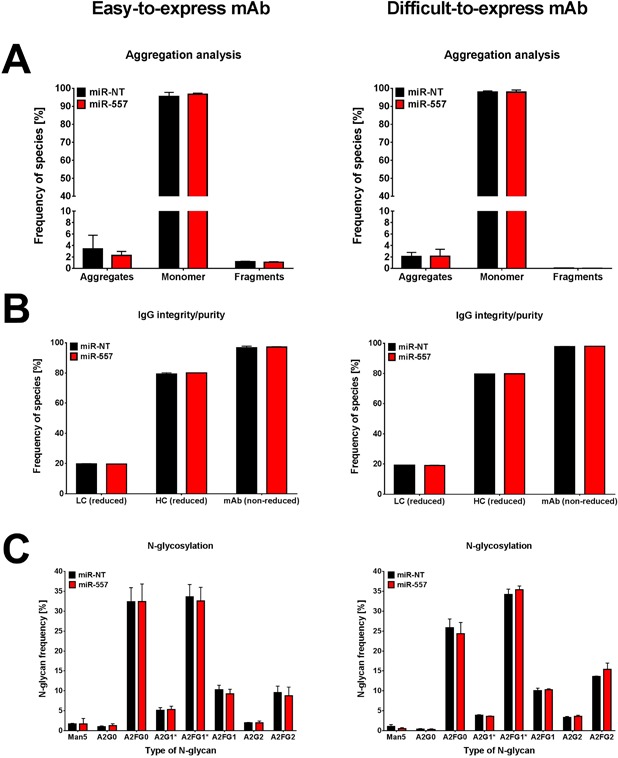
Analysis of product quality attributes of monoclonal antibodies produced by miRNA engineered CHO host cells. Data are presented for both easy (left panel) as well as difficult‐to‐express mAb (right panel). (A) Analysis and quantification of aggregates, fragments as well as monomers of mAbs produced by miR‐557 (red) compared to miR‐NT expressing control cells (black). Analysis was performed by Ultra‐Performance Size‐Exclusion Chromatography (UP‐SEC) using a priori Protein A purified antibodies. (B) Separation and quantification of antibody light (LC) and heavy chain (HC) as well as assembled mAb was achieved using capillary electrophoresis (CE) at either reducing or non‐reducing conditions to examine purity and integrity of produced antibodies. (C) Analysis and quantification of N‐linked glycan structures present on mAbs. Separation of N‐glycans was performed using CE using a priori Protein A purified antibodies. Samples for product quality analyses were taken from culture supernatant of ambr®15 fed‐batch cultivation runs at day of harvest. Data are presented as mean ± SD of three different cell clones (TOP 3 clones).
Discussion
Transient Transfection of miR‐557 Mimics Increases Protein Production in CHO Cells Independent of Molecule Type and Expression Level
Transient transfection of miRNA mimics represents a powerful method to elucidate miRNA function in mammalian cells and has recently been introduced to study phenotypic effects of specific miRNAs on CHO cells (Barron et al., 2011a; Fischer et al., 2014, 2015d; Stiefel et al., 2016; Strotbek et al., 2013). A previously conducted high‐throughput miRNA mimics screening has identified the primate specific microRNA miR‐557 to induce a hyperproductive phenotype in an antibody producing CHO DG44 cell line. In order to investigate if miR‐557 might also positively affect protein production of other CHO cell lines, we selected a subset of seven different recombinant CHO production cell lines and transiently transfected each cell line either with miR‐557 or non‐targeting control mimics. The seven CHO cell lines were selected such that they cover various CHO host cell backgrounds (CHO, DG44, and CHO‐K1), and selection systems (DHFR, GS), alternate molecule types (standard IgGs, bispecific IgGs, fusion proteins) of different complexity as well as various expression levels (high, medium, low producer). Noteworthy, we carefully optimized transfection conditions for each cell line in advance to ensure optimal transfection conditions since transient transfection of nucleic acids is heavily dependent on the cell type as well as the culture medium (Bertschinger et al., 2006; Eberhardy et al., 2009; Fischer et al., 2013; Geisse and Fux, 2009). Interestingly, transient introduction of miR‐557 increased recombinant protein production in all seven cell lines tested indicating that this miRNA might represent a universal productivity enhancing genetic element in CHO cells. Consequently, we sought to determine the effect of stable miR‐557 expression in recombinant CHO cells and its influence on an industrial CLD process.
Previous miRNA profiling studies suggested that miR‐557 is not expressed in CHO cells (Diendorfer et al., 2015; Hackl et al., 2011; Hammond et al., 2012; Hernandez Bort et al., 2012; Lin et al., 2011; Stiefel et al., 2016), which we could confirm via qRT‐PCR based quantification of mature miR‐557 in our stable negative control miRNA expressing CHO host cells. According to the latest release of the miRBase repository (Kozomara and Griffiths‐Jones, 2014), miR‐557 has yet only found to be expressed in four species comprising human, chimpanzee, rhesus macaque, and orangutan. Interestingly, although miR‐557 is not expressed in CHO cells it seems to have a substantial phenotypic influence on this cell type. It might be that during evolution rodents could have lost their genomic microRNA‐557 locus, but target genes of miR‐557 may still be present. Rescue experiments where miR‐557 is ectopically introduced back into CHO cells might restore an ancient functionality which may finally lead to increased protein biosynthesis. There are no reports available yet describing a specific function of miR‐557 in any tissue. Only very few reports have mentioned miR‐557 to be potentially linked to the development of cancer (Chen et al., 2013; Jiang et al., 2014; Katayama et al., 2012), but no validated target genes could be revealed so far for this miRNA.
In order to shed initial light on potentially regulated target genes and pathways by miR‐557, we have performed a computational target prediction analysis for miR‐557 using four different miRNA target prediction algorithms (miRWalk, miRDB, miRanda, TargetScan). Potential target genes, which were predicted in silico by each of the four algorithms (296 genes; Supplementary Table S1), were subjected to a functional classification analysis using the publically available PANTHER tool (Mi et al., 2017). The TOP5 regulated cellular pathways as well as the respective genes involved in these pathways are listed in Supplementary Table S2. Interestingly, mitogen‐activated protein kinase 9 (MAPK9) was found to be involved in four of the TOP5 potentially regulated pathways. MAPK9 belongs to stress‐activated protein kinases that play a crucial role in the cellular response to environmental stress (Chen et al., 1996). Among the many functions of MAPK9 in mammalian cells is also the inactivation of the transcription factor TIF‐IA, which results in a down‐regulation of ribosomal RNA (rRNA) synthesis (Mayer et al., 2005). As a potential target gene of miR‐557, knock‐down of MAPK9 by miR‐557 might inhibit inactivation of TIF‐IA resulting in an elevated generation of rRNA in CHO cells. Santoro et al. (2009) demonstrated that up‐regulation of rRNAs enhanced ribosome formation leading to increased recombinant protein production in CHO cells.
Furthermore, the regulation of the oxidative stress response was amongst the TOP5 regulated pathways. Elevated antibody production in CHO cells during fed‐batch cultivation requires increased oxidative metabolism (Templeton et al., 2013). Engineering of the oxidative metabolism using miRNAs has recently been described by Kelly et al. (2015), who decreased miR‐23 expression in CHO cells in order to enhance recombinant protein production. The authors demonstrated that miR‐23 is involved in the regulation of the oxidative metabolism and knock‐down of miR‐23 led to an increased mitochondrial activity presumably leading to a pro‐productive phenotype (Kelly et al., 2015). Due to the very limited information about miR‐557 function, it is rather speculative to hypothesize potential molecular mechanisms leading to the observed pro‐productive phenotype in CHO cells. We could demonstrate that the presence of miR‐557 increased production titers of both tested mAbs, but enhancing effects were found to be more pronounced with regard to the DTE proteins. The reason for such behavior might be that miR‐557 induces transcriptomic changes favoring the production of challenging molecules. However, it may also be conceivable that for DTE proteins the room for improvement simply is larger since these molecules are less close to the biological limit of cellular productivity compared to easy‐to‐express mAbs. Nonetheless, due to its obvious productivity enhancing effects in CHO cells miR‐557 remains an attractive target miRNA to be further investigated in more detail.
Stable Expression of miR‐557 Improves Bioprocess Performance Without Negatively Influencing Product Quality
Previous reports describing the functionality of specific miRNAs on CHO cell phenotypes via stable miRNA overexpression generally introduced these miRNAs into cell lines already producing a recombinant protein (Fischer et al., 2014, 2015d; Jadhav et al., 2014; Loh et al., 2014; Strotbek et al., 2013). Although these studies provided very interesting insights into the impact of miRNAs on protein production or cell growth in CHO cells, there is no evidence yet that modification of CHO host cells using miRNAs can improve overall cell line and process development. To close this gap, we established genetically modified CHO host cells constitutively expressing miR‐557 and compared them to control host cells expressing a non‐targeting miRNA. The generation of miRNA overexpressing CHO cells requires an antibiotic selection step to allow for stable genomic integration of the miRNA expression cassette. In this context, it was crucial to establish negative control host cells that have undergone a similar selection procedure to ensure accurate comparison during CLD processes. Comparison of newly developed negative control (miR‐NT) host cells with unmodified parental CHO host cells revealed that selected TOP clones exhibited highly comparable bioprocess performance in controlled fed‐batch cultivations (data not shown). This suggests that negative control miRNA expressing host cells show a competitive performance which further underscores the superiority of miR‐557 expressing CHO host cells.
For proof‐of‐concept studies, we used a commercially available vector system where a given miRNA expression cassette is located within the 3′UTR of a GFP gene. This vector has previously been reported to be functional for generating stable miRNA overexpressing CHO cells (Jadhav et al., 2012, 2014; Strotbek et al., 2013). The main advantage of such a system is that transfected cells can be analyzed and sorted for GFP‐positive cells via flow cytometry and miRNA expression can be easily monitored. Though reporter genes such as GFP are valuable tools for functional investigations on miRNA function they may possess disadvantages, when it comes to application in industrial manufacturing cell lines. Generation of industrial CHO host cell lines used for the commercial production of therapeutic proteins rather requires a vector system lacking reporter genes to avoid translational burden and to reduce host cell protein (HCP) complexity which is important for downstream purification processes. Consequently, future expression vectors for stable miRNA expression in CHO manufacturing host cell lines might be designed in a way that miRNAs are either co‐expressed with non‐reporter proteins or independently expressed from RNA polymerase III (e.g., U6 or H1) promoters similar to short‐hairpin RNA (shRNA) expression constructs (Zhou et al., 2008). This also includes vector set‐ups that facilitate co‐expression of multiple miRNAs since combined expression of diverse miRNAs might lead to regulation of different cellular pathways to induce a superior cell phenotype (Fischer et al., 2014).
Although current CLD platforms are able to generate production clones exceeding productivities of >3 g/L for a large number of IgG molecules, there is a substantial number of these classical IgGs that behave different and show considerably low productivity in CHO cells (Pybus et al., 2014). Furthermore, in recent years the number of complex but effective biologicals such as multi‐specific antibodies or fusion proteins has increased dramatically (Spiess et al., 2015). However, although these effective therapeutic proteins often promise a superior clinical benefit to address unmet medical needs, they are also frequently reported to be DTE for current CHO expression systems. Consequently, CHO production host cell lines have to be prepared for these future challenges in order to meet requirements for economic manufacturing processes. In the present study, we aimed at developing a novel genetically modified CHO host cell system suitable for enhanced production of DTE proteins. Toward this end, we investigated cell culture performance of miR‐557 expressing CHO cells using an easy‐as well as a DTE antibody candidate. Interestingly, comparison of final product titers of uncontrolled fed‐batch cultivations revealed that stable clones co‐expressing miR‐NT and the DTE mAb candidate could not outperform stable cell pools in terms of productivity (Figs. 4C and 5C), which is usually one of the main goals of SSC. This indicated that the selected DTE antibody candidate can really be ascertained as challenging molecule, since clones may only survive if they exhibit sufficient expression of the selection marker gene but only limited amount the antibody. In contrast, co‐expression of miR‐557 and the DTE antibody led to increased mAb titers and single clones were found to achieve higher product titers compared to stable cell pools. In controlled fed‐batch processes, production clones derived from miR‐557 expressing host cells actually achieved final product titers of approximately 1 g/L after 10 days in regard of the DTE mAb, which means a twofold increase compared to our previously performed CLD campaigns for this particular antibody. Thus, our findings suggest that presence of miR‐557 can substantially increase success in industrial CLD. Noteworthy, expression of miR‐557 had no impact on product quality attributes such as N‐glycosylation, aggregation, fragmentation, purity, and integrity of both antibodies used in this study. Further analyses will be necessary before implementing these novel host cell lines into routine drug development processes. For instance, as miRNAs generally fine‐tune the expression of hundreds of different genes in a cell, overexpression of miRNAs might also change HCP composition. In addition, as some miRNAs have been demonstrated to induce carcinogenesis and malignant transformation (Frixa et al., 2015), miRNA quantification studies in purified antibody solution need to be performed to prove depletion of the miRNA in order to prevent potential transmission to the drug product. However, since RNAs are extremely unstable it is expected that miRNAs released into culture supernatant will immediately be degraded by nucleases. Taken together our transient as well as comprehensive stable expression data indicate that miR‐557 is suitable to increase overall recombinant protein production in CHO cells which is especially beneficial for challenging proteins. Thus, we provide several lines of evidence that host cell engineering using the novel miRNA technology in industrial manufacturing CHO cell lines represents an attractive tool to overcome challenges of DTE therapeutic proteins.
S. Fischer, J. Fieder, S. Barden, M. Gamer, I. Gorr, H. Bradl, and P. Schulz are employees of Boehringer Ingelheim Pharma which develops and sells pharmaceuticals. Boehringer Ingelheim funded all of this research. The authors are grateful to Simone Heinrich, Stefanie Gessler, Donja Spiess, Thomas Krieg, and Stefanie Scheffold for cell culture work. Furthermore, we thank Renate Straub and Karin Rechtsteiner for ambr™15 cultivation support, Anja Ebe, Alexandra Ziegler, and Sophie Aftring for FACS sorting and Juergen Rapp, Jens Poesl, and Melanie Luz for their help in protein quantification. We are also grateful to Dr. Stephan Barden and Birgit Sonntag for mAb purification and product quality analytics.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure S1.
Supporting Table S1.
Supporting Table S2.
Supporting Information S1.
Conflict of interest: None.
References
- Barron N, Kumar N, Sanchez N, Doolan P, Clarke C, Meleady P, O'Sullivan F, Clynes M. 2011a. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR‐7. J Biotechnol 151(2):204–211. [DOI] [PubMed] [Google Scholar]
- Barron N, Sanchez N, Kelly P, Clynes M. 2011b. MicroRNAs: Tiny targets for engineering CHO cell phenotypes? Biotechnol Lett 33(1):11–21. [DOI] [PubMed] [Google Scholar]
- Bentley KJ, Gewert R, Harris WJ. 1998. Differential efficiency of expression of humanized antibodies in transient transfected mammalian cells. Hybridoma 17(6):559–567. [DOI] [PubMed] [Google Scholar]
- Bertschinger M, Backliwal G, Schertenleib A, Jordan M, Hacker DL, Wurm FM. 2006. Disassembly of polyethylenimine‐DNA particles in vitro: Implications for polyethylenimine‐mediated DNA delivery. J Control Release 116(1):96–104. [DOI] [PubMed] [Google Scholar]
- Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M, Teal CB, Brem RF, Stojadinovic A, Grinkemeyer M, McCaffrey TA, Man YG, Fu SW. 2013. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS ONE 8(1):e54213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Wang X, Templeton D, Davis RJ, Tan TH. 1996. The role of c‐Jun N‐terminal kinase (JNK) in apoptosis induced by ultraviolet C and gamma radiation. Duration of JNK activation may determine cell death and proliferation. J Biol Chem 271(50):31929–31936. [DOI] [PubMed] [Google Scholar]
- Diendorfer AB, Hackl M, Klanert G, Jadhav V, Reithofer M, Stiefel F, Hesse F, Grillari J, Borth N. 2015. Annotation of additional evolutionary conserved microRNAs in CHO cells from updated genomic data. Biotechnol Bioeng 112(7):1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druz A, Son YJ, Betenbaugh M, Shiloach J. 2013. Stable inhibition of mmu‐miR‐466h‐5p improves apoptosis resistance and protein production in CHO cells. Metab Eng 16:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Radzniak L, Liu Z. 2009. Iron (III) citrate inhibits polyethylenimine‐mediated transient transfection of Chinese hamster ovary cells in serum‐free medium. Cytotechnology 60:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerling VV, Fischer S, Stiefel F, Holzmann K, Handrick R, Hesse F, Horer M, Kochanek S, Otte K. 2015. Temperature‐sensitive miR‐483 is a conserved regulator of recombinant protein and viral vector production in mammalian cells. Biotechnol Bioeng 113(4):830–841. [DOI] [PubMed] [Google Scholar]
- Fischer S, Buck T, Wagner A, Ehrhart C, Giancaterino J, Mang S, Schad M, Mathias S, Aschrafi A, Handrick R, Otte K. 2014. A functional high‐content miRNA screen identifies miR‐30 family to boost recombinant protein production in CHO cells. Biotechnol J 9(10):1279–1292. [DOI] [PubMed] [Google Scholar]
- Fischer S, Handrick R, Aschrafi A, Otte K. 2015a. Unveiling the principle of microRNA‐mediated redundancy in cellular pathway regulation. RNA Biol 12(3):238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S, Handrick R, Otte K. 2015b. The art of CHO cell engineering: A comprehensive retrospect and future perspectives. Biotechnol Adv 33(8):1878–1896. [DOI] [PubMed] [Google Scholar]
- Fischer S, Mathias S, Schaz S, Emmerling VV, Buck T, Kleemann M, Hackl M, Grillari J, Aschrafi A, Handrick R, Otte K. 2015c. Enhanced protein production by microRNA‐30 family in CHO cells is mediated by the modulation of the ubiquitin pathway. J Biotechnol 212:32–43. [DOI] [PubMed] [Google Scholar]
- Fischer S, Paul AJ, Wagner A, Mathias S, Geiss M, Schandock F, Domnowski M, Zimmermann J, Handrick R, Hesse F, Otte K. 2015d. MiR‐2861 as novel HDAC5 inhibitor in CHO cells enhances productivity while maintaining product quality. Biotechnol Bioeng 112(10):2142–2153. [DOI] [PubMed] [Google Scholar]
- Fischer S, Wagner A, Kos A, Aschrafi A, Handrick R, Hannemann J, Otte K. 2013. Breaking limitations of complex culture media: Functional non‐viral miRNA delivery into pharmaceutical production cell lines. J Biotechnol 168(4):589–600. [DOI] [PubMed] [Google Scholar]
- Frixa T, Donzelli S, Blandino G. 2015. Oncogenic MicroRNAs: Key players in malignant transformation. Cancers (Basel) 7(4):2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisse S, Fux C. 2009. Recombinant protein production by transient gene transfer into Mammalian cells. Methods Enzymol 463:223–238. [DOI] [PubMed] [Google Scholar]
- Hackl M, Borth N, Grillari J. 2012a. MiRNAs‐pathway engineering of CHO cell factories that avoids translational burdening. Trends Biotechnol 30(8):405–406. [DOI] [PubMed] [Google Scholar]
- Hackl M, Jadhav V, Jakobi T, Rupp O, Brinkrolf K, Goesmann A, Puhler A, Noll T, Borth N, Grillari J. 2012b. Computational identification of microRNA gene loci and precursor microRNA sequences in CHO cell lines. J Biotechnol 158(3):151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl M, Jakobi T, Blom J, Doppmeier D, Brinkrolf K, Szczepanowski R, Bernhart SH, Honer Zu Siederdissen C, Bort JA, Wieser M, Kunert R, Jeffs S, Hofacker IL, Goesmann A, Pühler A, Borth N, Grillari J. 2011. Next‐generation sequencing of the Chinese hamster ovary microRNA transcriptome: Identification, annotation and profiling of microRNAs as targets for cellular engineering. J Biotechnol 153(1–2):62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S, Swanberg JC, Polson SW, Lee KH. 2012. Profiling conserved microRNA expression in recombinant CHO cell lines using Illumina sequencing. Biotechnol Bioeng 109(6):1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Bort JA, Hackl M, Hoflmayer H, Jadhav V, Harreither E, Kumar N, Ernst W, Grillari J, Borth N. 2012. Dynamic mRNA and miRNA profiling of CHO‐K1 suspension cell cultures. Biotechnol J 7(4):500–515. [DOI] [PubMed] [Google Scholar]
- Jadhav V, Hackl M, Bort JA, Wieser M, Harreither E, Kunert R, Borth N, Grillari J. 2012. A screening method to assess biological effects of microRNA overexpression in Chinese hamster ovary cells. Biotechnol Bioeng 109(6):1376–1385. [DOI] [PubMed] [Google Scholar]
- Jadhav V, Hackl M, Druz A, Shridhar S, Chung CY, Heffner KM, Kreil DP, Betenbaugh M, Shiloach J, Barron N, Grillari J, Borth N. 2013. CHO microRNA engineering is growing up: Recent successes and future challenges. Biotechnol Adv 31(8):1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav V, Hackl M, Klanert G, Hernandez Bort JA, Kunert R, Grillari J, Borth N. 2014. Stable overexpression of miR‐17 enhances recombinant protein production of CHO cells. J Biotechnol 175:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HB, Yang TJ, Lu P, Ma YJ. 2014. Gene expression profiling of gastric cancer. Eur Rev Med Pharmacol Sci 18(15):2109–2115. [PubMed] [Google Scholar]
- Johari YB, Estes SD, Alves CS, Sinacore MS, James DC. 2015. Integrated cell and process engineering for improved transient production of a “difficult‐to‐express” fusion protein by CHO cells. Biotechnol Bioeng 112(12):2527–2542. [DOI] [PubMed] [Google Scholar]
- Jost C, Pluckthun A. 2014. Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr Opin Struct Biol 27:102–112. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Maeda M, Miyaguchi K, Nemoto S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S, Tanaka H. 2012. Identification of pathogenesis‐related microRNAs in hepatocellular carcinoma by expression profiling. Oncol Lett 4(4):817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PS, Breen L, Gallagher C, Kelly S, Henry M, Lao NT, Meleady P, O'Gorman D, Clynes M, Barron N. 2015. Re‐programming CHO cell metabolism using miR‐23 tips the balance towards a highly productive phenotype. Biotechnol J 10(7):1029–1040. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths‐Jones S. 2014. MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue):D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Davis A, Bahr S, Borgschulte T, Achtien K, Kayser K. 2011. Profiling highly conserved microrna expression in recombinant IgG‐producing and parental Chinese hamster ovary cells. Biotechnol Prog 27(4):1163–1171. [DOI] [PubMed] [Google Scholar]
- Loh WP, Loo B, Zhou L, Zhang P, Lee DY, Yang Y, Lam KP. 2014. Overexpression of microRNAs enhances recombinant protein production in Chinese hamster ovary cells. Biotechnol J 9(9):1140–1151. [DOI] [PubMed] [Google Scholar]
- Mason M, Sweeney B, Cain K, Stephens P, Sharfstein ST. 2012. Identifying bottlenecks in transient and stable production of recombinant monoclonal‐antibody sequence variants in Chinese hamster ovary cells. Biotechnol Prog 28(3):846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Bierhoff H, Grummt I. 2005. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF‐IA and down‐regulates rRNA synthesis. Genes Dev 19(8):933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Katinger H, Grillari J. 2008. MicroRNAs as targets for engineering of CHO cell factories. Trends Biotechnol 26(7):359–365. [DOI] [PubMed] [Google Scholar]
- Pybus LP, Dean G, West NR, Smith A, Daramola O, Field R, Wilkinson SJ, James DC. 2014. Model‐directed engineering of “difficult‐to‐express” monoclonal antibody production by Chinese hamster ovary cells. Biotechnol Bioeng 111(2):372–385. [DOI] [PubMed] [Google Scholar]
- Sanchez N, Kelly P, Gallagher C, Lao NT, Clarke C, Clynes M, Barron N. 2014. CHO cell culture longevity and recombinant protein yield are enhanced by depletion of miR‐7 activity via sponge decoy vectors. Biotechnol J 9(3):396–404. [DOI] [PubMed] [Google Scholar]
- Santoro R, Lienemann P, Fussenegger M. 2009. Epigenetic engineering of ribosomal RNA genes enhances protein production. PLoS ONE 4(8):e6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D, Schulz P, Litzenburger T, Spitz J, Hoerer S, Blech M, Enenkel B, Studts JM, Garidel P, Karow AR. 2015. Boosting antibody developability through rational sequence optimization. MAbs 7(3):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Zhai Q, Carter PJ. 2015. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol 67(2 Pt A):95–106. [DOI] [PubMed] [Google Scholar]
- Stiefel F, Fischer S, Hackl M, Handrick R, Hesse F, Borth N, Otte K, Grillari J. 2015. Noncoding RNAs, post‐transcriptional RNA operons and Chinese hamster ovary cells. Pharm Bioprocess 3(3):227–247. [Google Scholar]
- Stiefel F, Fischer S, Sczyrba A, Otte K, Hesse F. 2016. MiRNA profiling of high, low and non‐producing CHO cells during biphasic fed‐batch cultivation reveals process relevant targets for host cell engineering. J Biotechnol 225:31–43. [DOI] [PubMed] [Google Scholar]
- Strotbek M, Florin L, Koenitzer J, Tolstrup A, Kaufmann H, Hausser A, Olayioye MA. 2013. Stable microRNA expression enhances therapeutic antibody productivity of Chinese hamster ovary cells. Metab Eng 20:157–166. [DOI] [PubMed] [Google Scholar]
- Templeton N, Dean J, Reddy P, Young JD. 2013. Peak antibody production is associated with increased oxidative metabolism in an industrially relevant fed‐batch CHO cell culture. Biotechnol Bioeng 110(7):2013–2024. [DOI] [PubMed] [Google Scholar]
- Zhou H, Huang C, Xia XG. 2008. A tightly regulated Pol III promoter for synthesis of miRNA genes in tandem. Biochim Biophys Acta 1779(11):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure S1.
Supporting Table S1.
Supporting Table S2.
Supporting Information S1.


