Summary
Plasmodium falciparum and Toxoplasma gondii are obligate intracellular parasites that belong to the phylum of Apicomplexa and cause major human diseases. Their access to an intracellular lifestyle is reliant on the coordinated release of proteins from the specialized apical organelles called micronemes and rhoptries. A specific phosphatidic acid effector, the acylated pleckstrin homology domain-containing protein (APH) plays a central role in microneme exocytosis and thus is essential for motility, cell entry, and egress. TgAPH is acylated on the surface of the micronemes and recruited to phosphatidic acid (PA)-enriched membranes. Here, we dissect the atomic details of APH PA-sensing hub and its functional interaction with phospholipid membranes. We unravel the key determinant of PA recognition for the first time and show that APH inserts into and clusters multiple phosphate head-groups at the bilayer binding surface.
Keywords: pleckstrin homology domain, phosphatidic acid, NMR, Toxoplasma gondii, Plasmodium falciparum, microneme, exocytosis, gliding motility, invasion, egress
Graphical Abstract
Highlights
-
•
Solution structures of APH from Toxoplasma gondii and Plasmodium falciparum
-
•
APH represents a new class of PH domain
-
•
APH phosphatidic acid binding site encompasses canonical and atypical sites
-
•
APH inserts into the bilayer and clusters multiple phosphate head-groups
High-resolution structures of a phosphatidic acid effector protein (APH) from both Plasmodium falciparum and Toxoplasma gondii parasites and its interaction with the lipid mediator unravel a new class of PH domain that initiates microneme-plasma membrane fusion by inserting and clustering phosphatidic acid within membranes.
Introduction
Apicomplexans form a group of parasitic protists that includes agents of major human diseases: Toxoplasma gondii responsible for toxoplasmosis (Robert-Gangneux and Darde, 2012) and Plasmodium species causing malaria (Bhatt et al., 2015). Among the five species of Plasmodium capable of infecting humans, Plasmodium falciparum is responsible for the most severe form of malaria, particularly in endemic areas of sub-Saharan Africa where ∼90% of global malaria-related deaths occur (Kim and Schneider, 2013). The intracellular lifestyle of apicomplexan parasites (Cowman and Crabb, 2006) is reliant on the actions of proteins released from specialized apical organelles, known as micronemes and rhoptries (Santos and Soldati-Favre, 2011). These apical secretory organelles critically contribute to gliding motility, invasion, and egress from infected cells. Notably, the micronemes secrete a perforin to egress from infected cells (Roiko and Carruthers, 2013). Several adhesins are also secreted to promote parasite attachment to the target cell and the formation of a moving junction between the cell and the actomyosin system, which drives the parasite inside the host-cell vacuole.
The signaling pathway leading to microneme secretion is complex and involves changes in potassium and cyclic nucleotide concentration levels that lead to an increase in parasite intracellular calcium levels (Brochet et al., 2014, Carruthers and Sibley, 1999, Moudy et al., 2001). Phosphoinositide-phospholipase C (PI-PLC) plays a central role in the signaling cascade leading to microneme secretion (Singh and Chitnis, 2012). PI-PLC catalyzes the conversion of PI(4,5)P2 into IP3 and diacylglycerol (DAG), which is further converted into phosphatidic acid (PA) via the activity of the specific diacylglycerol kinase 1 (DGK1) at the parasite plasma membrane, while IP3 is thought to stimulate a rise in cytosolic Ca2+ concentration (Bullen et al., 2016).
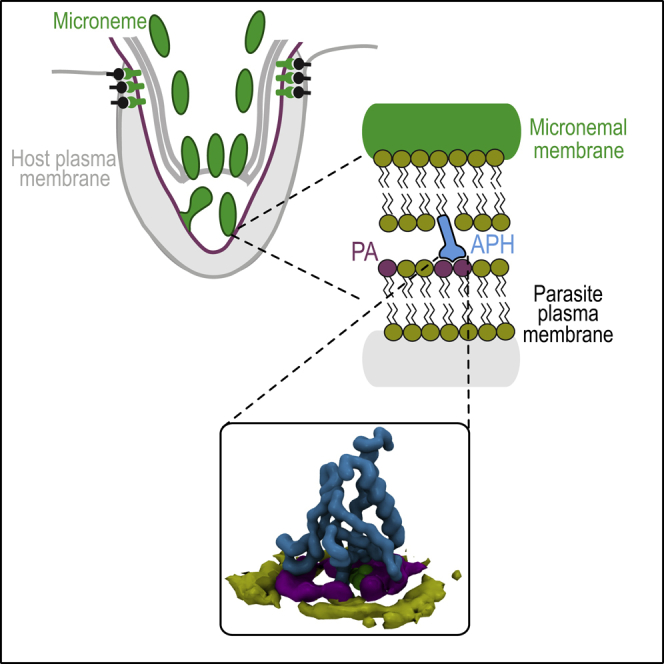
The discovery that changes in PA levels play an important role in controlling microneme exocytosis uncovered the identification of a novel PA sensor, conserved across the Apicomplexa and that possesses N-terminal lipid anchors and a predicted phospholipid binding domain (Bullen et al., 2016). This protein named acylated pleckstrin homology (PH) domain-containing protein (APH), is anchored at the surface of the micronemes via N-terminal myristoylation and palmitoylation. T. gondii and P. falciparum APH (TgAPH and PfAPH) bind selectively to PA both on PIP-strips and in liposome assays. It was proposed that this bipartite interaction tethers the microneme and plasma membranes together and participates in organelle fusion (Bullen et al., 2016) via the involvement of SNARE-like proteins (Figures 1A–1C), such as DOC2.1 (Farrell et al., 2012, Jean et al., 2014). The broader importance of PA in motility, invasion, and egress has been further highlighted by recent studies. The first is the discovery of the glideosome-associated connector protein (GAC), which links key microneme protein complexes to the actomyosin system and involves a specific interaction with PA via the C-terminal PH domain of GAC (Jacot et al., 2016). In a second example, new structural insight has been provided for the conserved protein CelTOS, which is a promising vaccine candidate (Pirahmadi et al., 2018). CelTOS is essential for parasite traversal of cells, and has been shown to bind to and disrupt PA-rich membranes (Jimah et al., 2016).
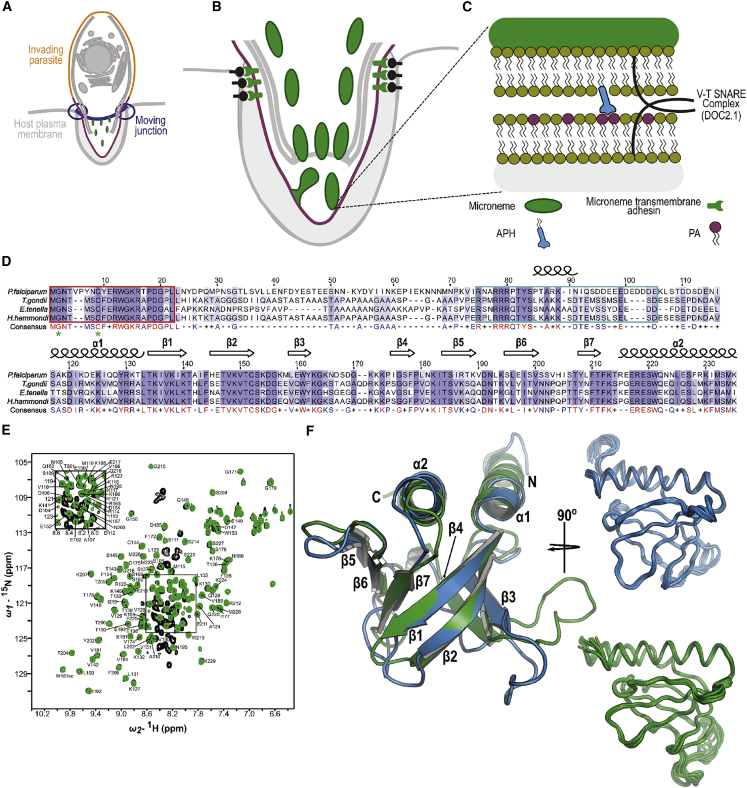
Figure 1.
Structural Characterization of PfAPH106-235 and TgAPH99-229 Reveals the C-Terminal Region of APH (APH) Adopts a Conserved Pleckstrin Homology Domain-like Fold
(A) Schematic representation of Toxoplasma parasite actively invading the host cell.
(B) Schematic representation of microneme fusion with parasite plasma membrane.
(C) Close-up of fusion event. APH embedded into the microneme surface via acylation interacts with PA accumulating on the inner leaflet of the plasma membrane, facilitating microneme exocytosis. PA is represented in purple, APH in light blue, micronemes and their contents in green.
(D) Multiple sequence alignment for APH full-length sequence from different apicomplexan species. Residues are colored in a purple spectrum according to the level of sequence identity, secondary structural elements are indicated, and numbering is shown for PfAPH. The consensus sequence is given below, invariant residues are colored red, highly conserved residues are colored blue, semi-conserved sequence identity is indicated by (+), and invariant residues are indicated by (−). The highly conserved 21 N-terminal residues required for targeting to the micronemal surface are highlighted by a red box, myristoylation (G2) and palmitoylation (C7) lipid anchor sites are indicated by green asterisks. A basic region within the linker sequence containing several conserved basic residues is highlighted by a cyan box.
(E) Overlay of 15N-labeled TgAPH22-229 (black) and TgAPH99-229 (green) 2D 1H-15N HSQC spectra. In comparison to TgAPH99-229, additional backbone amide peaks belonging to the linker region are visible in the TgAPH22-229 spectrum. There is expected to be an additional 77 backbone amide peaks in this linker region, but it is estimated only ∼54 peaks are visible. Residues that could be assigned in TgAPH99-229 are labeled, sc indicates resonances could be assigned to side chains (W161sc and W215sc).
(F) Left, aligned cartoon representations of the lowest-energy structures calculated for PfAPH106-235 (PDB: 6F24, blue) and TgAPH99-229 (PDB: 6F8E, green), the first 11 and 10 residues are omitted from PfAPH106-235 and TgAPH99-229 respectively as these were shown to be disordered. Right, ensembles of the ten lowest-energy structures calculated for PfAPH106-235 and TgAPH99-229.
The importance of this pathway suggests that it has potential as a target for therapeutic intervention, however the finer high-resolution mechanistic details are lacking. Furthermore, the mechanism by which the APH senses changes in the local PA concentration at the plasma membrane remains unclear. Here, we fill a gap of knowledge by elucidating the atomic resolution basis of the interaction between the apicomplexan PA effector, APH, and its lipid mediator PA in a variety of contexts, and by providing new atomic details into the initiation of microneme-plasma membrane fusion prior to release of the microneme contents.
Results
The Overall Atomic Structure of T. gondii and P. falciparum APH
Secondary structure predictions of APHs reveal a highly conserved mixed α/β domain at its C-terminus that is connected to the N-terminal acylation motifs via an extensive linker region (Drozdetskiy et al., 2015) (Figure 1D). Although the C-terminal half of APH possesses a structural organization consistent with a PH domain, predicted differences include an additional helical feature at its N-terminus and a shorter interstrand region between β1 and β2. Furthermore, a helical secondary structure is predicted within a charged portion of the APH linker (residues 85–91 in PfAPH) immediately upstream of the augmented PH domain. To provide further insight we compared 1D nuclear magnetic resonance (NMR) spectra for the full-length APH protein from T. gondii minus the acylation motif (TgAPH22-229) with a construct representing the structured PH domain within the C-terminal 99–229 residues (TgAPH99-229; Figure 1D). While the ordered PH domain (TgAPH99-229) is evident from the well-dispersed NMR resonance at high and low chemical shifts (Figure S1), the NMR spectrum of TgAPH22-229 is not consistent with the presence of an extensive disordered linker with over 20 alanine methyl resonances. It is therefore likely that many resonances for this region are broadened beyond detection due to conformational exchange on an intermediate timescale. Concurrently, only an estimated 54 out of an expected 77 backbone amide peaks belonging to the linker region, are visible in the heteronuclear single quantum coherence (HSQC) spectra for TgAPH22-229 when compared with TgAPH99-229 (Figure 1E). Absence of the additional linker region backbone amide resonances may be indicative of conformational exchange in this region. Interestingly, comparison between circular dichroism (CD) spectra for TgAPH22-229 and TgAPH99-229 indicate additional helical propensity within the N-terminal linker region (Figure S1). NMR spectra of recombinant produced PH domains from PfAPH (PfAPH106-235) and TgAPH (TgAPH99-229) were of excellent quality, so we determined the high-resolution solution structure of both proteins. These structures reveal an archetypal PH superfamily fold (Figure 1F; see Table 1 for structural statistics) (Lenoir et al., 2015), consisting of an open, seven-stranded β-barrel capped at one corner by a C-terminal α helix. Predicted differences to the classical PH fold, namely the β1-β2 loop and N-terminal α helix, are revealed by the experimental structures. The APH-specific N-terminal α helix packs against the C-terminus (Figure 1F), and the interstrand β1-β2 loop is much shorter and closed in APH compared with typical PH domains. These two features have potential functional implications.
Table 1.
NMR and Structural Validation Statistics for APH
| NMR-Derived Restraints | PfAPH PDB: 6F24 |
TgAPH PDB: 6F8E |
|---|---|---|
| Unambiguous Nuclear Overhauser Effect | ||
| Intra-residue | 945 | 946 |
| Sequential | 485 | 494 |
| Medium range (|i – j|) ≤ 4 | 215 | 279 |
| Long range (|i – j|) > 4 | 632 | 700 |
| Ambiguous NOE | 1,199 | 1,133 |
| Dihedral angle restraints (Φ/Ψ) | 230 | 234 |
| Structure Statistics | ||
| Violations | ||
| Number of dihedral angle violations >5° | 6.7 ± 1.0 | 2.2 ± 0.7 |
| Number of distance constraint violations >0.5Å | 0.30 ± 0.46 | 0.05 ± 0.22 |
| Deviation from idealized geometry | ||
| Bond length (Å) | 0.0040 ± 0.0001 | 0.0040 ± 0.0001 |
| Bond angle (o) | 0.57 ± 0.01 | 0.56 ± 0.01 |
| Average pairwise root-mean-square deviation r.m. SD for heavy atoms within secondary structures (Å) | 0.54 ± 0.07 | 0.52 ± 0.05 |
| Ramachandran plota | ||
| % In most favored positions | 91.0% ± 1.0% | 91.0% ± 1.0% |
| % In allowed regions | 98.0% ± 1.0% | 98.0% ± 1.0% |
| % In disallowed regions | 2.0% | 2.0% ± 1.0% |
Obtained from PDB NMR structure validation report.
The N-terminal helix of the PH domain extends to the linker region that connects to the microneme membrane anchor, and therefore this may play a role in signaling PA accumulation at the plasma membrane to the downstream membrane fusion machinery. Perhaps the most significant structural difference between APH and classic PH domains is the short β1-β2 loop, as this lies at the heart of the canonical phospholipid binding site and is usually longer and more open (Lenoir et al., 2015) (Figure S1). This striking difference indicates an altered mode of phospholipid binding for APH or a more restricted binding pocket to accommodate the small head group of PA. The similarity between PfAPH106-235 and TgAPH99-229 structures, and the high level of sequence conservation across the different apicomplexan species, suggest that this architecture applies to all apicomplexan APHs.
APH-PA Interface Overlaps with Canonical and Atypical Binding Surfaces
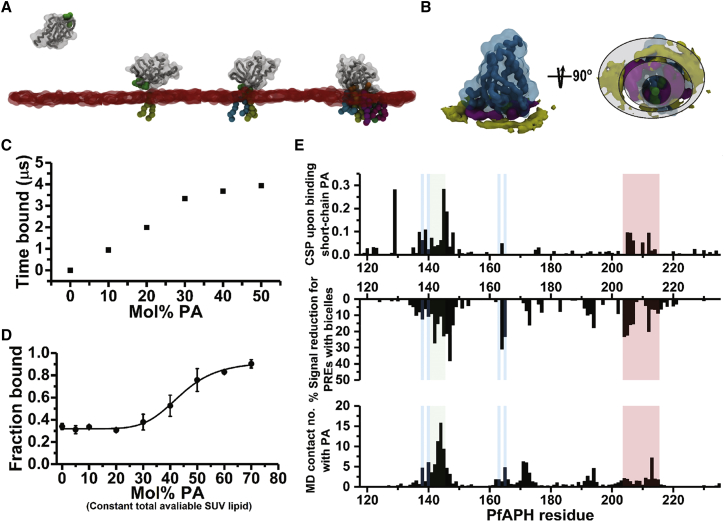
To delineate the binding site of the PA head group on the APH structure, 1H-15N HSQC NMR titration experiments with increasing molar ratios of short-chain PA, were performed first with 15N-labeled PfAPH106-235 due to its higher-quality spectra (Figures 2A and 2B). Chemical shift perturbation (CSP) of peaks upon addition of the PA ligand indicates a change in the chemical environment of the backbone amide group, and its likely proximity to the binding site. Small dose-dependent CSPs in fast exchange on the NMR timescale were observed for several peaks in the presence of short-chain PA. The location of these CSPs reveals a contiguous cluster around the β1 and β2 strands, suggesting that this region plays a role in recognition of PA (Figure 2C). Notably, one of the largest CSPs is observed for a lysine residue located on the β1 strand in PfAPH (K138), which represents the start of a conserved KxK motif. Prominent CSPs are also observed for residues that map to the β1-β2 loop region (T141 to H145 in PfAPH). Taken together, the NMR mapping of PA binding for PfAPH106-235 reveals a contiguous surface that overlaps with canonical and atypical binding sites identified in PH domains with specificity for other phospholipids (Figure S1).
Figure 2.
Mapping the APH:PA Interface
(A) Overlay of representative 2D 1H-15N HSQC spectra of PfAPH106-235 recorded upon titration with increasing molar ratios of short-chain PA. HSQC spectra are colored according to the molar ratio between 15N-labeled PfAPH106-235 and short-chain PA; black 1:0, green 1:1, blue 1:3, orange 1:7, purple 1:15.
(B) Plot of CSPs observed in (A) upon titration with 15-fold molar excess of short-chain PA, versus PfAPH106-235 sequence number. Residues that could not be assigned are indicated by a gray bar. Prominent CSPs are categorized as greater than 2σ from the mean noise (0.041 ppm), which is represented by a dotted line.
(C) CSPs mapped onto the structure of PfAPH106-235, colored in a 20-interval red spectrum. A more intense coloring indicates a greater CSP as each interval represents 0.5σ from the mean noise. Key residues clustered around the β1/2 strands and β3-β4 loop region are labeled, unassigned residues are colored dark gray.
(D) Representative 1H-15N HSQC spectra and 15N 1D profiles for PfAPH106-235 recorded in the presence of PA-enriched bicelles doped with and without a paramagnetic 5% PE-DTPA-Gd3+ lipid. PREs and therefore proximity to the PA-binding sites are indicated by a reduction in peak intensity.
(E) Plot of peak intensity reduction observed in (D) relative to the mean noise (61.80%), which is shown as the baseline, versus PfAPH106-235 sequence number.
(F) PREs mapped onto the structure of PfAPH106-235, residues are colored if greater than 1σ (yellow), 2σ (orange), or 3σ (red) from the mean noise, while unassigned residues are colored dark gray.
NMR studies using fast tumbling isotropic bicelles were initiated to gain further insight into the intermolecular interactions that occur in APH upon binding PA within the membrane. To limit the signal broadening of PfAPH106-235 resonances upon the addition of bicelles (∼100 kDa size range), high long-chain length lipids (1,2-dimyristoyl-sn-glycero-3-phosphocholine [DMPC] and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine [POPC] or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate [POPA])/short-chain length lipid (1,2-diheptanoyl-sn-glycero-3-phosphocholine [DHPC]) ratios, q = 0.33 bicelles (where q is the relative ratio), were employed to generate smaller bicelles. Specific CSPs were observed in the 1H-15N HSQC spectra of PfAPH106-235 upon the addition of bicelles with a bilayer enriched in POPA (Figure S2). Mapping these CSPs onto the structure of PfAPH106-235 reveals clusters around W161, β1-β2 (I143, F144 and H145), and β6-β7 (I205 to T207) loop regions, consistent with those identified in short-chain PA titrations.
Finally, to delineate the PA-binding surface more precisely, we performed NMR titration experiments with PA-enriched bicelles doped with a paramagnetic lipid (5% PE-DTPA-Gd3+ [1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid (gadolinium salt)]) to induce enhanced transverse relaxation for residues proximal to the binding surface. Following guidelines from previous studies, the distribution of the paramagnetic lipid is expected to be random at this lipid concentration and under these conditions (Koppisetti et al., 2014, Mazhab-Jafari et al., 2015). Paramagnetic relaxation enhancements (PREs) were quantified by the reduction in signal intensities in 1H-15N HSQC NMR spectra, when compared with the same experiment performed in the presence of diamagnetic bicelles. Although specific PREs were observed for amides in PfAPH106-235 (Figures 2D and 2E) that coincide with the CSP data, the PRE data also highlight the β3-β4 loop, which only showed very small chemical shifts changes in the short-chain PA titration. Interestingly, this region possesses a second conserved KxK motif that would likely be involved in PA recognition. Its increased prominence in the bicelle titration experiments could suggest that this motif is more specific for a PA bilayer context. The observed PA-binding site for PfAPH106-235 was delineated in identical NMR titration experiments for TgAPH99-229 (Figure S3).
While PIP-strip assays showed that both TgAPH and PfAPH bind specifically to PA, it was also suggested that APH may be capable of binding to PI(4,5)P2, albeit more weakly (Bullen et al., 2016). Although no other lipid specificity was suggested in these studies, we used NMR to test the possible PI(4,5)P2 binding. NMR titrations of PfAPH106-235 with PI(4,5)P2 did not induce CSPs, confirming the absence of a specific interaction (Figure S4). Dual specificity for lipids via distinct binding sites has been reported for several PH domains (Jian et al., 2015, Lai et al., 2013, Lucas and Cho, 2011). To assess whether APH is capable of dual phosphoinositide binding or perhaps more importantly, whether PA binding enhances recognition of a second phospholipid, NMR titration experiments were performed with increasing molar ratios of short-chain PI(4,5)P2 after saturation with PA (Figure S5). As expected, the PA-specific CSPs were observed in the PfAPH106-235 1H-15N HSQC spectra, however no further CSPs occurred with PI(4,5)P2. Taken together, these data indicate that the weak PI(4,5)P2 binding observed in PIP-strip assay is likely a result of non-specific binding (Bullen et al., 2016).
APH Binds Specifically to PA-Enriched Unilamellar Vesicles
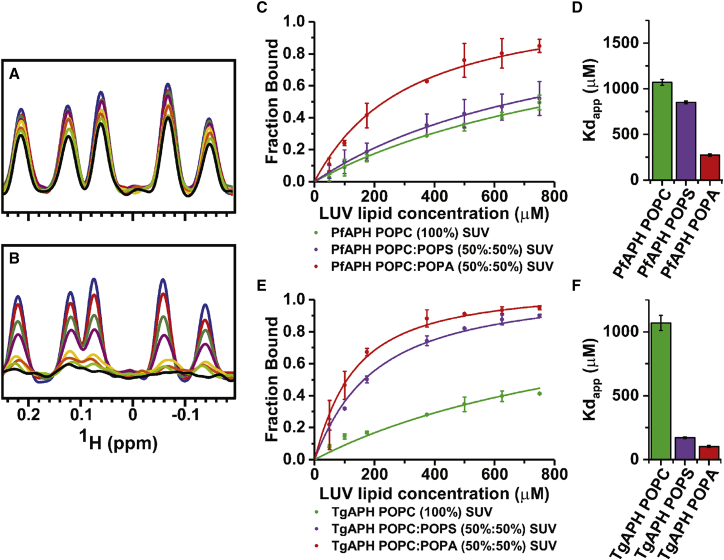
Binding experiments with recombinant APHs and short-chain PA enabled mapping of the PA head group interaction in a residue-specific manner. Notably, CSPs are small and binding to the short-chain PA is in the fast exchange regime on the NMR timescale, suggesting that the interaction, in this context, is weak (estimated to be >50 μM). To quantify the affinity and facilitate an assessment of site-directed mutants, we monitored the 1D 1H NMR spectrum for PfAPH106-235 or TgAPH99-229 following the addition of liposomes (Ceccon et al., 2013, Mercredi et al., 2016) (Figures 3 and S5). A loss in signal intensity can be interpreted as the formation of a large, NMR-invisible complex between APH and the liposomes. Only modest signal intensity losses are observed for PfAPH106-235 upon titration with large unilamellar vesicles (LUVs) composed solely of POPC (POPC LUVs; Figure 3A). In comparison, titration with LUVs composed of 50% POPA and 50% POPC (POPA LUVs) resulted in significant signal attenuation, with a complete loss of the PfAPH106-235 spectrum at high liposome concentrations (Figure 3B).
Figure 3.
APH Specifically Binds PA-Enriched Membranes
(A and B) PfAPH106-235 1D 1H NMR spectral region corresponding to the upfield-shifted methyl region (0.255 to −0.170 ppm) was monitored upon titration with increasing concentration of LUVs composed of (A) POPC (100%) or (B) POPC and POPA (50%:50%). PfAPH106-235:LUVs molar ratios: blue, free PfAPH106-235 in solution; red 1:2; green 1:4; purple 1:7; yellow 1:15; orange 1:20; lime 1:25; black 1:30.
(C) This region was monitored upon titration with variable LUV compositions (POPC [100%] green, POPC:POPS [50%:50%] purple, or POPC:POPA [50%:50%] red), integrated, expressed as the fraction of bound protein, and plotted against total lipid concentration to generate binding curves. Data are represented as mean ± 1σ.
(D) Apparent dissociation constants (Kdapp) for binding LUVs were calculated from fitting binding curves. Data are shown as mean ± 1σ for fitting curves.
(E and F) (E) and (F) are identical to (C) and (D), but for TgAPH99-229 using the downfield-shifted amide region (9.4–6.4 ppm).
Binding curves were generated from these data by integrating the upfield-shifted methyl region, plotting values against lipid concentration and fitting to a single-site binding isotherm. The apparent dissociation constant (Kdapp) for PfAPH106-235 binding LUVs containing 50% POPA is 275 ± 13 μM, almost 5-fold lower than the Kdapp for POPC LUVs (1070 μM ± 33 μM), which indicates that the presence of POPA enhances the interaction between PfAPH106-235 and LUVs (Figures 3C and 3D). To establish the general role of electrostatic interactions between LUVs and APH, titrations were repeated with LUVs in which PA was replaced with 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), which also possesses a negatively charged head group. A value of Kdapp of 850 ± 13 μM was obtained for PfAPH106-235 (Figures 3C and 3D). Although the observed trend in LUV binding affinity for PfAPH106-235 is also borne out in identical titration experiments with TgAPH99-229 (Figures 3E and 3F), the affinity TgAPH99-229 shows for POPS liposomes is higher than PfAPH106-235. The increased affinity of PfAPH106-235 or TgAPH99-229 for POPA LUVs over the similarly negatively charged POPS LUVs suggests interaction for PA or clusters of PA molecules is specific and not dictated by simple electrostatic attraction alone.
Twin KxK Motifs and the β1-β2 Loop Are Essential for PA Recognition
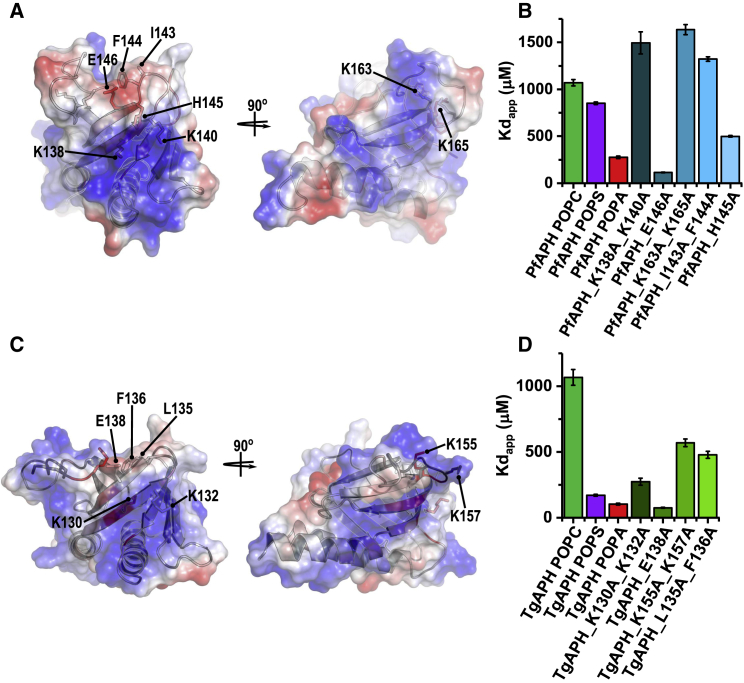
A series of alanine substitutions were generated in PfAPH106-235 and TgAPH99-229 (referred to with the mutation type and position hereafter) that target residues highlighted in the NMR-based PA titrations. The 1D NMR LUV assay was used to determine the influence of these mutations on PA binding. Mutation of K138 (PfAPH_K138A, Kdapp = 959 ± 35 μM) or K140 (PfAPH_K140A, Kdapp = 604 ± 29 μM) reduces PfAPH106-235 affinity for POPA LUVs compared with wild-type (data not shown). Mutation of both lysines (PfAPH_K138A_K140A, Kdapp = 1,494 ± 117 μM) has a more dramatic effect on binding, reducing the affinity of PfAPH106-235 for POPA LUVs beyond that for wild-type binding to neutral POPC LUVs (Figures 4A and 4B). Mutation of the second conserved KxK motif present in the β3-β4 loop (K163 and K165 in PfAPH) has a similarly detrimental effect on PA binding, increasing Kdapp to 1,636 ± 54 μM for the double-mutant PfAPH_K163A_K165A (Figures 4A and 4B). PfAPH106-235 affinity for POPA LUVs is also reduced when exposed hydrophobic side chains located at the tip of the β1-β2 loop are mutated; namely I143 and F144A (PfAPH_I143A_F144A, Kdapp = 1,322 ± 24 μM), and H145 (PfAPH_H145A, Kdapp = 499 ± 9 μM) (Figures 4A and 4B). Removal of a negative charge that disrupts the β1 strand KxK motif in the mutation of E146 to alanine, resulted in an increase in PfAPH106-235 affinity for POPA LUVs (PfAPH_E146A, Kdapp = 155 ± 4 μM). Identical LUV titration assays with TgAPH99-229 confirm that the PA-binding behavior is consistent with that observed for PfAPH106-235 (Figures 4C and 4D). It is noted that compared with PfAPH106-235, the solvent-exposed patch of basic charge present on the same face as the β3-β4 loop KxK motif, is extended in TgAPH99-229 (compare Figures 4A and 4C). Electrostatic attraction between this extended basic patch and negatively charged membranes may explain why TgAPH99-229 has a greater affinity for POPS LUVs.
Figure 4.
Conserved APH Basic Residues Mediate Binding to PA Within a Membrane Environment
(A–D) APH contains only two, highly conserved KxK motifs present on the β1 strand (K138 and K140 in PfAPH, K130 and K132 in TgAPH) and within the β3-β4 loop region (K163 and K165 in PfAPH, K155 and K157 in TgAPH). Coulombic colored surface representation of (A) PfAPH106-235 and (C) TgAPH99-229 reveals KxK motifs form patches of solvent-exposed basic charge. 1D NMR LUV titration experiments show that mutation of KxK motifs reduce (B) PfAPH106-235 and (D) TgAPH99-229 affinities for PA-enriched LUVs. A glutamate residue (E146 in PfAPH, E138 in TgAPH) present in the β2 strand disrupts the β1 strand KxK surface exposed basic charge (A and C). As measured by 1D NMR LUV titration experiments, mutation of this glutamate residue increases PfAPH106-235 and TgAPH99-229 affinity for PA-enriched LUVs (B and D).
Functional Characterization of Key Residues in TgAPH
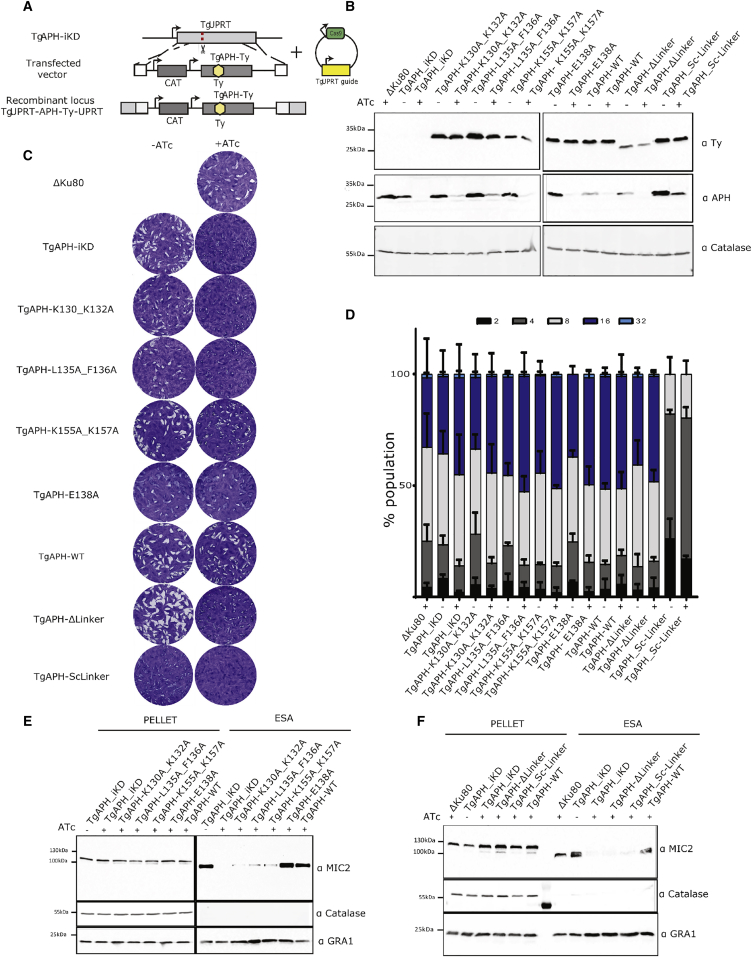
In vivo validation of the importance of APH residues was determined by complementation of T. gondii parasites due to the availability of a strain bearing a regulatable endogenous TgAPH (TgAPH-iKD). The tested second copies of TgAPH mutants containing an internal Ty tag epitope were targeted to the non-essential uracil phosphoribosyltransferase locus (UPRT) (Figure 5A). TgAPH mutants were generated in the two KxK PA-binding motifs (TgAPH-K130A_K132A and TgAPH-K155A_K157A), the hydrophobic motif at tip of the β1-β2 loop (TgAPH-L135A_F136A) and also in the negatively charged amino acid that disrupts the β1 strand KxK motif (TgAPH-E138A). The charged linker region upstream of the PH domain displays significant levels of sequence conservation among apicomplexan APH (Figure 1A). A 34-amino acid sequence within this linker region was subsequently deleted in TgAPH-Δlinker. To address whether features of the linker region other than its length are functionally important, this region was first deleted and then a subsequent mutation introduced a scrambled linker sequence (TgAPH-Sc-linker). Integration and expression of these APH mutants was confirmed by genomic PCR and western blot analysis (Figure 5B). Although, the second copies of APH are expressed at lower levels compared with the endogenous or wild-type copy, the expression levels between mutants are comparable. The TgAPH-Δlinker mutant leads to a modest decrease in protein expression, whereas the TgAPH-Sc-linker mutant shows a striking 5- to 10-fold increase in protein levels. No changes in expression levels from the second gene copies were observed upon TgAPH-iKD depletion with anhydrotetracycline (ATc).
Figure 5.
In Vivo Functional and Mutagenesis Studies of APH
(A) Schematic representation of APH-Ty mutant generation.
(B) Western blot analysis of endogenous and second copy TgAPH ± ATc 48 hr. Catalase provides a loading control.
(C) Plaque assay on human foreskin fibroblast monolayer 7 days ± ATc.
(D) Intracellular growth assays at 24 hr ± ATc treatment, with 24-hr pre-treatment. Data are presented as mean ± 1σ.
(E and F) Microneme secretion assay of mutants in the PH domain (E) and linker region (F). Extracellular secreted antigen (ESA) MIC2 was compared with parental strain ± ATc 48 hr. Catalase represents a loading control for parasite number and lysis, GRA1 represents a control for constitutive secretion.
None of the mutants within the PH domain or the linker deletion led to defects in intracellular growth rate (±ATc), nor did they affect the lytic cycle in the absence of ATc (Figures 5C and 5D). In contrast, the scrambled linker mutant showed a marked defect in the intracellular growth rate and lytic cycle (–ATc), which is likely a result of the high expression of this APH mutant coupled with a defect in its function. All other mutants expressed at lower levels compared with wild-type APH. Upon the depletion of TgAPH-iKD, TgAPH-WT and TgAPH-E138A were capable of restoring the lytic cycle, whereas the TgAPH-K155A_K157A partially complemented the phenotype by generating small plaques. In contrast, TgAPH-K130A_K132A, TgAPH-L135A_F136A, TgAPH-Δlinker, and TgAPH-Sc-linker exhibited severe defects comparable to TgAPH-iKD + ATc, indicating that these mutants are non- or poorly functional variants of TgAPH (Figure 5E).
Depletion of APH also leads to a defect in microneme secretion (Bullen et al., 2016), which was partially complemented by the various mutants. TgAPH-K130A_K132A, TgAPH-L135A_F136A, and TgAPH-K155A_K157A mutants showed limited ability to secrete micronemes, with TgAPH-K130A_K132A being the most severely affected (Figure 5E), which is consistent with the plaque assay data. No microneme secretion was observed in the presence of either TgAPH-Δlinker or TgAPH-Sc-linker mutants, which highlight the importance of the linker sequence (Figure 5F).
The APH-Phospholipid Binding Surface Accommodates Multiple PA Head-Groups
The presence of two conserved KxK motifs and mutagenesis data, indicating a key role in PA binding, raises the notion that multiple phosphate head-groups may be recognized by APH. To challenge this hypothesis further, we performed coarse-grained molecular dynamics (CG-MD) simulations in which the PfAPH PH domain (PfAPH106-235) was placed ∼9 nm away from an equilibrated PC:PA lipid bilayer of varying composition from 0% to 50% PA; 3 × 5 μs simulations at each lipid composition were performed in which PfAPH106-235 was free to diffuse and encounter the membrane (Figures 6 and S6). Similar techniques have been used to characterize the interactions of PH domains with PIP-containing membranes (Lai et al., 2013, Lumb et al., 2011, Yamamoto et al., 2016). In all simulations, PfAPH106-235 encountered the bilayer multiple times, forming transient complexes with the membrane surface (Figure S6), the frequency of which increases with PA concentration. Mapping the contacts between PfAPH106-235 and the lipid molecules over time at each lipid composition allowed us to probe features of binding to PA-enriched membrane.
Figure 6.
Coarse-Grained MD Simulation of APH Binding to PA-Enriched Membranes
(A) Snapshots from an individual binding series of PfAPH106-235 (gray) to a 10% PA membrane. The hydrophobic residues I143-F144-H145 that become anchored in the membrane are shown as a green surface. POPA residues within 6 Å of the protein surface are shown as spheres colored individually and the lipid head-groups are shown as a transparent red surface. The recruitment of POPA following the initial association is apparent in the final panel.
(B) Average occupancy of PA (magenta) and PC (yellow) head-groups averaged over five simulations of 50% PA membranes, PfAPH106-235 is shown in light blue. POPA is found to be preferentially in the first shell of lipids around the buried anchor residues I143-F144-H145 (green) whereas PC is found in the second annular layer. Rough lipid shell boundaries are indicated by gray-shaded circles. The protein backbone is shown as a gray trace.
(C) Relationship between average time (μs) bound to membrane and PA membrane enrichment for PfAPH106-235 coarse-grained MD simulation (5 μs total simulation time).
(D) Binding between PfAPH106-235 and a fixed concentration of LUVs (500 μM total available lipid) increasingly enriched with PA (Mol% PA). Hill plot analysis indicates PfAPH106-235 binds to PA in a positively cooperative manner.
(E) Comparison between coarse-grained MD simulation and NMR experiments probing binding between PfAPH106-235 and PA reveal three regions key to interaction with a PA-enriched membrane. Coarse-grained MD simulations indicate residues 99–110 (including β5-β6 loop) are involved in initial contact with a PA-enriched membrane (red). Hydrophobic residues located at the tip of the β1-β2 loop region (green, I143/F144) dip into the membrane. This anchoring is stabilized by electrostatic interaction between conserved charged residues including KxK motifs (blue, K138-K140 and K163-K165), and PA head-groups.
The simulations converge upon a stable binding mode of PfAPH106-235 on the membrane surface, which is consistent across the PA concentration range (Figures 6A and S6). In this bound state, the hydrophobic residues I143-F144-H145 in the β1-β2 loop penetrate the membrane leaflet surface, whereas the KxK motifs accommodate multiple, negatively charged PA head-groups. The APH membrane contact points identified from independent MD simulations are consistent with the binding interface highlighted in NMR mapping experiments (Figure 6E). At all concentrations of PA, upon binding of the protein, POPA molecules are recruited to the protein, with up to six POPA lipids present at the interface in 10% PA membranes (Figure S6). PA lipids cluster tightly in the first “shell” around the anchoring loop, with PC lipids displaced to form the second layer or “shell” around the protein (Figures 6B and S6). Furthermore, PA forms small clusters of dimers and trimers within the CG membrane (Figure S6).
Taken together, the experimental observation of at least two PA-binding sites on APH suggests that binding PA-enriched membranes may be cooperative. Initial encounter with a PA head group and β1-β2 loop insertion in the membrane leaflet could in turn enhance the affinity for a second PA molecule. To test this, we performed NMR binding experiments with PfAPH106-235, in which the proportions of PA within the LUV bilayer was increased, while the total concentration of LUVs was kept constant (Figure 6D). The fraction of bound APH increased sharply only when PA levels in the LUV were above 40% and began to plateau above 70% (Hill constant, n = 6.77 ± 0.72; goodness of fit R2 > 0.99). Bilayer binding rates from individual MD simulations can be calculated for membranes with increasing PA concentration (Figure 6C). Although the curve is shifted to lower concentrations, likely due to the limitations of a CG-MD model, the dependence of time bound on the PA concentration echoes the sigmoidal curve of the NMR binding data (Figure 6C).
Discussion
PH domains are ubiquitous in signal transduction pathways. The vast majority of the PH domains characterized to date bind phosphatidylinositol phosphates or inositol phosphate head-groups and subsequently target proteins to a specific endomembrane compartment (Lemmon, 2008). The structures of TgAPH99-229 and PfAPH106-235 reveal deviations from the canonical PH domain fold, with an N-terminal helix connected to the APH-microneme linker and a much shorter β1-β2 loop (Figure 1). The binding surface of the PA head-groups also encompasses both canonical and atypical binding sites of typical PH domains that target phosphatidylinositol/inositol phosphates (Figure 2). Canonical binding sites comprise a basic sequence within the β1-β2 loop, usually Kxn(K/R)xR that coordinates phosphates from PIP ligands. PH domains lacking this motif often use the opposite face of the β1-β2 strands and the intervening loop, which has been termed the atypical binding site and is observed for the β-spectrin and ArhGap9 PH domains (Ceccarelli et al., 2007, Yamamoto et al., 2016). Although the Kxn(K/R)xR motif is not present in any of the APH sequences, they harbor two conserved, but separated KxK motifs that are important for binding of PA-enriched membranes. The lysine side chains of the first motif (K138-K140 in PfAPH or K130-K132 in TgAPH) project toward the atypical binding surface on the upper side of the β1-β2 region. The second motif (K163-K165 in PfAPH or K155-K157 in TgAPH) delineates one edge of the canonical binding site, which is capped by the shorter and closed β1-β2 loop. Two further basic sequences are conserved in APH, namely RRR within the linker (R78-R79-R80 in PfAPH or R73-R74-R75 in TgAPH) and K/RxK in the β3-β4 loop (K171-K173 in PfAPH or R165-K167 in TgAPH), and these may play minor roles in membrane binding.
The true nature of PH domain interactions with membranes is far more complex than single phospholipid recognition. Recent structural studies on PH domains revealed that PIPs can bind to both canonical and atypical sites simultaneously (Jian et al., 2015, Vonkova et al., 2015), and often in a cooperative manner; for example, by the PH domain from the Arf GAP (Vonkova et al., 2015) and ASAP1 (Jian et al., 2015). Despite these advances, reports on PH PA binding are sparse and no structural insight is currently available. Mutagenesis of the nucleotide-exchange factor Son of Sevenless (Sos) implicated a role for two positively charged residues from an extended β3-β4 loop in PA binding and subsequent Ras activation. These observations raise the possibility that another phospholipid interaction may play a role in APH membrane engagement, i.e., in addition to PA, and the relationship could be cooperative. Our studies do not support additional phospholipid specificity for APH, but instead reveal a high selectivity for PA that is driven through cooperative binding to more than one PA lipid molecule. Two major PA-binding sites exist within the APH domain, which are represented by two conserved KxK motifs, with the first lying on the atypical face within β1 and the other at the end of β3. These motifs lie in distinct locations on the APH structure and are juxtaposed to the well-established canonical and atypical binding sites of PH domains. A similar behavior has been reported for the kindlin-3 and Brag2 PH domains, which are able to accommodate multiple PIP lipid head-groups (Karandur et al., 2017, Ni et al., 2017). The β1-β2 loop is also important for the membrane association of APH, with the surface exposed, bulky hydrophobic side chains from this region (I143-F144-H145 in PfAPH), inserting into the lipid bilayer. It is conceivable that membrane insertion of the β1-β2 loop occurs after initial encounter of a KxK motif with a PA head group and subsequent conformation change could facilitate increased binding to the bilayer. Similar conclusions have been postulated from structural and dynamic studies of PH domains from ACAP1 and Grp1 (Lumb et al., 2011, Pang et al., 2014). It is widely recognized that some PA-binding proteins respond to negative curvature stress (Putta et al., 2016). This property together with the multiple contacts of APH with PA head-groups and insertion of the hydrophobic loop may provide a mechanism to sense increased local PA concentration and subsequent negative curvature.
The presence of a basic region linker sequence between the PH domain and the microneme N-terminal anchor (residues R75-K89 in PfAPH and R70-K84 in TgAPH) is worth noting. The in vivo complementation data confirm that the APH linker is critical and plays a key functional role in triggering microneme secretion. The conserved basic “Rx2RRRx8RK” would be influenced by the proximity of the negatively charged membrane surface in membrane-bound APH. It is tempting to speculate that the PH domain together with the basic APH linker interact with the negatively charged bilayer and further stabilize binding to the PA-enriched plasma membrane. It is worth noting that in tandem BAR-PH domain proteins, such as ACAP1 or the ArfGAPs (Frost et al., 2009), an additional basic surface on the helical BAR domain enhances interaction of the PH domains with the membrane surface and induces curvature. Upon sensing PA at the parasitic plasma membrane, APH tethers the micronemal membrane in close proximity. This function is somewhat reminiscent of Num1, a protein that tethers mitochondria to the plasma membrane in budding yeast through a bipartite interaction (Ping et al., 2016). Num1 C-terminal PH domain binds PI(4,5)P2 at plasma membrane, whereas the N-terminal coiled coil domain preferentially binds cardiolipin at the mitochondrial outer membrane via basic residues. Similarities between the domain architecture of Num1 and APH may also suggest that the N-terminal linker region in APH plays a role in membrane binding.
The association of APH with the plasma membrane leading to the engagement of microneme fusion and exocytosis is poorly understood. Presumably, these events are not independent and are connected either via an APH-induced molecular signal or a direct interaction. Indeed, many PA-binding effectors are targeted through cooperative binding with additional protein cofactors (Lemmon, 2008). For example, Opi1 binds and senses changes in PA in the ER of yeast by binding to the ER protein Scs2 in addition to PA (Loewen et al., 2004). The engagement of APH molecules with the PA-enriched regions of the plasma membrane could provide a stable molecular scaffold for the subsequent recruitment of membrane fusion machinery, e.g., SNARE-like DOC2 proteins (Farrell et al., 2012).
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-APH | The Soldati-Favre Lab | N/A |
| α-Ty | The Soldati-Favre Lab | N/A |
| α-Catalase | The Soldati-Favre Lab | N/A |
| α-MIC2 | The Carruthers Lab | N/A |
| α-GAP45 | The Soldati-Favre Lab | N/A |
| α-GRA3 | The Dubremetz Lab | N/A |
| Peroxidase conjugated goat α-mouse/rabbit | ThermoFisher | Cat# 62-6520, Cat# 31460 |
| Alexa Fluor 680 conjugated goat α-rabbit | ThermoFisher | Cat# A-21109 |
| Alexa Fluor 488 conjugated goat α-mouse | ThermoFisher | Cat# A-11001 |
| Alexa Fluor 594 conjugated goat α-rabbit | ThermoFisher | Cat# R37117 |
| Bacterial and Virus Strains | ||
| XL10 Gold | Stratagene | Cat# 200315 |
| E. coli DH5α | NEB | Cat# C2987I |
| E. coli BL21 (DE3) | NEB | Cat# C2527I |
| Deposited Data | ||
| Solution structure of PfAPH106-235 | This paper | PDB: 6F24 |
| Solution structure of TgAPH99-229 | This paper | PDB: 6F8E |
| Experimental Models: Organism/Strains | ||
| Human foreskin fibroblasts (HFFs) | Igcstandards | Cat# ATTC-112Sk |
| Toxoplasma gondii: RHΔKu80 | (Fox et al., 2009, Huynh and Carruthers, 2009) | N/A |
| E. coli: pNIC28a-Bsa4_PfAPH106-235 | This paper | N/A |
| E. coli: pNIC28a-Bsa4_TgAPH99-229 | This paper | N/A |
| E. coli: pNIC28a-Bsa4_TgAPH22-229 | This paper | N/A |
| Oligonucleotides | ||
| Primers used for APH expression and functional characterisation see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Plasmids used for expression of APH and functional studies see Table S1. | This paper | N/A |
| Software and Algorithms | ||
| Sequence Manipulation Suite: Shuffle Protein | GenScript | https://www.genscript.com/sms2/shuffle_protein.html |
| PSI-PRED | (McGuffin et al., 2000) | http://bioinf.cs.ucl.ac.uk/psipred/ |
| Pymol | Version 1.3 (DeLano Scientific LLC/ Schrödinger) | https://pymol.org/ |
| Prism | Version 7, GraphPad Software | https://www.graphpad.com |
| OriginPro | 2017 version, OriginLab | https://www.originlab.com/index.aspx?go=PRODUCTS/Origin |
| Topspin | Version 3.5, Bruker | https://www.bruker.com/products/mr/nmr/nmr-software/nmr-software/topspin/overview.html |
| MARS | (Jung and Zweckstetter, 2004) | http://www3.mpibpc.mpg.de/groups/zweckstetter/_links/software_mars.htm |
| TALOS+ | (Shen et al., 2009) | https://spin.niddk.nih.gov/bax/software/TALOS/ |
| NMRView | NMRviewJ/In house version | http://www.onemoonscientific.com/nmrviewj/ |
| Aria/CNS | Versions 2.3 and 1.1(Rieping et al., 2007) | http://aria.pasteur.fr/ |
| Gromacs | Versions 4.6 and 5, (Hess et al., 2008) | www.gromacs.org |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Steve Matthews (s.j.matthews@imperial.ac.uk).
Experimental Model and Subject Details
HFF Cell Culture
Human foreskin fibroblasts (HFFs) were grown at 37°C, 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM; GIBCO, Invitrogen) supplemented with 2 mM glutamine, 5% foetal calf serum and 25 μg/ml gentamicin.
Toxoplasma gondii Cell Culture
RHΔKu80 were grown at 37°C, 5%CO2 in confluent human foreskin fibroblasts (HFFs) maintained in DMEM, supplemented with 2 mM glutamine, 5% foetal calf serum and 25 μg/ml gentamicin. Tet-inducible gene expression was regulated with 1 μg/ml anhydrotetracycline (ATc) (Meissner et al., 2001).
Escherichia coli DH5α Cell Culture
Transformed DH5α strains were grown in LB media or plated onto LB agar supplemented with 50μg/ml Kanamycin (Sigma), and grown at 37°C.
Escherichia coli BL21 Cell Culture
Transformed BL21 strains were grown at 37°C in either LB media or M9 media supplemented with 15NH4Cl and/or 13C-glucose until OD600 reached 0.8 units. Media was supplemented with 50μg/ml Kanamycin (Sigma). Expression was induced at 18°C by the addition of 0.5mM IPTG (Sigma) for PfAPH106-235/TgAPH99-229, or 0.25mM for TgAPH22-229.
Method Details
PfAPH and TgAPH Cloning, Expression and Purification for Structural Studies
Based on secondary structure prediction (PSI-PRED), the sequence corresponding to the C-terminal pleckstrin-homology domain was amplified from full length, codon optimised PfAPH and TgAPH genes (PfAPH and TgAPH) and cloned into an pNIC28a-Bsa4 vector containing an TEV cleavable N-terminal-(His)6 tag fusion, using LIC methods, to generate pNIC28a-Bsa4_PfAPH106-235 and pNIC28a-Bsa4_TgAPH99-229 (see Table S1). TgAPH22-229 was amplified from the full length codon optimised gene, excluding the conserved acylation site corresponding to the first N-terminal 21 residues. PfAPH106-235 mutants, K138A_K140A/I143A_F144A/H145A/E146A and K163A_K165A, and TgAPH99-229 mutants K130A_K132A/L135A_F136A/E138A and K155A_K157A were generated using Q5 site-directed mutagenesis kits (NEB) using pNIC28a-Bsa4_PfAPH106-235 and pNIC28a-Bsa4_TgAPH99-229 vectors as respective templates (see Table S2). DH5α E.coli (NEB) were used for cloning.
Vectors were transformed into an E.coli BL21 strain (NEB) and grown as stated in experimental models. Cells were lysed and clarified by centrifugation at 17,000rpm for 35mins. Supernatants were initially purified by nickel-affinity chromatography followed by TEV cleavage during overnight dialysis to remove the N-terminal-(His)6 fusion tag. Cleaved protein was further purified by gel filtration using a Superdex-75 column (GE healthcare) pre-equilibrated in 10mM HEPES, 0.3M NaCl, 2mM TCEP, pH 6.5 (PfAPH106-235), 50mM HEPES, 150mM NaCl, 2mM TCEP, pH 7 (TgAPH99-229) buffer, or 10mM HEPES, 300mM NaCl, 2mM TCEP, pH7 (TgAPH22-229). Expression and purification of PfAPH and TgAPH mutants followed the same procedure as wild-type protein. Like wild-type protein, the folding status of each mutant was verified by 1D 1H-NMR.
PfAPH106-235 and TgAPH99-229 Short-Chain Phosphatidic Acid and PI(4,5)P21H-15N HSQC Titration Experiments
550μl NMR samples were prepared with purified 15N-labelled protein (250 μM final concentration) and D20 added (10% v/v). Short-chain PA (1,2-dihexanoyl-sn-glycero-3-phosphate, Avanti lipids) and PI(4,5)P2 (1,2-dioctanoyl-sn-glycero-3-phospho-(1'-myo-inositol-4',5'-bisphosphate, Avanti lipids) was initially dissolved in chloroform. Chloroform was removed by evaporation under a stream of N2 to leave a lipid film, which was left to dry overnight. Dried lipid was rehydrated with gel filtration buffer to generate a concentrated lipid stock (40mM). 2D 1H-15N HSQC spectra were recorded for protein alone and protein titrated with increasing molar ratios of short-PA or PI(4,5)P2 from the concentrated stock.
Large Unilamellar Vesicle (LUV) Preparation
POPA (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate), POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine) and POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) were obtained commercially (Avanti) as chloroform dissolved lipids. Volumes of lipids were pipetted into glass vials,chloroform removed by evaporation under a stream of N2 to leave a lipid film, and residual chloroform removed by desiccation overnight. Dried lipids were re-suspended in gel filtration buffer through shaking (1500rpm) at room temperature for 2hrs to generate a cloudy solution. To form LUVs, the re-suspension was sonicated using a probe tip sonicator until transparent and then centrifuged at 17,000rpm to removed titanium debris and large or multilamellar vesicles. LUVs were prepared at an 8mM total lipid concentration and used within 48hrs of preparation. LUVs had a typical hydrodynamic diameter of between 80-100 nm, which was measured by dynamic light scattering (Malvern, Zetasizer Nano S DLS analyser).
1D 1H-NMR LUV Titration Experiments
550μl NMR samples were prepared with purified protein (50μM final concentration) and D20 (10% v/v). For each titration at the specified large unilamellar vesicle (LUV) concentration, a LUV preparation was added to the NMR sample from a concentration stock (8mM), and mixed (see Table S3). 1D 1H-NMR spectra were recorded after each titration and overlaid using Topspin 3.5 software (Bruker).
Bicelle Preparation
Bicelle lipids DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine), DHPC (1,2-diheptanoyl-sn-glycero-3-phosphocholine), POPA (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate), POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and PE-DTPA-Gd3+ (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid (gadolinium salt)) were obtained commercially (Avanti) as chloroform dissolved lipids. Volumes of long chain lipids (DMPC, POPA, POPC or PE-DTPA-Gd3+, depending on bicelle composition) and short-chain lipid (DHPC) were separately pipetted into glass vials. Chloroform was removed by evaporation under a stream of N2 to leave a lipid film, and residual chloroform removed by desiccation overnight. Long-chain lipids were re-suspended in gel filtration buffer through shaking (1500rpm) at room temperature for 2hrs to generate a cloudy solution. Large unilamellar vesicles (LUVs) were generated from re-suspended long-chain lipids using methods previously described. To prepare isotropic bicelles with a q value of 0.33 (q = [Long-chain lipids]/[DHPC]), dried DHPC was re-suspended with long-chain lipid LUVs through vortexing, and the solution subjected to 10 freeze-thaw cycles between liquid nitrogen and a 60°C water bath. Bicelles were generated using a 40mM total lipid concentration, and in accordance with previously published results (Koppisetti et al., 2014), had an average hydrodynamic diameter of ∼10nm which was measured using dynamic-light scattering (Malvern, Zetasizer Nano S DLS analyser). The long-chain lipid composition of POPA enriched bicelles contained varying molar ratios of DMPC and POPA, whilst POPC enriched bicelles contained a 50%:50% DMPC:POPC long-chained lipid composition. Bicelles doped with PE-DTPA-Gd3+ had a 50%:45%:5% POPA:POPC:PE-DTPA-Gd3+ long-chained lipid composition. Bicelles were used within 24 hrs of preparation.
PfAPH106-235 and TgAPH99-229 Bicelle HSQC Titration and PRE Experiments
Prepared bicelles (30mM final total lipid concentration) were diluted with 15N-labelled protein (50μM final concentration) and D20 (10% v/v), to generate a 550μl NMR sample. Separate NMR samples were prepared for each bicelle composition, including samples containing bicelles increasingly enriched with POPA. 2D 1H-15N HSQC spectra were recorded for protein alone and in the presence of bicelles. Spectra were overlaid and combined chemical shift-perturbations values were calculated. For studies with paramagnetic probes, separate NMR samples were prepared with PA-enriched bicelles doped with and without PE-DPTA-Gd3+. Separate 2D 1H-15N HSQC spectra were recorded for protein in the presence of non-doped or doped bicelles.
PfAPH106-235 and TgAPH99-229 NMR Resonance Assignment and Structure Calculation
500μl samples of purified 15N/13C-PfAPH106-235 (700μM) or 15N/15C-TgAPH99-229 (830μM) were prepared and D20 added (10% v/v). All NMR spectra were acquired at 298K on Bruker Avance-III DRX 800 and Avance-III 600 spectrometers. An initial 1D 1H NMR spectra and 2D 1H-15N HSQC spectra were acquired prior to and between acquisition of 3D-NMR experiments used for backbone and side chain assignment, to assess protein folding and the quality of the sample. Triple resonance HNCA, HNCACB, HNCO and HN(CO)CA spectra were recorded and analysed to obtain backbone assignments. Linking assigned backbone chemical shifts was performed automatically using MARS (Jung and Zweckstetter, 2004) which incorporates PSI-PRED secondary structure prediction (McGuffin et al., 2000). Triple resonance HBHA(CO)NH, H(CCO)NH and CC(CO)NH and HCCH-TOCSY spectra were recorded for use in side-chain chemical shift assignment. 15N-NOESY and 13C-NOESY spectra were recorded, peaks picked, and peak files used as distance restraints in structural calculation. Chemical shift assignment and analysis was performed using an in-house version of NMRview. Dihedral angles were calculated using TALOS+ (Shen et al., 2009) and used as restraints in structural calculations. Automatic NOE assignment and structural calculation were performed using Aria 2.3/CNS 1.1 software (Rieping et al., 2007). A set of 100 structures were calculated in the final iteration and the 10 lowest-energy structures were refined in water. Structure ensembles for PfAPH106-235 and TgAPH99-229 have been deposited in the PDB under accession codes 6F24 and 6F8E, respectively. The medoid structure from ensembles is represented in figures using PyMOL.
Circular Dichroism
Purified TgAPH99-229 and TgAPH22-229 were dialysed into 10mM HEPES, 150mM NaF, 1mM TCEP, pH7 buffer overnight at 4°C, and then diluted with 10mM HEPES, 150mM NaF, pH7 buffer to 40μM and 30μM respectively for use in circular dichroism (CD). 200μl samples were loaded into a quartz 100-QS cuvette with a 1mm path length, and CD was performed on a Chirascan circular dichroism spectrometer (Applied Photophysics) at 20°C, wavelength 200 to 260nm, 5s scan length per point, 5 repeats.
Cloning of DNA Constructs for In-Vivo Studies
All amplifications were performed with either KOD polymerase (Novagen) or Q5 polymerase (New England Biolabs). RNA was isolated using TRIzol extraction. Total cDNA was generated by RT-PCR using the Superscript II reverse transcriptase (Invitrogen) according to manufacturer’s protocol. Primers used are listed in the Key Resources Table above.
APH gRNA/Cas9 Vector
Specific gRNA/Cas9 vector used for the generation of APH-iKD was made using the Q5 site-directed mutagenesis kit (New England Biolabs) with 6326-4883 and pSAG1::CAS9-GFP-U6::sgUPRT as a template (Shen et al., 2014).
APH Complementation
pT8-N21-Ty-APH-BleO (Bullen et al., 2016) was digested with EcoRI-PacI and ligated into 5'UPRT-pT8-MycGFPPfMyoAtail-Ty-3'UPRT (Jacot et al., 2016), pTub5-CAT was then digested with SpeI-ApaI and inserted into the intermediate plasmid generating 5'UPRT-CAT-pT8-N21-Ty-APH -3'UPRT. The modified APH variants were generated via Q5 mutagenesis of 5'UPRT-CAT-pT8-N21-Ty-APH-3'UPRT. The constructs and primers were used as follows, 5'UPRT-CAT-pT8-N21-Ty-APH-K130A+K132A-3'UPRT(6339-6529), 5'UPRT-CAT-pT8-N21-Ty-APH-L135A+F136A-3'UPRT (6341-6342), 5'UPRT-CAT-pT8-N21-Ty-APH-K155A+K157A-3'UPRT (6343-6344), 5'UPRT-CAT-pT8-N21-Ty-APH-E138A-3'UPRT (7327-7368), 5'UPRT-CAT-pT8-N21-Ty-APH-Δ-linker-3'UPRT(6423-6424). 5'UPRT-CAT-pT8-N21-Ty-APH-Sc-linker-3'UPRT was generated via triple ligation of amplicons (2170-7400) ClaI-XmaI, (7399-4749) XmaI-NotI and inserted into 5'UPRT-CAT-pT8-N21-Ty-APH -3'UPRT ClaI-NotI. The scrambled amino acid sequence was generated using the Shuffle protein program - Genscript.
Parasite Transfection and Selection of Stable Transfectants
T. gondii tachyzoites were transfected by electroporation as previously described (Soldati and Boothroyd, 1993). TgAPH-iKD strain was generated via transfection of RHΔKu80 (here referred as ΔKu80) (Fox et al., 2009, Huynh and Carruthers, 2009) with 30μg of pSAG1::CAS9-GFP-U6::sgAPH vector along with purified KOD PCR amplicon using primers 6324-6325 with iKD-GAC-DHFR (Jacot et al., 2016) as the template. Resistant parasites were selected using pyrimethamine (1 μg/ml). TgAPH-iKD strain was transfected with 5μg pSAG1::CAS9-GFP-U6::sgUPRT and 30μg of one of the TgAPH complementation plasmids (digested KpnI-NotI), refer to list above. Resistant parasites were selected using chloramphenicol (20μM). Parasites were cloned by limiting dilution in 96 well plates and plates and analysed for the integration and expression of the transgenes by PCR and Western blot, respectively.
Western Blot Analysis
3-4mL of parasites were lysed in 80μL RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 50mM Tris pH 7.5) using standard procedures and suspended to 120μL final volume with SDS–PAGE loading buffer (50mM Tris-HCl pH 6.8, 10% glycerol, 2mM EDTA, 2% SDS, 0.05% bromophenol blue, 100mM DTT) under reducing conditions. This suspension was subjected to 5min boiling at 95°C and two sonication cycles. SDS-PAGE was performed using standard methods, between 5-15μL of parasites were loaded per well. Separated proteins were transferred to nitrocellulose membranes and probed with appropriate antibodies in 5-10mL of 5% non-fat milk powder in 0.05%Tween20-PBS. Bound secondary peroxidase conjugated antibodies were visualized using either the ECL system (GE healthcare) or SuperSignal (Pierce).
Microneme Secretion Assay
3-4mL of freshly egressed parasites ± ATc 48hrs were resuspended in equal volume intracellular (IC) buffer (5mM NaCl, 142mM KCl, 1mM MgCl2, 2mM EGTA, 5.6mM glucose, 25mM HEPES, pH to 7.2 with KOH) prior to pelleting at 1050 rpm, 10 minutes. Pellets were subsequently washed in 500μL IC buffer and re-pelleted. Pellets were resuspended in 100μL of serum-free media and incubated with 2% EtOH for 30min at 37°C. Parasites were pelleted at 1000g, 5min at 4°C. The supernatant was subsequently transferred to new Eppendorf tubes and re-pelleted at 2000g, 5min at 4°C. Final supernatant (ESA - excreted secreted antigens) and pellet fractions were resuspended in 120μL SDS sample buffer final volume and subjected to 5min boiling at 95°C and two sonication cycles, prior to immunoblotting.
Plaque Assay
HFF monolayers were infected with 100μL of serially diluted parasites (1/100, 1/1000 and 1/10000) and allowed to develop for 7 days ± ATc. Plaques were fixed in 200μL of 4% paraformaldehyde, 0.05% glutaraldehyde (PAF-Glu), 10 minutes quenched in 600μL of 0.1M glycine-PBS and subsequently stained with 200μL Crystal Violet (Sigma-Aldrich), 10min. Data are representative of three independent biological experiments.
T. gondii Growth Assay
20μL of freshly egressed parasites ± ATc 24hrs were inoculated onto HFF coated coverslips. 24hrs post-infection the parasites were fixed with 200μL PAF-Glu for 20min, quenched in 600μL 0.1M glycine-PBS. Growth was assessed via immunofluorescence assay staining for both GAP45 (1/10000) and GRA3 (1/2000). 100 vacuoles were counted for three independent experiments. Data presented is mean value ± SD of experiments.
Immunofluorescence Assay
Previously fixed cells were permeabilized 20min in 100μL 0.2%Triton-PBS, blocked for 20min in 100μL 2%BSA-PBS. 100μL of primary antibodies (diluted as required in PBS) were incubated for 1hr, washed 3 times in 500μL PBS, followed by a 1hr incubation of 100μL of secondary antibodies and washed as previously. Coverslips were mounted onto slides with 3-5μL DAPI-Fluromount G (SouthernBiotech).
Molecular Dynamics Simulations
Simulations were performed using gromacs 4.6 and gromacs 5 (www.gromacs.org) with GPU acceleration (Hess et al., 2008). The lowest-energy PfAPH NMR model was simulated using the GROMOS56a3 force field (Oostenbrink et al., 2004) in 0.15 M NaCl for 100 ns. Multiple frames from the final 75 ns of this simulation were used to generate MARTINI version 2.2 (http://md.chem.rug.nl/) (de Jong et al., 2013) coarse-grained PfAPH parameters with the martinize.py script, using an elastic network for structured regions with a 1 nm cutoff and a force constant of 500 kJ mol–1 nm–2. A 300-lipid POPC membrane was generated by self-assembly(Scott et al., 2008) and individual lipids from each leaflet were randomly converted to POPA in order to generate symmetric mixed PC:PA bilayers of the following compositions: 0%, 10%, 20%, 30%, 40% and 50% POPA (Koldso et al., 2014). The protein was then centered at 9 nm from the membrane centre-of-mass and randomly rotated in x, y, and z dimensions to generate 5 separate starting points for each lipid composition. Simulations were performed at 310 K using the V-rescale algorithm and 1 tau using the Parrinello-Rahman barostat (Bussi et al., 2007) with semiisotropic coupling. Visualisation used Pymol (http://pymol.org) and VMD (Humphrey et al., 1996). Lipid density isosurfaces of phosphate particles in the reference frame of the protein were generated using the Volmap plugin of VMD. Lipid contacts were calculated between each residue of the protein and the phosphate headgroup particles of POPA and POPC using a cutoff of 1.0 nm. Lipid contact analysis was performed as described elsewhere (Hedger et al., 2016) using scripts from Heidi Koldsoe (D.E.Shaw Research).
Quantification and Statistical Analysis
The coordinates of the final ensembles of PfAPH106-235 and TgAPH99-229 structures are deposited at the Protein Data Bank Europe (https://www.ebi.ac.uk/pdbe/) under the accession codes 6F24 and 6F8E respectively. PfAPH106-235 and TgAPH99-229 assigned chemical shifts are also deposited at the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) under the accession numbers 34202 and 34216 respectively.
Data and Software Availability
HSQC Titration Analysis
Spectra were overlaid and chemical shifts measured for assigned backbone resonances using NMRview software. All combined chemical shift perturbation values were calculated using ((Δ1H chemical shift)2 +(0.2Δ15N chemical shift))1/2 (Williamson, 2013). Mean noise was calculated using an iterative method (Williamson, 2013).
PRE Analysis
Peak intensities for assigned backbone amide resonances were measured using NMRview software. Signal reduction was obtained from (I∗/Io), where I∗ and Io are equal to peak intensities in the presence of doped and non-doped bicelles respectively, and expressed as a percentage. Signal reduction was subtracted from mean noise to obtain relative signal reduction.
1D 1H-NMR LUV Titration Analysis
Using Topspin 3.5 software (Bruker), peaks in the region corresponding to amide (9.5 to 6.4ppm) and aliphatic methyl (0.255 to -0.175ppm) groups were integrated for TgAPH99-229 and PfAPH106-2351D 1H-NMR spectra respectively, to exclude resonances from lipids and unstructured protein regions. The fraction of bound protein is expressed as 1-I/I0, where I is the integral of protein NMR signal for a given total available lipid concentration (ALC) and I0 is the integral of protein NMR signal when ALC = 0 (protein alone). Total available lipid is calculated as half the total lipid concentration added to account for the inaccessible lipid present in the LUV inner leaflet. OriginPro software was used to plot fraction of bound protein against ALC and fit non-linear binding isotherms to estimate apparent dissociation constants (Kdapp) according to (Ceccon et al., 2013):
PC represents the total protein concentration and Bmax is a fixed constant. Titrations with varying LUV compositions were replicated in triplicate whilst titrations with POPA enriched LUVs for TgAPH99-229 or PfAPH106-235 mutants were replicated in duplicate. Error bars for binding curves represent1σ from the mean for replicates, whilst error bars for calculated Kdapp represent 1σ from the mean for fitting binding curves.
Circular Diochroism
From 5 repeats, spectra were averaged, corrected for baseline contributions, and the net spectra smoothed with a Savitsky–Golay filter (window 2).
Acknowledgments
The authors would like to thank the Wellcome Trust (PhD program; equipment grant no. 085464 awarded to S.M.) and The Leverhulme Trust (grant RPG-2018-107). This research was supported by the Swiss National Science Foundation 310030B_166678 (to D.S.-F.). D.J.D is supported by IZRJZ3_164183 (to Dr. Karine Frenal).
Author Contributions
N.D., D.J.D., and S.L.R designed and conducted experiments, wrote the paper, and constructed figures. P.M.H., T.B., and S.B. conducted experiments. B.L. contributed to solving NMR structures. D.S.-F. and S.M. designed experiments, wrote the paper, and secured funding.
Declaration of Interests
The authors declare no competing financial interests.
Published: June 14, 2018
Footnotes
Supplemental Information includes six figures and three tables and can be found with this article online at https://doi.org/10.1016/j.str.2018.05.001.
Contributor Information
Dominique Soldati-Favre, Email: dominique.soldati-favre@unige.ch.
Steve Matthews, Email: s.j.matthews@imperial.ac.uk.
Supporting Citations
The following references appear in the Supplemental Information: Kim et al., 1993.
Supplemental Information
References
- Bhatt S., Weiss D.J., Cameron E., Bisanzio D., Mappin B., Dalrymple U., Battle K.E., Moyes C.L., Henry A., Eckhoff P.A. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet M., Collins M.O., Smith T.K., Thompson E., Sebastian S., Volkmann K., Schwach F., Chappell L., Gomes A.R., Berriman M. Phosphoinositide metabolism links cGMP-dependent protein kinase G to essential Ca2+ signals at key decision points in the life cycle of malaria parasites. PLoS Biol. 2014;12:e1001806. doi: 10.1371/journal.pbio.1001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen H.E., Jia Y., Yamaryo-Botte Y., Bisio H., Zhang O., Jemelin N.K., Marq J.B., Carruthers V., Botte C.Y., Soldati-Favre D. Phosphatidic acid-mediated signaling regulates microneme secretion in toxoplasma. Cell Host Microbe. 2016;19:349–360. doi: 10.1016/j.chom.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Carruthers V.B., Sibley L.D. Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol. Microbiol. 1999;31:421–428. doi: 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- Ceccarelli D.F.J., Blasutig I.M., Goudreault M., Li Z., Ruston J., Pawson T., Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Ceccon A., D'Onofrio M., Zanzoni S., Longo D.L., Aime S., Molinari H., Assfalg M. NMR investigation of the equilibrium partitioning of a water-soluble bile salt protein carrier to phospholipid vesicles. Proteins. 2013;81:1776–1791. doi: 10.1002/prot.24329. [DOI] [PubMed] [Google Scholar]
- Cowman A.F., Crabb B.S. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- de Jong D.H., Singh G., Bennett W.F., Arnarez C., Wassenaar T.A., Schafer L.V., Periole X., Tieleman D.P., Marrink S.J. Improved parameters for the Martini Coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell A., Thirugnanam S., Lorestani A., Dvorin J.D., Eidell K.P., Ferguson D.J., Anderson-White B.R., Duraisingh M.T., Marth G.T., Gubbels M.J. A DOC2 protein identified by mutational profiling is essential for apicomplexan parasite exocytosis. Science. 2012;335:218–221. doi: 10.1126/science.1210829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B.A., Ristuccia J.G., Gigley J.P., Bzik D.J. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot. Cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A., Unger V.M., De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedger G., Rouse S.L., Domanski J., Chavent M., Koldso H., Sansom M.S. Lipid-loving ANTs: molecular simulations of cardiolipin interactions and the organization of the adenine nucleotide translocase in model mitochondrial membranes. Biochemistry. 2016;55:6238–6249. doi: 10.1021/acs.biochem.6b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-38. [DOI] [PubMed] [Google Scholar]
- Huynh M.H., Carruthers V.B. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D., Tosetti N., Pires I., Stock J., Graindorge A., Hung Y.F., Han H., Tewari R., Kursula I., Soldati-Favre D. An apicomplexan actin-binding protein serves as a connector and lipid sensor to coordinate motility and invasion. Cell Host Microbe. 2016;20:731–743. doi: 10.1016/j.chom.2016.10.020. [DOI] [PubMed] [Google Scholar]
- Jean S., Zapata-Jenks M.A., Farley J.M., Tracy E., Mayer D.C. Plasmodium falciparum double C2 domain protein, PfDOC2, binds to calcium when associated with membranes. Exp. Parasitol. 2014;144:91–95. doi: 10.1016/j.exppara.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Jian X., Tang W.K., Zhai P., Roy N.S., Luo R., Gruschus J.M., Yohe M.E., Chen P.W., Li Y., Byrd R.A. Molecular basis for cooperative binding of anionic phospholipids to the PH domain of the Arf GAP ASAP1. Structure. 2015;23:1977–1988. doi: 10.1016/j.str.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimah J.R., Salinas N.D., Sala-Rabanal M., Jones N.G., Sibley L.D., Nichols C.G., Schlesinger P.H., Tolia N.H. Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption. Elife. 2016;5:e20621. doi: 10.7554/eLife.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.S., Zweckstetter M. Mars – robust automatic backbone assignment of proteins. J. Biomol. NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- Karandur D., Nawrotek A., Kuriyan J., Cherfils J. Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane. Proc. Natl. Acad. Sci. USA. 2017;114:11416–11421. doi: 10.1073/pnas.1707970114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schneider K.A. Evolution of drug resistance in malaria parasite populations. Nat. Educ. Knowl. 2013;4:6. [Google Scholar]
- Kim K., Soldati D., Boothroyd J.C. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science. 1993;262:911–914. doi: 10.1126/science.8235614. [DOI] [PubMed] [Google Scholar]
- Koldso H., Shorthouse D., Helie J., Sansom M.S. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. 2014;10:e1003911. doi: 10.1371/journal.pcbi.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppisetti R.K., Fulcher Y.G., Jurkevich A., Prior S.H., Xu J., Lenoir M., Overduin M., Van Doren S.R. Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12. Nat. Commun. 2014;5:5552. doi: 10.1038/ncomms6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-L., Srivastava A., Pilling C., Chase A.R., Falke J.J., Voth G.A. Molecular mechanism of membrane binding of the GRP1 PH domain. J. Mol. Biol. 2013;425:3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lenoir M., Kufareva I., Abagyan R., Overduin M. Membrane and protein interactions of the pleckstrin homology domain superfamily. Membranes. 2015;5:646–663. doi: 10.3390/membranes5040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., Levine T.P. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Lucas N., Cho W. Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 2011;286:41265–41272. doi: 10.1074/jbc.M111.300806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb C.N., He J., Xue Y., Stansfeld P.J., Stahelin R.V., Kutateladze T.G., Sansom M.S.P. Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhab-Jafari M.T., Marshall C.B., Smith M.J., Gasmi-Seabrook G.M.C., Stathopulos P.B., Inagaki F., Kay L.E., Neel B.G., Ikura M. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. USA. 2015;112:6625. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Meissner M., Brecht S., Bujard H., Soldati D. Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res. 2001;29:E115. doi: 10.1093/nar/29.22.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercredi P.Y., Bucca N., Loeliger B., Gaines C.R., Mehta M., Bhargava P., Tedbury P.R., Charlier L., Floquet N., Muriaux D. Structural and molecular determinants of membrane binding by the HIV-1 matrix protein. J. Mol. Biol. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudy R., Manning T.J., Beckers C.J. The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J. Biol. Chem. 2001;276:41492–41501. doi: 10.1074/jbc.M106154200. [DOI] [PubMed] [Google Scholar]
- Ni T., Kalli A.C., Naughton F.B., Yates L.A., Naneh O., Kozorog M., Anderluh G., Sansom M.S.P., Gilbert R.J.C. Structure and lipid-binding properties of the kindlin-3 pleckstrin homology domain. Biochem. J. 2017;474:539–556. doi: 10.1042/BCJ20160791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink C., Villa A., Mark A.E., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Pang X., Fan J., Zhang Y., Zhang K., Gao B., Ma J., Li J., Deng Y., Zhou Q., Egelman E.H. A PH domain in ACAP1 possesses key features of the BAR domain in promoting membrane curvature. Dev. Cell. 2014;31:73–86. doi: 10.1016/j.devcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping H.A., Kraft L.M., Chen W., Nilles A.E., Lackner L.L. Num1 anchors mitochondria to the plasma membrane via two domains with different lipid binding specificities. J. Cell Biol. 2016;213:513–524. doi: 10.1083/jcb.201511021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirahmadi S., Zakeri S., Mehrizi A.A., Djadid N.D. Analysis of genetic diversity and population structure of gene encoding cell-traversal protein for ookinetes and sporozoites (CelTOS) vaccine candidate antigen in global Plasmodium falciparum populations. Infect. Genet. Evol. 2018;59:113–125. doi: 10.1016/j.meegid.2018.01.023. [DOI] [PubMed] [Google Scholar]
- Putta P., Rankenberg J., Korver R.A., van Wijk R., Munnik T., Testerink C., Kooijman E.E. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta. 2016;1858:2709–2716. doi: 10.1016/j.bbamem.2016.07.014. [DOI] [PubMed] [Google Scholar]
- Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T.E., Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Robert-Gangneux F., Darde M.L. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin. Microbiol. Rev. 2012;25:264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiko M.S., Carruthers V.B. Functional dissection of Toxoplasma gondii perforin-like protein 1 reveals a dual domain mode of membrane binding for cytolysis and parasite egress. J. Biol. Chem. 2013;288:8712–8725. doi: 10.1074/jbc.M113.450932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.M., Soldati-Favre D. Invasion factors are coupled to key signalling events leading to the establishment of infection in apicomplexan parasites. Cell. Microbiol. 2011;13:787–796. doi: 10.1111/j.1462-5822.2011.01585.x. [DOI] [PubMed] [Google Scholar]
- Scott K.A., Bond P.J., Ivetac A., Chetwynd A.P., Khalid S., Sansom M.S. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Brown K.M., Lee T.D., Sibley L.D. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio. 2014;5 doi: 10.1128/mBio.01114-14. e01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Chitnis C.E. Signalling mechanisms involved in apical organelle discharge during host cell invasion by apicomplexan parasites. Microbes Infect. 2012;14:820–824. doi: 10.1016/j.micinf.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Soldati D., Boothroyd J.C. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science. 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- Vonkova I., Saliba A.-E., Deghou S., Anand K., Ceschia S., Doerks T., Galih A., Kugler K.G., Maeda K., Rybin V. Lipid cooperativity as a general membrane-recruitment principle for PH domains. Cell Rep. 2015;12:1519–1530. doi: 10.1016/j.celrep.2015.07.054. [DOI] [PubMed] [Google Scholar]
- Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. NMR Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Yamamoto E., Kalli A.C., Yasuoka K., Sansom M.S.P. Interactions of Pleckstrin homology domains with membranes: adding back the bilayer via high-throughput molecular dynamics. Structure. 2016;24:1421–1431. doi: 10.1016/j.str.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.