Summary
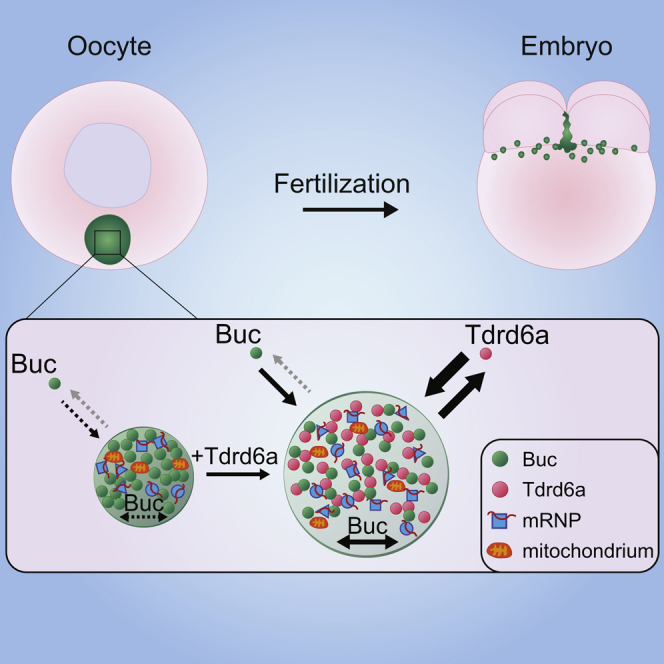
Phase separation represents an important form of subcellular compartmentalization. However, relatively little is known about how the formation or disassembly of such compartments is regulated. In zebrafish, the Balbiani body (Bb) and the germ plasm (Gp) are intimately linked phase-separated structures essential for germ cell specification and home to many germ cell-specific mRNAs and proteins. Throughout development, these structures occur as a single large aggregate (Bb), which disperses throughout oogenesis and upon fertilization accumulates again into relatively large assemblies (Gp). Formation of the Bb requires Bucky ball (Buc), a protein with prion-like properties. We found that the multi-tudor domain-containing protein Tdrd6a interacts with Buc, affecting its mobility and aggregation properties. Importantly, lack of this regulatory interaction leads to significant defects in germ cell development. Our work presents insights into how prion-like protein aggregations can be regulated and highlights the biological relevance of such regulatory events.
Keywords: phase separation, Tdrd6, Tudor domain, Bucky ball, germ plasm, Balbiani body, zebrafish, arginine methylation, prion-like domain, primordial germ cell
Graphical Abstract
Highlights
-
•
Tdrd6a is required for Bucky ball mobility within aggregates, and for PGC formation
-
•
Maternal Tdrd6a coordinates transcript deposition into future PGCs
-
•
A dimethylated tri-RG motif in Bucky ball mediates interaction with Tdrd6a
-
•
The tri-RG motif is essential for Balbiani body and germ cell formation
Zebrafish Balbiani body and germ plasm are related phase-separated structures. Roovers, Kaaij et al. show that Tdrd6a is required for their formation and mobility through interaction with dimethylated arginines in the prion-like protein Bucky ball, revealing a role for Tudor domain-methylated arginine interactions in in vivo phase separation modulation.
Introduction
Phase-separating mechanisms have been acknowledged as important aspects of cell biology. After the initial description of the liquid-like behavior of P granules, peri-nuclear RNA-rich protein aggregates in the Caenorhabditis elegans germline and many other RNA-containing granules have been shown to have similar properties (Brangwynne et al., 2009, Brangwynne et al., 2011, Kroschwald et al., 2015). Important players in the formation of these structures are proteins containing intrinsically disordered regions (IDRs) and/or prion-like domains (PrDs) (Kato et al., 2012, Kroschwald et al., 2015). Such proteins have the propensity to self-aggregate and potentially trigger other proteins to phase separate as well (Prusiner, 1998, Shorter and Lindquist, 2005). In many ways, biologically functional protein assemblies such as P granules resemble pathogenic protein-aggregation states. It has been suggested that such disease-causing aggregations are an extreme manifestation of an abundantly used mechanism to form membrane-less compartments (Shin and Brangwynne, 2017). This suggests that mechanisms are in place that prevent healthy, functional aggregates to transform into pathological forms.
In many organisms, germ cell fate is imposed on cells through the cytoplasmic inheritance of P granule-like structures, called germ plasm (Gp) (Ikenishi, 1998, Raz, 2003). In zebrafish, Gp originates from an evolutionary conserved electron-dense aggregate in the oocyte, called the Balbiani body (Bb) (Kloc et al., 2004). The mRNAs enriched in the Bb and Gp are often germline-specific, and in zebrafish, these include vasa, nanos3, and dazl (Hashimoto et al., 2004, Köprunner et al., 2001, Yoon et al., 1997). Depletion of single Gp mRNAs can have detrimental effects on primordial germ cell (PGC) numbers, showing that individual Gp components are important for PGC specification and survival (Köprunner et al., 2001, Slaidina and Lehmann, 2017, Tzung et al., 2015, Weidinger et al., 2003).
Bucky ball (Buc) is a protein known to play a key role in the formation of the Bb in zebrafish (Bontems et al., 2009, Marlow and Mullins, 2008). Overexpression of Buc in zygotes revealed that Buc is sufficient to induce ectopic PGCs, suggesting it is also involved in the formation of the Bb-related Gp structure (Bontems et al., 2009). Buc contains a PrD, and elegant studies on its homolog in Xenopus (Xvelo) have demonstrated that these proteins self-aggregate into membrane-less organelles that display amyloid-like features (Boke et al., 2016).
Core Piwi-interacting RNA (piRNA) pathway components, such as Ziwi in zebrafish and Aub in Drosophila, are present in the Gp as well (Harris and Macdonald, 2001, Houwing, 2009). Furthermore, it has been shown in Drosophila that piRNA pathway components inherited via the Gp are essential for transposon silencing in the offspring (Brennecke et al., 2008), and piRNA-mRNA interactions have been proposed to drive mRNA localization to Gp (Barckmann et al., 2015, Vourekas et al., 2016). Many proteins involved in the piRNA pathway have been identified through genetic and biochemical approaches including multi-Tudor domain-containing proteins (Tdrds) (Siomi et al., 2010). Tdrds play important roles in the formation of nuage, a peri-nuclear protein-RNA aggregate that associates closely with mitochondria. For some Tdrds, it has been shown that they bind to symmetrically dimethylated arginine (sDMA) residues on their interaction partners. In zebrafish, for instance, the interaction between Tdrd1 and the Piwi protein Zili is mediated via a specific sDMA site in Zili (Huang et al., 2011).
One of the Tdrds that has received relatively little attention is Tdrd6, the closest vertebrate homolog to Drosophila Tudor (Tud). Tud has been shown to interact with Piwi proteins Aub and Ago3 and plays a role in the localization of Aub to Gp and polar granule formation (Kirino et al., 2010, Nishida et al., 2009, Thomson and Lasko, 2004). In mice, TDRD6 plays a role in establishing the chromatoid body, a testis-specific structure that resembles Gp, and the localization of piRNA pathway components to this body (Vasileva et al., 2009). In addition, it is involved in spliceosome assembly in primary spermatocytes (Akpınar et al., 2017). However, a specific molecular function of Tdrd6 or Tud has thus far not been demonstrated.
We show that Tdrd6a is required for coordinated loading of essential Gp components into PGCs through fine-tuning of the aggregating properties and mobility of the Bb organizer Buc. The Tdrd6a-Buc interaction represents one of the few documented cases that demonstrate how the aggregation of a prion-like protein is regulated in vivo. We speculate that similar phase separation-regulating mechanisms may act in other cell types as well.
Results
Tdrd6a Is Gonad Specific and Localizes to Nuage, the Bb, and Gp
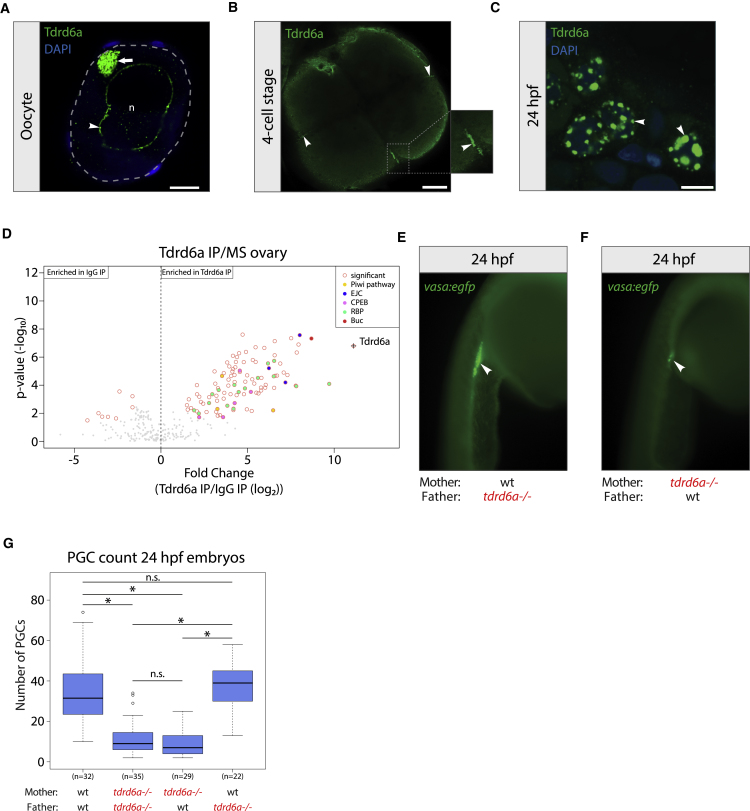
The zebrafish genome encodes three Tdrd6 paralogs: tdrd6a–c. In this study, we focused on tdrd6a. Tdrd6a contains seven Tudor domains and is 2,117 amino acids in length (Figure S1A). Germline-specific expression of tdrd6a was validated by RT-PCR (Figure S1B). Immunohistochemistry (IHC) confirmed that Tdrd6a is expressed in the ovary, where it localizes to nuage (Figure 1A, arrowhead) and to the Bb (Figure 1A, arrow). Tdrd6a is also maternally provided and localizes to the Gp in 4-cell stage embryos (Figure 1B, arrowheads). 24 hours post fertilization (hpf), Tdrd6a is restricted to PGCs, where it again localizes to nuage (Figure 1C, arrowheads). We confirmed the identity of the Tdrd6a-containing structures using established markers for the nuage, Bb and the Gp, using both IHC and localization of transgenic Tdrd6a-mCherry (Figure S1C). These results demonstrate that Tdrd6a is maternally contributed and localizes to three conserved and related structures involved in germline specification and maintenance: the Bb, Gp, and nuage.
Figure 1.
Tdrd6a Is Germline Specific and Required for PGC Formation
(A) IHC for Tdrd6a in oocytes. Arrowhead and arrow indicate Tdrd6a staining in the nuage and Bb, respectively. Gray dashed line outlines the cell, n = nucleus. Scale bar, 10 μm.
(B) IHC for Tdrd6a in 4-cell stage embryos. Arrows indicate Tdrd6a localization to the Gp. Scale bar, 100 μm.
(C) Tdrd6a localizes to peri-nuclear nuage granules (arrowheads) in PGCs at 24 hpf. Scale bar, 7.5 μm.
(D) MS of Tdrd6a IPs on an ovary, compared to IgG (immunoglobulin G) control.
(E and F) 24 hpf embryos derived from wt (E) or tdrd6a mutant mothers (F) in a vasa:egfp background. Arrowheads indicate the PGCs.
(G) Quantification of PGC numbers in 24 hpf embryos from the crosses indicated on the x axis (∗ indicates p value < 0.0001, n.s. = non-significant, calculated by Wilcoxon test).
See also Figures S1 and S2.
Identification and Characterization of a tdrd6a Mutant Allele
We isolated a tdrd6a allele harboring a premature stop codon (Q158X) from an ENU mutagenized library (Wienholds, 2002). Western blot analysis confirmed loss of Tdrd6a in homozygous mutant animals (Figure S1D). Tdrd6a−/− oocytes showed complete loss of Tdrd6a staining in peri-nuclear nuage (Figure S1E, arrowhead) and Gp in 4-cell stage embryos (Figure S1F, arrowheads). Some residual staining remained in the Bb in tdrd6a mutants (Figure S1E, arrow); however, a strong Tdrd6a-related Bb phenotype (see later) and the presence of a Tdrd6a-mCherry transgene in both nuage and the Bb suggest that this is due to cross reactivity of the antibody in IHC. Homozygous zygotic (Z) and maternal-zygotic (MZ) tdrd6a mutants are fertile, indicating that Tdrd6a is not essential for fertility. We conclude that tdrd6aQ158X represents a strong loss-of-function allele.
Tdrd6a Does Not Affect piRNAs
Next, we performed a Tdrd6a immunoprecipitation (IP) on ovary lysates, followed by label-free quantitative mass spectrometry (Figure 1D). Besides Tdrd6a, we found strong enrichments for several complexes containing RNA-binding proteins (RBPs), including the Exon Junction Complex (EJC) and the cytoplasmic polyadenylation element binding protein complex (CPEB). In addition, we identified the Piwi pathway components Ziwi, Zili, and Tdrd7. Finally, we found that Buc was highly enriched.
Given the interaction with Ziwi and Zili, we probed for a role of Tdrd6a in the piRNA pathway. We first validated the Tdrd6a interaction with Ziwi and Zili (Figure S2A). Despite these interactions, small RNA (smRNA) sequencing of total ovary did not show significant differences between piRNAs of tdrd6a+/− and tdrd6a−/− animals (Figures S2B–S2D). We only observed a small but significant reduction in the typical antisense bias for piRNAs mapping to retrotransposons (Figure S2E). When we roughly divided oocytes into early (⌀ < 300 μm) and later stages (⌀ > 300 μm), we noticed that this represents a defect in accumulation of antisense piRNAs during early oogenesis only (Figures S2F–S2J). In conclusion, while Tdrd6a associates with Ziwi and Zili, its absence barely affects piRNA populations.
Tdrd6a Affects PGC Formation
MZ tdrd6a mutants have a strong tendency to develop into males. Since the amount of PGCs can have an impact on sex determination in zebrafish (Tzung et al., 2015), we examined the effect of Tdrd6a on PGC formation. In both wild-type (wt) and MZ tdrd6a−/− embryos, PGCs marked by the vasa:egfp transgene (Krøvel and Olsen, 2002) were at the genital ridge at 24 hpf (Figures 1E and 1F, arrowhead). However, we observed a significant reduction in PGC number in the offspring from tdrd6a−/− females, irrespective of the genotype of the father (Figure 1G).
Tdrd6a Affects Coordinated Loading of Gp mRNAs into PGCs
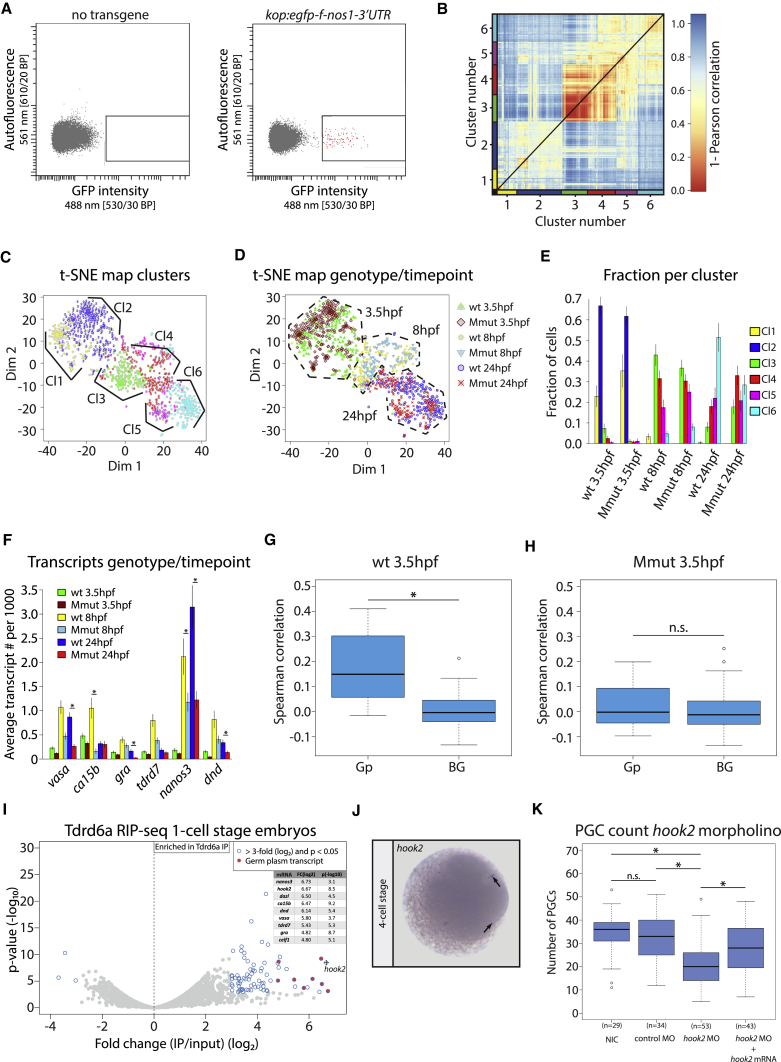
To learn more about the underlying cause of the PGC defect, we performed single-cell RNA-sequencing (scRNA-seq) on PGCs isolated from embryos spawned by tdrd6a+/− (wt) and tdrd6a−/− (Mmut) mothers, both crossed with tdrd6a+/+ males. PGCs were marked using the kop:egfp-f-nos1-3ʹ UTR transgene (Blaser et al., 2005) and isolated by fluorescence-activated cell sorting (FACS) (Figure 2A). Three time points were analyzed: (1) when PGCs can be first identified using transgenic GFP expression (3.5 hpf), (2) during migration of the PGCs (8 hpf), and (3) when the PGCs have reached the genital ridge (24 hpf).
Figure 2.
Single Cell RNA-Seq Analysis Reveals that Maternal Tdrd6a Mediates Positive Correlation of Loading of Gp-Residing mRNAs into PGCs
(A) Flow cytometry plots of the sort strategy used in this study. Representative FACS plots of embryos 8 hpf without or with the kop:egfp transgene are shown. Positive events are indicated in red.
(B) Heatmap indicating transcriptome distances of ∼1,100 PGCs computed as 1-Pearson’s correlation coefficient. K-medoids clustering identified six clusters, which are color coded on the x and y axes.
(C) Similar as in (B) but now visualized in a t-SNE map. Clusters identified by k-medoids clustering are color coded as in (B).
(D) t-SNE map highlighting the genotype and developmental time point of the individual PGCs as indicated.
(E) Barplot displaying the fraction of cells per clusters identified in (B) for the different genotype-developmental time combinations. Error bars were derived from error propagation.
(F) Barplot showing the average transcript counts per 1,000 transcripts per cell of six Gp transcripts in all six different genotype-developmental time point combinations, as indicated. Error bars represent the SEM (∗ indicates p value < 0.01, calculated by negative binominal statistics and corrected for multiple testing [Benjamini–Hochberg]).
(G and H) Boxplots displaying the Gp-Gp and BG-BG correlations in wt and Mmut embryos, respectively (∗ indicates p value < 0.001, n.s. = non-significant, calculated by Wilcoxon test).
(I) Volcano plot displaying the fold difference between Tdrd6a RIP-seq and input on the x axis (average of three biological replicates). y axis: p value belonging to the observed differences between Tdrd6a RIP-seq and input. Listed are the values of enriched Gp transcripts.
(J) ISH against hook2 at the 4-cell stage. Arrows indicate Gp.
(K) Quantification of PGC numbers observed in embryos in morpholino knockdown (MO KD) injection experiment. NIC = non-injected control; the control MO targets the fus transcript; the hook2 mRNA contained mismatches at the hook2 MO target site and rescues the KD (∗ indicates p value < 0.01, n.s. = non-significant, calculated by Wilcoxon test).
See also Figure S3.
Roughly 1,100 PGCs were sequenced and analyzed using RaceID2 (Figure S3A) (Grün et al., 2014, Grün et al., 2016, Hashimshony et al., 2012). Representation of the pairwise distances of the single cell transcriptomes in a heatmap revealed two main clusters, which can be further subdivided into clusters 1 and 2 and clusters 3–6 by k-medoids clustering (Figure 2B). Representation of this data in t-distributed stochastic neighbor embedding (t-SNE) maps (Van Der Maaten and Hinton, 2008) revealed that clusters 1 and 2 predominantly harbor 3.5 hpf old PGCs, whereas clusters 3–6 consist of PGCs from 8 hpf and 24 hpf (Figures 2C and 2D). Consistent with this, the pluripotency gene nanog is selectively expressed in clusters 1 and 2 (Figure S3B) (Takahashi and Yamanaka, 2006). In contrast, the rps gene family, which has been shown to be upregulated after the maternal-to-zygotic transition (MZT) (Siddiqui et al., 2012), is expressed in clusters 3–6 (Figure S3C). No strong differences between genotypes could be observed for 3.5 hpf and 8 hpf PGCs (Figure 2E). However, a significant fraction of 24 hpf Mmut PGCs was enriched in cluster 4 (Figures 2E and S3D), which is dominated by wt PGCs of 8 hpf, suggesting that PGCs lacking maternal Tdrd6a experience developmental delay between 8 hpf and 24 hpf.
Since individual Gp transcripts can influence PGC numbers (Köprunner et al., 2001, Tzung et al., 2015, Weidinger et al., 2003), we tested if PGCs lacking Tdrd6a generally have lower Gp mRNA levels. Of the 8 known zebrafish Gp transcripts (Hashimoto et al., 2004, Köprunner et al., 2001, Strasser et al., 2008, Wang et al., 2013, Weidinger et al., 2003, Yoon et al., 1997), 6 transcripts passed our filtering criteria (see STAR Methods). While at 8 hpf and 24 hpf PGCs lacking Tdrd6a indeed tended to have significantly fewer Gp mRNAs than wt, at 3.5 hpf, no significant difference was found (Figure 2F). In line with this, bulk RNA-seq at the 1-cell stage did not reveal significant effects on mRNA levels (Figures S3E and S3F). Hence, the reduction in PGC number observed upon loss of maternal Tdrd6a most likely is not due to an overall reduction of Gp transcripts provided by the mother.
We then computed all pairwise correlations between the individual Gp mRNAs in wt PGCs at 3.5 hpf and compared these to pairwise correlations of non-Gp background (BG) mRNAs (see STAR Methods). This revealed a general positive correlation for Gp mRNAs in wt PGCs (Figures 2G and S3G), indicating that relatively fixed ratios of individual Gp transcripts are loaded into PGCs. Strikingly, in the absence of Tdrd6a, this positive correlation is completely lost (Figures 2H and S3G). Together, these data show that the stoichiometry of Gp mRNAs in single PGCs is tightly controlled and that this depends on maternally provided Tdrd6a.
Tdrd6a Interacts with Known Gp mRNAs
Since Tdrd6a is required for correct loading of Gp transcripts into PGCs, we next explored whether Tdrd6a interacts with Gp-residing mRNAs through RNA-IP followed by sequencing (RIP-seq). Strikingly, all known Gp mRNAs were strongly enriched in the Tdrd6a RIP-seq compared to input (Figure 2I). We validated these findings using Tdrd6a RIP-qPCR for the Gp markers vasa, dazl, and nanos3, revealing between a 50- and a 100-fold enrichment in the Tdrd6a RIPs (Figure S3H). The mRNA that was most strongly enriched in the RIP-seq was hook2 (Figure 2I). Hook2 is an unknown Gp component in zebrafish but reported to be present in Xenopus Gp (Owens et al., 2017). Interestingly, in our scRNA-seq data, hook2 behaves similar to other Gp markers and also displays the typical Tdrd6a-dependent positive correlation with other Gp transcripts (Figures S3I and S3J). Indeed, in situ hybridization (ISH) confirmed the presence of hook2 in zebrafish Gp (Figure 2J). Finally, translation inhibition morpholino (MO) injections revealed that hook2 affects PGC numbers (Figure 2K), substantiating that hook2 is a bona fide Gp component.
It has been reported that in Drosophila, the PIWI protein Aub plays a role in regulating Gp mRNA stability and localization (Barckmann et al., 2015, Vourekas et al., 2016). In analogy, we performed Ziwi RIP-qPCR experiments on 1-cell stage embryos for vasa, dazl, and nanos3, using the same experimental conditions used for the Tdrd6a RIP-qPCR experiment. The enrichment values for the tested mRNAs were all below 3-fold (Figure S3K), while western blot confirmed that the IPs were successful (Figure S3L). These enrichment values for Ziwi are in sharp contrast to the values obtained in Tdrd6a RIPs, indicating that Ziwi-messenger ribonucleoprotein (mRNP) interactions are not very prominent, or stable in these experiments.
Tdrd6a Affects Bb Organization
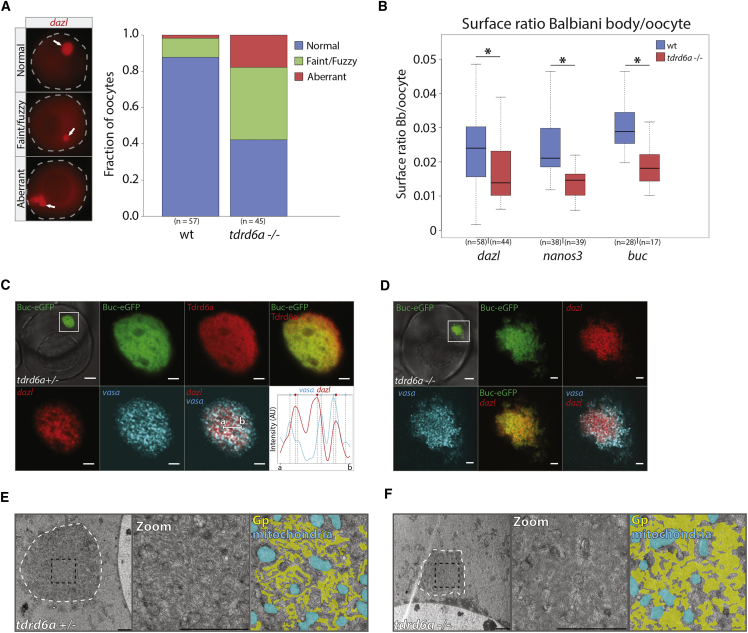
We next probed for Bb integrity in the presence and absence of Tdrd6a by doing whole mount fluorescence ISH (FISH) on oocytes against dazl. In tdrd6a mutant oocytes, the Bb often appears to be smaller relative to the entire oocyte, lacking a well-defined edge or even being further distorted. We quantified these defects by classifying the observed structural abnormalities (Figure 3A) and calculating the size ratio between the Bb and the oocyte using various probes (Figure 3B).
Figure 3.
Tdrd6a Is Required for Bb Integrity
(A) Quantification of Bb phenotypes as indicated based on dazl FISH on oocytes (examples indicated on the left, arrows indicate Bb).
(B) Surface ratio of Bbs in wt versus tdrd6a mutant oocytes (∗ indicates p value < 0.001, Wilcoxon test).
(C and D) Confocal images of Buc-eGFP-positive oocytes in tdrd6a+/− (C) and tdrd6a−/− (D) background. IHC for Tdrd6a and double smFISH was performed and displayed as indicated. Dazl and vasa signals typically do not overlap, illustrated in the line graph. Intensity for dazl (red) and vasa (cyan) signals over line a-b (see overlay), with vertical lines indicating fluorescence peaks per smFISH signal (highlighted by colored circle on top) showing transcript peaks are in a separate phase.
(E and F) Electron micrographs of Bbs of tdrd6a+/− (E) and tdrd6a−/− (F) oocytes (white dashed line). The zoom (black dashed square) is shown with (right) and without (middle) overlays that mark the Gp (yellow) and mitochondria (cyan). Scale bars, 10 μm (overview C and D), 2 μm (zoom) (C and D), and 2 μm (E and F).
See also Figure S4.
We extended these experiments by combining double single-molecule FISH (smFISH) with IHC for Tdrd6a in a Buc-eGFP background (Riemer et al., 2015). In the Bb, Buc-eGFP and Tdrd6a form a continuous structure in which Gp mRNAs are embedded (Figure 3C). SmFISH shows that different Gp transcripts display diverse sub-localization within the Bb. The dazl signal is found as a rather compact core in the Bb, whereas vasa is found more throughout the entire Bb (Figure 3C). Interestingly, the smFISH signals do not overlap with each other but rather form transcript-specific networks (Figure 3C, line graph). In tdrd6a mutant oocytes, the Buc-eGFP signal is more irregular (Figure 3D). Gp-transcripts still localize to the Bb, indicating that Tdrd6a is not essential for these transcripts to accumulate in the Bb (Figure 3D).
Electron microscopy (EM) revealed that the electron-dense structures in the Bb display a heterogeneous, fibrillary appearance (Figures 3E and S4A, yellow overlays). In contrast, Bbs without Tdrd6a have larger and more homogenous electron-dense areas than with Tdrd6a (Figures 3F and S3B, yellow overlays). A more widely conserved function of the Bb is mitochondrial selection, which is therefore highly represented in the Bb (Bilinski et al., 2017). We found that mitochondria still accumulate in the Bb in the absence of Tdrd6a (Figures 3E, 3F, S4A, and S4B, cyan overlays). In conclusion, Tdrd6a is required for the overall organization of the Bb, even though mRNAs and mitochondria are still present.
Tdrd6a Is Required for Merging Particles with Distinct mRNA Content into Mature Gp Structures in the Embryo
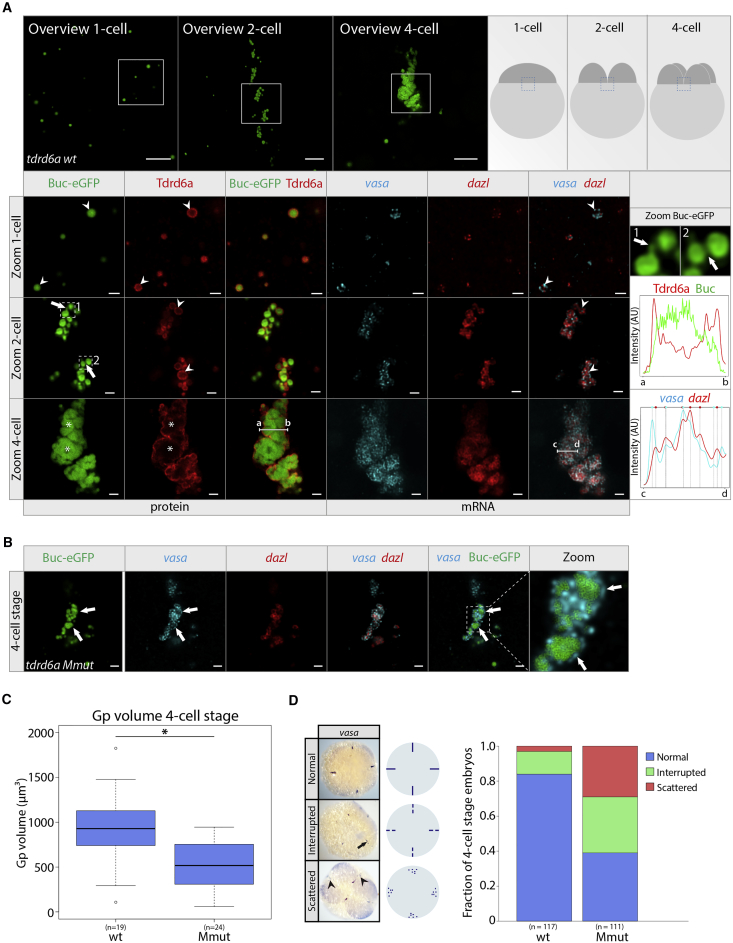
In late oogenesis, the Bb disperses into fragments at the vegetal cortex of the oocyte. Upon fertilization, these Buc-containing assemblies accumulate at the cleavage planes to form larger Gp structures (Riemer et al., 2015). Using smFISH and IHC, we found that in 1-cell stage embryos, Buc and Tdrd6a form isolated particles, decorated with discrete mRNA foci at their periphery (Figure 4A, arrowheads). Buc forms the core of the Gp particles, whereas the Tdrd6a signal is predominantly found at the edge (Figure 4A, arrowheads). As in the Bb, transcript signals do not overlap. At the 2-cell stage, the smaller Buc-Tdrd6a units organize themselves along the cleavage planes and start to cluster together (Figure 4A). The Buc signal often bridges individual granules (Figure 4A, arrows). Furthermore, Tdrd6a appears to be localized around the Buc-assemblies, similar to the mRNA (Figure 4A, arrowheads).
Figure 4.
Tdrd6a Is Required for Gp Integrity
(A) Confocal images of Buc-eGFP-positive embryos from tdrd6a+/− mothers, at 1-, 2-, and 4-cell stages, focusing on Gp as schematically indicated. In zoom of the Gp, IHC for Tdrd6a and double smFISH was performed and displayed as indicated. Arrowheads indicate mRNA and Tdrd6a that is peripherally localized on the Gp granule; arrows indicate Buc-eGFP bridges (zooms 1 and 2 on the right). Asterisks mark mature Gp, containing fused Buc-eGFP and mRNA networks inside the structure. Line graphs display intensity for Buc (green) versus Tdrd6a (red) signals over line a-b and dazl (red) versus vasa (cyan) intensity over line c-d, with vertical lines indicating fluorescence peaks per smFISH signal (highlighted by colored circle on top).
(B) Confocal images of Buc-eGFP-positive Gp of a 4-cell stage embryo from a tdrd6a−/− mother (Mmut). SmFISH was performed and displayed as indicated. Arrows indicate areas where Buc-eGFP has fused, but mRNA remains peripherally localized.
(C) Boxplot representing volumes of the Buc-eGFP signal at the cleavage planes of wt and Mmut 4-cell stage embryos. The largest Gp fragment of each embryo was measured (also see Figure S5B) after 3D reconstruction in Imaris. (∗ indicates p value < 0.001, Wilcoxon test).
(D) Quantification of Gp phenotypes of 4-cell stage embryos using an ISH against vasa. Scale bars, 10 μm (overview A), 2 μm (zoom) (A), and 2 μm (B).
See also Figure S5.
The Gp grows further toward the 4-cell stage into a larger structure, in which the Tdrd6a signal surrounds the Buc signal (Figure 4A). We also observe that in these parts of the Gp transcripts have mostly moved inward, forming large intermingled networks (Figure 4A, line graph). Overall, the smFISH signals for different mRNAs are very well mixed within the larger Gp structure, but areas of overall enrichment for one or the other mRNA can still be observed. We note that structures similar to the internal smFISH signal were found using antibody-mediated FISH (Figure S5A), suggesting that the peripheral Tdrd6a signal on Gp does not result from issues related to general antibody penetration into the structure.
In embryos lacking Tdrd6a, mRNAs still associate with Buc particles (Figure 4B), showing that like in the Bb, Tdrd6a is not required for this association. However, without Tdrd6a the Gp structure fails to grow and remains relatively small and highly fragmented (Figures 4C, 4D, and S5B). We do observe some apparent fusion of Buc particles, but typically, also in these cases, mRNA remains at the periphery (Figure 4B, arrows). These observations lead us to propose that Gp forms through the ongoing accumulation of small granules, containing Buc, Tdrd6a, and mRNPs. Tdrd6a contributes to the accumulation of these granules and for the mRNP particles to move into the Buc structure, where they intermingle and form networks with mRNPs of the same kind. We speculate that it is the lack of Gp growth that ultimately results in the above described Gp mRNA defects we see in PGCs lacking Tdrd6a.
Tdrd6a Interacts with Buc via Symmetrically Dimethylated Arginines
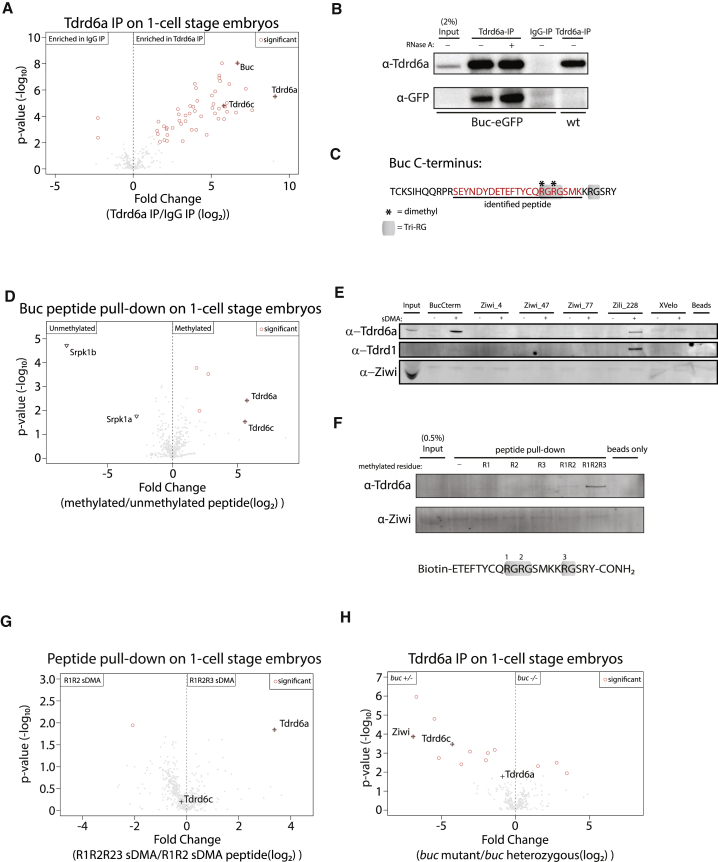
The IP-mass spectrometry (MS) experiments on ovary extracts identified Buc as a strong interactor of Tdrd6a (Figure 1D). We also found Buc, as well as the close Tdrd6a paralog Tdrd6c, to be among the strongest interactors of Tdrd6a in freshly laid embryos (Figure 5A). We verified the Buc-Tdrd6a interaction on western blot and show resistance to RNase A treatment (Figure 5B). Tdrds often bind sDMA residues in a binding partner (Siomi et al., 2010). Indeed, analysis of our MS results identified two dimethylated arginine residues within the C terminus of Buc, residing in a tri-RG (RG[X0-4]RG[X0-4]RG) motif (Figure 5C) (Thandapani et al., 2013). In order to test their relevance for interaction with Tdrd6a, we performed pull-down experiments using biotinylated peptides covering these arginines in either an sDMA- or non-methylated state followed by MS. In the pull-down using the methylated Buc-peptide, Tdrd6a was highly enriched (Figure 5D). Interestingly, another Tdrd6 paralog, Tdrd6c, was also among the few enriched proteins. The pull-down with the non-methylated peptide showed enrichment for two members of the serine-arginine protein kinase family, Srpk1a and Srpk1b (Figure 5D), confirming that this pull-down was also successful and revealing potential additional post-translational regulation of Buc besides sDMAs. To test the specificity of the Buc peptide for Tdrd6a, we repeated the pull-down using peptides derived from Ziwi, Zili, and the analogous C-terminal region of the Xenopus Buc homolog XVelo. Clear enrichment of Tdrd6a was found using the methylated Buc peptide, but not using the non-methylated Buc peptide (Figure 5E). We do see some affinity of Tdrd6a for the methylated Zili peptide previously shown to interact with Tdrd1 (Huang et al., 2011). Since Zili is not maternally provided, this affinity could be biologically relevant in ovarian nuage where Zili interacts with Tdrd6a (Figure S2A). Tdrd1 displayed affinity only for the methylated Zili228 peptide, as shown before (Huang et al., 2011), and did not interact with the sDMAs residing in the Buc C terminus (Figure 5E). Moreover, we found that all three sDMAs on the Buc peptide are required for Tdrd6a interaction (Figure 5F). Interestingly, MS analysis demonstrated that Tdrd6c does bind to both the di- and tri-methylated Buc peptides (Figure 5G). Lastly, Tdrd6a IP/MS from buc+/− and buc−/− embryos showed that without Buc, Tdrd6c (and also Ziwi) is lost from Tdrd6a IPs (Figure 5H). We conclude that both Tdrd6a and Tdrd6c specifically interact with sDMA-modified Buc and that they may be responsible for recruitment of different protein complexes to Gp.
Figure 5.
Tdrd6a and Buc Interact via sDMAs in the C Terminus of Buc
(A) Volcano plot of Tdrd6a IP compared to IgG IP on embryo extracts, followed by MS.
(B) Confirmation of Tdrd6a co-IP with Buc using the Buc-eGFP transgenic line.
(C) C terminus of Buc with the identified dimethylated peptide underlined. Asterisk indicates residues that were found to be dimethylated by MS. Three RG sites together form a tri-RG motif, indicated in gray.
(D) Volcano plot of peptide pull-down on embryo extracts followed by MS. On the “Methylated” peptide, all 3 RG motifs were symmetrically dimethylated.
(E) Peptide pull-down followed by western blot for multiple methylated (sDMA) and non-methylated peptides derived from proteins known to contain sDMA modifications and the Buc homolog XVelo on ovary extracts. Listed are all peptides used.
(F) Peptide pull-down of Buc C terminus peptides with different methylation states on embryo extracts.
(G) MS of pull-downs of double and triple sDMA modified peptides.
(H) MS of Tdrd6a IP in the buc+/− compared to buc−/− background. Tdrd6c and Ziwi are specifically enriched in the buc+/− background, indicating that they require the presence of Buc to associate with Tdrd6a.
Tdrd6a Affects the Aggregation Behavior of Buc
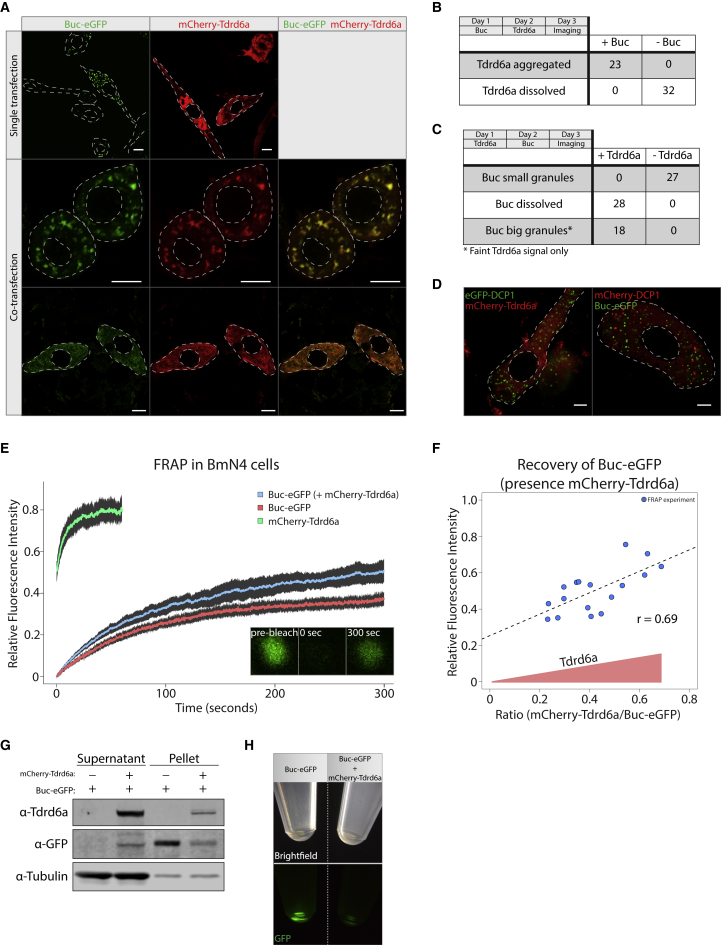
It has been shown previously that XVelo is a protein with an N-terminal PrD that has a tendency to self-aggregate. We showed that Buc aggregation, illustrated by detailed Bb and Gp imaging, is highly regulated in vivo and affected by Tdrd6a. We reasoned that Tdrd6a might be involved in spatiotemporal regulation of Buc aggregation. We first tested this in a heterologous cell culture system, using silkworm-derived BmN4 cells. These are of ovarian origin and cultured at 27°C, the same temperature at which zebrafish are kept, thereby mimicking natural conditions for Tdrd6a and Buc. IP-MS experiments on transfected Buc-eGFP revealed the same arginine methylation on Buc as observed in zebrafish (data not shown). Expression of Buc in BmN4 cells results in abundant, cytoplasmic, small granules (Figure 6A). In contrast, Tdrd6a displays a ubiquitous cytoplasmic signal (Figure 6A). Upon co-transfection, we observe two possible outcomes: the presence of both Tdrd6a and Buc either results in co-localization in enlarged, cytoplasmic aggregates with a broad variety in size (Figures 6A, middle row, and S6A) or in diffuse cytoplasmic localization of both proteins (Figure 6A, bottom row). We then performed consecutive transfection rather than co-transfection and quantified protein behavior. If we first transfect Buc, followed by Tdrd6a the next day, we always observed enlarged granules that are positive for both Buc and Tdrd6a (Figure 6B). When the order of transfection is reversed, Buc mostly localizes throughout the cytoplasm (Figure 6C). Only when the Tdrd6a signal is low, Buc seems to be able to form enlarged granules. This mutual effect between Buc and Tdrd6a is specific, since co-transfection of Buc and Tdrd6a with Dcp1, a P-body marker, leaves both proteins unaffected (Figure 6D). These results demonstrate that Tdrd6a can either stimulate the accumulation of Buc into larger granules or prevent its aggregation altogether.
Figure 6.
Tdrd6a Stimulates Buc Mobility in BmN4 Cells
(A) Localization of Buc-eGFP and mCherry-Tdrd6a in BmN4 cells in a single transfection (upper panel) and when they are co-transfected (middle and bottom). Co-transfected BmN4 cells displaying enlarged Buc-eGFP granules to which mCherry-Tdrd6a co-localizes (middle) or dissolved Buc-eGFP (bottom). Scale bars, 10 μm.
(B) Quantification of localization of Tdrd6a transfected 1 day after Buc.
(C) Quantification of localization of Buc transfected 1 day after Tdrd6a.
(D) Co-transfection of Dcp1-mCherry with Buc-eGFP (left) or Dcp1-eGFP with mCherry-Tdrd6a (right). Scale bars, 5 μm.
(E) FRAP recovery curves of mCherry-Tdrd6a and Buc-eGFP (with or without the presence of mCherry-Tdrd6a as indicated). Fluorescence intensity is the calculated fraction of the pre-bleach intensity and is plotted with the 95% confidence interval.
(F) FRAP recovery of Buc-eGFP plotted against increasing relative amounts of mCherry-Tdrd6a present in the bleached granule.
(G) Western blot for GFP and Tdrd6a on transfected BmN4 cell lysates and corresponding pellets.
(H) Pellets of lysates of Buc-EGFP-expressing BmN4 cells in the presence or absence of Tdrd6a as indicated.
See also Figure S6.
We investigated the properties of these granules in more detail using fluorescence recovery after photobleaching (FRAP). Tdrd6a recovers rapidly upon bleaching of Buc-Tdrd6a double-positive granules, reflecting a high mobility in and out of the granule (Figure 6E, n = 17). Buc alone only recovers up to ∼35% (n = 17) of the initial fluorescence intensity. Interestingly, Buc recovery increases to ∼55% (n = 17) in the presence of Tdrd6a (Figure 6E). Quantification of the FRAP experiments shows that this increase in Buc recovery in the presence of Tdrd6a is significant (Figure S6B). However, we did observe a rather broad distribution in recovery in Tdrd6a-Buc double-positive granules (Figure S6B). We hypothesized that this variation in Buc recovery could be due to differences in relative Buc and Tdrd6a concentrations in the granules that were studied. Hence, we normalized the protein amounts in the FRAP experiments by calibrating relative fluorescence using an mCherry-eGFP construct (Figure S6C). This revealed that the more Tdrd6a is present in a granule, the better Buc can recover (Figure 6F). Furthermore, without Tdrd6a Buc-eGFP cannot be detected in the soluble fraction of BmN4 lysates and is predominantly found in the pellet (Figures 6G and 6H). In contrast, in the presence of Tdrd6a, significant amounts of Buc-eGFP were soluble (Figure 6G). We conclude that Tdrd6a positively stimulates Buc mobility and solubility and that this can contribute to growth of Buc granules.
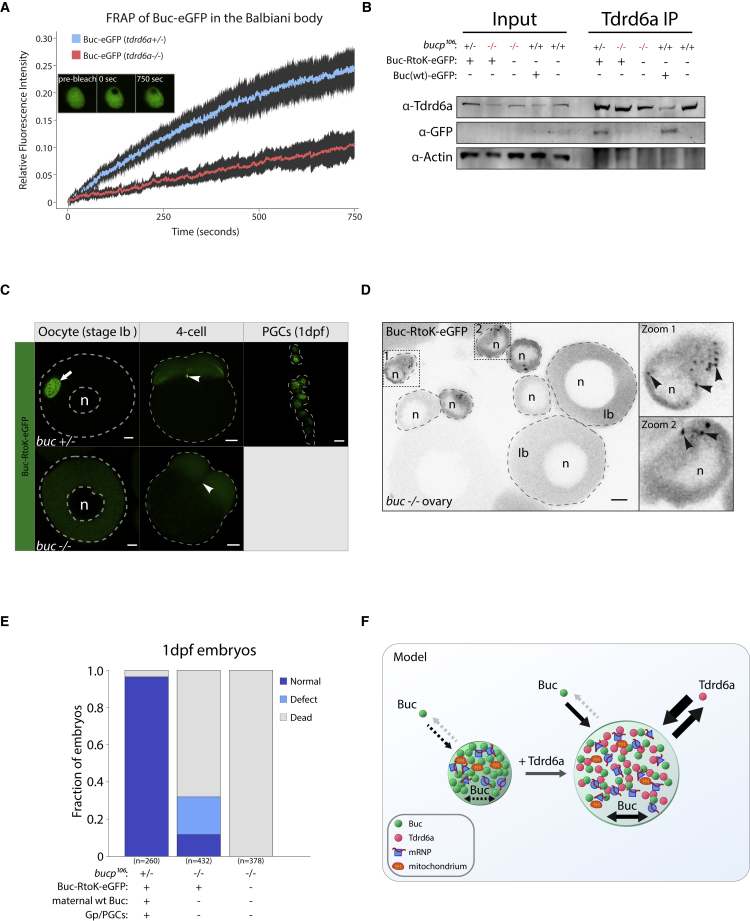
The tri-RG Motif of Buc Is Required for Bb Formation
We then aimed to test the in vivo relevance of the FRAP results and the Tdrd6a-Buc interaction data we describe in Figure 5. First, we performed FRAP on the Bb of Buc-eGFP-positive oocytes in tdrd6a+/− and −/− backgrounds. These studies showed a remarkable decrease of mobility of Buc within the Bb in the absence of Tdrd6a (Figure 7A). Second, we created a line that expresses a modified version of Buc-eGFP in which we replaced the arginine residues in the C-terminal tri-RG motif by lysines (Buc-RtoK). In the presence of wt Buc, Buc-RtoK can interact with Tdrd6a (Figure 7B), can be incorporated into the Bb and Gp, and is found in PGCs 1 day post fertilization (dpf) (Figure 7C). In contrast, in the absence of wt Buc, Buc-RtoK does not interact with Tdrd6a (Figure 7B) and fails to form a Bb in stage-Ib oocytes (Figures 7C and S7A). Tdrd6a still localizes to nuage in these oocytes (Figure S7A). Furthermore, in the absence of wt Buc, Buc-RtoK embryos neither have Gp at the 4-cell stage cleavage planes nor form PGCs (Figure 7C). Buc-RtoK alone does form small granules in early stage-I oocytes (ø < ∼30 μm), but these are detached from the nucleus and never progress to form a Bb (Figure 7D, arrowheads). Despite the absence of a Bb, Buc-RtoK embryos are polarized and are partly viable (Figures 7E and S7B), indicating that Buc-RtoK can partly rescue the buc phenotype and that polarization is Bb-independent. However, most embryos do show severe defects in cell division and/or subsequent development (Figures 7E and S7B).
Figure 7.
Tdrd6a Stimulates Buc-eGFP Mobility In Vivo
(A) FRAP recovery curves of Buc-eGFP in tdrd6a heterozygous or mutant Balbiani bodies. Fluorescence intensity is the calculated fraction of the pre-bleach intensity and plotted with a 95% confidence interval.
(B) Tdrd6a IPs probed for the indicated proteins by western blot. Bucp106 = buc loss-of-function allele. Note that Buc-eGFP is typically very hard to detect in total lysates.
(C) Localization of Buc-RtoK-eGFP in the buc+/− and buc−/− background. Arrow indicates Bb, arrowheads indicate Gp (buc+/−) or where Gp should be (buc−/−). Scale bars for oocyte and 1dpf, 10 μm. Scale bar for 4-cell, 100 μm. n = nucleus.
(D) Overview of buc−/− ovary (whole mount) positive for Buc-RtoK-eGFP. Zooms 1 and 2 are examples of stage-I oocytes ø < ∼30 μm, containing small Buc-RtoK-positive granules (arrowheads). These granules are never detected in stage-Ib oocytes, where Buc-RtoK is diffusely cytoplasmic. Scale bar, 10 μm, n = nucleus.
(E) Quantification of progeny viability at 1 dpf spawned by mothers with background as indicated, crossed with wt males.
(F) Model of Buc-containing granules, with or without Tdrd6a. Arrows indicate movement in and out of the structure or mobility within the structure itself.
See also Figure S7.
In conclusion, our data show that Tdrd6a and its interaction with arginine-methylated Buc affect the aggregation behavior of Buc-containing structures by stimulating their growth, heterogeneity, and mobility (Figure 7F), both in cell culture as well as in vivo, and that this is directly relevant for germ cell formation and embryonic development.
Discussion
Proteins such as Tdrd6a, with multiple Tudor domains in tandem, are well known to act in germ cells, in particular in smRNA pathways and their organization in peri-nuclear granules. Their precise molecular functions, however, are far from resolved. Other highly abundant components of germ cells are proteins with low-complexity regions and/or PrDs, such as Buc. Other examples are MUT-16 and MEG proteins in C. elegans, and Xvelo in Xenopus, which nucleate a variety of subcellular aggregates (Boke et al., 2016, Phillips et al., 2012, Wang et al., 2014). However, insights into how their aggregation behavior is regulated remain scarce. We demonstrate that Tdrd6a regulates the aggregation of Buc. More specifically, it promotes solubility and mobility of Buc, and thereby growth of Buc aggregates into larger structures containing well-determined amounts of germ cell-specifying mRNPs and other Gp components. Various aspects related to our findings will be further discussed here.
Tdrd6a Does Not Affect piRNA Generation
Even though the Piwi proteins appear to interact with Tdrd6a in ovary extracts, lack of Tdrd6a does not have an effect on piRNA accumulation. Given the intimate connection between piRNA biogenesis and function, a mechanistic role for Tdrd6a in the piRNA pathway does not seem likely. Our results suggest, however, that Tdrd6c, instead of Tdrd6a, is the more relevant interaction partner for the piRNA pathway. Analysis of tdrd6c mutants will be required to clarify this.
Molecular Basis behind the PGC Phenotype of tdrd6a Mutants
We observed that Gp arises from the continuous merging of smaller Buc-Tdrd6a granules in embryos. In these granules, Buc is found at the core, while we detected Tdrd6a mainly at the periphery. We note, however, that at present, we cannot be certain that this apparent substructure is real, or whether it represents an experimental artefact due to very high local Tdrd6a concentrations surrounding the Gp structure.
The individual Buc-Tdrd6a granules in 1–2 cell embryos display discrete mRNA foci at their circumference. Interestingly, these foci move more internally and start to form networks when larger assemblies arise. Homotypic assemblies of mRNPs have been described recently in Drosophila, where it has been demonstrated that Gp mRNAs initially form homogenous mRNP granules, followed by fusion into heterogeneous mRNP aggregates, in which the quantities of Gp mRNAs positively correlate (Little et al., 2015, Trcek et al., 2015). In zebrafish, we could infer that mRNA quantities in mature Gp positively correlate as well and that Tdrd6a is required for this.
Why does this correlation between Gp mRNAs depend on Tdrd6a? In absence of Tdrd6a, mostly small incomplete Gp-like structures are found. Given that each Tdrd6a-Buc granule at the 1–2 cell stage only carries a limited number of individual mRNPs, sufficiently large numbers of Buc-Tdrd6a granules need to accumulate to attain the ratios as found on all granules combined. Without Tdrd6a, these numbers may not be reached. Since single Gp mRNAs can have a strong impact on PGC formation, such unstable ratios may directly relate to the observed PGC specification and/or maintenance defects.
mRNP Recruitment and Organization in Gp
Single Buc-containing particles contain individual foci of various Gp transcripts at their periphery. We show that this does not require Tdrd6a. How then are these transcripts recruited? Intrinsic properties, such as primary sequence or secondary structure of the mRNPs, may play a role (Knaut et al., 2002, Köprunner et al., 2001, Trcek et al., 2015). Furthermore, the fact that we identify the cytoplasmic EJC complex and the CPEB complex in our Tdrd6a interactome may reveal an additional aspect: mRNPs that have not undergone translation could be prone to be incorporated into Gp-related structures. Indeed, these complexes have been demonstrated to play a role in translational control and/or Gp transcript localization in Drosophila and Xenopus (Hachet and Ephrussi, 2004, Minshall et al., 2007, Nelson et al., 2004).
In more enlarged Gp structures, we observe bigger transcript networks, each consisting of single types of mRNA that spread throughout the Gp. Possibly, intrinsic properties of mRNPs trigger such network formation when local concentrations are sufficiently high. Indeed, the intrinsic tendency of transcripts of the same kind to cluster is a phenomenon that has been suggested previously in Drosophila (Little et al., 2015, Trcek et al., 2015). These larger homotypic structures may subsequently be further stabilized by their continued interaction with the growing Buc-containing structure, in which they intermingle with other homotypic networks. Our data show that Tdrd6a is required for this higher level organization of Gp mRNPs. Whether this results from its effect on Buc or on mRNPs directly cannot be distinguished at present.
How Does Tdrd6a Regulate Buc Aggregation?
We describe potentially contradicting effects of Tdrd6a on Buc behavior: on the one hand, Tdrd6a promotes Buc solubility and/or mobility, and, on the other hand, Tdrd6a drives the formation of larger Buc aggregates. Based on our cell culture experiments, we speculate that the effect of Tdrd6a on Buc may critically depend on relative and absolute concentrations of both proteins. Possibly, the multi-tudor domain organization of Tdrd6a/c allows it to increase local Buc concentrations, and hence Buc aggregation behavior. But at the same time, the high mobility of Tdrd6a, and possibly also Tdrd6c, may drive constant remodeling of Buc aggregates and prevent the formation of too rigid or too many condensed Buc aggregates and allow fusion and/or growth of Buc aggregates. To address these possibilities, in vitro systems will need to be established, such that protein-protein interactions and aggregation behavior can be studied in much greater detail.
Regardless of the exact mechanisms, our work reveals that post-translational modifications (PTMs) can play an important role in how aggregations are regulated: loss of Buc arginine methylation, and hence Tdrd6a interaction, severely affects Buc behavior in vivo. In fact, the RtoK mutations in Buc result in a much more severe phenotype than that observed in tdrd6a mutants. We consider it likely that this is caused by additional loss of Tdrd6c interactions with Buc. Possibly, additional PTMs and their dynamics are involved in the complex aggregation behavior that Buc displays in vivo but also in other scenarios. For instance, during early embryogenesis in C. elegans, phosphorylation and dephosphorylation of MEG-1/3 (maternal effect germ-cell defective 1 and 3) control P granule disassembly and assembly, respectively (Wang et al., 2014). Since kinases were identified in the non-modified Buc peptide pull-down, it is tempting to speculate that besides arginine dimethylation, phosphorylation may also regulate Buc aggregation dynamics.
The Bb Is Not Required for Generating Oocyte Polarity
We found that Buc-RtoK can rescue the oocyte polarization defect of buc mutants, even though a Bb never forms. This shows that the Bb as such is not essential for oocyte polarity establishment and that Buc may have a Bb-independent role that helps to maintain or establish polarity. Nevertheless, we did observe that Buc-RtoK could not fully rescue the loss of endogenous Buc because many embryos did not develop properly. This is unlikely to be due to tag interference since wt transgenic Buc that also carries a GFP tag at its C terminus can fully rescue (Riemer et al., 2015). Therefore, Buc and/or the Bb may play important roles downstream of polarity establishment in the oocyte as well.
Buc-Tdrd6a Interaction as a Model for Regulated Protein Aggregation
Phase separation of proteins with IDRs has been recognized as a research field of major importance. It represents a pivotal type of compartmentalization, which mediates diverse cellular processes. There appears to be a wide range of aggregation states, spanning from liquid-like droplets to almost solid aggregations (Brangwynne et al., 2011, Patel et al., 2015, Shin and Brangwynne, 2017). It has been proposed that pathogenic protein aggregates, such as those found in Alzheimer disease or amyotrophic lateral sclerosis (ALS), represent a detrimental state of normally occurring protein aggregation. Hence, knowledge about how aggregation states can be regulated in vivo will be directly relevant to the understanding of these types of disease. Since Buc aggregation is very dynamic during zebrafish oogenesis and embryogenesis, it represents a powerful model to study the spatiotemporal regulation of protein aggregation, both by trans-acting factors as well as PTMs.
Analogous to previous studies, Buc typically behaves like a “scaffold,” recovering slowly and only partially. Tdrd6a recovery is typical for a granule “client,” displaying rapid, near complete recovery, indicating high mobility in and out of the Buc-aggregate (Woodruff et al., 2017). This may mean that in other scenarios in which Tdrd6a-like proteins have been described to affect aggregations, such as for example, the chromatoid body in mammalian spermatocytes or peri-nuclear nuage, scaffold proteins such as Buc are still to be discovered. Alternatively, well-known proteins may in fact act as such scaffolds. For instance, Piwi proteins typically have rather long and seemingly unstructured N-terminal tails, and also other well-studied proteins, like Vasa, contain disordered regions and can phase separate in vitro (Nott et al., 2015). Indeed, these proteins are rich in RG motifs that could be sites of aggregation modulation by Tdrd proteins.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Tdrd6a | This paper Eurogentec | Epitope: QAVVHEPESEKEKRD |
| Rat polyclonal anti-Ziwi | This paper Eurogentec | Epitope: QLVGRGRQKPAPGAM |
| Rabbit polyclonal anti-Zili | Houwing et al. (2007) | N/A |
| Rabbit polyclonal anti-Tdrd1 | Huang et al. (2011) | N/A |
| Sh-anti-DIG | Roche | Cat# 11333089001; RRID: AB_514496 |
| Ms-anti-GFP (B-2) | Santa Cruz | Cat# Sc9996; RRID: AB_627695 |
| Anti-Rb-Alexa405 | Abcam | Cat# ab175651 |
| Anti-Rb-Alexa647 | Abcam | Cat# ab150075 |
| Anti-Rt-Alexa488 | Abcam | Cat# ab150153 |
| Anti-Sh-Alexa555 | Invitrogen | Cat# A21436; RRID: AB_2535857 |
| Anti-Rb-IRDye | LI-COR | Cat# 926-32211; RRID: AB_621843 |
| Anti-Rt-IRDye | LI-COR | Cat# 926-68076; RRID: AB_10956590 |
| Anti-Ms-IRDye | LI-COR | Cat# 926-68070; RRID: AB_10956588 |
| Anti-Ms-HRP | Cell Signaling Technology | Cat# 7076; RRID: AB_330924 |
| Anti-Rb-HRP | Cell Signaling Technology | Cat# 7074; RRID: AB_2099233 |
| Anti-DIG-AP Fab fragments | Roche | Cat# 11093274910; RRID: AB_514497 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Buc_R1 | Peptide Specialty Laboratories GmbH | Biotin – ETEFTYCQ(sDMA)GRGSMKKRGSRY – CONH2 |
| Buc_R2 | Peptide Specialty Laboratories GmbH | Biotin – ETEFTYCQRG(sDMA)GSMKKRGSRY – CONH2 |
| Buc_R3 | Peptide Specialty Laboratories GmbH | Biotin – ETEFTYCQRGRGSMKK(sDMA)GSRY – CONH2 |
| Buc_R12 | Peptide Specialty Laboratories GmbH | Biotin – ETEFTYCQ(sDMA)G(sDMA)GSMKKRGSRY – CONH2 |
| Buc_Cterm | Peptide Specialty Laboratories GmbH | Biotin – ETEFTYCQ(sDMA)G(sDMA)GSMKK(sDMA)GSRY – CONH2 |
| Ziwi_4 | Peptide Specialty Laboratories GmbH | H2N-MTG(sDMA)ARARSRGRGRGQEP(BiotinC6) - CONH2 |
| Ziwi_47 | Peptide Specialty Laboratories GmbH | Biotin - EGQLVG(sDMA)GRQKPAPGAMS - CONH2 |
| Ziwi_77 | Peptide Specialty Laboratories GmbH | Biotin - KIGE(sDMA)GGRRRDFHDSG - CONH2 |
| Zili_228 | Peptide Specialty Laboratories GmbH | H2N - G(sDMA)GFTGFGRAAMPHMTVK(BiotinC6) - CONH2 |
| XVelo | Peptide Specialty Laboratories GmbH | Biotin - RSFLY(sDMA)GHGLQK(sDMA)GTKKKGLN - CONH2 |
| TRIzol | Thermo Fisher | Cat# 15596018 |
| TRIzol LS | Thermo Fisher | Cat# 10296010 |
| TrypLE Express (1x ) | Life Technologies | Cat# 12605036 |
| Pronase | Sigma-Aldrich | Cat# P5147 |
| MOPS buffer | Thermo Fisher | Cat# NP0001 |
| 4%-12% NuPage NOVEX gradient gel | Thermo Fisher | Cat# NP0321 |
| NuPAGE LDS sample buffer 4x | Thermo Fisher | Cat# NP0007 |
| Paraformaldehyde | Sigma-Aldrich | Cat# 441244 |
| PBS | Gibco | Cat# 14190-094 |
| Immobilon-FL PVDF membrane | Merck | Cat# IPFL00010 |
| Osmium Tetroxide, crystalline, highest Purity 99,95% | ScienceService | Cat# E19120 |
| EMbed-812 Kit with DMP | ScienceService | Cat# E14120-DMP |
| Glutaraldehyde, 25% aqueous solution, EM grade | ScienceService | Cat# E16210 |
| Paraformaldehyde, 20% aqueous solution, EM grade | ScienceService | Cat# E15713 |
| Sodium cacodylate trihydrate | Sigma-Aldrich | Cat# 20840 |
| 4-Nitro blue tetrazolium chloride (NBT) | Roche | Cat# 11383213001 |
| 5-Bromo-4-chloro-3-indolyl phosphate (BCIP) | Roche | Cat# 11383221001 |
| Yeast RNA | Sigma-Aldrich | Cat# R6625 |
| Heparin | Sigma-Aldrich | Cat# H4784 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A7906 |
| Dextran sulfate | Sigma-Aldrich | Cat# 42867-5G |
| Vanadyl-ribonucleoside complex | NEB | Cat# S1402S |
| ProLong™ Gold Antifade Mountant | Thermo Fisher | Cat# P10144 |
| Formamide | Ambion | Cat# AM9342 |
| Triton-X100 | Sigma-Aldrich | Cat# T8787 |
| Tween20 | Sigma-Aldrich | Cat# P1379 |
| cOmplete Mini, EDTA-free protease inhibitor cocktail Tablets | Roche | Cat# 11836170001 |
| Dynabeads protein G | Invitrogen | Cat# 10004D |
| Streptavidin magnetic beads | Thermo Fisher | Cat# 65001 |
| Glycoblue | Invitrogen | Cat# AM9515 |
| Acetonitrile | Sigma-Aldrich | Cat# 271004 |
| ReproSil-Pur 120 C18-AQ 1.9μm | Dr. Maisch GmbH | Cat# r119.aq. |
| Rhodamine B Dextran | Sigma-Aldrich | Cat# R9379-100MG |
| DIG labelling mix | Merck | Cat# 11277073910 |
| IPL-41 insect medium | Gibco | Cat# 11405057 |
| 8-well μ-slides | ibidi | Cat# 80826 |
| 9μl X-tremeGENE™ HP | Roche | Cat# 6365779001 |
| DpnI | New England Biolabs | Cat# R0176L |
| BP clonase II | Thermo Fisher | Cat# 11789020 |
| LR clonase II plus | Thermo Fisher | Cat# 12538120 |
| GFP-Trap Agarose | Chromotek | Cat# gta-100 |
| Critical Commercial Assays | ||
| Sp6 mMESSAGE MACHINE kit | Invitrogen | Cat# AM1340 |
| Poly(A) tailing kit | Invitrogen | Cat# AM1350 |
| Bioanalyzer Small RNA assay | Agilent | Cat# 5067-1548 |
| Bioanalyzer High Sensitivity DNA assay | Agilent | Cat# 5067-4626 |
| NEBNext® Small RNA Library Prep Set for Illumina | New England Biolabs | Cat# E7330 |
| DNA 300 assay kit for Labchip XT | Caliper | Cat# PN 760601 |
| Qubit dsDNA HS Assay | Life Technologies | Cat# Q32851 |
| M-MLV reverse transcriptase, RNase H point mutant | Promega | Cat# M3681 |
| iQ SYBR Green supermix | BioRad | Cat# 1708880 |
| QIAquick PCR Purification Kit | Qiagen | Cat# 28106 |
| Ovation RNA-seq System V2 | NuGEN | Cat# 7102 |
| TruSeq DNA Sample Prep Kit | Illumina | Cat# 15026486 |
| Deposited Data | ||
| Raw and processed RNA-seq data | This paper | GEO: GSE79285 |
| Mass spectrometry data | This paper | ProteomeXchange ID: PXD008322 |
| Danio rerio (Zebrafish), Zv9 (GCA_000002035.2) | Zebrafish genome project | https://support.illumina.com/sequencing/sequencing_software/igenome.html |
| Uniprot/Trembl Danio rerio fasta | The UniProt Consortium | www.uniprot.org |
| smFISH probes | This paper; Mendeley Data | https://doi.org/10.17632/f9ckhbrvpd.1 |
| Raw Confocal, Western blot and EM data | This paper; Mendeley Data | https://doi.org/10.17632/f9ckhbrvpd.1 |
| Experimental Models: Cell Lines | ||
| BmN4 | Laboratory of Ramesh Pillai, Université de Genève | Bombyx mori ovary derived cell line |
| Experimental Models: Organisms/Strains | ||
| Zebrafish:tdrd6aQ185 | This paper | N/A |
| Zebrafish:bucp106 | Bontems et al. (2009) | N/A |
| Zebrafish: Tg(buc:buc-RtoK-egfp) | This paper | N/A |
| Zebrafish: Tg(buc:buc-wt-egfp) | Riemer et al. (2015) | N/A |
| Zebrafish: Tg(ziwi:tdrd6a-mcherry-polyA) | This paper | N/A |
| Zebrafish: Tg(kop:egfp-f-nos1-3’UTR) | Köprunner et al. (2001) | N/A |
| Zebrafish: Tg(vasa:egfp) | Krøvel and Olsen (2002) | N/A |
| Oligonucleotides | ||
| Fus_MO | GeneTools | GCCCATAATGATTTCACGGCATCTT |
| hook2_MO | GeneTools | GCTGATGTTTATTCAGGCTCATGGT |
| Tdrd6aQ185seq _F | N/A | GCCAATGCCTTACCACTATC |
| Tdrd6aQ185seq_genotype_R | N/A | CACTTGCCTCTGAATTCTTC |
| bucp106seq_F | N/A | TCTCCCCAAAGGGAGAACTCCATTG |
| bucp106seq_R | Sequence without transgene | GTTTAACATTTTAAACTGCTCAACATACCTCTG |
| Bucp106seqUTR_R | Sequence in presence of transgene | GTG TCC ATG TGT ACA TTT ATA GTG AAG TGC |
| hook2_mismatch_SP6_F | N/A | CATACGATTTAGGTGACACTATAGACAATGTCTTTAAACAAG CACCAACTGAGCGACTCTTTATTTATCTGGCTG |
| hook2_R | N/A | TCA TCG GGG CTG CAG GCG |
| Tdrd6a_attF | N/A | GGGG ACAAGTTTGTACAAAAAAGCAGGCT CCACC ATG TGC TCC ATT CCG GGA CTC CC |
| Tdrd6a_attR | N/A | GGGG AC CAC TTT GTA CAA GAA AGC TGG GTG ATC ACG CTT TTC TTT TTC ACT CTC GG |
| RtoKmut_GFPstart_F | N/A | P-GGCTCAAGATACGGCGGAAGCGGCATGGTGAGCAAG GGCGAGGAG |
| RtoKmut_R | N/A | CTT TTT CTT CAT AGA ACC TTT GCC CTT CTG GCA GTA GGC |
| BmDcp1 Fwd (BamHI) | N/A | AGT GGA TCC cAT GGC TGA CAC CGG GTT ACG |
| BmDcp1 Rev (NotI) | N/A | GGT GCG GCC GCT TAT GAC ACA GAA AAT GCT TTT TCT G |
| eGFP_F(BamHI) | N/A | AGT GGA TCC cAT GGT GAG CAA GGG CGA G |
| eGFP_R(BamHI) | N/A | GGT GGA TCC GCC TTG TAC AGC TCG TCC ATG CC |
| DrTdrd6a (NotI) Fwd | N/A | AGT GCG GCC GCC ATG TGC TCC ATT CCG GGA C |
| DrTdrd6a (XbaI) Rev | N/A | GGT TCT AGA CTA ATC ACG CTT TTC TTT TTC ACT C |
| Buc_F | N/A | P-GAAGGAATAAATAACAATTCACAACCAATGG |
| Buc_R | N/A | GGG TAG GCC ATG GTG TAA GCT TG G TAT CTT GAG CCT CTT TTC TTC ATA GAA C |
| pBEMBL_R | N/A | GGC AGC CTC GAG CGG TGG |
| qpcr_nanos_F | N/A | GGCTTTTTCTCTTCTCCAATTCATCCTTTC |
| qpcr_nanos_R | N/A | GAGACTCCAGCAGCGCGGC |
| qpcr_dazl_F | N/A | CGGCGGTATTGATATGAAGGTGGATGAG |
| qpcr_dazl_R | N/A | GGAGATGACACTGACCGAGAACTTCG |
| qpcr_vasa_F | N/A | GGTCGTGGAAAGATTGGCCTG |
| qpcr_vasa_R | N/A | CAGCAGCCATTCTTTGAATATCTTC |
| qpcr_bactin_F | N/A | GACCCAGACATCAGGGAGTGATGG |
| qpcr_bactin_R | N/A | GGTCTCGAACATGATCTGTGTCATCTTC |
| Recombinant DNA | ||
| pBEMBL-NHA-Buc-eGFP | This paper, backbone from Xiol et al. (2012) | N/A |
| pBEMBL-NHA-mCherry-DrTdrd6a | This paper, backbone from Xiol et al. (2012) | N/A |
| pBEMBL-NHA-mCherry-DCP1 | This paper, backbone from Xiol et al. (2012) | N/A |
| pBEMBL-NHA-eGFP-DCP1 | This paper, backbone from Xiol et al. (2012) | N/A |
| pME_tdrd6a | This paper | N/A |
| P5E_pziwi | Leu and Draper (2010) | N/A |
| P3E_mcherry-polyA | Tol2 kit | http://tol2kit.genetics.utah.edu/index.php/Main_Page |
| Tol2CG2 | Tol2 kit | http://tol2kit.genetics.utah.edu/index.php/Main_Page |
| Tol2CG2_pziwi-tdrd6a-mcherrypA | This paper | N/A |
| Software and Algorithms | ||
| MaxQuant v.1.5.2.8 | Cox and Mann (2008) | http://www.coxdocs.org/doku.php?id=:maxquant:start |
| cutadapt | [https://doi.org/10.14806/ej.17.1.200] | https://cutadapt.readthedocs.io/ |
| fastq quality_filter | N/A | http://hannonlab.cshl.edu/fastx_toolkit/ |
| seqtk trimfq | N/A | https://github.com/lh3/seqtk |
| Bowtie v0.12.8 | http://genomebiology.com/2009/10/3/R25 | http://bowtie-bio.sourceforge.net/index.shtml |
| TopHat | Trapnell et al. (2009) | http://cole-trapnell-lab.github.io/projects/tophat/ |
| DESeq | Anders and Huber (2010) | https://www.bioconductor.org/packages/release/bioc/html/DESeq.html |
| RaceID | Grün et al. (2015) | https://github.com/dgrun/RaceID |
| TrackMate | Tinevez et al. (2017) | https://github.com/fiji/TrackMate/releases/tag/TrackMate_-3.5.3 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, René F. Ketting (r.ketting@imb.de).
Experimental Model and Subject Details
Zebrafish Lines
Zebrafish strains were housed at the Institute of Molecular Biology in Mainz and bred and maintained under standard conditions (26-28oC room and water temperature and lighting conditions in cycles of 14:10 hours light:dark) as described by (Westerfield, 1995). Larvae < 5 days post fertilization were kept in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28oC. The tdrd6aQ185X/+ mutant allele zebrafish was derived from ENU mutagenized libraries using target-selected mutagenesis as described before (Wienholds, 2002). Animals carrying tdrd6aQ185X/+ were out crossed against wt fish (AB and Tue), and the following pre-existing lines: kop:egfp-f-nos1-3’UTR, vasa:egfp or buc:buc-egfp-buc3’UTR transgenic fish (Köprunner et al., 2001, Krøvel and Olsen, 2004, Riemer et al., 2015) and subsequently incrossed to obtain tdrd6aQ185X/Q185X offspring. All experiments were conducted according to the European animal welfare law and approved and licensed by the ministry of Rhineland-Palatinate.
Cell Culture
BmN4 cells were a kind donation of the laboratory of Ramesh Pillai. BmN4 cells were cultured at 27oC in IPL-41 (Gibco) medium supplemented with 10%FBS (Gibco) and 0.5% Pen-Strep.
Method Details
Genotyping
For genotyping, the DNA was extracted form caudal fin tissue, amputated from anesthetized fish. The primers used to amplify and sequence the tdrd6aQ185 allele were Tdrd6aQ185seq_F: GCCAATGCCTTACCACTATC and Tdrd6aQ185seq_genotype_R CACTTGCCTCTGAATTCTTC. The lesion induces a truncation after amino acid Q185. This residue precedes the epitope used for immunization. The bucp106 allele was amplified with bucp106seq_F: TCT CCC CAA AGG GAG AAC TCC ATT G and bucp106seq_R: GTT TAA CAT TTT AAA CTG CTC AAC ATA CCT CTG and sequenced with the reverse oligo.
Tdrd6a Antibody
Tdrd6a antibodies were raised in rabbits with the synthetic peptide H2N-QAVVHEPESEKEKRD-CONH2. Antisera were subsequently purified against the synthetic peptide (Eurogentec).
Whole Mount Colorimetric In Situ Hybridization Embryos
Embryos were collected and fixed at 4-cell stage in 4% PFA/PBS ON at 4oC. Next, they were washed with PBST and dechorionated using forceps, followed by storage in MeOH at least ON at -20oC. Upon rehydration, embryos were blocked in Hyb+ (50% de-ionized formamide, 5xSSC, 0.1%Tween-20, 5mg/ml yeast RNA, 50μg/ml heparin) for 2 hrs at 70 oC. Next, samples were incubated ON at 70 oC with DIG-labelled probe in Hyb+. After probe removal, samples were washed at 70 oC: 2 x 20 minutes in Hyb- (Hyb+ without yeast RNA and heparin), 2 x 20 minutes in 2xSSCT, 2 x 20 minutes in 0.2xSSCT. Samples were washed twice in TBST at RT and blocked 1 hour in 10% BSA. Next, samples were incubated with 1:2000 anti-DIG-AP Fab fragments (Roche) in 10% BSA ON at 4oC. Next day, they were washed 3 x 20 min. in TBST at RT and 2 x 10 min. in AP-staining buffer (100mM NaCl, 100mM Tris pH9.5, 50mM MgCl2, 0.1% Tween-20), followed by incubation with NBT (4.5μl/ml AP staining buffer; Roche) and BCIP (3.5μl/ml AP staining buffer; Roche) to stain the embryos.
Whole Mount Fluorescent In Situ Hybridization
Ovary tissue was put in OR2 medium (82mM NaCl, 2mM KCl, 1mM MgCl2, 5mM HEPES pH7.5) and filtered through a 300μm mesh to collect only stage I-III oocytes. These oocytes were fixed in 4% PFA/PBS 3 hours at RT, followed by dehydration in MeOH and storage ON at -20oC. The same procedure was followed as described above for colorimetric ISH, only after blocking in 10% BSA, samples were incubated with 1:1000 sheep-anti-DIG (Roche) in 10% BSA ON at 4oC. After 3 x 20 min. washing in TBST, samples were incubated with 1:500 anti-sheep-Alexa555 (Invitrogen) for 1 hr/RT. Samples were washed 3 x 10 min. in TBST and mounted in 80% glycerol, followed by imaging under DM6000 Leica microscope.
Whole Mount Double smFISH and IHC
PFA fixed oocytes/embryos were collected and prepared as described above and were incubated overnight with 1:100 anti-Tdrd6a and 1:100 of both smFISH probes (Stellaris™ custom design, Quasar 570 (vasa) and Quasar 670 (dazl) labelled) stocks (12.5μM in TE buffer) in hybridization buffer (10% dextran sulfate, 10% formamide, 1mg/mL tRNA, 0.02%BSA, 2mM vanadyl-ribonucleoside complex (NEB S1402S) in 2xSSC) at 30oC. Next day, wash 15 minutes in wash buffer (10% formamide, 2xSSC) and incubate in 1:500 anti-rabbit alexa-405 for 30 minutes in wash buffer. Then wash 2 x 15 minutes in wash buffer and mount in ProLong™ Gold Antifade Mountant (ThermoFisher).
Peptide Pull-Down
Peptides were synthesized by Peptide Specialty Laboratories GmbH. 20μg peptide in 500μL IP buffer (25 mM Tris pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton-X100, 1mM DTT, protease inhibitor) was pre-incubated with streptavidin-coupled magnetic beads for 30 minutes at RT, rotating. 20μL resin (ThermoFisher 65001) was used per pulldown. Then, the respective lysate was added to the washed beads and incubated for 1hr at 4oC while rotating. The beads were then washed with wash buffer (25mM Tris pH 7.5, 300mM NaCl, 1.5mM MgCl2, 1mM DTT) and either used for Western blot analysis or MS.
FRAP
FRAP was performed on a TCS SP5 Leica confocal microscope, equipped with a FRAP-booster, using a 63x oil objective with an NA of 1.4 (BmN4 cells) or a 63x water objective with an NA of 1.2 (Bb). In BmN4 cells, entire granules were bleached in a fixed region of 1.5μm ø and recovery was followed for 1500 frames (0.2s/frame). Bbs were bleached partially in a fixed region of 2.5μm ø and recovery was followed for 1500 frames (0.5s/frame). Regions (pre-- and post-bleach) were tracked using TrackMate (Tinevez et al., 2017). 10 pre-bleach frames were recorded and after background subtraction, the average intensity was used as pre-bleach intensity. Post-bleach frames were background subtracted and to make replicates comparable, post-bleach frame #1 of each measurement was set to 0 and corresponding pre-bleach intensity was corrected for this. Normalization of Buc-eGFP and mCherry-Tdrd6a was performed by plotting intensities of mCherry-eGFP using the same microscope settings as for the FRAP. Intensities were plotted after background subtraction and the resulting curve was used to calculate protein ratios using the initial/pre-bleach intensities of each experiment.
Immunohistochemistry
Samples were prepared as described for the ISH above and either embedded in paraffin and sectioned (ovary Figures 1A and S7A) or used whole mount. The samples were blocked in block buffer (2% sheep serum, 2%BSA in PBS with 0.05% Tween (PBST)) for 1 hour at RT and next incubated with the primary antibody in block buffer overnight at 4oC. The next day, samples were washed 3 x 5 minutes in PBST and incubated with the secondary antibody in block buffer for 1 hour at RT. The samples were washed 4 x 15 minutes in PBST and mounted in ProLong™ Gold Antifade Mountant prior to imaging. Rb-anti-Tdrd6a antibody was used 1:100, rat-anti-Ziwi was used 1:100. Anti-rabbit alexa-647 (Abcam, ab150075) and anti-rat alexa-488 (Abcam ab150153) was used 1:500.
Confocal Imaging
Samples were imaged using a TCS SP5 Leica confocal microscope using a 10x dry objective (NA 0.3), 40x oil (NA 1.3), 63x oil (NA of 1.4) or a 63x water objective (NA 1.2). The following figures were deconvolved using the Huygens software: Figures 3C, 3D, 4A, 4B, 6A, 6D, 7D, S1C, S4A, and S6A
Immunoprecipitation
Per IP the following amounts of sample was used: 2 testis lobes, stage I-III oocytes from 1 female or 40 1-cell stage embryos (remove all E3 prior to addition of lysis/IP buffer). Samples were taken up in 650 μL lysis/IP buffer (25 mM Tris pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton-X100, 1mM DTT, protease inhibitor) and homogenized with a micropestle followed by sonication for 3 x 30 seconds at low power. The oocyte and testis samples were spun down 10 minutes at 12,000 x g at 4oC and the supernatant was used for IP. The embryo samples were filtered through a 70 micron mesh to remove the chorion fragments and the filtrate was used for IP directly. The Tdrd6a antibody was used at a dilution of 1 to 100 and incubated for 2 hours at 4oC while rotating. Then 30 μl washed Dynabeads protein G were added and incubated with the sample for another 45 minutes at 4oC. Next, 3 washes were performed with wash buffer (25 mM Tris pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 1mM DTT). For pulldown of the GFP-tagged constructs, 25μl GFP-Trap (GFP-Trap_A, Chromotek) per lysate was used and incubated for 1 hour at 4oC, followed by 3 washes with wash buffer.
For q-RT-PCR and RIP-seq analysis, Dynabeads were eluted in TRIzol LS (ThermoFisher) for RNA isolation. For western blot and mass spectrometry analysis, Dynabeads or GFP-Trap beads were eluted in NuPAGE LDS sample buffer (ThermoFisher).
Western Blot
Samples were heated to 95oC for 5 minutes prior to loading on a 4%-12% NuPage NOVEX gradient gel (ThermoFisher) and blotted on an Immobilon-FL PVDF membrane (Merck) overnight at 15V, RT. The membranes were incubated next day with primary antibodies (Rb-α-Tdrd6a 1:1000, Rt-α-Ziwi 1:1000, Rb-α-Tdrd1 1:500, Rb-α-Zili 1:10,000, Ms-α-GFP (Santa Cruz) 1:1000 ) and upon washing incubated with 1:10,000 800CW IRDye α-Rb and IRDye 680RD α-Rt (LI-COR) and imaged on an Odyssey CLx imaging system (LI-COR).
Library Construction and High-Throughput Sequencing
Total RNA was subjected to 15% TBE-urea gel for size selection of 15– 35 nt. This excised gel fraction was eluted in 0.3 M NaCl for N16 h and precipitated with 100% isopropanol and Glycoblue for 1h at −20 °C. The precipitated RNA pellet was washed once with 75% ethanol and dis- solved in nuclease-free water. The purified RNA fraction was confirmed by Bioanalyzer Small RNA assay (Agilent). Library preparation was based on the NEBNext® Small RNA Library Prep Set for Illumina® (New England Biolabs) with minor modifications. To counteract ligation bias and to remove PCR duplicates, small RNA was first ligated to the 3′ adapter and then the 5′ adapter, both of which contained four random bases at the 5′ and 3′ end, respectively. Adapters with random bases were chemically synthesized by Bioo Scientific. Adapter-ligated RNA was reverse-transcribed and PCR amplified for 14 cycles using index primers. The PCR amplified cDNA construct was checked on the Bioanalyzer (Agilent) using High Sensitivity DNA assay. We performed a size selection of the small RNA library on LabChip XT instrument (PerkinElmer) using the DNA 300 assay kit. All libraries were pooled to obtain 10 nM, which was denatured to 9 or 10 pmol with 5% PhiX spiked-in and sequenced as single-read for 50 cycles on an Illumina MiSeq or HiSeq 2500 instrument in either rapid or high-output mode.
Bioinformatic Analysis
The quality of raw sequenced reads was accessed with FastQC, Illumina adapters were then removed with cutadapt (-O 8 -m 26 -M 38), reads with low-quality calls were filtered out with fastq quality_filter ( -q 20 -p 100 -Q 33). Using information from unique molecule identifiers (UMIs) added during library preparation, reads with the same sequence (including UMIs) were collapsed to removed putative PCR duplicates using a custom script. Prior to mapping, UMIs were trimmed (seqtk trimfq) and library quality re-assessed with FastQC. Reads were aligned against the Zebrafish (_Danio rerio) genome assembly Zv9 with bowtie v0.12.8 (—tryhard —best —strata —chunkmbs 256 -v 1 -M 5).
The locations of transposable elements were downloaded from the UCSC genome browser (repeat masker track, Zv9) and used to select reads reads mapping to either RNA (SINE, LINE and LTR) or DNA transposons. The strength of the ping-pong cycle was assessed as the 5’ overlap of reads in opposite strands (Brennecke et al., 2007) and the Z-scores scores were calculated as Z-score = (P10-M)/SD, where P10 is the number of read pairs with an offset of 10 bases, M the mean of read pairs with 1-9 and 11-30 bases, and SD the standard deviation.
LC-MS/MS
Mass Spectrometry Sample Preparation
Proteins were heated to 80°C for 10 min prior to loading on a 4%-12% NuPage NOVEX gradient gel (Life Technologies). The proteins were separated using MOPS buffer (ThermoFisher) at 170V for 10 min. In-gel digestion was performed essentially as previously described (Shevchenko et al., 2006). Peptides were desalted on StageTips and stored on them until MS measurement.
Mass Spectrometry Measurement
Peptides were eluted from StageTips with 80 percent ACN/0.5% formic acid. The mixture was separated using an EASY-nLC1000 with a reversed phase column (25 cm, 75 μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 1.9 μm (Dr. Maisch GmbH)) mounted directly at a Q Exactive Plus mass spectrometer (ThermoFisher). A 88 minute gradient of 2% to 40% acetonitrile at a flow of 225 nl/min was combined with a wash-out of 95% acetonitrile in an overall 105 min instrument method. Spray voltage was set to ca. 2.4 kV. The instrument performed a top10 data-dependent acquisition with up to 10 HCD fragmentations per MS full scan (70k resolution, 300-1650 m/z).
MS Analysis
The raw files were processed with MaxQuant v.1.5.2.8 and searched with the incorporated Andromeda search engine against a Uniprot/Trembl Danio rerio fasta file (58,793 entries) (Cox and Mann, 2008). Carbamidomethylation was set as fixed modification while methionine oxidation, protein N-acetylation, phosphorylation and lysine/arginine dimethylation were considered as variable modifications. The search was performed with an initial mass tolerance of 7ppm mass accuracy for the precursor and 20 ppm for the MS/MS spectra in the HCD fragmentation mode. Standard settings were applied except match between runs and the LFQ quantitations were activated. Search results were filtered at a false discovery rate of 0.01 on protein and peptide level.
Data Analysis
For statistical analysis of the MS data, protein groups identified by site, known contaminants and reverse hits were excluded. The dataset was further filtered for at least 2 peptide identifications (at least 1 unique and 1 razor) per protein group. Missing LFQ values were imputed using lower values of a beta distribution derived from the measured values. Provided LFQ values were log2 transformed and for the volcano plot, the mean of the LFQ intensity of the Tdrd6a IP subtracted with the mean of the LFQ intensity of IgG IP (x-axis) against the p-value from a Welch t-test between both groups (y-axis) were plotted.
Fluorescence-Activated Cell Sorting (FACS)
Embryos were collected at the stage of interest in 20 ml E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) in a glass beaker. 500μl 10mg/ml pronase was added to remove the chorions. The chorions were washed away with E3 and the embryos were taken up in 1 mL TrypLE Express (1x) (Life Technologies) and dissociated with the help of a syringe. The samples were added to 6 mL E3, 5 μL 1M EDTA and 0.5 mL FCS was added. Samples were centrifuged for 5 minutes at 1000g. Supernatant was removed and cells were taken up in PBS. Cells were pipetted through a cell strainer and 10 μL Dnase was added. Prior to sorting, DAPI was added. Single GFP-positive cells were sorted using a BD FACSAria III (Becton Dickinson) with an 85 μm nozzle in 96 well plates containing 150 μL TRIzol Reagent (ThermoFisher).
CEL-Seq Library Preparation
After sorting, single cell TRIzol extractions were performed and the dried RNA pellet was resuspended in primer solution, containing the 5’ Illumina adapter, a cell specific barcode, a unique molecular identifier (UMI), the T7 promoter and a poly T stretch. The RNA-primer solution was briefly denatured, cooled on ice and subsequently first strand synthesis mix was added. After the first strand synthesis reaction the DNA:RNA hybrids were converted into dsDNA, cleaned up and subsequently o/n in vitro transcription was performed. The obtained RNA was thereafter fragmented and Illumina compatible libraries were made using the TruSeq small RNA sample prep kit. (Grün et al., 2014).
Quantification of Transcript Abundance
Paired end read processing obtained by CEL-Seq was essentially done as described with minor modifications (Grün et al., 2015). The transcriptome of all Ensembl genes (version Zv9) was downloaded and all isoforms were merged into single genes. We aligned the reads with BWA using standard settings to the improved Ensembl transcriptome as described (Junker et al., 2014). The right read was mapped to the gene models whereas the left mate, containing the UMI, was used for quantification. Barcode frequencies were converted based on binominal statistics into transcript count.
RaceID2
RaceID2 was used with default settings. The lower limit of transcript counts per cell we used was 1100 transcripts per cell. Thereafter the transcript counts of all cells passing this threshold were down sampled to 1100 transcripts per cell. The data described in Figures S3I and S3J was obtained from cells down sampled to 1750 transcripts to be able to pick up more lowly expressed genes as well. K-medoids clustering was performed similarly as the k-means clustering as described (Grün et al., 2015), but optimal cluster number was not determined by gap statistics, but by determining the saturation point of the within cluster dispersion (Grün et al., 2016). For t-SNE-map generation a seed of 2500 was used. Standard deviations on the individual Gp correlations were obtained by bootstrapping (n=100).
Detection of Differentially Expressed Genes in scRNA-Seq Data
To identify differentially expressed genes we applied an approach akin to previously published method (Anders and Huber, 2010). First, the down-sampled version computed by RaceID2 was used as input, in order to compare two subsets of cells from the same dataset. A p-value for a significant difference in mean expression of a gene between the two subsets was computed using DESeq as described in (Anders and Huber, 2010) while using the RaceID background model (Grün et al., 2015) to estimate the dispersion parameter of the negative binomial expression distribution within the two subgroups. These p-values were corrected for multiple testing by the Benjamini-Hochberg method.
Background Correlation Model and Random Cell Generation
To compute the expected Spearman correlations values of transcripts with a particular average expression we ranked the genes in our dataset according to their average expression. For every Gp pair for which we computed the pairwise correlation we took the three genes above and below the two Gp genes in question from the ranked list and computed all their pairwise correlations. The pairwise correlation between Gp markers was done with all cells at 3.5hpf except one, which was discarded after manually checking of atypical Gp expression levels. This particular cell expressed tdrd7 > 200-fold higher than the average cell in the 3.5hpf dataset and was considered a technical artifact.
Generation of random cell was done by randomly ascribing Gp counts derived from cells at the 3.5hpf timepoint and we subsequently computed the pairwise correlation of the Gp mRNAs in these hypothetical cells.
qPCR
From total RNA from input and Tdrd6a IP samples, cDNA was synthesized with M-MLV reverse transcriptase (RNase H point mutant, Promega), using random hexamers (Promega). qPCR was performed using iQ SYBR Green supermix (BioRad) on a CFX384 Real-Time thermal cycler (Bio Rad). All oligos were tested for linearity prior to the experiment.
RIP-Seq
RNA was isolated from input samples and Tdrd6a IP experiments on freshly laid embryos by TRIzol extraction according to manufacturer’s instructions. 1.5 ng of immunoprecipitated or input material as total RNA was amplified to cDNA with Ovation RNA-seq System V2 (NuGEN) followed by purification with QIAquick PCR Purification Kit (Qiagen). The purified full-length cDNA was sheared to around 200 bp by a focused-ultrasonicator (Covaris). 1 μg of sheared cDNA was end-repaired and adapter-ligated following manufacture’s instruction of TruSeq DNA Sample Prep Kit (Illumina). Size selection of adapter-ligated cDNA between 200-400 bp was done by LabChip XT (PerkinElmer). This size-selected fraction was PCR amplified for 3 cycles before pooling for paired-end sequencing on HiSeq 2500 for 50 bp read length. Around 30 million reads per sample were available for processing. Reads were aligned to Zv9 using TopHat with default settings. DESeq was used to compute significance of observed differences between Tdrd6a RIP and input counts per gene (Anders and Huber, 2010). Only genes with on average more than 50 RPM were used for further analysis.
Morpholino Knockdown and PGC Quantification
A morpholino was designed antisense to the region containing the start-codon of hook2 (GeneTools). 1nl was injected into 1-cell stage pvasa:egfp positive embryos at a concentration of 0.5mM and co-injected with 1/10th volume of rhodamine dextran. This was also performed using a morpholino targeting the fus transcript at the same concentration. Rescue experiments were performed by mixing 200ng/μL final concentration in the MO injection mix. Hook2 mRNA was amplified using an oligo containing an Sp6 promoter and mismatches at the MO binding site. mRNA was synthesized using the Sp6 mMESSAGE MACHINE kit (Invitrogen), followed by poly-A-tailing using the poly(A) tailing kit (Invitrogen). At 24hpf, the larvae were dechorionated and fixed in 4%PFA/PBS so GFP positive PGCs could be counted at the same developmental stage. 1dpf, embryos were dechorionated and fixed in 4%PFA/PBS and observed through Leica stereo microscope for counting PGCs.
Electron Microscopy
Gonads of 5wpf Tdrd6a Mut and WT fish were fixed with half-strength Karnovsky fixative (pH 7.4, Karnovsky, 1965) and postfixed with 1% OsO4 in 0.1M Cacodylate buffer (pH7.4), dehydrated in an Acetone series and embedded in EPON (Karnovsky, 1965). Semi-thin sections (1.5μm) were cut on a Reichert Ultracut 2040 and a Butler diamond knife (Diatome) until the desired area in the gonad was reached. Ultra-thin sections (90nm) were cut on a Reicher Ultracut E and collected on pioloform coated copper slot grids, dried and stained with leadcitrate under oxygen free conditions for 2 minutes. Sections were examined with a Zeiss LIBRA 120.
BmN4 Cell Transfection
For imaging of transfected BmN4 cells, cells were grown overnight in 8-well μ-slides (ibidi 80826). A total of 300ng construct mix (co-transfection 300ng total or single transfection 150ng plus 150ng empty vector) was incubated with 0.9μl X-tremeGENE™ HP (Roche) filled up to 30μL with insect medium without additives for 30 minutes prior to transfection.
Cloning
Tdrd6a-mCherry-polyA Construct
The transgene was made using Tol2 kit for multisite Gateway cloning. The tdrd6a CDS was amplified and cloned into pDonr221 using BP clonase in order to obtain pME_tdrd6a. Next, an LR reaction was performed using p5E_pziwi (ziwi promoter), pME_tdrd6a, p3E_mCherry-polyA and tol2CG2 (pDest) (Kwan et al., 2007, Leu and Draper, 2010). Injected embryos were screened for cmlc2:gfp to create a line. The expression pattern of this transgene was limited to oocytes up to stage Ib to early stage II (data not shown).
Buc-RtoK-eGFP Mutant Construct
The Buc construct used to make the Buc-eGFP line (Riemer et al., 2015) was modified with PCR in order to obtain the RtoK mutated version. First, the plasmid was digested with NotI and SalI and the resulting 4064 bp fragment was subcloned in pre-digested pCS2+. Arginine codons were replaced by Lysine codons using RtoKmut_GFPstart_F (phosphorylated): GGCTCA AGATACGGCGGAAGCGGCATGGTGAGCAAGGGCGAGGAG and RtoKmut_R: CTT TTT CTT CAT AGA ACC TTT GCC CTT CTG GCA GTA GGC. This also causes loss of intron 6, however, since the other introns remained unaffected and the transgene was translated and could localize normally in the presence of wt Buc, this does not seem to affect the functionality of the transgene. After the PCR, the original plasmid was digested with DpnI and the PCR fragment was circularized by ligation and sequenced. Next, the mutated fragment was ligated back into the pre-digested buc-egfp plasmid. The mutated construct was injected into wt embyros together with tol2 transposase and adult females were screened for GFP-positive gonads. The F0 females were then outcrossed in order to obtain a stable line, prior to crossing it with the bucp106 allele.
Cloning of the BmN4 Expression Constructs
Dcp1 was amplified and digested with BamHI and NotI and ligated into pBEMBL-NHA. Next, mCherry and eGFP were amplified and ligated N-terminally into the BamHI site in order to create mCherry-Dcp1 and eGFP-DCP1 respectively. Tdrd6a was amplified and digested with NotI and XbaI and ligated into pre-digested pBEMBL-NHA-mCherry. Buc was amplified using Buc F, which was 5’ phosphorylated, and Buc_R, containing an overhang for the pBEMBL-NHA-eGFP. Next, the amplicon was mixed with pBEMBL-NHA-eGFP and together with the pBEMBL_R oligo, a PCR was performed on the entire plasmid. Next, the original backbone was digested with DpnI and the linear PCR product was ligated (and thereby circularized) in order to obtain pBEMBL-NHA-Buc-eGFP.
Quantification and Statistical Analysis
Quantification of the fluorescent Western blot was done using the Image Studio software from LI-COR.
For the qPCR, standard deviations of 2 biological replicates of triplicate measurements were calculated. P-values were calculated using a two-sided Student’s t-test on the ΔCt values.
The surface of the 2D information of the Buc granules in the BmN4 cells was calculated in Fiji. First, single plane confocal images (1 Airy unit) were binarised using the Otsu method. Next, the surface of the granules was measured using ‘Analyze Particles’ (size: > 0.1μm2).
All p-values for the boxplots were calculated using the two-sided Wilcoxon test, in order to compare significant differences between two populations.
Data and Software Availability
Raw and processed RNA-seq data files have been deposited in the NCBI Gene Expression Omnibus (GEO) under the accession number GEO: GSE79285.
Mass spectrometry data has been submitted to ProteomeXchange: PXD0088322.
Acknowledgments
We thank the members of our laboratory for fruitful discussions. Yasmin El Sherif is thanked for extensive experimental support. We thank the Cuppen group (Hubrecht) for identifying the tdrd6aQ158X allele. We would like to thank Jeroen Krijsgveld and the proteomics core facility at EMBL for performing initial proteomics analysis and Eugene Berezikov (Hubrecht/ERIBA) for initial bioinformatics support. We thank the following IMB Core Facilities for their contributions and valuable services: genomics, microscopy, bioinformatics, flow cytometry, and the media lab. In particular, we thank Mária Hanulová for support in FRAP experiments. We thank Edward Lemke for critical reading of the manuscript. This work was supported by the Rhineland Palatinate Forschungsschwerpunkt GeneRED, a Marie Curie fellowship 623119 (L.J.T.K.), the ERC (ERC-StG202819, R.F.K., and ERC-AdG294325-GeneNoiseControl, A.v.O.), and Vici award 724.011.001 (R.F.K.) from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO).
Author Contributions
E.F.R., L.J.T.K., and R.F.K. conceived the study and designed experiments. L.J.T.K. performed scRNA-seq experiments and data analysis, RIP-seq analysis, and some of the IP experiments. E.F.R. performed most other experiments. S. Redl performed electron microscopy experiments, with assistance of W.S. A.W.B. performed BmN4 cell culture experiments. A.M.d.J.D. performed smRNA-seq analysis. H.Y.H. performed initial analyses of the tdrd6a mutant and Tdrd6a localization. S.Riemer and R.D. provided the wt Buc-eGFP construct and transgenic line. K.W. performed scRNA-seq experiments, and D.G. and A.v.O devised scRNA-seq data analyses. C.T.H. optimized RIP-seq protocols, and F.B. performed MS analysis. R.F.K. supervised the project. L.J.T.K., E.F.R., and R.F.K. wrote the paper with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: August 6, 2018
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.devcel.2018.07.009.
Supplemental Information
References
- Akpınar M., Lesche M., Fanourgakis G., Fu J., Anastassiadis K., Dahl A., Jessberger R. TDRD6 mediates early steps of spliceosome maturation in primary spermatocytes. PLoS Genet. 2017;13:e1006660. doi: 10.1371/journal.pgen.1006660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B., Pierson S., Dufourt J., Papin C., Armenise C., Port F., Grentzinger T., Chambeyron S., Baronian G., Desvignes J.P. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015;12:1205–1216. doi: 10.1016/j.celrep.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinski S.M., Kloc M., Tworzydlo W. Selection of mitochondria in female germline cells: is Balbiani body implicated in this process? J. Assist. Reprod. Genet. 2017;34:1405–1412. doi: 10.1007/s10815-017-1006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H., Eisenbeiss S., Neumann M., Reichman-Fried M., Thisse B., Thisse C., Raz E. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci. 2005;118:4027–4038. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S.P., Alberti S., Drechsel D., Hyman A.A., Mitchison T.J. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F., Stein A., Marlow F., Lyautey J., Gupta T., Mullins M.C., Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Curr. Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Grün D., Kester L., van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- Grün D., Muraro M.J., Boisset J.C., Wiebrands K., Lyubimova A., Dharmadhikari G., van den Born M., van Es J., Jansen E., Clevers H. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Harris A.N., Macdonald P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Maegawa S., Nagai T., Yamaha E., Suzuki H., Yasuda K., Inoue K. Localized maternal factors are required for zebrafish germ cell formation. Dev. Biol. 2004;268:152–161. doi: 10.1016/j.ydbio.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Wagner F., Sher N., Yanai I. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Houwing S. Utrecht University Repository; 2009. Piwi-piRNA complexes in the zebrafish germline. Dissertation. [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H.Y., Houwing S., Kaaij L.J.T., Meppelink A., Redl S., Gauci S., Vos H., Draper B.W., Moens C.B., Burgering B.M. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011;30:3298–3308. doi: 10.1038/emboj.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenishi K. Germ plasm in Caenorhabditis elegans, Drosophila and Xenopus. Dev. Growth Differ. 1998;40:1–10. doi: 10.1046/j.1440-169x.1998.t01-4-00001.x. [DOI] [PubMed] [Google Scholar]
- Junker J.P., Noël E.S., Guryev V., Peterson K.A., Shah G., Huisken J., McMahon A.P., Berezikov E., Bakkers J., Van Oudenaarden A. Genome-wide RNA tomography in the zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- Karnovsky M.J. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 1965;27:137A. [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y., Vourekas A., Sayed N., de Lima Alves F., Thomson T., Lasko P., Rappsilber J., Jongens T.A., Mourelatos Z. Arginine methylation of aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Bilinski S., Etkin L.D. The Balbiani body and germ cell determinants: 150 years later. Curr. Top. Dev. Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- Knaut H., Steinbeisser H., Schwarz H., Nüsslein-Volhard C. An evolutionary conserved region in the vasa 3’UTR targets RNA translation to the germ cells in the zebrafish. Curr. Biol. 2002;12:454–466. doi: 10.1016/s0960-9822(02)00723-6. [DOI] [PubMed] [Google Scholar]
- Köprunner M., Thisse C., Thisse B., Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krøvel A.V., Olsen L.C. Sexual dimorphic expression pattern of a splice variant of zebrafish vasa during gonadal development. Dev. Biol. 2004;271:190–197. doi: 10.1016/j.ydbio.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Krøvel A.V., Olsen L.C. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech. Dev. 2002;116:141–150. doi: 10.1016/s0925-4773(02)00154-5. [DOI] [PubMed] [Google Scholar]
- Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Leu D.H., Draper B.W. The ziwi promoter drives germline-specific gene expression in zebrafish. Dev. Dyn. 2010;239:2714–2721. doi: 10.1002/dvdy.22404. [DOI] [PubMed] [Google Scholar]
- Little S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F., Gavis E.R. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 2015;17:558–568. doi: 10.1038/ncb3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L.J.P., Hinton G.E. Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Marlow F.L., Mullins M.C. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev. Biol. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Reiter M.H., Weil D., Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Nelson M.R., Leidal A.M., Smibert C.A. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K.M., Okada T.N., Kawamura T., Mituyama T., Kawamura Y., Inagaki S., Huang H., Chen D., Kodama T., Siomi H. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D.A., Butler A.M., Aguero T.H., Newman K.M., Van Booven D., King M.L. High-throughput analysis reveals novel maternal germline RNAs crucial for primordial germ cell preservation and proper migration. Development. 2017;144:292–304. doi: 10.1242/dev.139220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Montgomery T.A., Breen P.C., Ruvkun G. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26:1433–1444. doi: 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E. Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- Riemer S., Bontems F., Krishnakumar P., Gömann J., Dosch R. A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish. Gene Expr. Patterns. 2015;18:44–52. doi: 10.1016/j.gep.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Siddiqui N.U., Li X., Luo H., Karaiskakis A., Hou H., Kislinger T., Westwood J.T., Morris Q., Lipshitz H.D. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13:R11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M.C., Mannen T., Siomi H. How does the royal family of Tudor rule the piwi-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaidina M., Lehmann R. Quantitative differences in a single maternal factor determine survival probabilities among drosophila germ cells. Curr. Biol. 2017;27:291–297. doi: 10.1016/j.cub.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser M.J., Mackenzie N.C., Dumstrei K., Nakkrasae L.-I.I., Stebler J., Raz E. Control over the morphology and segregation of zebrafish germ cell granules during embryonic development. BMC Dev. Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thandapani P., O’Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- Thomson T., Lasko P. Drosophila Tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T., Grosch M., York A., Shroff H., Lionnet T., Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzung K.W., Goto R., Saju J.M., Sreenivasan R., Saito T., Arai K., Yamaha E., Hossain M.S., Calvert M.E.K., Orbán L. Early depletion of primordial germ cells in zebrafish promotes testis formation. Stem Cell Reports. 2015;4:61–73. doi: 10.1016/j.stemcr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A., Tiedau D., Firooznia A., Müller-Reichert T., Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A., Alexiou P., Vrettos N., Maragkakis M., Mourelatos Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature. 2016;531:390–394. doi: 10.1038/nature17150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Teng Y., Xie Y., Wang B., Leng Y., Shu H., Deng F. Characterization of the carbonic anhydrases 15b expressed in PGCs during early zebrafish development. Theriogenology. 2013;79:443–452. doi: 10.1016/j.theriogenology.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3:e04591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., Thisse C., Thisse B., Raz E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Westerfield M. Third Edition. University Oregon Press; 1995. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- Wienholds E. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077.e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Xiol J., Cora E., Koglgruber R., Chuma S., Subramanian S., Hosokawa M., Reuter M., Yang Z., Berninger P., Palencia A. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell. 2012;47:970–979. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Yoon C., Kawakami K., Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.