Abstract
Arhgap21 is a member of the Rho GTPase activating protein (RhoGAP) family, which function as negative regulators of Rho GTPases. Arhgap21 has been implicated in adhesion and migration of cancer cells. However, the role of Arhgap21 has never been investigated in hematopoietic cells. Herein, we evaluated functional aspects of hematopoietic stem and progenitor cells (HSPC) using a haploinsufficient (Arhgap21+/−) mouse. Our results show that Arhgap21+/− mice have an increased frequency of phenotypic HSC, impaired ability to form progenitor colonies in vitro and decreased hematopoietic engraftment in vivo, along with a decrease in LSK cell frequency during serial bone marrow transplantation. Arhgap21+/− hematopoietic progenitor cells have impaired adhesion and enhanced mobilization of immature LSK and myeloid progenitors. Arhgap21+/− mice also exhibit reduced erythroid commitment and differentiation, which was recapitulated in human primary cells, in which knockdown of ARHGAP21 in CMP and MEP resulted in decreased erythroid commitment. Finally, we observed enhanced RhoC activity in the bone marrow cells of Arhgap21+/− mice, indicating that Arhgap21 functions in hematopoiesis may be at least partially mediated by RhoC inactivation.
Keywords: Arhgap21, Hematopoiesis, Erythroid cells, Hematopoietic stem and progenitor cells, Fate decision
1. Introduction
Hematopoiesis is a complex process initiated by the differentiation of hematopoietic stem cell (HSC) through progressive stages of lineage commitment (Sun et al., 2014; Yu et al., 2016). HSC functions are finely regulated by intrinsic and extrinsic factors, which prevent their exhaustion and allow hematopoiesis to be sustained throughout the life (Baryawno et al., 2017).
Hematopoietic stem and progenitor cells (HSPCs) are constantly undergoing fate-decisions, including quiescence vs. proliferation, self-renewal vs. differentiation, lineage commitment, and mobilization vs. adhesion. These processes are regulated by growth factors, cell-cell interactions, transcriptional networks, and epigenetics, many of which lead to cytoskeletal rearrangements (Nayak et al., 2013; Narla & Mohandas, 2016).
Rho GTPases are central regulators of cytoskeletal dynamics (Ridley, 2015) that cycle between an inactive GDP-bound and an active GTP-bound state. This cycle is tightly controlled by regulatory proteins, such as RhoGEFs and RhoGAPs, which respectively catalyze Rho activation and inactivation (Infante & Ridley, 2013). Despite efforts to understand the participation of Rho GTPases, such as Cdc42 and RhoA, in hematopoiesis, there are few studies regarding the role of RhoC and its regulators (GEFs and GAPs) in this system.
ARHGAP21 is a RhoGAP protein (Basseres et al., 2002) that contains a PDZ and a pleckstrin homology (PH) domain in addition to the RhoGAP domain.(Basseres et al., 2002; Dubois et al., 2005) ARHGAP21 has been shown RhoGAP activity for Cdc42,(Dubois et al., 2005; Bigarella et al., 2009) RhoA and RhoC (Lazarini et al., 2013) and is thought to integrate signals from multiple pathways. Our group has previously identified the participation of ARHGAP21 in cell adhesion and migration of solid tumor cell lines, and described an increase of ARHGAP21 mRNA expression during erythroid differentiation of primary human CD34+ cells (Bigarella et al., 2009; Lazarini et al., 2013; Barcellos et al., 2013).
Here we investigate the role of Arhgap21 in hematopoiesis using a heterozygous knockout mouse model. We show that reduction of Arhgap21 levels leads to changes in the relative frequencies of hematopoietic stem and progenitor cell populations, and mobilization of immature progenitor and myeloid cells. Using both murine and human primary cells, we observed that ARHGAP21 is important for erythroid commitment of common myeloid progenitor (CMP) and megakaryocyte-erythroid progenitor (MEP) cells. To provide mechanistic insight, we show that there is increased RhoC activity (but not Cdc42 or RhoA) in the bone marrow, and decreased fibronectin adhesion in vitro, both of which likely play a role in causing the hematopoietic defects observed. These results provide strong evidence that Arhgap21 participates in cellular processes related to the maintenance of hematopoiesis, mainly through the regulation of RhoC activity.
2. Methods
2.1. Generation and genotyping of Arhgap21 haploinsufficient mice (Arhgap21+/−)
An embryonic stem cell line containing vector insertion in the Arhgap21 gene was obtained from the GeneTrap consortium (Gene Bank Accession number: CG784642) and injected into blastocysts of C57/Bl6 mice. Chimeras were genotyped for genomic insertion of the β-Geo cassette (Fig. S1A) and backcrossed with wild-type C57/Bl6 mice for 10 generations before performing experiments. Arhgap21−/− mice were embryonic lethal at E8. The reasons for embryonc lethality at 8 days post-conception are currently under investigation. Because hematopoietic stem cells emerge in the aortogonad-mesonephros region at E10.5, which occurs after Arhgap21−/− embryos have died, we have characterized the hematopoietic compartment of the haplo-insufficient mice.
Arhgap21+/− mice were genotyped by PCR, using DNA extracted from tail and primers targeting the β-Geo cassette (β-Geo forward: GGCGCCTCATGAATATTAACC; β-Geo reverse: CACTCCAACCTCCGCAAA CTC). All procedures were approved by the Ethics Committee for Experimental Research at the University of Campinas.
2.2. Isolation of bone marrow cells
Bone marrow cells were isolated by crushing the femurs, tibias and humerus of 6–10 week old mice. Cells were passed through a 70 μM strainer and red blood cells were lysed with lysis solution (155 mM NH4Cl, 10 mM NaHCO3, 0.1 mM EDTA). For histology, femurs were fixed in 10% formalin and embedded in paraffin, sectioned and placed on silanized slides followed by hematoxylin and eosin staining. Five random high-powered fields from stained slides were captured at 10× objective magnification and visualized for manual counting for mega-karyocytes, using ImageJ (http://imagej.nih.gov/ij/).
2.3. Real time PCR
RNA was purified with Illustra RNAspin Mini Kit (GE Healthcare Life Sciences, UK) and reverse transcribed with RevertAid H minus First Strand cDNA synthesis Kit (ThermoScientific, Inc., USA). Real time quantitative PCR was carried out as previously described (Xavier-Ferrucio et al., 2015), in an Eppendorf MasterCycler using SYBR green master mix (ThermoScientific, Inc., USA). Gene expression was determined, using specific primers: murine Arhgap21 (NM_001128084) Arhgap21 forward: GAGGAAAGCTTCAAGCACCA, Arhgap21 reverse: GATGACAGC AGATCAGGAA; Hprt forward: GGGGGCTATAAGTTCTTTGCT and HPRT reverse: GGCCTGTATCCAACACTTCG; human ARHGAP21 (NM_020824) ARHGAP21 forward: CAATGGATACCATATTTGTTAAGCAAGTT, ARHGAP21 reverse: CACTTTCTCCATTGACTTTTATAATTCG, HPRT forward: TTGCTTTCCTTGGTCAGGCA and HPRT reverse: TTCGTGGGGTC CTTTTCACC.
2.4. Flow cytometric analysis
Bone marrow, spleen or peripheral blood cells were incubated with specific antibodies for 15 min at room temperature in order to characterize the following hematopoietic populations: HSC: Lineage markers (CD4, CD8, B220, Ter119, Gr1, Mac1) conjugated to FITC, Sca1-PerCP, c-Kit-APC-Cy7, CD150-PE and CD48-PE-Cy7; LSK: Lineage markers conjugated to FITC, Sca1-PE and c-Kit-APC; erythroid committed cells: Ter119-FITC; myeloid cells: Gr1-PE, Mac1-APC; PreMegE, MkP and ErP: Lineage markers conjugated to PeCy7, Sca1-BV421, c-Kit-APC, CD150-PE, FcgR-APCR700, CD41-BV510 and CD105-BB515. Ter119-FITC and CD71-PE staining accessed the erythroid differentiation stages. Gating of murine BM subpopulations was performed as in Pronk et al. (2007).
Expression of CXCR-4, α4β1 and α5β1 integrins in hematopoietic progenitor cells was evaluated by staining total bone marrow cells with APC-conjugated lineage markers (Lineage APC cocktail) and CXCR-4-PE, CD49d/CD29-PE or CD49e/CD29-PE. Unless specified, all antibodies were from BD Biosciences. For intracellular assessment of CXCR-4, cells were fixed with 4% paraformaldehyde, permeabilized in PBS containing 0.2% BSA, 0.1% azide and 0.5% saponin. Fluorescence analysis was performed with a FACSCalibur or FACS Canto (Becton–Dickinson) and analyzed with FlowJo software (TreeStar Inc.)
2.5. Murine colony-formation assay
A total of 5 × 104 murine bone marrow cells were cultured in 1 mL of methylcellulose medium supplemented with growth factors (M3434, StemCell Technologies). After 10 days at 37 °C under 5% CO2 and high humidity, the numbers of granulocyte–macrophage colony forming units (CFU-GM) and burst forming unit–erythroid (BFU-E) were assessed. Secondary plating was performed to determine sequential clonogenicity and the number of colonies (CFU-GM) was counted after an additional 10 days.
2.6. CFU-S assay
The colony-forming unit in spleen (CFU-S) assay was performed by transplanting 105 bone marrow cells from wild type or Arhgap21+/− mice into 9.5Cy irradiated wild-type mice. After 12 days, the spleens were fixed with Bouin’s fixative and CFU-S were scored.
2.7. In vitro hematopoietic progenitor cell adhesion
Lineage depleted (Lin−) cells were plated on retronectin-coated dishes and incubated in DMEM (Mediatech, Herndon, VA) containing 1% BSA and 100 ng/mL CXCL12 (Peprotech, NJ, USA) for 1 h. Plates were then washed two times with cold PBS to remove non-adherent cells. The amount of adherent cells was assessed with CCK-8 reagent (Sigma-Aldrich) with optical density (OD) measured at 570 nm using an automated plate reader. The following formula was used to calculate the percentage of adherent cells: % adherent cells = OD adherent cells/OD total cells × 100.
2.8. Serial bone marrow transplantation
Serial bone marrow transplantation was performed to evaluate the engraftment of long-term HSCs. Briefly, 106 CD45.2+ bone marrow cells from wild type or Arhgap21+/− mice were transplanted into 9.5Gy irradiated B6·SJL/BoyJ (CD45.1+, Jackson Labs) recipient mice. After 16 weeks, primary recipients were sacrificed and 106 cells bone marrow cells were used for secondary transplantation into additional 9.5Gy irradiated B6·SJL/BoyJ. Donor reconstitution (CD45.2+ cells) was monitored monthly in peripheral blood and the contribution of each lineage to the reconstituted bone marrow was accessed in the end of week 16 (32 weeks total).
2.9. Human cell source, preparation, and FACS
Human granulocyte colony-stimulating factor (G-CSF) mobilized peripheral blood was CD34+ enriched (CliniMACS; Miltenyi), stained with Lineage cocktail-BV510 (BD), CD34-BV421 (Biolegend), CD38-PECF594(BD), CD45Ra-BV711, CD135-PE (Biolegend), CD36-PerCPCy5.5 (BD), CD110-APC (BD) and CD41a-APCH7(BD) antibodies. Human MEP (Lin−CD34+CD45Ra−CD135−CD38midCD110+CD36−-CD41a−), ErP (Lin−CD34+CD45Ra−CD135−CD38highCD110−), MkP (Lin−CD34+CD45Ra−CD135−CD38midCD110+CD36−CD41a+) and CMP (Lin−CD34+CD45Ra−CD135+) were sorted on a FACSAria, as previously described (Sanada et al., 2016).
2.10. ARHGAP21 vector and virus production
Lentiviral vector pLV[shRNA]-EGFP:T2A:Puro-U6 was purchased from VectorBuilder (Cyagen Biosciences) and customized with short hairpin RNA (shRNA) against ARHGAP21 or against a scramble control (shSc). The sequences of the shRNAs against human ARHGAP21 were ShARHGAP21#1: GGATCTAATTAGTCGAAGA and ShARHGAP21#2: GCACAGAGATGCTACCGAA. For lentivirus production, 80% confluent 293T cells were transfected with pLV[shRNA]-EGFP:T2A:Puro-U6 shARHGAP21–1/2 (or shScramble), Gag/VS VG, and Gag/Rev. with FuGENE HD (Promega). Virus was collected 24, 36, 48, and 60 h after transfection, and each collection was 0.45-mm filtered, concentrated using Amicon filters (Millipore), and frozen in aliquots at −80 °C.
2.11. Transduction and colony formation assays
Sorted MEPs or CMPs were cultured in expansion media (100 ng/mL recombinant hFLT3 ligand (hFLT3, 100 ng/mL recombinant human stem cell factor SCF, 20 ng/mL, recombinant human interleukin IL-3, 20 ng/mL recombinant human interleukin 6 IL-6, and 20 ng/mL recombinant human trombopoetin hTPO, all from ConnStem) in StemSpan Serum Free Expansion Medium (Stem Cell Technologies) for 16 h, transduced with lentivirus and plated in colony formation assays 48 h after transduction. For the Dual Mk/E colony assay (performed as previously described Sanada et al., 2016), 250 transduced MEPs were plated in 2 plates (125 cells/plate) of MegaCult C Medium plus Lipids (Stem Cell Technologies) with 0.5 μg/mL of puromycin (Sigma Aldrich), 3.0 U/mL rhEPO (recombinant human erythropoietin), 10 ng/mL rhIL-3, 10 ng/mL rhIL-6, 25 ng/mL rhSCF, and 50 ng/mL rhTPO. All cytokines were from ConnStem except hEPO (Amgen). After 12–14 days, the colonies were stained overnight with CD41-PE (Biolegend) and GlyA-APC (BD) (1:100 dilution) and assessed by fluorescence microscopy (DMI6000B, Leica). Colonies were scored based on GFP+ (transduced) and on GlyA and CD41a staining as megakaryocyte only (CFU-Mk), erythroid only burst forming unit (BFU-E), or megakaryocyte/erythroid (CFU-Mk/E). Transduced CMPs were also plated in Methocult (StemCell Technologies) in the presence of 2.5 μg/mL of puromycin (Sigma Aldrich) and scored according to manufacturer’s instructions.
2.12. Rho GTPase activity assay
The pull down assay protocol was performed as previously described (Pellegrin & Mellor, 2008). Briefly, 1 × 107 bone marrow cells from WT and Arhgap21+/− mice were lysed in RIPA buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% SDS, 1% NP-40 and 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 1 μg/mL leupeptin) at 4–6 °C. Lysates were cleared by centrifuging 20 min at 15,000g, 4 °C. The cleared lysate was incubated with Rhotekin-GST or Wasp-GST beads overnight in rotation at 4 °C. The beads were washed 4 times in 4–6 °C 1× lysis buffer. Proteins eluted from beads and the input samples were analyzed by SDS-PAGE. The blots were probed with anti-Cdc42, anti-RhoA, anti-RhoC (all from Cell Signaling) and anti-actin antibodies (Santa Cruz). Wasp-GST beads were kindly provided by Prof. Anne Ridley (King’s College London) and Rhotekin-GST beads were from Cytoskeleton.
2.13. Statistical analysis
Data are expressed as mean ± standard deviation. Statistical analyses were performed using the Student’s t-test or Anova with a p value ≤ 0.05 considered statistically significant. The statistical significance of all data from figures is presented in the respective legend.
3. Results
3.1. Alterations in HSPC and myeloid compartments of Arhgap21+/− mice
The role of Arhgap21 in hematopoiesis was systematically investigated in Arhgap21 haploinsufficient mice. Arhgap21+/− mice are born from Arhgap21+/− x WT crossed mating pairs with the expected Mendelian distribution (Supplementary Table 1) and live in normal conditions of animal care, with a lifespan similar to wild type. Bone marrow cells from Arhgap21+/− mice presented a 50% reduction of Arhgap21 mRNA and protein levels compared to those from WT mice (Fig. S1B and C).
Cell blood count revealed a small but statistically significant decrease in the numbers of red blood cells (RBC), platelets and hemoglobin levels and an increased number of white blood cells (WBC) and neutrophils in Arhgap21+/− mice; (all p < 0.05). No changes were observed in lymphocyte numbers (Table 1) and similar percentages of B220+ B lymphocytes and T lymphocytes (CD4+, CD8+ or CD4+CD8+ cells) were respectively detected in the bone marrow in the thymus of ARHGAP21−/− and WT mice (Fig. S2A). Substitute ARHGAP21−/− for Arhgap21+/−
Table 1.
Hematologic parameters of WT and Arhgap21+/− mice.
| Hematologic Parameters | WT | Arhgap21+/− | p value |
|---|---|---|---|
| WBC (× 103 cells/μL) | 9.6 ± 1.82 | 12.4 ± 1.8 | 0.018 |
| Lymphocytes (cells/μL) | 7884 ± 1587 | 9842 ± 1392 | 0.09 |
| Neutrophils (cells/μL) | 1715 ± 189.7 | 2265 ± 157.6 | 0.04 |
| RBC (× 106 cells/μL) | 10.93 ± 0.34 | 10.42 ± 0.28 | 0.015 |
| Hemoglobin (g/dL) | 16.27 ± 0.36 | 15.63 ± 0.35 | 0.0087 |
| Hematocrit (%) | 55.0 ± 1.73 | 53.5 ± 1.51 | ≥0.05 |
| MCV (fL) | 50.04 ± 0.48 | 51.05 ± 0.88 | 0.024 |
| Platelets (105 cells/μL) | 1.84 ± 0.05 | 1.74 ± 0.07 | 0.020 |
| MPV (fL) | 7.05 ± 0.12 | 7.2 ± 0.08 | 0.042 |
Blood samples were collected from 9 WT and 8 Arhgap21+/− mice (8 wk. old littermates). Counts were performed by automated hematology analyzer (Poch 100iv Diff; Sysmex; Roche). RBC: red blood cell count; MCV: mean cell volume; WBC: White blood cell count; MPV: mean platelet volume.
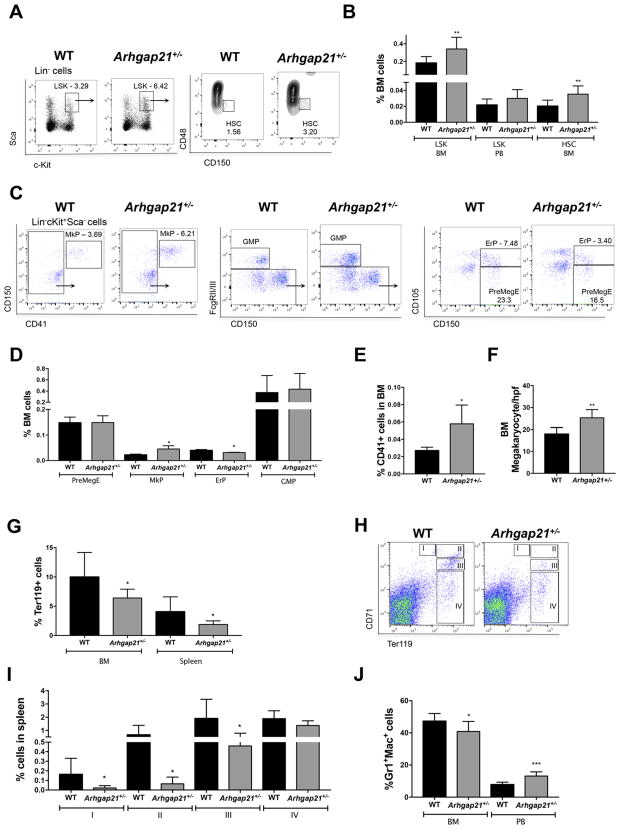
The bone marrow cellularity and the frequency of Lin− hematopoietic progenitors in WT and Arhgap21+/− mice were similar (Fig. S2 B–C). However, there was a statistically significant increase in the frequency of both Lin−Sca+cKit+ (LSK) and Lin−Sca+cKit+CD48−CD150+ (hematopoietic stem cells, (HSC)) in the bone marrow of Arhgap21+/− mice (Fig. 1A–B).
Fig. 1.
HSPC and myeloid compartments are altered in Arhgap21+/− mice. (A–B) Flow cytometry analysis show increased frequency of Lin−Sca-1+c-Kit+(LSK) and of Lin−Sca-1+c-Kit+CD150+CD48− (LSK CD150+CD48−) cells in bone marrow of Arhgap21+/− mice (WT n = 6 and Arhgap21+/− n = 4; LSK BM P = 0.04; HSC P = 0.03). (C–D) Flow cytometry based on Pronk et al. (2007) to quantify MkP (Lin− FcgRII/III− cKit+ Sca− CD150+ CD41+), ErP (Lin− FcgRII/III− cKit+ Sca− CD150+ CD41− CD105+), PreMegE (Lin− FcgRII/III− cKit+ Sca− CD150+ CD41− CD105−) and GMP (Lin− FcgRII/III+ cKit+ Sca− CD150− CD41−) shows no change in GMP or PreMegE frequency, an increase of MkP (P = 0.02) and a decrease of ErP (P = 0.02) (WT n = 5 and Arhgap21+/− n = 3). Confirming the megakaryocytic over erythroid change, there was an increase in (E) CD41+ cells by FACS analysis (P = 0.03) and in (F) megakaryocyte number assessed by morphology (P = 0.002) in 5 high power fields (hpf) of hematoxylin eosin stained bone marrow sections of each mouse (WT n = 8 and Arhgap21+/− n = 5). There was a decrease in (G) Ter119+ cells in bone marrow (P = 0.01) and spleen (P = 0.02). (H–I) Flow cytometric characterization of erythroid differentiation showing a decreased frequency Arhgap21+/− splenic proerythroblasts (I, P = 0.03), basophilic erythroblasts (II, P = 0.03), and late basophilic/polychromatophilic erythroblasts (III, P = 0.01), but no difference in the late subtypes of the erythroid differentiation such as orthochromatophilic erythroblasts (IV, WT n = 6 and Arhgap21+/− n = 9). (J) Flow cytometry results showing reduced frequency of Gr1+/Mac-1+ cells in bone marrow (P = 0.04) and increased frequency in peripheral blood (P = 0.0009) of Arhgap21+/− mice (WT n = 6 and Arhgap21+/− n = 10).
The subtle changes in RBC and platelets led us to investigate the progenitor cells that give rise to both megakaryocytes and erythroid cells. Flow cytometric analysis showed no difference in the frequency of granulocyte monocyte progenitors (GMP) or PreMegE. However, an increased frequency of megakaryocyte progenitors (MkP, P = 0.02) and a decreased frequency of erythroid progenitors (ErP, P = 0.02), was observed in Arhgap21+/− mice, suggesting that the PreMegEs could be biased toward the megakaryocytic lineage (Fig. 1C–D). This result was further corroborated by the 2-fold increase in the frequency of megakaryocytic committed cells (CD41+, P = 0.04) and in the megakaryocyte number (P = 0.002) from Arhgap21+/− bone marrow (Fig. 1E–F). Consistent with a potential effect on MEP fate, there was a 2-fold decrease of erythroid committed (Ter119+) cells in both the bone marrow (P = 0.01) and spleen (P = 0.02) of Arhgap21+/− mice (Fig. 1G).
In order to better characterize the erythroid differentiation, we further dissected terminal erythroid differentiation. (Koulnis et al., 2011; Liu et al., 2006; Socolovsky et al., 2001) While there were no statistically significant differences in the bone marrow of the haploinsufficient mice (Fig. S2D) compared to WT, there were differences in the spleen. We observed a decreased frequency of proerythroblasts (Ter119medCD71high, I), basophilic erythroblasts (Ter119highCD71high, II), and late basophilic/polychromatophilic erythroblasts (Ter119highCD71med, III) in the spleen of Arhgap21+/− mice, but no difference in the late subtypes of the erythroid differentiation such as orthochromatophilic erythroblasts (Ter119highCD71low, IV; Fig. 1H–I), which suggests that there is some compensatory process to prevent anemia in the haploinsufficient mice.
Within the myeloid compartment, no changes were observed in the percentage of the granulocyte/monocyte progenitor (GMP, Fig. 1C–D), however there was a 10% decrease in Gr+Mac-1+ myeloid cell frequency in Arhgap21+/− bone marrow concomitant with 5% increase in frequency of myeloid cells in the peripheral blood of the same mice (Fig. 1J). This finding underlies the increase in the absolute leukocyte and neutrophil numbers in the peripheral blood (Table 1). Taken together, these results show that the myeloid compartment is altered in Arhgap21+/− mice perhaps due to enhanced myeloid egress from the marrow.
3.2. Enhanced HSCP egress in Arhgap21+/− mice may be due to lower adhesion
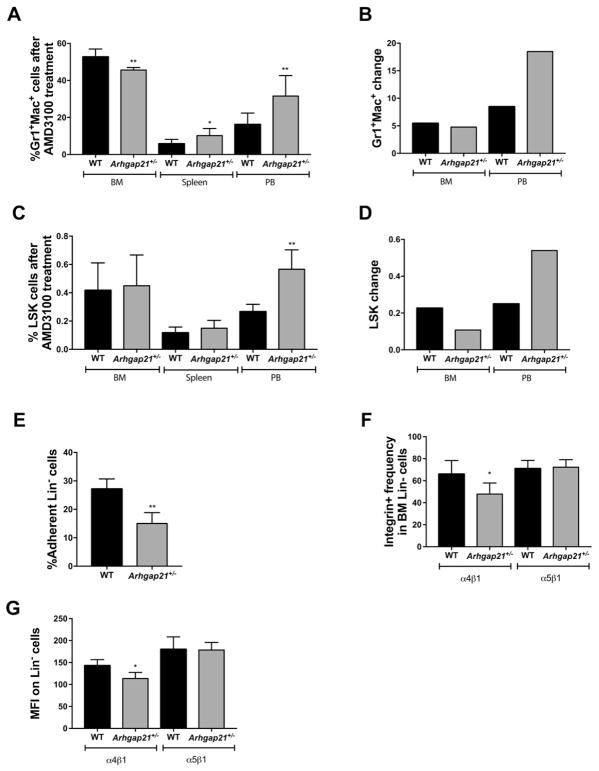
In order to investigate whether the increased number of myeloid cells in the peripheral blood and apparent loss from the bone marrow, was due to enhanced egress, we induced HSPC mobilization using AMD3100, a CXCR4 antagonist that mobilizes hematopoietic stem/progenitor cells from the marrow (Winkler et al., 2012).
After 1 h of treatment, myeloid cells (Gr-1+Mac-1+) from Arhgap21+/− mice maintain their decreased frequency of Gr-1+Mac-1+ in the bone marrow compared to WT, and showed a significantly increased frequency in peripheral blood compared to WT (Fig. 2A, P = 0.001). After normalizing for the frequency of Gr-1+Mac-1+ cells at baseline for each genotype (Fig. 1J, no AMD3100 treatment), the mobilization of Gr-1+Mac-1+ from the BM to the peripheral blood was 30% higher in Arhgap21+/− compared to WT mice (Fig. 2B). In both genotypes, AMD3100 increased the percentage of LSK cells in the BM, with both reaching approximately 0.4%. The frequency of LSK cells was increased in the peripheral blood of both genotypes (Fig. 2C). However, the increase in Arhgap21+/− mice was significantly higher with a 17% greater increase in mobilization for Arhgap21+/− vs. controls after normalization to baseline levels (Fig. 2D).
Fig. 2.
Arhgap21 deficiency in progenitor cells leads to increased mobilization and defective adhesion. (A–D) Hematopoietic cells were analyzed by flow cytometry 1 h after AMD3100 treatment. (A) Bone marrow from Arhgap21+/− mice exhibited a reduction of Gr-1+/Mac-1+ cells compared to WT (P = 0.001), whereas the frequencies of Gr-1+/Mac-1+ cells in spleen (P = 0.02) and peripheral blood (P = 0.007) were increased. (B) The change in the frequency of Gr-1+Mac-1+ cells before and after AMD3100 for each genotype is shown (after AMD3100 treatment as shown in Fig. 2A) minus before AMD3100 treatment as in Fig. 1J). (C) The frequency of LSK cells was not significantly altered in the bone marrow and spleen, whereas the frequency of LSK was increased in peripheral blood (P = 0.0005) of Arhgap21+/− mice after AMD3100 treatment (WT n = 7 and Arhgap21+/− n = 7). (D) The change in the frequency of LSK cells before and after AMD3100 for each genotype is shown ((after AMD3100 treatment as shown in Fig. 2C) minus before AMD treatment as in Fig. 1B). (E) Adhesion to fibronectin was impaired in Lin− cells obtained from Arhgap21+/− mice (WT n = 5 and Arhgap21+/− n = 5; P = 0.003). (F) Arhgap21+/− mice showed a reduction of Lin− cells expressing CD49d/CD29 (a4b1 integrin) in bone marrow (P = 0.04), but no difference in the frequency of Lin− cells expressing CD49e/CD29 (a5b1 integrin). (G) Similarly, the mean fluorescence intensity of α4β1 integrin in Lin− Arhgap21+/− cells were also decreased (P = 0.02) when compared to WT.
To investigate whether the enhanced mobilization was associated with a change in CXCR-4 expression, Lin− bone marrow cells were fractionated and membrane-associated and intracellular CXCR-4 expression was evaluated by flow cytometry. As shown in Fig. S3, the frequency of Lin− cells expressing CXCR-4 in the membrane (A), or in the intracellular compartment (B) did not differ between WT and Arhgap21+/− mice.
We next determined whether the higher level of Arhgap21+/− myeloid cells at baseline might be due to decreased adhesion of hematopoietic cells in the BM environment. To investigate in vitro the role of Arhgap21 in the adhesion of hematopoietic progenitor cells, we used a recombinant fibronectin that has binding sites for α4β1 and α5β1 integrins, important molecules for hematopoietic progenitor cell adhesion to the bone marrow niche (Klein et al., 2015). We observed that adhesion of Arhgap21+/−Lin− hematopoietic progenitor cells to fibronectin was significantly reduced compared to WT progenitors (Fig. 2E, P = 0.003). Consistent with this, we observed a decrease in the frequency (P = 0.04) and in the mean fluorescence intensity (P = 0.03) of α4β1+ on Lin− bone marrow cells of Arhgap21+/− mice with no difference in Lin− α5β1+ cells (Fig. 2F–G).
Together, these results suggest that reduced levels of Arhgap21 promote the egress of myeloid cells and HSPC, and that this may be due to defective adhesion to the bone marrow niche.
3.3. Impaired function of Arhgap21+/− HSCP in vitro and in vivo
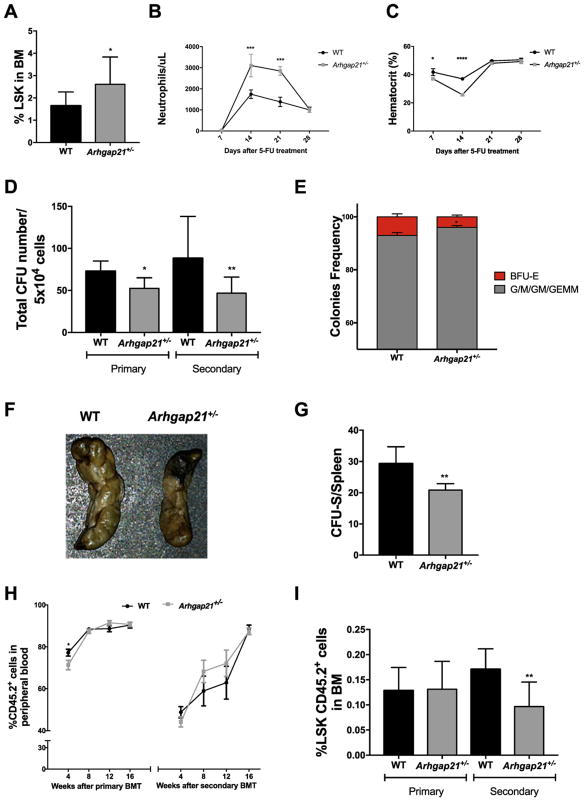
In order to confirm the increased frequency of HSPC, we challenged WT and Arhgap21+/− mice with 5-FU (5-fluorouracil), which depletes cycling cells. Hematologic recovery after 5-FU depends on both the quantity and function of the surviving HSPC population. Arhgap21+/− mice recovered a higher number of neutrophils in peripheral blood compared to WT (Fig. 3B). In addition, Arhgap21+/− mice presented an increased frequency of LSK cells in the bone marrow 28 days after 5-FU treatment, suggesting that their higher hematologic recovery may have been due to increased HSPC (Fig. 3A). Interestingly, the hematocrit of Arhgap21+/− mice was significantly reduced 7 (P = 0.03) and 14 (P < 0.0001) days after 5-FU treatment compared to WT, suggesting that the erythroid defects observed under steady state resulted in slower erythroid recovery that resolved by day 28 (Fig. 3C).
Fig. 3.
Impaired function of Arhgap21+/− hematopoietic stem and progenitor cells in vitro and in vivo. (A-C) Short-term reconstitution was evaluated by hematopoietic recovery after 5-FU treatment. (A) We observed a higher frequency of Lin−Sca+cKit+ (LSK) cells 28 days after 5-FU treatment (P = 0.04). (B–C) Peripheral blood was collected 7, 14, 21 and 28 days after treatment and blood cell count was performed by automated hematology analyzer. (B) There is an increase in the neutrophil number of Arhgap21 mice 14 (P = 0.0005) and 21(P = 0.0002) days after 5-FU treatment. (C) Despite an increase in the LSK frequency, there is a decrease in erythroid reconstitution 7 (P = 0.03) and 14 days (P < 0.0001) after the 5-FU treatment. (D-E) Bone marrow cells from wild type and Arhgap21+/− mice were cultured in methylcellulose media supplemented with cytokines for myeloid differentiation and the total number of colonies was scored after ten days of incubation. (D) Colonies were then replated in methylcellulose and secondary CFU formation was accessed after an additional 10 days (Primary: P = 0.007, Secondary: P = 0.02). (E) When analyzed by frequency, there is a decrease of BFU-E percentage in the haploinsufficient mice (P = 0.03), suggesting that the erythroid compartment is the most affected by the reduction of Arhgap21. (F-G) For CFU-S, bone marrow cells were injected into lethally irradiated WT mice, and spleen colonies counted after 12 days. (F) Macroscopic view of CFU-S in recipients that received cells from WT and Arhgap21+/− mice. (G) Quantification of CFU-S colonies showing reduced number of CFU-S in mice that received Arhgap21+/− cells (P = 0.002). (H–I) Serial bone marrow transplantation was performed using bone marrow cells from Arhgap21+/− and WT mice transplanted into lethally irradiated CD45.1 mice. Chimerism (CD45.2+) in the peripheral blood of the recipients was examined at indicated times after primary and secondary transplantation, showing a slight reduction of donor cells in PB of Arhgap21+/− mice, 4 weeks after primary transplant (P = 0.04). (I) Donor derived (CD45.2+) immature LSK cells, which were analyzed in bone marrow 16 weeks after primary and secondary transplantation by flow cytometry, are reduced in recipient mice that received Arhgap21+/− bone marrow in secondary transplantation (P = 0.001). Minimum of 8 WT and 8 Arhgap21+/− animals used in each experiment.
Despite the increased amount of HSPC by FACS phenotype, the total number of colonies forming units (CFU-GEMM, CFU-G/M (combined number of CFU-GM, CFU-G, CFU-M) and BFU-E) in the bone marrow was decreased in Arhgap21+/− mice compared to WT (Fig. 3D). The mean ± SD of colonies was as follows: CFU-G/M: (WT: 66.7 ± 5.2 and Arhgap21+/−: 48.1 ± 4.7, P = 0.02); CFU-GEMM: (WT: 2.1 ± 0.36 and Arhgap21+/−: 1 ± 0.2, P = 0.03); and BFU-E: (WT: 5.5 ± 0.7 and Arhgap21+/−: 3.2 ± 0.5, P = 0.04). This result was further evidenced after secondary plating (Fig. 3D). We also assessed the ratio of erythroid (BFU-E) vs. myeloid (CFU-GM/GEMM) colonies between WT and Arhgap21+/−. Interestingly, when analyzed by frequency, there is a decrease of BFU-E percentage in the haploinsufficient mice (P = 0.03), suggesting that the erythroid compartment is more affected by the reduction of Arhgap21. The decrease in BFU-Es in Arhgap21+/− mice was more dramatic than other CFU subtypes tested (Fig. 3E).
Consistent with the decreased number of functional progenitors assessed in vitro, Arhgap21+/− mice had fewer CFU-S than WT mice. WT and Arhgap21+/− mice bone marrow cells generated on average 29.3 and 20.8 CFU-S per 105 injected cells, respectively (P = 0.002, Fig. 3F–G). Thus, although an increase in HSPC number was observed, progenitor cells from Arhgap21+/− have decreased ability to produce colonies in vitro and in vivo.
To better understand the role of Arhgap21 in HSC function, the capacity of Arhgap21+/− HSC to long-term reconstitute hematopoiesis was analyzed in a serial noncompetitive transplantation using CD45.1+ lethally irradiated recipients. CD45.2+ Arhgap21+/− bone marrow cells were able to fully reconstitute irradiated mice, as assessed by peripheral blood chimerism, although a slight reduction of Arhgap21+/− derived cells compared to WT was observed 4 weeks after primary transplantation (Fig. 3H). No significant differences in the frequency of Arhgap21+/− cells in the peripheral blood were observed after 8, 12 and 16 weeks of the transplant.
Sixteen weeks after the primary bone marrow transplant, recipient bone marrow cells were analyzed by flow cytometry and transplanted into secondary lethally irradiated recipients. Some immunophenotype characteristics previously observed in Arhgap21+/− mice were recapitulated in the primary recipients, such as decreased myeloid cells in the bone marrow (Supp. Fig. 4A). After primary bone marrow reconstitution, the frequency of donor derived LSK cells was similar in the recipients of WT and Arhgap21+/− cells, however, 16 weeks after the secondary transplant, the frequency of LSK cells in the bone marrow of recipients of Arhgap21+/− cells was significantly decreased compared to recipients of WT cells (Fig. 3I). This progressive reduction of LSK cell frequency with serial transplantation despite the increased frequency of phenotypic HSC observed in Arhgap21+/− bone marrow at baseline suggests that the engraftment of long-term self-renewing Arhgap21+/− HSC may be impaired.
3.4. ARHGAP21 knockdown impairs erythroid commitment in human primary cells
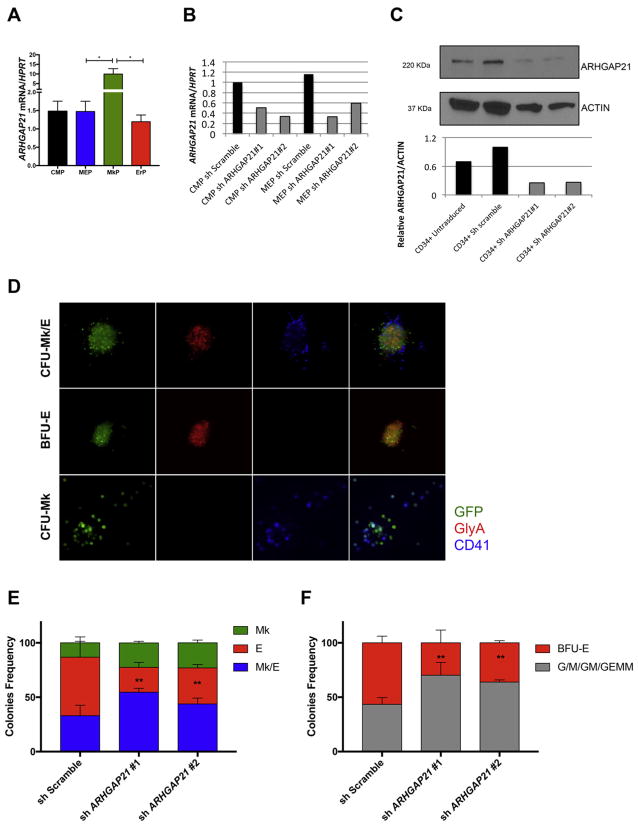
We next sought to investigate possible alterations caused by ARHGAP21 downregulation in primary human cells. ARHGAP21 gene expression was higher in FACS-sorted MkP (Lin−CD34+ CD45Ra−CD135−CD38midCD110+CD36−CD41a+) than in CMP (Lin−CD34+ CD45Ra− CD135+), MEP (Lin−CD34+ CD45Ra−CD135−CD38mid CD110+CD36−CD41a−) and ErP (Lin−CD34+CD45Ra−CD135−CD38highCD110−, Fig. 4A purified accordantly to our recent publication.(Sanada et al., 2016) We knocked down ARHGAP21 expression in MEP and CMP using two different shRNAs against ARHGAP21 that each gave >60% inhibition of ARHGAP21 mRNA and protein (Fig. 4B–C). To assess the effect of ARHGAP21 knockdown on the Mk vs. E fate decision, MEPs were plated in the CFU-Mk/E assay 48 h after transduction in the presence of selective antibiotic. After 12 days, colonies were stained with CD41 and GlyA and scored for CFU-Mk/E, CFU-Mk and BFU-E using fluorescence microscopy (Fig. 4D). Despite the increase of ARHGAP21 in the MkP population, the knockdown of ARHGAP21 increased CFU-Mk and CFU-Mk/E at the expense of BFU-E (P = 0.002 in two-way Anova, Fig. 4E). These data are consistent with the results showing an increased frequency of MkPs and decreased frequency of ErPs in Arhgap21+/− mice, and support the hypothesis that ARHGAP21 plays a role in the MEP fate decision by inhibiting megakaryocytic and promoting erythroid commitment.
Fig. 4.
ARHGAP21 knockdown impairs erythroid commitment of human CMP and MEP. (A) Real-time PCR analysis of ARHGAP21 mRNA expression of FACS-sorted CMP, MEP, MkP and ErP (from 3 different donors) show upregulation of the transcript in MkP when compared to MEP (P = 0.04) and ErP (P = 0.04). (B–C) Knockdown of ARHGAP21 in CMP and MEP cells showing decreased (B) mRNA and (C) protein expression 24 h after transduction. (D) Improved dual CFU-Mk/E assay to determine Mk/E potential of transduced MEPs. Cells are plated in a collagen-based colony assay with EPO, TPO, SCF, IL-3, and IL-6 to promote growth of megakaryocyte and erythroid colonies. After 12 days of culture, colonies are stained for CD41a (Mk specific, false colored as blue) and GlyA (E specific, false colored as red) and checked for transduction efficiency (GFP+, green). Representative images of different colony types are shown. Typical CFU-Mk/E stains for both GlyA and CD41, BFU-E stains for GlyA only and CFU-Mk stains for CD41a only. Both Dual Mk/E of MEPs (E) and methylcellulose of CMPs (F) assays show decreased BFU-E when ARHGAP21 is knocked down (P = 0.002 in Anova tests for both assays). Results are presented as average + SD from 3 independent experiments.
We also observed a significant reduction of BFU-E colonies in CMPs with ARHGAP21 knockdown in the methylcellulose assay (Fig. 4F). Overall, these results suggest that Arhgap21 haploinsufficiency leads to a reduction of erythroid commitment in both mouse and human systems.
3.5. RhoC activity is enhanced in Arhgap21+/− mice
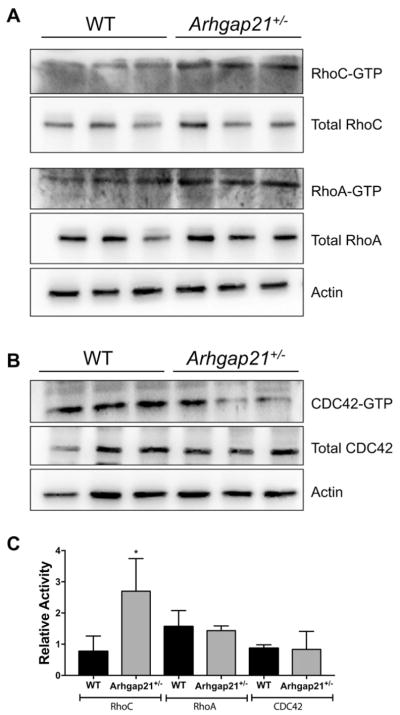
Given the important role of Rho GTPases in HSPC functions (Nayak et al., 2013) and previous publications showing that Arhgap21 can inhibit RhoA, RhoC and Cdc42 activities, (Dubois et al., 2005; Bigarella et al., 2009; Lazarini et al., 2013; Barcellos et al., 2013) we wondered if these Rho GTPases could be downstream targets affected in the Arhgap21+/− mice. Pull-down assays showed that Arhgap21+/− bone marrow cells exhibited enhanced RhoC activity, but no changes in RhoA or Cdc42 activities (Fig. 5A–C) suggesting that Arhgap21 functions as a RhoGAP to decrease RhoC activity in the bone marrow.
Fig. 5.
RhoC activity is increased in Arhgap21+/− mice. (A-B) Active Rho A, RhoC and CDC42 was determined by GST-pull down assay from WT and Arhgap21+/− BM cells lysates. (A) Representative Western blots of Rhotekin-GST pull down (respective upper panel), assessment of total RhoA and RhoC (respective intermediate panels) and Actin (lowest panel). (B) Representative Western blots of PAK-GST pull down (upper panel), assessment of total CDC42 (intermediate panels) and Actin (lowest panel). (C) Quantification of western blot band intensities from GST-pull down assays normalized to the total amount of the respective RhoGTPase showing significant (P = 0.04) increase in RhoC activity.
4. Discussion
ARHGAP21 has been shown to negatively regulate Cdc42, RhoA and RhoC in cell lines.(Dubois et al., 2005; Bigarella et al., 2009; Lazarini et al., 2013; Barcellos et al., 2013) In the present study, we describe the functional role of this RhoGAP in hematopoiesis, using Arhgap21 haploinsufficient mice, which showed decreased Arhgap21 expression associated with increased RhoC activity in bone marrow cells.
Rho GTPases have been extensively studied in hematopoiesis and are implicated in HPSC cellular process that require cytoskeleton rearrengements, such as differentiation, migration, adhesion, engrafment, survival, self-renewal and aging.(Mulloy et al., 2010; Florian et al., 2012; Wang et al., 2006; Yang et al., 2001; Yang et al., 2007) These data demonstrate the importance of studying the role of Rho GTPase regulators in hematopoiesis.
We first investigated the HSC pool in bone marrow of Arhgap21+/− mice and its ability to recover after myeloablative stress. Our results suggest a deregulation of HSC production in Arhgap21+/− mice, as they exhibited an increased frequency of both short and long-term HSC in bone marrow, which in turn may have contributed to the better recovery of the hematopoietic system after stress. Besides the increased frequency of HSC and faster recovery ability, the bone marrow of Arhgap21+/− mice exhibited a reduction in functional hematopoietic progenitors, assessed by the number of CFU and CFU-S, indicating that the progenitors might be short-lived.
Previous studies showed that ARHGAP21 plays an essential role in both migration and adhesion of non-hematopoietic cells (Bigarella et al., 2009; Lazarini et al., 2013; Barcellos et al., 2013). Herein, we showed that this could be also applied to the hematopoietic system, since we observed defective adhesion to fibronectin of Lin− progenitor cells and a reduction of α4β1 (VLA4) integrin expression on bone marrow cells of haplo insufficient mice. VLA-4 binds to fibronectin and to VCAM-1 (Vascular cell adhesion-1), mediating the interaction between hematopoietic progenitors and the bone marrow microenvironment (Gur-Cohen et al., 2015). The inhibition of this integrin leads to egress of hematopoietic cells from bone marrow (Spiegel et al., 2008). Thus, Arhgap21 downregulation may contribute to the disruption of fibronectin-adhesion, stimulating the mobilization of hematopoietic progenitors from bone marrow through the reduction of VLA-4 expression.
Several steps are involved in the reconstitution of hematopoietic system after bone marrow transplantation: adhesion, proliferation and engraftment. Afterwards, HSC must renew their compartment in bone marrow, ensuring the maintenance of their pool for the proper repopulation of hematopoietic cells (Morrison & Scadden, 2014; Mendez-Ferrer et al., 2015). Although Arhgap21+/− mice showed normal repopulation of hematopoietic system, Arhgap21+/− HSC could not sustain the long-term reconstitution at the same levels as WT HSC, indicated by their lower frequency after secondary transplantation. Our results show that Arhgap21 does not alter initial repopulation ability of hematopoietic system; however, Arhgap21 appears to be important for the HSC long-term reconstitution, perhaps by regulating HSC self-renewal.
Analysis of the erythroid and megakaryocytic lineages in Arhgap21+/− bone marrow indicated a reduced number of erythroid progenitors (BFU-E in colony formation and ErP by flow cytometry) along with increased frequency of MkP and megakaryocytic-committed (CD41+) cells, suggesting that the downregulation of Arhgap21 could bias the MEP fate decision toward the megakaryocytic lineage. In order to test this observation, one needs to evaluate a truly bipotent MEP in assays with single cell resolution, but with high capacity so that hundreds of cells can be assayed simultaneously to assess not only the fate decision of this purified population, but also the molecular mechanisms on which their function relies (Narla & Mohandas, 2016; Sanada et al., 2016). Taking advantage of our recently published strategies to enrich for human MEP, MkP and ErP (Sanada et al., 2016) along with the development of a novel assay to quantify at the single cell level the MEP fate decision, our hypothesis of a Mk-biased MEP under Arhgap21 inhibition in mice was confirmed in human primary cells.
In addition to the fate decision process, after erythroid commitment, Arhgap21 also affects the later stages of erythroid differentiation as we observed a decreased number of BFU-E colonies and Ter119+ cells in the bone marrow and spleen. By flow cytometry, we observed a decreased frequency of proerythroblasts, basophilic erythroblasts, and late basophilic/polychromatophilic erythroblasts in the spleen of Arhgap21+/− mice, without affecting late subtypes of the erythroid differentiation such as orthochromatophilic erythroblasts, which suggests that there is some compensatory process to prevent anemia in the haploinsufficient mice. This process was further evidenced when the mice were challenged with 5-FU treatment. While presenting a higher neutrophil reconstitution, the same haploinsufficient mice showed a reduced hematocrit 7 and 14 days after the treatment that reached the same levels as the WT by day 28. A previous study from our group demonstrated that ARHGAP21 expression increases during erythroid differentiation of human CD34+ cells, indicating that ARHGAP21 might play a role in erythroid differentiation (Basseres et al., 2002).
Different from RhoA and CDC42, the role of RhoC in hematopoiesis remains largely unknown. RhoC is known to participate in cancer cell proliferation, migration and metastasis (Guan et al., 2017; Vega et al., 2011). Interestingly, RhoC methylation and expression levels were associated with overall survival in acute myeloid leukemia, supporting not only a prognostic but also a biologic relevance of RhoC that could be acting as an oncogene (Marcucci et al., 2014). RhoC was also described to significantly lower the expression levels of the core stem cell transcription factors, Nanog, Sox2 and Oct3/4 through reducing levels of phosphorylated STAT3 via the IL-6/JAK signaling pathway (Islam et al., 2014). Here we described Arhgap21 as a new regulator of RhoC in the hematopoietic system.
In conclusion, the establishment and maintenance of the hematopoiesis involve restrict control of the renewal and differentiation of HSCs together with their migration and retention in specialized stem-cell niches. This is the first study implicating Arhgap21 in hematopoiesis and our results include Arhgap21 as a new modulator of hematopoietic cell mobilization, adhesion, and erythroid commitment, supporting the specificity of different RhoGAPs in orchestrating hematopoietic homeostasis. Our results indicate that Arhgap21 functions in hematopoiesis may be at least partially mediated by RhoC, providing new insights about the function of this Rho GTPase in hematopoiesis and stimulating further investigations.
Supplementary Material
Acknowledgments
Funding
This work was supported by FAPESP (2009/08908-6, 2010/50682-2 and 2011/51959-0), CNPq (INCT-Sangue), the National Institutes of Health (DK086267 and DK094934) and the Yale Cooperative Center of Excellence in Hematology (1U54DK106857).
We thank IPEN (Instituto de Pesquisa Energéticas e Nucleares), especially Elisabeth Somessari and Carlos Silvei, for performing irradiation for bone marrow transplantion. We also would like to thank Dr. Anne Ridley for helpful discussions and reagents, and Dr. Stefan Bohlander and the members of his laboratory for helping us to develop the bone marrow transplantation assay.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2017.11.014.
References
- Barcellos KS, Bigarella CL, Wagner MV, Vieira KP, Lazarini M, Langford PR, Machado-Neto JA, Call SG, Staley DM, Chung JY, Hansen MD, Saad ST. ARHGAP21 protein, a new partner of alpha-tubulin involved in cell-cell adhesion formation and essential for epithelial-mesenchymal transition. J Biol Chem. 2013;288:2179–2189. doi: 10.1074/jbc.M112.432716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N, Severe N, Scadden DT. Hematopoiesis: reconciling historic controversies about the niche. Cell Stem Cell. 2017;20:590–592. doi: 10.1016/j.stem.2017.03.025. [DOI] [PubMed] [Google Scholar]
- Basseres DS, Tizzei EV, Duarte AA, Costa FF, Saad ST. ARHGAP10, a novel human gene coding for a potentially cytoskeletal Rho-GTPase activating protein. Biochem Biophys Res Commun. 2002;294:579–585. doi: 10.1016/S0006-291X(02)00514-4. [DOI] [PubMed] [Google Scholar]
- Bigarella CL, Borges L, Costa FF, Saad ST. ARHGAP21 modulates FAK activity and impairs glioblastoma cell migration. Biochim Biophys Acta. 2009;1793:806–816. doi: 10.1016/j.bbamcr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Dubois T, Paleotti O, Mironov AA, Fraisier V, Stradal TE, De Matteis MA, Franco M, Chavrier P. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–364. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- Florian MC, Dorr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, Filippi MD, Hasenberg A, Gunzer M, Scharffetter-Kochanek K, Zheng Y, Geiger H. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Chen S, Zhao Y. The role of RhoC in malignant tumor invasion, metastasis and targeted therapy. Histol Histopathol. 2017:11915. doi: 10.14670/HH-11-915. [DOI] [PubMed] [Google Scholar]
- Gur-Cohen S, Itkin T, Chakrabarty S, Graf C, Kollet O, Ludin A, Golan K, Kalinkovich A, Ledergor G, Wong E, Niemeyer E, Porat Z, Erez A, Sagi I, Esmon CT, Ruf W, Lapidot T. PAR1 signaling regulates the retention and recruitment of EPCR-expressing bone marrow hematopoietic stem cells. Nat Med. 2015;21:1307–1317. doi: 10.1038/nm.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante E, Ridley AJ. Roles of Rho GTPases in leucocyte and leukaemia cell transendothelial migration. Philos Trans R Soc Lond Ser B Biol Sci. 2013;368 doi: 10.1098/rstb.2013.0013. 20130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M, Sharma S, Teknos TN. RhoC regulates cancer stem cells in head and neck squamous cell carcinoma by overexpressing IL-6 and phosphorylation of STAT3. PLoS One. 2014;9:e88527. doi: 10.1371/journal.pone.0088527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Schmal O, Aicher WK. Matrix metalloproteinases in stem cell mobilization. Matrix Biol. 2015;44–46:175–183. doi: 10.1016/j.matbio.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Koulnis M, Pop R, Porpiglia E, Shearstone JR, Hidalgo D, Socolovsky M. Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J Vis Exp. 2011 doi: 10.3791/2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarini M, Traina F, Machado-Neto JA, Barcellos KS, Moreira YB, Brandao MM, Verjovski-Almeida S, Ridley AJ, Saad ST. ARHGAP21 is a RhoGAP for RhoA and RhoC with a role in proliferation and migration of prostate adenocarcinoma cells. Biochim Biophys Acta. 2013;1832:365–374. doi: 10.1016/j.bbadis.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pop R, Sadegh C, Brugnara C, Haase VH, Socolovsky M. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 2006;108:123–133. doi: 10.1182/blood-2005-11-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, Kohlschmidt J, Mrozek K, Wu YZ, Bucci D, Curfman JP, Whitman SP, Eisfeld AK, Mendler JH, Schwind S, Becker H, Bar C, Carroll AJ, Baer MR, Wetzler M, Carter TH, Powell BL, Kolitz JE, Byrd JC, Plass C, Garzon R, Caligiuri MA, Stone RM, Volinia S, Bundschuh R, Bloomfield CD. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32:548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Scadden DT, Sanchez-Aguilera A. Bone marrow stem cells: current and emerging concepts. Ann N Y Acad Sci. 2015;1335:32–44. doi: 10.1111/nyas.12641. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy JC, Cancelas JA, Filippi MD, Kalfa TA, Guo F, Zheng Y. Rho GTPases in hematopoiesis and hemopathies. Blood. 2010;115:936–947. doi: 10.1182/blood-2009-09-198127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Mohandas N. The road not taken? Blood. 2016;128:886–888. doi: 10.1182/blood-2016-07-722413. [DOI] [PubMed] [Google Scholar]
- Nayak RC, Chang KH, Vaitinadin NS, Cancelas JA. Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol Rev. 2013;256:255–268. doi: 10.1111/imr.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. Rho GTPase activation assays. Curr Protoc Cell Biol. 2008;Chapter 14(Unit 14):18. doi: 10.1002/0471143030.cb1408s38. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada C, Xavier-Ferrucio J, Lu YC, Min E, Zhang PX, Zou S, Kang E, Zhang M, Zerafati G, Gallagher PG, Krause DS. Adult human megakaryocyte-erythroid progenitors are in the CD34+CD38mid fraction. Blood. 2016;128:923–933. doi: 10.1182/blood-2016-01-693705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Spiegel A, Zcharia E, Vagima Y, Itkin T, Kalinkovich A, Dar A, Kollet O, Netzer N, Golan K, Shafat I, Ilan N, Nagler A, Vlodavsky I, Lapidot T. Heparanase regulates retention and proliferation of primitive Sca-1+/c-Kit+/Lin- cells via modulation of the bone marrow microenvironment. Blood. 2008;111:4934–4943. doi: 10.1182/blood-2007-10-116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol. 2011;193:655–665. doi: 10.1083/jcb.201011038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang L, Filippi MD, Williams DA, Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler IG, Pettit AR, Raggatt LJ, Jacobsen RN, Forristal CE, Barbier V, Nowlan B, Cisterne A, Bendall LJ, Sims NA, Levesque JP. Hematopoietic stem cell mobilizing agents G-CSF, cyclophosphamide or AMD3100 have distinct mechanisms of action on bone marrow HSC niches and bone formation. Leukemia. 2012;26:1594–1601. doi: 10.1038/leu.2012.17. [DOI] [PubMed] [Google Scholar]
- Xavier-Ferrucio JM, Pericole FV, Lopes MR, Latuf-Filho P, Barcellos KS, Dias AI, de Campos PM, Traina F, Vassallo J, Saad ST, Favaro P. Abnormal Hedgehog pathway in myelodysplastic syndrome and its impact on patients’ outcome. Haematologica. 2015;100:e491–493. doi: 10.3324/haematol.2015.124040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FC, Atkinson SJ, Gu Y, Borneo JB, Roberts AW, Zheng Y, Pennington J, Williams DA. Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc Natl Acad Sci U S A. 2001;98:5614–5618. doi: 10.1073/pnas.101546898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci U S A. 2007;104:5091–5096. doi: 10.1073/pnas.0610819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VW, Yusuf RZ, Oki T, Wu J, Saez B, Wang X, Cook C, Baryawno N, Ziller MJ, Lee E, Gu H, Meissner A, Lin CP, Kharchenko PV, Scadden DT. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell. 2016;167:1310–1322. e1317. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.