SUMMARY
Using integrative genomic analysis of 360 metastatic castration-resistant prostate cancer (mCRPC) samples, we identified a novel subtype of prostate cancer typified by biallelic loss of CDK12 that is mutually exclusive with tumors driven by DNA repair deficiency, ETS fusions, and SPOP mutations. CDK12 loss is enriched in mCRPC relative to clinically-localized disease and characterized by focal tandem duplications (FTDs) that lead to increased gene fusions and marked differential gene expression. FTDs associated with CDK12 loss result in highly recurrent gains at loci of genes involved in the cell cycle and DNA replication. CDK12-mutant cases are baseline diploid and do not exhibit DNA mutational signatures linked to defects in homologous recombination. CDK12-mutant cases are associated with elevated neoantigen burden ensuing from fusion-induced chimeric open reading frames and increased tumor T cell infiltration/clonal expansion. CDK12 inactivation thereby defines a distinct class of mCRPC that may benefit from immune checkpoint immunotherapy.
INTRODUCTION
Comprehensive genomic analyses have substantially furthered our understanding of primary prostate cancer (PCa) and metastatic castration-resistant prostate cancer (mCRPC) (Barbieri et al., 2012; Beltran et al., 2016; Fraser et al., 2017; Grasso et al., 2012; Robinson et al., 2015; The Cancer Genome Atlas Research Network, 2015). These studies have discovered common genetic drivers of prostate cancer, such as fusions of ETS genes (Tomlins et al., 2005), amplification of AR, and loss of CDKN2A, PTEN, RB1, SPOP, and TP53 (Robinson et al., 2015). Integrative genomic studies have further delineated distinct molecular subtypes in primary and metastatic prostate cancer and specific molecular pathways that contribute to prostate cancer onset and progression, including AR, WNT, and PI3K/AKT/MTOR signaling (Barbieri et al., 2012; Beltran et al., 2016; Robinson et al., 2017; The Cancer Genome Atlas Research Network, 2015).
This knowledge is being actively translated into promising drug targets. Recently, recurrent germline and somatic mutations in genes involved in DNA repair provided a rationale for the use of poly ADP ribose polymerase (PARP) and immune checkpoint inhibitors in homologous recombination-deficient (HRD) and mismatch repair-deficient (MMRD) metastatic prostate cancer, respectively (Le et al., 2015; Mateo et al., 2015; Robinson et al., 2015). Intriguingly, in both cases, the genomic instability engendered by the deficiency becomes a “double-edged sword”. On one hand, it is the mechanism by which the tumor generates secondary oncogenic drivers, while on the other, it makes the tumor susceptible to a specific therapy. For example, cancer cells with MMRD have a high mutation burden that generates tumor neoantigens, thereby making the patients favorable candidates for intervention with immunotherapies (Le et al., 2015).
CDK12 is a cyclin-dependent kinase that associates with its activating partner, cyclin K, to form a heterodimeric complex that regulates several critical cellular processes (Blazek et al., 2011; Cheng et al., 2012). CDK12 consists of different functional domains: a centrally-located kinase domain, several RS (arginine/serine) motifs near the N-terminus, and a proline-rich motif (PRM) which can function as a binding site for additional proteins (Ko et al., 2001). CDK12 directly regulates transcription by phosphorylating serine residues of the hepta-peptide repeats (YSPTSPS) within the C-terminal domain of RNA polymerase II essential for transcriptional elongation (Bartkowiak et al., 2010; Blazek et al., 2011; Cheng et al., 2012). Multiple studies have also suggested a role for CDK12 in controlling genomic stability through regulation of genes involved in the DNA damage response (ATR, BRCA1, FANCD2, FANCI, etc.) (Blazek et al., 2011; Juan et al., 2016). Depletion or loss of function of CDK12 have further been observed to sensitize ovarian cancer cells to PARP inhibitors through defects in HR (Bajrami et al., 2014; Ekumi et al., 2015; Joshi et al., 2014).
In previous studies, we found recurrent CDK12 mutations in metastatic prostate cancer (Robinson et al., 2015), while similar observations were later made in serous ovarian tumors (Popova et al., 2016). Herein, we delineate a novel genetically unstable subtype of mCRPC associated with biallelic inactivation of CDK12. We show that CDK12-mutants are genetically, transcriptionally, and phenotypically distinct from HRD and MMRD tumors. Further, we identify that CDK12-mutant tumors have synthetic genetic dependencies and a characteristic immunophenotype, which provide candidate targets for precision therapy.
RESULTS
CDK12 mutations are enriched in cases of mCRPC
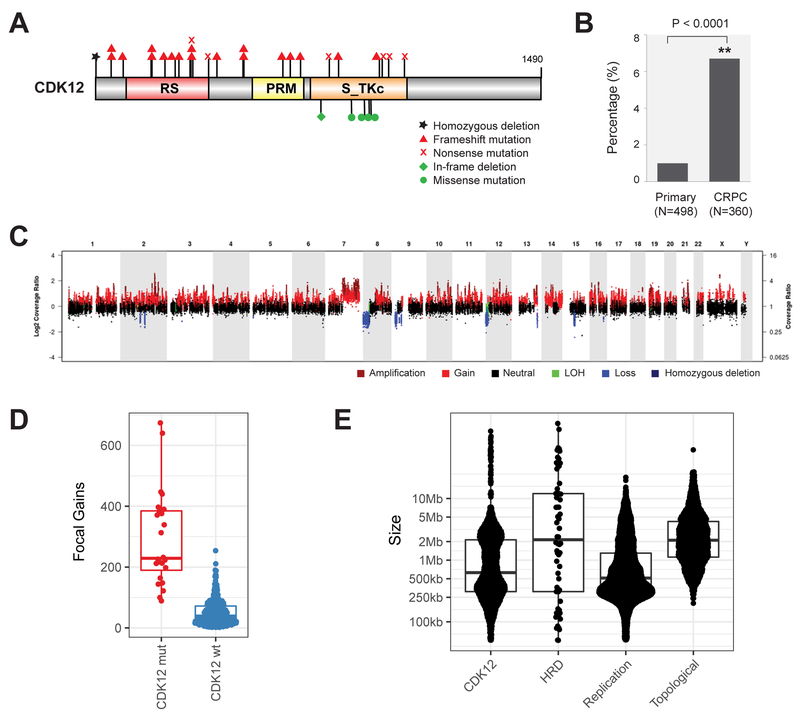
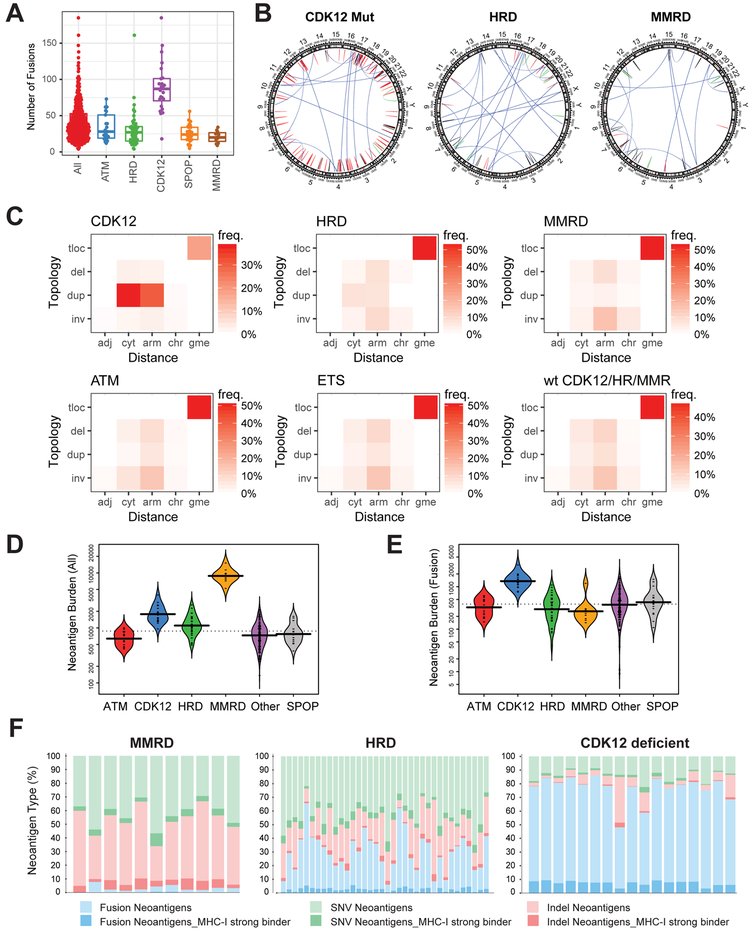
We previously reported that 4.7% of mCRPC patients harbored biallelic aberrations of CDK12. To confirm this observation, we have compiled an extended multi-site metastatic prostate cancer cohort of 360 patients (CRPC360), comprising SU2C (Robinson et al., 2015), MI-OncoSeq (Robinson et al., 2017), and UMich rapid autopsy cases (Grasso et al., 2012) (Table S1), a majority of which have matched whole-exome and transcriptome data (Table S2). The combined data sets were reanalyzed using the MI-Oncoseq workflow (Robinson et al., 2017), producing harmonized call sets of somatic, germline, and structural variants. We also analyzed, using the MI-Oncoseq workflow, sequence data from 498 cases of primary prostate cancer in the TCGA (The Cancer Genome Atlas) dataset. We detected aberrations of CDK12 in 25/360 of mCRPC patients (6.9%), 95% CI [4.6%,10.2%] (Figure 1A). This is significantly higher than in primary PCa, 6/498 patients (1.2%) (Figure 1B and Table S3) (p<0.0001 Fisher exact test). Examination of data across additional primary and metastatic prostate cancer datasets revealed a similar difference in the frequency of biallelic CDK12 mutations between primary and metastatic cancer (Table S4) (Abida et al., 2017; Beltran et al., 2016; Fraser et al., 2017; Kumar et al., 2016). CRPC genomes are more highly mutated than those of localized tumors; however, the magnitude of the increased mutation rate is not sufficient to explain the increased frequency of biallelic loss of CDK12. The majority of CDK12 mutations (83%) were truncating and resulted in the loss of the kinase domain. Missense mutations were clustered around conserved residues in the kinase domain (Figure S1). All patients showed biallelic inactivation of CDK12. CDK12 has been shown to have a very low tolerability for germline loss-of-function variants (Juan et al., 2016), and, consistently, no germline aberrations were detected in our cohort (Table S5).
Figure 1. Biallelic loss of CDK12 is enriched in mCRPC and results in focal tandem duplications.
(A) Schematic of mutations in CDK12.
(B) Increased frequency of CDK12 loss in metastatic castration-resistant prostate cancer (CRPC) compared to primary disease.
(C) Characteristic pattern of genomic instability found in all cases with CDK12 loss. Copy gains are indicated in shades of red. LOH, loss of heterozygosity.
(D) Number of focal copy gains (< 8Mb) by CDK12 mutational status, as determined by whole-exome analysis.
(E) Size of copy gains (tandem duplications), as ascertained by whole-genome sequencing of index cases with CDK12 mutations (CDK12) and homologous recombination deficiency (HRD). Sizes of replication domains and topological domains in normal tissues are shown for comparison.
See also Figures S1-S3 and Tables S1-S5.
CDK12-mutant tumors are baseline diploid with an excess of focal tandem duplications
A significant increase in genomic instability is a hallmark of metastatic tumors (Negrini et al., 2010). While primary prostate cancers are largely diploid, metastatic tumors often show extensive LOH, aneuploidies, and a significant increase in mutational burden (Robinson et al., 2017). We examined the landscape of CDK12-mutated mCRPC cases and observed a distinctive genomic landscape (Figures 1C and S2), similar to that identified in a subset of ovarian cancers (Popova et al., 2016). The prototypical CDK12-mutant tumor was baseline diploid and had few arm-level copy-number aberrations except gain of 8q, but notably, hundreds of focal copy-number gains were dispersed across the genome. While focal gains were present on all chromosomes within a sample, other focal events, such as high-level amplifications or deletions, were rare or absent. CDK12 biallelic inactivation was strongly associated with this form of genomic instability (p < .00001, Fisher exact test). All cases with CDK12 inactivation, and only cases with CDK12 mutation, exhibited this form of genome instability in both the metastatic and primary cohorts (Figure S2 and Table S3). No other genes were positively associated with this genome instability. ETS fusions and PTEN mutations were depleted in cases with CDK12 mutations (p<.00001 for both, Fisher exact test). None of the CDK12-mutated tumors exhibited a neuroendocrine phenotype.
The genomic phenotype of CDK12-mutant tumors was compared to other cases in the CRPC360 cohort, particularly those associated with frequent primary genetic drivers (PGDs) of prostate cancer: ATM mutations, HRD, SPOP mutations, and MMRD. Like CDK12-mutant cases, SPOP- and MMRD-driven tumors were mostly diploid, while a large subset of ATM- and HRD-driven tumors showed large-scale aneuploidy (Figure S3A). The high number of focal gains was consistently observed in CDK12-mutant cases compared to those in the cohort with wild-type CDK12 (Figure 1D). Detection of genomic structural variants (SV) from whole-genome sequencing (WGS) data confirmed that the gains were focal tandem duplications (FTDs) (Figure S3B) and enriched in gene-dense regions (Figure S3C). Strikingly, comparison of CDK12-mutant and HRD index cases revealed a bimodal distribution of FTD sizes in CDK12-mutant, but not HRD, tumors (Figure S3D). The modes of this distribution were consistent with the sizes of replication domains (RD), but not topological domains (TD) (Figure 1E). Specifically, the ~2.4Mb peak was close to the mode of the early/late RDs, while the ~0.4Mb peak matched the size of transitional RDs (Hiratani et al., 2008) (Figure S3E). Breakpoint sequence assembly revealed that FTDs were enclosed by error-prone junctions indicative of a non-homologous end joining (NHEJ)-mediated repair process (Figure S3F). We refer to these events as CDK12-associated FTDs (CDK12-FTDs) to distinguish them from BRCA-dependent events and focal amplifications.
CDK12-mutants represent a specific class of prostate cancer with a distinct transcriptional phenotype
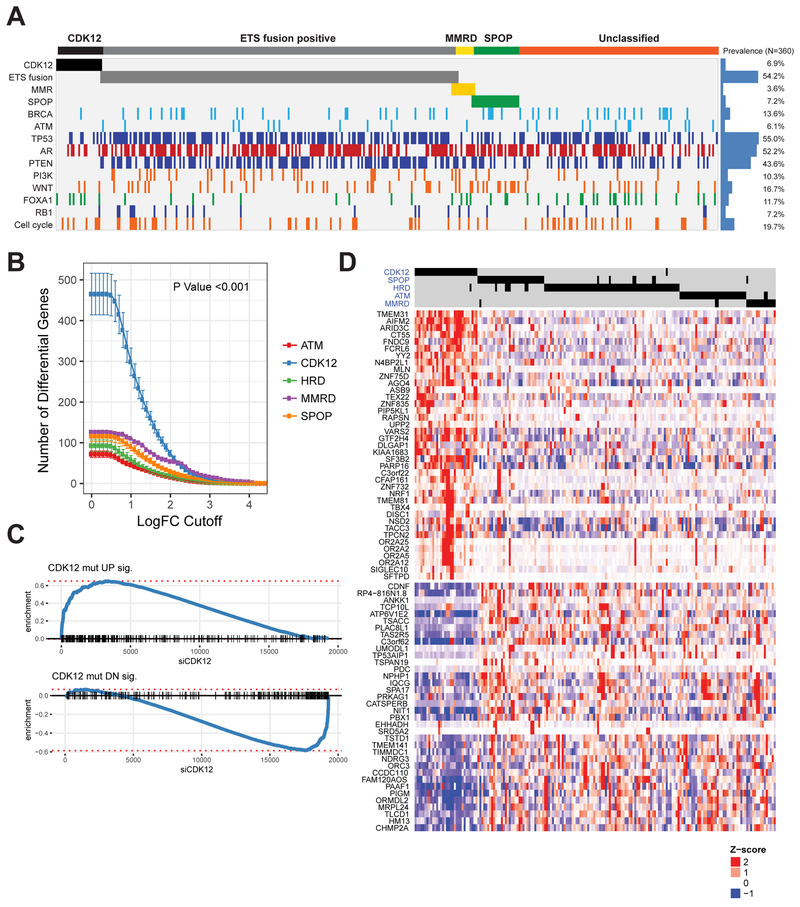
We next tested for genetic associations between CDK12 loss and the most frequent PGDs of prostate cancer to determine whether CDK12-mutant cases were a unique class of mCRPC. Strikingly, CDK12 aberrations were mutually exclusive with all of the PGDs analyzed (ETS fusions, SPOP mutations, HRD, ATM mutations, and MMRD) (Figure 2A).
Figure 2. CDK12-mutant prostate cancer is a novel molecular subtype of mCRPC.
(A) Mutual exclusivity of CDK12 loss, ETS fusions, mismatch repair deficiency (MMRD), SPOP mutations, and homologous recombination deficiency (HRD).
(B) Number of significantly differentially expressed genes (DEGs) for the prostate tumors with different primary genetic drivers.
(C) Enrichment plot for signatures of up- (top) and downregulated (bottom) genes in CDK12 mutant tumors. Genes are ranked by their fold change following siCDK12 knockdown in LNCaP cells, with CDK12-loss signature genes indicated as black dashes. The increased relative frequency (enrichment score) of genes at either end of this spectrum is shown as a blue line.
(D) Heatmap of the top DEGs in CDK12-mutant prostate cancer. Differential expression for all samples (columns) in this heatmap is relative to tumors that are wild-type for primary genetic drivers of prostate cancer (as in B).
Several of the established prostate cancer PGDs have been associated with characteristic gene expression profiles (Herschkowitz et al., 2008; Parikh et al., 2014; Saal et al., 2007). We hypothesized that CDK12 loss may similarly constrain a specific transcriptional phenotype. To test this, we compared the expression profiles of mCRPC cases with aberrations in specific PGDs or CDK12 to a reference set of cases (n=92) that were wild-type for all the PGDs, including CDK12 (PGD-WT). Interestingly, we found that CDK12 aberrations were associated with the highest number of differentially expressed genes (DEGs) (Figure S4A), independent of differences in the number of cases for each PGD, and across a wide range of effect-size (Figure 2B), and p-value cutoffs. The most up- (e.g. AIFM2, ARID3C, TBX4) or downregulated (e.g. TSACC, CDNF, ABCC12) genes have not been previously studied in the context of prostate cancer (Figure S4B). To establish a causal link between this transcriptional phenotype and loss of CDK12, we performed a siRNA-mediated knockdown experiment in LNCaP cells. Depletion of CDK12 at the RNA and protein levels resulted in growth arrest (Figures S4C-E) and profound transcriptional changes. In addition, DEGs associated with CDK12 mutations in patients were almost perfectly recapitulated in vitro (Figure 2C), which allowed us to define a transcriptional signature of CDK12-loss in mCRPC (Table S6).
While most CDK12-mutants retained active androgen receptor (AR) signaling (Figure S4F) (Beltran et al., 2016), their expression signature was distinct from the equivalent signatures for the other PGDs (Figures 2D and S4G). Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) across the MSigDB (Liberzon et al., 2015) revealed significantly perturbed curated gene sets (Figure S4H). The most prominently altered were those related to oxidative phosphorylation (down), inflammatory response (up), hormone receptor signaling (down), and epithelial dedifferentiation (down). To understand this further, we delineated a core set of 28 genes downregulated in both metaplastic and stem-like breast cancer (i.e. two of the most significant gene sets). Strikingly, the majority of those genes were significantly downregulated in CDK12-mutant mCRPC (Figure S4I). Although the shift from oxidative to glycolytic metabolism (Warburg effect) is a hallmark of many cancer types (Vander Heiden et al., 2009), it is not a characteristic of most prostate cancers (Cutruzzola et al., 2017).
CDK12-mutant tumors display characteristic copy-number and mutational signatures distinct from DNA repair-deficient prostate cancer
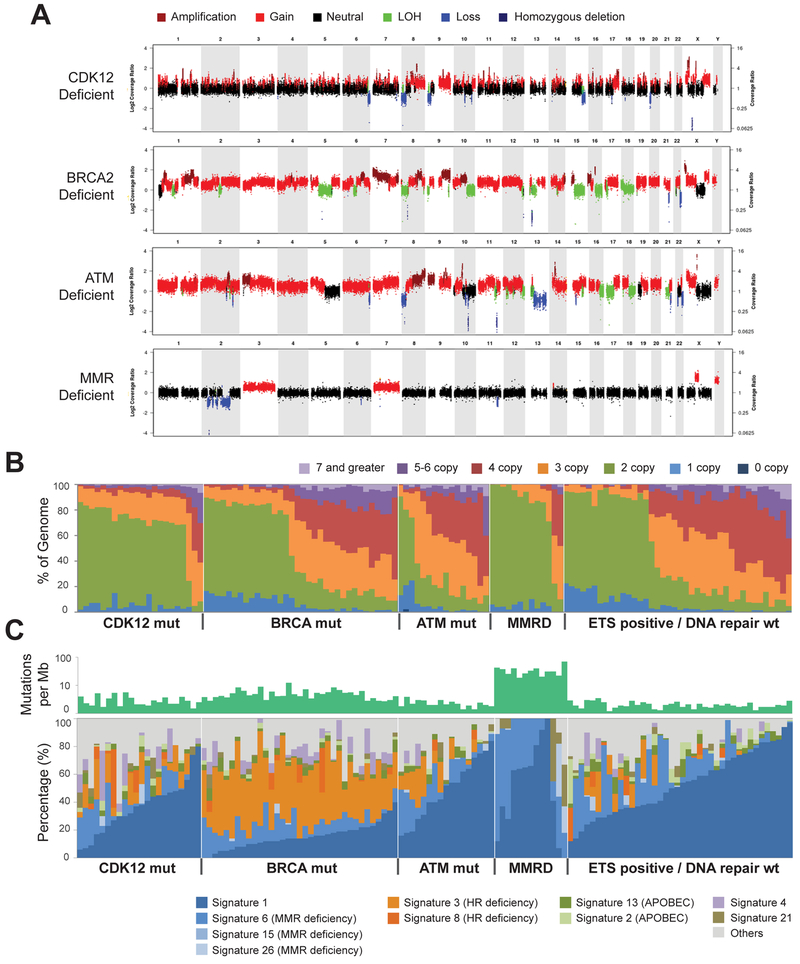
Previous studies suggested that CDK12 is involved in controlling genomic stability through regulation of HR or other DNA damage response effectors (Blazek et al., 2011; Ekumi et al., 2015; Joshi et al., 2014; Juan et al., 2016). Our CRPC360 transcriptional data also showed a unique signature for CDK12-mutant tumors (Figure 2D). Large-scale copy-number gains were evident in the BRCA2- and ATM-deficient cases, as compared to CDK12-mutant or MMRD cases (Figure 3A). To quantitate and contrast the CDK12-mutant pattern with the other PGDs on a larger scale, we tallied absolute copy-number levels from whole-exome sequencing (WES) data across the entire CRPC360 cohort (Figure 3B). These analyses showed that BRCA and ATM mutated, as well as ETS fusion-positive, tumors had the highest percentage of copy-number gains, while the majority of CDK12-mutant and MMRD tumors did not exhibit changes in ploidy (Figure 3B).
Figure 3. CDK12 loss results in a distinct pattern of genomic instability.
(A) Representative copy-number plots for prostate tumors with deficiencies in key DNA damage response or repair pathways.
(B) Spectrum of copy-number aberrations in tumors with distinct genetic drivers.
(C) Spectrum of inferred mutational signatures in tumors with distinct genetic drivers.
See also Figure S4.
Genomic signatures are a powerful approach to study the mutagenic imprints of environmental and genetic factors. To determine if loss of CDK12 activity is associated with a distinct signature, we computed mutational burden as well as mutational signature across various genetic drivers (Figure 3C). As expected, MMRD cases had the highest mutational burden and a signature consistent with microsatellite instability (signature 6) (Alexandrov et al., 2013). HRD tumors had the next highest mutational burden, and BRCA-loss was associated with an evident signature 3 (Polak et al., 2017). The remaining PGDs, including CDK12, had a baseline level of SNVs and were dominated by age-related 5-methylcytosine deamination (signature 1). Combined, these data support that the CDK12-mutant subtype is distinct from either the HRD or MMRD type of prostate cancer. In particular, CDK12-mutants are different from tumors with HRD, which was previously presumed to be the pathway through which CDK12 regulated genomic stability. Notably, the expression of BRCA1 or BRCA2 was not affected by CDK12 mutational status (Figure S4J) and neither was the expression of other genes encoding long transcripts and cognate proteins (Figure S4K), a class previously suggested to be regulated by CDK12 (Blazek et al., 2011).
CDK12-FTDs result in highly recurrent gains of genes involved in the cell cycle and DNA replication
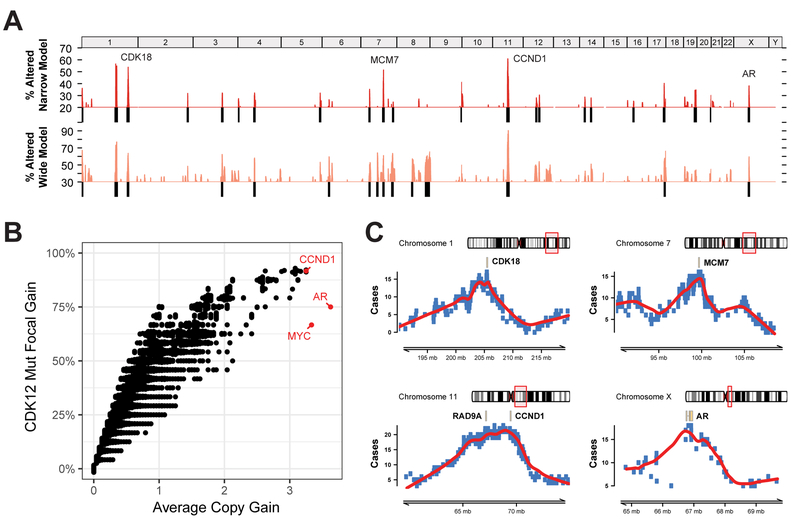
The large number of FTDs present in all CDK12-mutant tumors introduces the possibility of detecting synthetic genetic dependencies or epistasis. One approach is to look for loci with recurrent CDK12-FTDs at the cohort level. To identify such genomic regions, we developed a Monte Carlo null model to simulate the expected distribution of FTD recurrences, given their number and size. We applied both stringent (2 Mb, “narrow”) and relaxed (8 Mb, “wide”) definitions (Figures S5A-B). Using both models, we detected a total of 27 loci with recurrent focal gains at false-discovery rates of 3.5% and 5%, respectively (Figure 4A and S5C). Indicative of strong positive selection, several of these loci showed copy-number gains in almost all CDK12-mutant cases (Figure S5C). Strikingly, their recurrence was significantly lower in CDK12 wild-type tumors, which suggests a synthetic dependency (Figure S5D). As a prominent exception, the MYC and AR loci (Figure S5E) were recurrently amplified, regardless of CDK12 status, which underscores their fundamental role in prostate cancer. Although most of the CDK12-FTDs result in the gain of one additional copy (Figure 3B), we observed that the most recurrent genes also had the highest copy-number gains (MYC, AR, CCND1), suggestive of gene dosage selective pressure (Figure 4B).
Figure 4. Recurrence of focal tandem duplications (FTDs) associated with CDK12 loss.
(A) Genome-wide frequency (percentage of CDK12-mutant patients) of FTDs based on a narrow (<2Mb) and wide (<8Mb) definition of focality.
(B) FTD recurrence and average copy-number gain of FTDs at the individual gene level. Genes with the highest average copy-number are highlighted in red.
(C) Delineation of minimal common regions (MCR) for loci with the most recurrent gains specific to CDK12-loss tumors. Genes related to the cell cycle are highlighted in each MCR. The AR locus is presented as a positive control.
See also Figures S5-S6.
The delineation of minimal common regions (MCR) is an established strategy to identify genetic targets that are subject to positive selection and, hence, responsible for the recurrent copy-number aberrations (Mermel et al., 2011). In order to nominate such candidate genes in CDK12-mutant mCRPC, we summarized FTDs into MCRs at the most recurrent loci (Figure 4C). The AR locus, whose MCR was centered on the AR gene as expected, represents a positive control for this approach. Of the recurrent loci, two were consolidated into a narrow MCR harboring a single candidate gene (MCM7 and CDK18), while one required further prioritization. The chr11_q13.2 locus is characterized by high gene density and the presence of RAD9A and CCND1, all of which could contribute to the FTD recurrence of this region. CCND1 was also associated with the highest copy-number gains, comparable in magnitude with amplifications at the MYC and AR loci (Figure 4B). Strikingly, candidate genes under positive selection, MCM7, RAD9A, CDK18, and CCND1, have crucial roles in DNA replication and genome stability. Amplifications of MYC and AR are among the most recurrent genetic events in mCRPC and not specific to CDK12-mutants. Correspondingly, their molecular functions are pleiotropic; both regulate the cell cycle (Bretones et al., 2015; Yuan et al., 2006), and contribute independently to proliferation of prostate cancer cells (Bernard et al., 2003).
CDK12-FTDs induce expression in a dosage dependent and independent manner
In order to better understand some of the functional consequences of CDK12-FTDs, we interrogated both global and gene-specific associations between copy-number and expression levels. To assess global effects of CDK12-FTDs, we probed changes in average expression levels associated with the focal increases in copy-number (Figures S5F-G). We observed a significant increase in the number of DEGs at each absolute copy-number level (Figure S5F). To demonstrate the feasibility of identifying gene-specific effects given our sample size, we interrogated the expression of three genes associated with the highest average copy-number gains and high recurrence: CCND1, MYC, and AR (Figure 4B). A significant dose dependent relationship for CCND1 and AR, but not MYC, was observed (Figure S6A). We expanded this analysis to other cancer-related genes and identified similar trends for key oncogenes in the MAPK, AKT, and MTOR pathways (Figure S6B). Strikingly, dosage dependence was much less robust for receptor tyrosine kinases (RTK), which were dominated by singleton expression outliers (Figure S6C). A global analysis was performed to determine the contribution of CDK12-FTDs to the prevalence of expression outliers. Overall, outliers were more frequent in CDK12-FTDs, and their frequency increased with copy-number gains (0.5% to 4%) (Figure S5G).
Mutant CDK12 prostate cancers exhibit a unique structural signature characterized by increased gene fusions
Transcriptome sequencing data were used to delineate signatures of structural genomic instability across the different classes of PGDs. Interestingly, as shown in Figure 5A, CDK12-mutant tumors had the highest fusion burden, consistent with the large number of focal copy-number events (Figure 1D) and their enrichment in gene-rich regions (Figure S3C). The prototypical CDK12-mutant case exhibited a large number of fusions (Figure 5A) generated by tandem duplications and relatively fewer by translocations, inversions, or deletions (Figure 5B). This contrasts with HRD and MMRD tumors, which have a significantly lower fusion burden dominated by translocations. Next, we devised “fusion-grams” to quantitatively compare signatures of structural variants between the varying prostate cancer classifications (Figure 5C). In a fusion-gram, structural variants are classified according to the observed distance and topology of their breakpoints (i.e. deletion, duplication, inversion, translocation). For CDK12-mutant tumors, the majority of fusions (70%) were classified as duplications within a cytoband or chromosome arm. All other PGDs had signatures dominated by translocations (~49%) and fewer overall duplications (11%) than deletions (18%) or inversions (22%), further supporting the uniqueness of CDK12-mutant PCa.
Figure 5. Signatures of structural variation and neoantigen presentation in CDK12-mutant tumors.
(A) Total number of detected gene fusions for prostate tumors with different genetic drivers.
(B) Representative examples of circos plots showing the pattern structural variation in tumors with major types of genomic instability. Structural variants (SVs) detected from RNA-seq are classified into translocations, deletions, duplications, and inversions based on the topology of the breakpoints. Color code: blue - translocations, red - duplication, green - inversion, black - deletion.
(C) Classification of SVs based on the topology and distance between the breakpoints. adj – breakpoints in adjacent loci, cyt – in same cytoband, arm – on same chromosome arm, gme – genomic translocation, inv – inversion, dup – duplication, del – deletion, tloc – translocation. Heatmap color indicates frequency of a SV class across all index cases. (numbers of patients: CDK12 = 24, HRD = 47, MMRD = 11, ATM = 21, ETS = 190, WT = 31).
(D-E) Antigen burden in tumors with distinct types of genetic instability. Overall burden based on single nucleotide variants, insertions/deletions, and fusions is shown in D. Fusion-specific burden is shown in E.
(F) Distribution of neoantigens based on genetic variant type and predicted MHC class-I (MHC-I) binding affinity.
See also Figure S6.
Since CDK12-FTDs are associated with expression outliers (Figure S5G) and a large number of gene fusions (Figure 5A), we hypothesized that some of those events are potential secondary genetic cancer drivers. We searched for candidate driver events where a chromosomal aberration resulted in either outlier expression of an oncogene or formation of a likely oncogenic gene fusion. In addition to the singleton outlier RTKs (Figure S6C), we found two cases of BRAF fusions (KIAA1549-BRAF and HIPK2-BRAF) generated as a result of a CDK12-FTD (Figures S6D-E). While we, and others, have previously reported BRAF fusions in prostate cancer (Palanisamy et al., 2010), duplications involving the KIAA1549-HIPK2-BRAF locus have thus far been noted as hallmarks of pilocytic astrocytoma (Yu et al., 2009). Surprisingly, we also found a promoter hijacking event leading to outlier expression of ETV1 (Figure S6F). However, not all secondary events could be inferred as direct consequences of CDK12-FTDs. For example, we found a translocation leading to extremely high expression of full-length FGFR2 (Figure S6G). Importantly, FGFR fusions can be found in many solid tumors and are compelling targets for precision therapy (Wu et al., 2013).
CDK12-mutant tumors are characterized by increased gene fusion-induced neoantigen open reading frames
Tumor immunogenicity is associated with mutational burden and neoantigen load (Le et al., 2015). We reasoned that gene fusions and their chimeric protein products yield significant numbers of neoantigens in CDK12-mutant tumors. We carried out comprehensive prediction of novel peptides from mutation and fusion calls (STAR Methods section) and found that MMRD, HRD, and CDK12-mutant tumors had a significantly higher neoantigen burden compared to other mCRPC molecular subtypes (Figure 5D). Strikingly, the mutational mechanism by which the neoantigens were generated was specific to each subtype. While neoantigens in MMRD and HRD tumors were formed by indels and SNVs, fusions contributed most of the novel epitopes in CDK12-mutant mCRPC (Figures 5E-F). The calculated neoantigen burden from fusions was the highest in CDK12-mutant tumors among the other PGDs (Figure 5E). Importantly, these analyses also identified neoantigens with strong MHC class-I binding affinities that are predicted to be candidate epitopes for immunotherapy (Figure 5F).
CDK12-mutant tumors show high immune infiltration and imprints of immune evasion
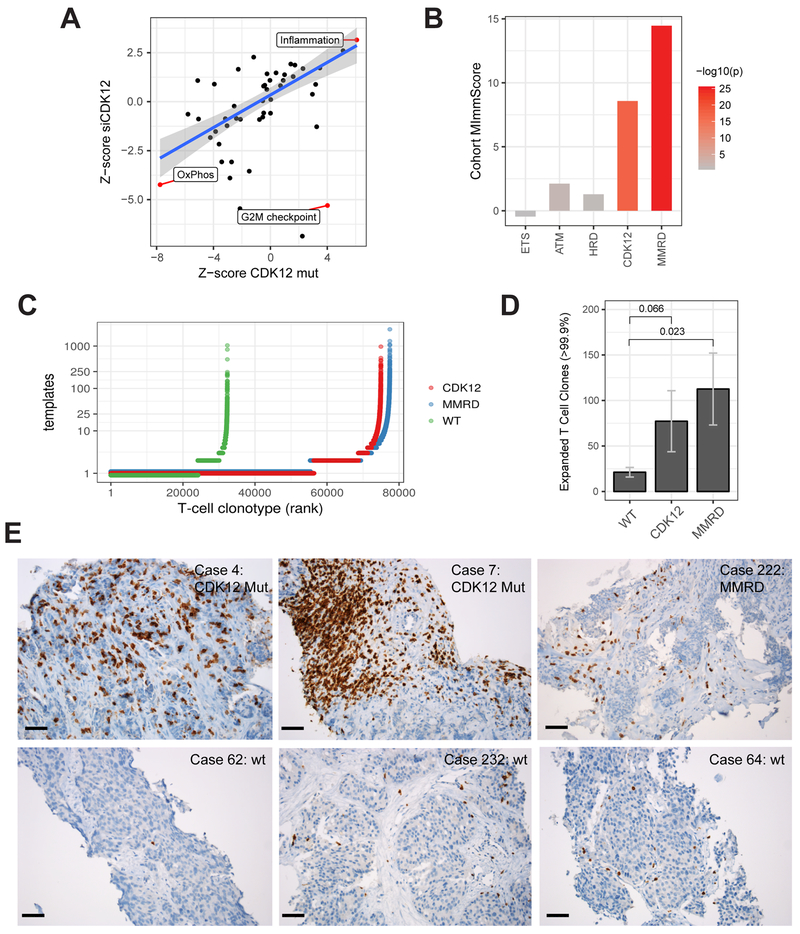
We found significant activation of the cancer inflammatory hallmark gene-set in CDK12-mutant tumors and LNCaP cells transfected with siRNA to CDK12 (Figure 6A). Compared to wild-type tumors (PGD-WT, see above), CDK12-mutant cases showed increased expression of chemokines and their receptors (Figure S7A). Overall, we observed reduced or low expression of chemokines that can recruit regulatory T cells (CCL17, CCL20, CCL22) (Curiel et al., 2004; Zou, 2006) and an increase in chemokines that support dendritic cell migration into the tumor microenvironment (CCL21, CCL25). Interestingly, certain direct pro-tumor chemokines, including CCL18 and CXCL8 (Nagarsheth et al., 2017), were enriched in patients with CDK12 mutations. To determine whether this immune phenotype was specific to CDK12-mutant tumors, we contrasted the activation of the top signatures across genetically unstable mCRPC subtypes. Strikingly, only MMRD and CDK12-mutant tumors showed robust activation of chemokine signaling/inflammatory response and high immune infiltration as estimated by the cohort MImmScore (Robinson et al., 2017) (Figures 6B and S7B). Taken together, these data indicate that CDK12-mutant tumors are immunogenic and infiltrated by leukocytes but evolve chemokine-mediated mechanisms of immune evasion.
Figure 6. Immunogenomic properties of CDK12-mutant tumors.
(A) Differential expression of MSigDB cancer hallmark gene-sets in CDK12-mutant patients and in LNCaP cells depleted with CDK12 by siRNA. Highlighted hallmarks are significant (FDR < 0.05, limma moderated t-test).
(B) Levels of global immune infiltration across prostate tumors with distinct genetic drivers compared to genetically stable (PGD wild-type) tumors. The “Cohort MImmScore” is defined as the gene-set enrichment Z-score and p-value based on Random-Set test and moderated cohort DE log2 fold-changes.
(C) Overview of T cell clonotypes across CDK12-mutant (n=10), MMRD (n=10), and WT (n=10) tumors. T cell clonotypes (i.e. identical CDR3 sequences) are ranked by their frequency (number of templates). CDK12-mutant and MMRD tumors show, overall, an increase in the total number of T cells (X-axis), and higher levels of clonal expansion (Y-axis).
(D) Comparison of clonal expansion between immunogenic (MMRD, CDK12) and wild-type mCRPC tumors (t-test). Expanded clones are defined as those with the highest number of clonal expansion (estimated number of templates > 99.9 percentile across all cohorts; n > 12).
(E) Immunohistochemistry (IHC) performed on formalin-fixed paraffin-embedded tumor sections using anti-CD3 antibody. Six representative cases are shown, including two CDK12-mutant tumors, one MMRD tumor, and three tumors which are wild type for CDK12, MMR genes, and HR genes. Black bar: 50 μm.
See also Figure S7.
Antigen recognition by T cells leads to their clonal expansion. To detect whether increased neoantigen burden was mirrored by an increase in T cell clonality (McGranahan et al., 2016), we performed T cell repertoire analysis using TCRb sequencing on a set of 60 tumors across all molecular subtypes (n=10 per group). We found that, compared to genomically stable tumors, CDK12-mutant tumors showed higher overall levels of T cell infiltration (Figure 6C) and larger numbers of expanded T cell clones (Figure 6D), regardless of the template cutoff used (Figure S7C). To confirm these trends, we performed T cell repertoire profiling of RNA-seq data (Bolotin et al., 2015). First, we established that RNA and DNA-based estimates of T cell infiltration were in agreement (Figure S7D). We found that relative to wild-type cases (PGD-WT), MMRD, HRD, and CDK12-mutant tumors all had a significant increase in both the number of detected T cell clones (Figure S7E) and the total number of CDR3 sequences (Figure S7F). Importantly, immunohistochemical (IHC) staining of CD3 on representative index cases further confirmed the presence of tumor-infiltrating T cells in a subset of CDK12-mutant tumors (Figure 6E).
Pilot clinical study to determine CDK12-mutant prostate cancer response to checkpoint inhibitor immunotherapy
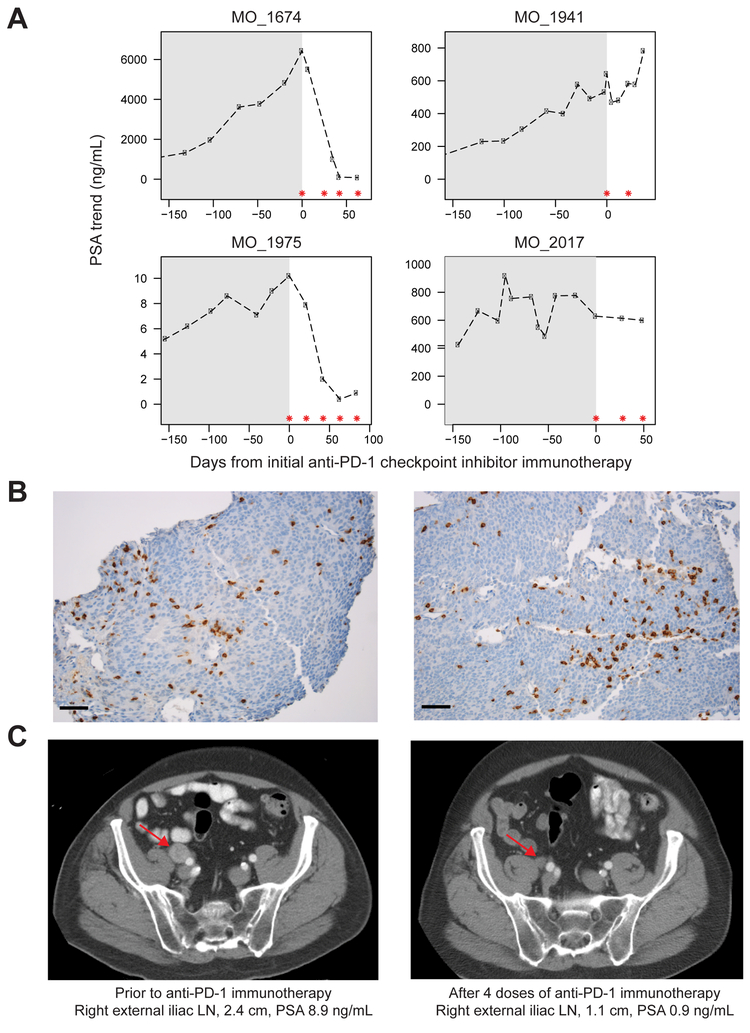
Of eleven CDK12 mutant patients identified in the MI-Oncoseq program, a total of five late stage, pre-treated mCRPC patients had some exposure to immunotherapy in the form of the immune checkpoint inhibitor anti-PD1. One patient received one dose of anti-PD1 as part of combination therapy on a clinical trial and was excluded, as he did not receive anti-PD1 monotherapy and could, therefore, not be compared to the other treated patients. Detailed prostate-specific antigen (PSA) response data are presented on the four patients treated with anti-PD1 monotherapy for whom we also have associated clinical data and detailed sequencing information (Figure 7A). Strikingly, two of the four patients had an exceptional response in terms of PSA decline. This was surprising as checkpoint inhibitor immunotherapy has typically not been efficacious in prostate cancer, with the exception of patients with mismatch repair defects (Le et al., 2015).
Figure 7. Response of CDK12-mutant patients to anti-PD1 checkpoint inhibitor immunotherapy.
(A) PSA levels of four CDK12-mutant prostate cancer patients treated with anti-PD1 monotherapy. Gray shading represents PSA levels prior to anti-PD-1 therapy. Asterisks indicate anti-PD1 doses of 200 mg IV.
(B) Representative CD3 IHC images of metastatic lymph node biopsies of patient MO_1975 prior to anti-PD1 treatment. Cells exhibited membranous and cytoplasmic staining of CD3, highlighting the presence of T lymphocytes. Black bar: 50 μm.
(C) CT imaging of patient MO_1975 pre- and post-immunotherapy treatment. Arrows indicate metastatic lymph node.
One patient (MO_1674) was treated with anti-PD1 checkpoint inhibitor immunotherapy and displayed a marked PSA response after four doses of therapy, but eventually succumbed to multisystem organ failure, possibly due to anti-PD1 induced pneumonitis (Nishino et al., 2015). Patient MO_1941 received only two doses of anti-PD1 with a subsequently rising PSA and is deceased. Two patients are still alive on active therapy (MO_2017, MO_1975). Patient MO_2017 had heavily pre-treated disease, with prior disease progression on abiraterone, enzalutamide, docetaxel, and cabazitaxel. Pre-treatment PSA prior to initiation of immunotherapy was 628.8 ng/mL with a modest improvement in PSA after three doses of anti-PD-1, and subsequent PSA decline to 599.2 ng/mL. Patient MO_1975 had a Gleason 9 metastatic prostatic adenocarcinoma and prior lymph node progression on abiraterone and enzalutamide. Evaluation of a metastatic lymph node biopsy demonstrated robust CD3 staining by IHC (Figure 7B). To date, the patient has received five doses of anti-PD1 with a significant PSA decrement (Figure 7A), as well as marked decline in pelvic lymph node disease burden (Figure 7C). These early clinical results support the hypothesis that metastatic prostate cancer patients who harbor biallelic CDK12 loss may have a higher likelihood of response to immunotherapy than an unselected metastatic prostate cancer population. Further study in the context of a clinical trial is warranted.
DISCUSSION
In this report, we comprehensively characterized biallelic loss of CDK12 as a novel PGD of prostate cancer. Importantly, through an integrative genomic approach, we demonstrate that CDK12 mutations are mutually exclusive with other PGDs, such as SPOP mutations and ETS fusions. CDK12-mutant tumors present unique characteristics at the genetic, transcriptomic, and immunophenotypic levels, and have the potential to be therapeutically targeted.
At the genetic level, CDK12-mutant tumors show a characteristic pattern of genomic instability. Previous findings, primarily from cell-based assays, suggested that CDK12 impacts genome stability through defects in HR (Bajrami et al., 2014; Blazek et al., 2011; Ekumi et al., 2015; Joshi et al., 2014; Juan et al., 2016). However, our data, and the observations made previously in CDK12-mutant ovarian tumors (Popova et al., 2016), are inconsistent with that model. In contrast to HRD mCRPC tumors, which are characterized by translocations and aneuploidies, CDK12-mutant tumors are diploid with a large number of focal tandem duplications (CDK12-FTD) and few translocations. CDK12-mutant cases also lack mutational signatures of HRD (Figure 3C) and maintain the expression levels of BRCA1 and BRCA2 (Figure S4J). Overall, the genomic phenotypes of HR and CDK12 deficiency are clearly distinct.
Several lines of evidence indicate that CDK12-FTDs are a result of aberrant DNA re-replication during S-phase: (i) CDK12-FTDs have a characteristic bimodal size distribution which matches the length of replication domains but not topologically-associated domains or HR defects; (ii) CDK12-mutant cases have a synthetic dependency on aberrations in genes involved in DNA replication: MCM7, RAD9A, CCND1, and CDK18; (iii) CDK12-FTDs result most frequently in the gain of one additional copy, consistent with the firing of an additional origin of replication; (iv) knockdown of CDK12 results in growth arrest (Figure S4E). It remains unknown whether FTDs are generated through a one-time catastrophic event, a slow ongoing mutational process, or rescue of the phenotype by one of the recurrent gains.
At the transcriptional level, CDK12-mutant tumors are associated with over 300 DEGs, which makes them the most prominent molecular subtype in our analysis (Figures 2B and S4A). Perhaps most importantly for translational purposes, CDK12 mutant cases exhibit a characteristic immunophenotype. CDK12-mutant tumors show high overall immune infiltration (Figure 6B), increased levels of tumor-infiltrating lymphocytes (Figures 6C-E, S7E), and altered chemokine signaling.
This immunological phenotype may be influenced by the elevated neoantigen burden in CDK12-mutant tumors. While single-nucleotide variants (SNVs) and indels are the main source of neoantigens in MMRD and HRD tumors, neoantigens in CDK12-mutant tumors are mostly from FTD-induced fusions. Although the detection of neoantigens from fusions is still at an early stage, the present study is, to our knowledge, the first to demonstrate the analytical value of neoantigen prediction from RNA-seq data. Fusions are analogous to indels in that they can generate neoantigens through in-frame and frameshift mechanisms. The latter, often referred to as neo-ORFs (Hacohen et al., 2013), are particularly interesting because they generate completely novel epitopes that are potentially highly immunogenic. In line with this possibility, high levels of CCL21 and CCL25 may mediate dendritic cell tumor trafficking and neoantigen-specific T cell clonal expansion (Chan et al., 1999; Gosling et al., 2000; Vicari et al., 1997).
A large number of studies have established the complex and important roles of immunity in the development and progression of prostate cancer (Strasner and Karin, 2015). These findings gave rise to a number of clinical trials for several classes of immunotherapeutics, which have been met with mixed results. For example, a phase 3 trial comparing ipilimumab (anti-CTLA4 immune checkpoint inhibitor) with placebo failed in patients with mCRPC (Kwon et al., 2014). A plausible explanation is in the genetics of prostate cancer. Compared to other tumors, prostate cancer has a low mutation rate, few neoantigens, and, consequently, is less visible to the adaptive immune system. In spite of that, exceptional responses to anti-CTLA4 (Cabel et al., 2017) and anti-PD1 (Graff et al., 2016) treatment have been observed clinically. These findings clearly show that strategies are needed to identify those patients that will benefit from immunotherapy. Taken together, our data suggest that CDK12-mutant prostate cancer is intrinsically immunogenic (Sharma et al., 2017), and CDK12 mutations may identify a subset of patients where immunotherapy would be efficacious. Indeed, we observed an exceptional response (PSA decline) with anti-PD1 monotherapy in two out of four mCRPC patients in this study (Figure 7A). Furthermore, identification of CDK12 mutation-associated neoantigens may help in the design of personalized tumor vaccines. The immune phenotype of CDK12-mutated tumors may also broadly suggest a combinational strategy for prostate cancer treatment involving inhibition of CDK12 and immune checkpoint blockade.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arul M. Chinnaiyan (arul@med.umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
LNCaP (male, prostate carcinoma) and HeLaS3 (female, cervical adenocarcinoma) cell lines were obtained from the American Type Culture Collection. LNCaP cells were cultured in RPMI1640 medium, and HeLaS3 cells were cultured in Ham’s F-12K medium, both supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin (Invitrogen). Cell lines were maintained at 37°C in a 5% CO2 cell culture incubator. Cell lines were genotyped to confirm their identity at the University of Michigan Sequencing Core and tested routinely for Mycoplasma contamination.
Human subjects and patient inclusion
Sequencing of clinical samples was approved by the Institutional Review Board of the University of Michigan (Michigan Oncology Sequencing Protocol, MI-ONCOSEQ, IRB # HUM00046018, HUM00067928, HUM00056496). Patients with clinical evidence of metastatic castration-resistant prostate cancer (mCRPC) that could be feasibly accessed by image-guided biopsy were eligible for inclusion. Consecutive cases from SU2C, mCRPC enrolled in Mi-Oncoseq, and the University of Michigan rapid autopsy series, with at least 25% tumor content as determined by post-sequencing analysis of zygosity shift and copy-number adjusted variant allele fraction using the Mi-Oncoseq clinical analysis pipeline, were included in this study (see Table S1 for source cohort). All patients provided written informed consent to obtain fresh tumor biopsies and to perform comprehensive molecular profiling of tumor and germline exomes and tumor transcriptomes.
METHOD DETAILS
Kinase domain alignment
Alignment of the kinase domains of 30 members of the human CDK and MAPK families of protein kinases were performed using BLASTp followed by visualizations using NCBI Multiple Sequence Alignment Viewer 1.6.0 with no master sequence set. Amino acid residues were shaded by conservation using NCBI Multiple Sequence Alignment Viewer 1.6.0 using frequency based differences, with highly conserved residues shaded red, moderately conserved residues shaded blue, and nonconserved residues shaded gray (https://www.ncbi.nlm.nih.gov/projects/msaviewer/#).
siRNA-mediated knockdown of CDK12
For the CDK12 knockdown experiment, a pooled ON-TARGETplus siRNA targeting CDK12 (Dharmacon/ GE Healthcare) was transfected into LNCaP cells using Oligofectamine (Life Sciences). To ensure an efficient knockdown of CDK12, cells were transfected again with the same siRNA 48 hours later (48-hr time point), and incubated for another 24 hours (72-hr time point). Scrambled siRNA was used as a negative control (ON-TARGETplus Non-targeting Pool, Dharmacon/ GE Healthcare). For CDK12 protein detection, cells were lysed in RIPA buffer containing protease inhibitor cocktail (Pierce). Expression of CDK12 protein was measured by Western blotting using anti-CDK12 antibody (Cell Signaling). For the cell proliferation assay, LNCaP cells were trypsinized 72 hours post-transfection, and plated in triplicate in 24-well plates. The cells were incubated at 37°C and 5% CO2 atmosphere using the IncuCyte live-cell imaging system (Essen Biosciences). Cell proliferation was assessed by kinetic imaging confluence measurements at 3-hour time intervals.
Immunostaining of T lymphocytes
Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded tumor tissue sections using CONFIRM anti-CD3 (2GV6) rabbit monoclonal antibody (Ventana Medical Systems). IHC was carried out using an automated protocol developed for the Benchmark XT automated slide staining system and detected using the UltraView Universal DAB detection kit (Ventana Medical Systems). Hematoxylin II (Ventana-Roche) was used as counterstain. Human tonsil sections were used as the positive control. CD3-positive T lymphocytes exhibited membranous and cytoplasmic staining.
Integrative clinical sequencing
Integrative clinical sequencing was performed using standard protocols in our Clinical Laboratory Improvement Amendments (CLIA) compliant sequencing lab (Robinson et al., 2015; Robinson et al., 2017). In brief, tumor genomic DNA and total RNA were purified from the same sample using the AllPrep DNA/RNA/miRNA kit (Qiagen). Matched normal genomic DNA from blood, buccal swab, or saliva was isolated using the DNeasy Blood & Tissue Kit (Qiagen). RNA sequencing was performed by exome-capture transcriptome platform (Cieslik et al., 2015). Exome libraries of matched pairs of tumor/normal DNAs were prepared as described before (Robinson et al., 2015; Robinson et al., 2017), using the Agilent SureSelect Human All Exon v4 platform (Agilent). All the samples were sequenced on the Illumina HiSeq 2000 or HiSeq 2500 (Illumina Inc) in paired-end mode. The primary base call files were converted into FASTQ sequence files using the bcl2fastq converter tool bcl2fastq-1.8.4 in the CASAVA 1.8 pipeline.
T-cell receptor β repertoire deep sequencing
Amplification and sequencing of [TCRB / IGH / IGKL / TCRAD / TCRG] CDR3 was performed using the immunoSEQ Platform (Adaptive Biotechnologies). Same DNA aliquot obtained from frozen tumor tissues was used as for the exome sequencing. The immunoSEQ Platform combines multiplex PCR with high throughput sequencing and a sophisticated bioinformatics pipeline for [TCRB / IGH / IGKL / TCRAD / TCRG] CDR3 analysis that includes internal PCR amplification controls. PCR reactions were performed on 60 mCRPC tumor samples with 2 µg of DNA, and PCR fragments were sequenced on the Illumina MiSeq. Computational analysis of sequencing data, including the estimation of the total number of templates, identification, and clonotypes was performed using the vendor-supplied analysis portal.
QUANTIFICATION AND STATISTICAL ANALYSIS
Whole-genome sequencing data analysis
The bcbio-nextgen pipeline version 1.0.3 was used for the initial steps of tumor whole-genome data analysis. Paired-end reads were aligned to the GRCh38 reference using BWA (bcbio default settings), and structural variant calling was done using LUMPY (Layer et al., 2014) (bcbio default settings), with the following post-filtering criteria: “(SR>=1 & PE>=1 & SU>=7) & (abs(SVLEN)>5e4) & DP<1000 & FILTER==“PASS”“. The following settings were chosen to minimize the number of expected germline variants: (FDR<0.05 for germline status for both deletions and duplications). Replication domain sizes for normal tissues were obtained from GSE53984, and transactivation domain sizes for prostate cancer cell lines were obtained from GSE73782.
Exome data analysis
The FASTQ sequence files from whole exome libraries were processed through an in-house pipeline constructed for analysis of paired tumor/normal data. The sequencing reads were aligned to the GRCh37 reference genome using Novoalign (version 3.02.08) (Novocraft) and converted into BAM files using SAMtools (version 0.1.19). Sorting, indexing, and duplicate marking of BAM files used Novosort (version 1.03.02). Mutation analysis was performed using freebayes (version 1.0.1) and pindel (version 0.2.5b9). Variants were annotated to RefSeq (via the UCSC genome browser, retrieved on 8/22/2016), as well as COSMIC v79, dbSNP v146, ExAC v0.3, and 1000 Genomes phase 3 databases using snpEff and snpSift (version 4.1g). SNVs and indels were called as somatic if they were present with at least 6 variant reads and 5% allelic fraction in the tumor sample, and present at no more than 2% allelic fraction in the normal sample with at least 20X coverage; additionally, the ratio of variant allelic fractions between tumor and normal samples was required to be at least six in order to avoid sequencing and alignment artifacts at low allelic fractions. Minimum thresholds were increased for indels observed to be recurrent across a pool of hundreds of platform- and protocol-matched normal samples. Specifically, for each such indel, a logistic regression model was used to model variant and total read counts across the normal pool using PCR duplication rate as a covariate, and the results of this model were used to estimate a predicted number of variant reads (and therefore allelic fraction) for this indel in the sample of interest, treating the total observed coverage at this genomic position as fixed. The variant read count and allelic fraction thresholds were increased by these respective predicted values. This filter eliminates most recurrent indel artifacts without affecting our ability to detect variants in homopolymer regions from tumors exhibiting microsatellite instability. Germline variants were called using ten variant reads and 20% allelic fraction as minimum thresholds, and were classified as rare if they had less than 1% observed population frequency in both the 1000 Genomes and ExAC databases.
Exome data was analyzed for copy number aberrations and loss of heterozygosity by jointly segmenting B-allele frequencies and log2-transformed tumor/normal coverage ratios across targeted regions using the DNAcopy (version 1.48.0) implementation of the Circular Binary Segmentation algorithm. The Expectation-Maximization Algorithm was used to jointly estimate tumor purity and classify regions by copy number status. Additive adjustments were made to the log2-transformed coverage ratios to allow for the possibility of non-diploid tumor genomes; the adjustment resulting in the best fit to the data using minimum mean-squared error was chosen automatically and manually overridden if necessary.
Assignment of pathway status
For pathway status depicted in Figure 2A, the following criteria were applied: (1) TP53, RB1, PTEN, and ATM cases with biallelic inactivation by mutation, copy loss, copy neutral LOH, gene fusion or known pathogenic germline allele were scored as mutant for that pathway; (2) For BRCA pathway, biallelic inactivations of BRCA2, BRCA1, PALB2, or RAD51B/C were scored as mutant; (3) For PI3K pathway activation, activating mutations or amplifications of PIK3CA, PIK3CB, truncating or iSH2 mutations in PIK3R1, or known activating mutations in AKT1 were included; (4) For WNT pathway activation, biallelic inactivation of APC, ZNRF3, or RNF43, recurrent activating mutations of CTNNB1, or fusions and overexpression of RSPO family ligands were included; (5) For cell cycle aberrations, amplifications of CCND1, CCND2, CCND3, CCNE1, and CDK4, or biallelic inactivations of RB1, CDKN2A, CDKN1B, and CDKN2C were included. For all genes, amplification was defined as an absolute copy-number of seven or more.
RNA-seq data analysis
RNA-seq data processing, including quality control, read trimming, alignment, and expression quantification by read counting, was carried out as described previously (Robinson et al., 2017), using our standard clinical RNA-seq pipeline “CRISP” (available at https://github.com/mctp/rnascape-bootstrap) and our toolkit for the comprehensive detection of chimeric RNAs “CODAC” (available at https://github.com/mctp/codac). Both pipelines were run with default settings for paired-end RNA-seq data of at least 75bp. The only changes were made for unstranded transcriptome libraries sequenced at the Broad Institute, for which quantification using “featureCounts” (Liao et al., 2014) was used in unstranded mode “-s0”. Briefly, three separate alignment passes (STAR 2.4.0g1) against the GRCh38 (hg38) reference with known splice-junctions provided by the (Gencode 27) are made for the purposes of expression quantification and fusion discovery. The first pass is a standard paired-end alignment followed by gene expression quantification. The second and third pass are for the purpose of gene fusion discovery and enable STAR’s chimeric alignment mode (chimSegmentMin: 10, chimJunctionOverhangMin: 1, alignIntronMax: 150000, chimScoreMin: 1). Fusion detection was also carried out using CODAC with default parameters to balance sensitivity and specificity (annotation preset:balanced). CODAC uses MOTR v2 a custom reference transcriptome based on a subset of Gencode 27. Fusion-Grams were prepared using CODAC (v 3.2.2) based on its standard prediction of topology (inversion, duplication, deletion, translocation), and distance (adjacent – breakpoints in two directly adjacent loci, cytoband – breakpoints within the same cytoband based on UCSC genome browser, arm – breakpoints within the same chromosome arm).
Differential expression analysis
All differential expression analyses were done using limma R-package (Smyth, 2005), with the default settings for the “voom” (Law et al., 2014), “lmFit”, “eBayes”, and “topTable” functions. The contrasts were designed as follows. First, a set of “all wild-type” samples were identified. These samples were wild-type (WT) for mutations in all primary genetic drivers (PGDs) of prostate cancer, i.e. ETS fusions, homologous recombination deficiency (BRCA1/2, PALB2, etc.), ATM mutations, mismatch repair deficiency, SPOP mutations, and CDK12 mutations. These samples were formed a baseline to which all other groups were compared. Next, we constructed separate design matrices with coefficients for each of the primary genetic drivers, in addition to coefficients for TP53 status, different biopsy sites (bone marrow, lymph node, soft tissue), and type of RNA-seq library (capture RNA-seq vs polyA RNA-seq). For example, CDK12-mutant samples were contrasted with the wild-type samples, with separate coefficients for TP53 status, library type, etc. This allowed us to estimate the log fold-changes and adjusted p-values associated with each of the genetic drivers and some of the confounding variables (technical i.e. library type, and biological e.g. biopsy site, TP53 mutation status). Liver biopsies were excluded from this analysis because of the large variability in the expression of liver-specific genes in these biopsies. These estimated moderated log fold-changes and adjusted (FDR) p-values were used in all of the other downstream analyses.
To estimate the number of differentially expressed genes (DEGs) associated with each PGD (Figure 2B), we had to correct for the fact that we had different statistical power to detect those differences for different groups, since the number of samples are much higher among certain groups, e.g. for ETS-positive prostate cancer than for SPOP-mutant prostate cancer. Hence, we followed a sampling approach where we selected a random set of 13 samples (equal to the size of the smallest category, mismatch repair-deficient), and carried out the differential expression analysis as described before. We repeated this analysis 32 times to generate estimates of the average number of DEGs. We plot the number of DEGs, given a fixed p-value, as a function of absolute logFC cutoff.
Pathway and gene set enrichment analyses
All enrichment analyses have been carried out using the Random-Set approach (Newton et al., 2007) using the shrunken log fold changes estimated above. Gene signatures were obtained from the MSigDB (Liberzon et al., 2015), and the collection of pathway gene sets curated by SABiosciences (SABiosciences- a QIAGEN company, Oct 17 2017). Identifiers (entrez gene ids, gene symbols) were mapped onto Ensemble gene_id’s using Bioconductor and biomaRt (Durinck et al., 2005). If necessary, outdated gene symbols were corrected using HGNChelper (Waldron and Riester, 2017). The AR signaling score (Figure S4F) was computed using the signature by Beltran et al. (Beltran et al., 2016). Briefly, gene expression levels were converted into percentiles across the whole cohort. These percentiles were transformed using the quantile function for the normal distribution “qnorm” in R. For each sample these “inverse-normal” scores were summed to obtain the raw AR signaling score. Given that expression of AR targets strongly depends on tumor content, we constructed a linear model (R: lm), with tumor contents as a covariate and the raw score as a dependent variable. The final “AR signaling scores” were computed as the residual i.e. “raw score – predicted”. The cohort MImmScore is the cohort-level generalization of the MImmScore, as described previously (Robinson et al., 2017). It is based on the same set of immune system-related genes, but rather than scoring the immunological activity of one sample versus all other samples (MImmScore), it scores the immunological activity in one cohort vs. the WT cohort (as described above). The moderated fold-changes (see section: Differential expression analysis) and the Yoshihara et al. gene set are used as input to the Random-Set method (Yoshihara et al., 2013). The resulting Z-scores and adjusted p-values are shown in Figure 6B. Hallmark (Figure 6A) and immune pathway analyses (Figure S7B) were based on the Hallmark sets from MsigDB and the SABiosciences gene sets. For Figure S7B, activity scores were computed as “Z-score * -log10(p-value)” based on the Z-scores and p-values from the Random-Set method. The intersection of genes in the LIEN_BREAST_CARCINOMA_METAPLASTIC_VS_DUCTAL_DN and LIM_MAMMARY_STEM_CELL_DN signatures was designated as “Stem and Metaplastic dn” in Figure S4I.
Mutation signature analysis
Mutation signature analysis was performed by interpreting the set of somatic mutations in the context of 30 known mutational signatures from the COSMIC database (http://cancer.sanger.ac.uk/cosmic/signatures). The empirical distribution of the set of trinucleotide changes around somatic single nucleotide variants was extracted for each sample using the Bioconductor SomaticSignatures package, version 2.10.0 (Gehring et al., 2015). The R package quadprog, version 1.5–5 (Turlach and Weingessel, 2013), was then used to estimate a set of 30 non-negative weights each representing the contribution of a known COSMIC signature to the observed set of trinucleotide changes. Results were visualized using the plotMutationSpectrum function from the SomaticSignatures package.
CDK12 mutation frequency analysis
Using estimates of 2.3 Mutations/Mb in CRPC and 0.95 Mutations/Mb in localized tumors (determined from cohorts sequenced and analyzed uniformly here), we expect the rate of CDK12 mutations to increase by about 2.5-fold in CRPC. Using an empirical distribution of mutation rates for 277 localized prostate tumors, scaled to a median of 2.3 Mutations/Mb to reflect this increase, we sampled with replacement from this distribution 498 times (the size of the localized cohort), and simulated a number of mutations from the CDK12 locus (0.0045 Mb) using a Poisson distribution and computed the number of samples with one or two simulated CDK12 mutations. Across 1000 such iterations, we found a mean of 6 samples with single mutations in CDK12 and 0.06 samples with two or more mutations in CDK12; the maximum number of samples with two or more mutations in CDK12 across the 1000 simulations was 1. Therefore, even if the mutation rate in the localized cohort was inflated to reflect the observed mutation rate in CRPC, we would expect at most 1/498 (0.2%) extra double hits, far less than the difference observed between localized and CRPC samples.
CDK12-FTD recurrence analysis
To identify regions recurrently amplified in CDK12-mutant cases, we first developed a random model to estimate number of peaks at any genomic region controlling for differences in gene density (since our copy-number calls are based on whole-exome sequencing data). First, we determined the sizes of all copy gains relative to the baseline copy-number; these events included all regions with three+ copies and regions with two copies on X. Next, we filtered all CNVs for focal tandem duplications (FTDs) using a narrow (<2Mb) and wide (<10Mb) cutoff, resulting in two separate sets of FTDs in each sample. We developed two independent null models (background models) based on the two sets of FTDs (i.e. the narrow and the wider FTDs) in both the CDK12-mutant and CDK12-wild-type sets of cases. The overall statistical procedure was to: 1) sample random peaks (i.e. generate the same number of peaks as in any of the four input sets (narrow CDK12-wt, narrow CDK12-mut, wide CDK12-wt, wide CDK12-mut)- if a peak overlapped a region that is not covered by our capture kit, it was randomized again; 2) compute coverage at all loci in the genome; 3) compute how many loci are covered by more than a given number of random peaks. This procedure was repeated 800 times for each of the four sets of peaks. This allowed us to determine what the average (across all 800 randomization) number of loci was which were covered by a least given number of peaks, i.e. the expected number of false-positive calls. Based on these models, we chose cutoffs (i.e. the minimum number of peaks) that define a region as significant based on a pre-defined empirical false-discovery rate (i.e. the expected proportion of false-positive calls among all calls). Finally, regions exceeding the predefined threshold were merged into a contiguous peak based on a distance threshold of 1Mb. Regions significant in the CDK12-mutant cases (i.e. narrow CDK12-mut, wide CDK12-mut) were also subsequently merged to define a final set of loci with recurrent (narrow or wide) gains in CDK12-mutant cases.
Copy-number expression aggregation
When aggregating copy-number and expression at the gene level, we defined 100kb windows centered around the canonical promoter for each gene. We overlapped those promoter regions with the copy-number segments and assigned each gene to exactly one segment. If a promoter region overlapped multiple segments, we chose the one with the higher copy-number. To analyze expression differences in each sample, we followed a strategy very similar to the one above (Differential expression analysis section). We contrasted each individual CDK12-mutant sample with the all-wild-type group; therefore, for each gene in each sample, we computed a shrunken log fold change (relative to the all-wild-type group) and p-value (based on the variance estimate in the all-wild-type group). The following thresholds were used to compute the number of genes meeting differential expression criteria: Differentially Expressed Gene: Nominal p-value < 0.1. Outlier Expressed Gene: p-value < 1e-3 and log fold change > 3.322 and RPKM > 4 and percentile > 0.95.
Structural variant and fusion-gram analysis
Fusion-grams were plotted using data directly from the CODAC chimeric RNA discovery pipeline (see above), which includes gene-gene fusions as well as a number of types of truncating gene fusions. All of these events were categorized into broad classes of likely duplications, deletions, inversions, and translocations, based on the topology of their breakpoints, and also based on the distance between the breakpoints from GRCh38 cytobands and loci adjacency. To compute a fusion-gram, the frequency of events within a given class combination (distance x topology) was determined relative to the total number of events across all samples of a genetic subtype (e.g. CDK12-mutant cases). Similarly, to create fusion circos plots, we have color coded the CODAC variants based on the inferred topology of the breakpoints. To create circos plots that are representative both in terms of the number of structural variants and their topology within each genetic class, we first combined all of the structural variants across all cases within a group, and then sampled a random set of structural variants proportional to their average number.
HLA-typing analysis
PHLAT (Version 1.0) was used to determine the HLA haplotype of individuals for MHCI (HLA-A, HLA-B, HLA-C) at four-digit resolution using exome sequencing data from the patient’s matched normal sample.
Integrative in silico neoantigen translation
Mutation analysis from exome sequencing of patient’s matched tumor and normal pair along with fusion analysis from patient’s transcriptome sequencing was carried out. Somatic mutations from single/dinucleotide variants as well as small insertion/deletions from the cohort were used to identify the specific amino acid coding change. Missense mutations with >1 RPKM expression were selected and processed using Annovar (Version 07.16.17) and in-house perl script to get 17-mer amino acid neopeptides. Mutations with start-loss, stop-gain, and splice sites were excluded from the analysis. Indels and fusions with >1RPKM expression were selected. Inframe, indel, and fusion neopeptides of 17-mer length were created in the similar way as missense mutations. Frameshifts, indels, and fusions create novel open reading frames producing several neoantigenic peptides that are highly distinct from self. These frameshift peptides were generated until a stop codon was hit, or we reached the read evidence. Neopeptides created from indels and fusions with length less than 9-mer or with an immediate stop codon were excluded from further analysis.
IEDB peptide binding prediction
All of the neopeptides from single mutations, dinucleotides, small insertion/deletions, and fusions were than used to assess MHCI binding using the IEDB_recommended parameter from Immune Epitope Database (IEDB) (Version IEDB_MHC-2.17) and predicted high affinity MHCI binding neopeptide against patient autologous haplotypes. All neopeptides with an IEDB percentile rank <2 were considered as high affinity binding epitopes.
T cell repertoire analysis from RNA-seq data
Repertoire analysis was carried out using MiXCR (Bolotin et al., 2015) using the recommended workflow and setting for RNA-seq data, i.e. “-g -s hsa -p rna-seq -OallowPartialAlignments=true”, and two rounds of “assemblePartial” followed by “extendAlignments” and “assemble”. MiXCR was run on all unmapped reads, paired-end reads mapped to the T cell receptor loci. The number of reads mapped to the T cell receptor loci and normalized to the number of aligned reads and the number of different CDR3 sequences were used as the TCR CDR3 cpms and TCR clones. To verify the accuracy of this approach, we compared the RNA-based estimates to TCRb DNA-based sequencing and found them in excellent agreement (Figure S7D).
Statistical Analysis
Fisher exact tests were performed for CDK12 mutation incidence in CRPC vs. primary prostate cancer in the Results section and Figure 1B; n = 360 for CRPC and n = 498 for primary tumors. Fisher exact tests were performed for CDK12 mutation status vs. PTEN mutation status and CDK12 incidence vs. ETS fusion status in the Results section; n = 360. A t-test was used to evaluate expanded T cell clone values in differing subclasses of CRPC in Figure 6D; n = 10 for each subclass.
DATA AND SOFTWARE AVAILABILITY
Sequencing data can be obtained from the Database of Genotypes and Phenotypes (dbGaP) under accession numbers phs000673.v2.p1, phs000915.v1.p1, and phs000554.v1.p1. Mutation calls and clinical annotation of the SU2C-PCF mCRPC cases are also available at the cBioPortal for analysis and visualization: http://www.cbioportal.org/study?id=prad_p1000. All custom analysis software used in this study is publicly available on github at https://github.com/mctp/rnascape-bootstrap, https://github.com/mcieslik-mctp/codac, https://github.com/mcieslik-mctp/, https://github.com/milaboratory/mixcr, and https://github.com/mctp/.
Supplementary Material
Alignment of the kinase domains of CDK12 and CDK subfamily kinases. Related to Figure 1A. Highly conserved residues are in red, semi-conserved residues are in blue, and divergent residues are in grey. Missense mutations identified in CDK12 are indicated by arrowheads.
Copy-number plots of CDK12-mutant tumors. Related to Figure 1. Gene copy-number landscape was assessed by whole-exome sequencing matched to germline. Chromosomes are numbered above each plot. Copy-number changes are indicated by different colors. LOH, loss of heterozygosity. Representative mCRPC cases are shown in (A), and primary prostate cancer cases are shown in (B).
Genetic instability of CDK12-mutant tumors. Related to Figure 1. (A) Ploidy of tumors associated with distinct primary genetic drivers of prostate cancer. (B) Fusion-gram inferred from structural variants detected by whole-genome sequencing. (C) Density of genes within and outside focal tandem duplications (FTDs). (D) Size of FTDs of example cases of tumors with aberrations in CDK12 and homologous recombination deficiency (HRD). (E) Size of FTDs of tumors with mutant CDK12 or HRD compared with the size of topological domains or replication domains (transitional, early, or late). (F) Distribution of the number of inserted or deleted based at tandem duplication breakpoints.
Transcriptional characteristics of CDK12-mutant tumors. Related to Figure 2, Figure 3, and Table S6. (A) Number of differentially expressed genes (DEGs) in prostate tumors with common primary genetic drivers relative to tumors with no aberrations in any of those genes. (B) Volcano plot of DEGs in CDK12-mutant tumors. The most significant and differential genes are highlighted. (C) Depletion of CDK12 protein expression in LNCaP-CDK12 KD cells. CDK12 was knocked down by siRNA in LNCaP cells. (D) Volcano plot of DEGs in LNCaP-CDK12 KD cells, demonstrating the magnitude and significance of the CDK12 knockdown. (E) Effect of CDK12 knockdown on cell proliferation in LNCaP cells. (F) AR signaling in prostate tumors with common primary genetic drivers. The cumulative score is based on the expression of known AR targets. (G) Overlap between top 200 most DEGs for each of the genetic molecular subtypes of prostate cancer. (H) Most significant pathways and signatures from the MSigDB associated with CDK12 loss. (I) Differential expression of genes common to the “Metaplastic Breast dn” and “Mammary Stem Cell dn” signatures from (H). (J) Expression of BRCA1 and BRCA2 across genetic subtypes of prostate cancer is shown. (K) Role of CDK12 in the transcription of long transcripts. Lengths of differentially expressed genes across genetic subtypes of prostate cancer are shown.
Recurrence of CDK12-associated FTDs (CDK12-FTDs) and effect on expression/upregulation of genes within CDK12-FTDs. Related to Figure 4. (A-B) Empirical model to call genomic regions with recurrent focal tandem duplications. Number of loci (putative peaks, Y-axis) called at a given recurrence threshold (X-axis) are shown. Red line indicates the observed (empirical) distribution. Black boxplots indicate the observed number of sites at a given cutoff generated by placing the peaks randomly across the genome. Dotted line indicates a cutoff which achieves the indicated false-discovery rate i.e. number of expected false positives. (A) narrow model (peaks <2Mb). (B) wide model (peaks <8Mb). (C) Copy-number aberrations across loci with the most recurrent CDK12-FTDs and all CDK12-mutant mCRPC cases. (D) Genome-wide frequency (percentage of CDK12 wild-type patients) of FTDs based on a narrow (<2Mb) and wide (<8Mb) definition of focality. (E) Frequency of CDK12-FTDs at the most recurrent loci in CDK12-mutant and wild-type tumors. (F) Effect of CDK12-FTDs on the frequency of differential expression. (G) Dose-independent effect of CDK12-FTDs on the frequency of gene expression outliers.
Effect of CDK12-FTDS on the expression of select genes. Related to Figure 4 and Figure 5. (A) Genes with the highest average copy-number gains in CDK12-mutant tumors. (B) Genes associated with oncogenic signaling pathways (e.g. MAPK, AKT, MTOR). (C) Oncogenic tyrosine kinases. </p/> (D-G) Schematic diagram of driver gene fusions identified in CDK12-deficient cases. KIAA1549-BRAF fusion is shown in D, HIPK2-BRAF fusion is shown in E, BX117927-ETV1 fusion is shown in F, and AX747630-FGFR2 fusion is shown in G.
Immunophenotypic characteristics of CDK12-mutant tumors. Related to Figure 6. (A) Differential expression of chemokines and receptors in CDK12-mutant tumors. (B) Activity score for the most significant immune-related pathways across genetically unstable types of prostate cancer. (C) Measurement of expanded T cell clones using different template cutoffs. (D) RNA-seq and DNA-based (Adaptive) estimation of T cell infiltration in tumors. Total number of reads (RNA-seq) and estimated templates (Adaptive) is plotted for T cell CDR3 sequences. (E) Number of distinct T cell clones (based on unique CDR3 sequences) from RNA-seq data. (F) Number of T cell receptor CDR3 sequences (counts per million of aligned reads) from RNA-seq data.
Supplementary Material
Case descriptions and genetic events depicted in Figure 2A. Related to Figure 1 and Figure 2.
mutation details in metastatic and primary prostate cancer. Related to Figure 1B.
mutation incidence in sequenced prostate cancer cohorts. Related to Figure 1B.
Putative pathogenic germline alleles in the CRPC360 case cohort. Related to Figure 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-CDK12 | Cell Signaling | 11973 |
| Rabbit monoclonal anti-CD3 (2GV6) | Roche | 790–4341 |
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| Tumor/ Normal tissues from prostate cancer patients | University of Michigan MI-ONCOSEQ collection | See STAR Methods and Table S1 |
| Tumor/ Normal tissues from prostate cancer patients | University of Michigan Rapid autopsy program | See STAR Methods and Table S1 |
| Tumor/ Normal tissues from prostate cancer patients | SU2C-PCF, Multiple tissue source sites | See STAR Methods and Table S1 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Actinomycin D | Sigma-Aldrich | A1410–10MG |
| RQ1 RNase-Free DNase | Promega | M6101 |
| Superscript II Reverse Transcriptase | Invitrogen | 18064–071 |
| RNase H | Invitrogen | 18021–071 |
| DNA Polymerase I | New England Biolabs | M0209L |
| USER Enzyme | New England Biolabs | M5505L |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | M0530L |
| Critical Commercial Assays | ||
| AllPrep DNA/RNA/miRNA Universal Kit | Qiagen | 80224 |
| KAPA Hyper Prep Kit for Illumina | Kapa Biosystems | KK8504 |
| SureSelect XT Human All Exon V4 library | Agilent Technologies | 5190–4632 |
| SureSelectXT Reagent kit | Agilent Technologies | G9611B |
| RNA 6000 Nano kit | Agilent Technologies | 5067–1511 |
| DNA 1000 kit | Agilent Technologies | 5067–1504 |
| QIAGEN Multiplex PCR Kit | Qiagen | 206143 |
| immunoSEQ hsTCRB Kit | Adaptive Biotechnologies | ISK10101 |
| Deposited Data | ||
| BAM files of mCRPC in Mi-Oncoseq program, University of Michigan Clinical Sequencing Exploratory Research (CSER) | Robinson et al., 2017 | dbGaP (phs000673.v2.p1) |
| BAM files of the SU2C-PCF CRPC150 cohort | Robinson et al., 2015 | dbGaP (phs000915.v1.p1) |
| Mutation calls and clinical annotation of the SU2C-PCF CRPC150 and extended cohort | Robinson et al., 2015 | cBio portal,http://www.cbioportal.org/study?id=prad_p1000 |
| BAM files of mCRPC in Rapid autopsy cohort at the University of Michigan | Grasso et al., 2012 | dbGAP (phs000554.v1.p1) |
| Experimental Models: Cell Lines | ||
| LNCaP | ATCC | CRL-1740 |
| HeLaS3 | ATCC | CCL-2.2 |
| Experimental Models: Organisms/Strains | ||
| N/A | ||
| Oligonucleotides | ||
| NEBNext Multiplex Oligos for Illumina | New England Biolabs | E7535L |
| NEBNext Multiplex Oligos for Illumina Index Set 2 | New England Biolabs | E7500L |
| Random Primers | Invitrogen | 48190–011 |
| ON-TARGETplus CDK12 siRNA | GE Healthcare | L-004031–00-0005 |
| Recombinant DNA | ||
| N/A | ||
| Software and Algorithms | ||
| NCBI Multiple Sequence Alignment Viewer | NCBI | https://www.ncbi.nlm.nih.gov/projects/msaviewer/# |
| CRISPR Design | Zhang Lab, MIT 2017 | http://crispr.mit.edu |
| MiXCR | Bolotin et al., 2015 | https://github.com/milaboratory/mixcr |
| GenomicRanges | Lawrence et al., 2013 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| Clinical RNA-seq Pipeline (CRISP) | This paper and Robinson et al., 2017 | https://github.com/mcieslik-mctp/bootstrap-rnascape |
| Comprehensive Detection and Analysis of Chimeras (CODAC) | This paper and Robinson et al., 2017 | https://github.com/mcieslik-mctp/codac |
| Ggplot2 | http://ggplot2.org/book/ | https://cran.r-project.org/web/packages/ggplot2/index.html |
| DNACopy | Olshen et al., 2004 | http://bioconductor.org/packages/release/bioc/html/DNAcopy.html |
| biomaRt | Durinck et al., 2005 | https://bioconductor.org/packages/release/bioc/html/biomaRt.html |
| HGNChelper | Waldron and Riester, 2017 | https://www.rdocumentation.org/packages/HGNChelper/versions/0.3.4 |
| fgsea | Sergushichev et al., 2016 | https://github.com/ctlab/fgsea |
| edgeR | Robinson et al., 2010 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| limma | Ritchie et al., 2015 | http://bioconductor.org/packages/release/bioc/html/limma.html |
| Novoalign | Novocraft | http://www.novocraft.com/products/novoalign |
| Picard | Broad Institute | https://github.com/broadinstitute/picard |
| Freebayes | https://github.com/ekg/freebayes | https://github.com/ekg/freebayes |
| Pindel | https://github.com/genome/pindel | https://github.com/genome/pindel |
| SnpEff | http://snpeff.sourceforge.net | http://snpeff.sourceforge.net |
| SnpSift | http://snpeff.sourceforge.net/SnpSift.html | http://snpeff.sourceforge.net/SnpSift.html |
| Other | ||
| SeqCap EZ HE-Oligo Kit A | Roche | 06777287001 |
| SeqCap EZ HE-Oligo Kit B | Roche | 06777317001 |
| Agencourt RNAClean XP | Beckman Coulter | A63987 |
| AMPURE XP beads | Beckman Coulter | A63882 |
| Dynabeads MyOne Streptavidin T1 | Invitrogen | 65602 |
ACKNOWLEDGMENTS
We gratefully acknowledge all patients who participated. We thank Stephanie Ellison, Ph.D., for assistance in preparing this manuscript and Fuzon Chung for CDK12 knockdown in cell lines. We also acknowledge the efforts of the MI-Oncoseq team. This work was supported by the Prostate Cancer Foundation (PCF), StandUp2 Cancer (SU2C)-Prostate Cancer Foundation Prostate Dream Team Grant SU2C-AACR-DT0712, Department of Defense (DOD) Grant W81XWH-15–1-0562, Early Detection Research Network Grant U01 CA214170, and Prostate SPORE Grants P50 CA186786 and P50 CA097186. M.C. is supported by a PCF Young Investigator Grant and a DOD Prostate Cancer Research Program Idea Development Award PC160429. A.M.C. is a Howard Hughes Medical Institute Investigator, Taubman Scholar, and American Cancer Society Professor.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, Danila D, Rathkopf D, Morris M, Slovin S, et al. (2017). Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, Campbell J, Sims D, Rafiq R, Hooper S, et al. (2014). Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res 74, 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, White TA, Stojanov P, Van Allen E, Stransky N, et al. (2012). Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, and Greenleaf AL (2010). CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24, 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BVSK, Varambally S, et al. (2016). Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Pourtier-Manzanedo A, Gil J, and Beach DH (2003). Myc confers androgen-independent prostate cancer cell growth. J Clin Invest 112, 1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, and Peterlin BM (2011). The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25, 2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat Meth 12, 380–381. [DOI] [PubMed] [Google Scholar]
- Bretones G, Delgado MD, and Leon J (2015). Myc and cell cycle control. Biochim Biophys Acta 1849, 506–516. [DOI] [PubMed] [Google Scholar]
- Cabel L, Loir E, Gravis G, Lavaud P, Massard C, Albiges L, Baciarello G, Loriot Y, and Fizazi K (2017). Long-term complete remission with ipilimumab in metastatic castrate-resistant prostate cancer: case report of two patients. J Immunother Cancer 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan VWF, Kothakota S, Rohan MC, Panganiban-Lustan L, Gardner JP, Wachowicz MS, Winter JA, and Williams LT (1999). Secondary lymphoid-tissue chemokine (SLC) is chemotactic for mature dendritic cells. Blood 93, 3610–3616. [PubMed] [Google Scholar]
- Cheng SW, Kuzyk MA, Moradian A, Ichu TA, Chang VC, Tien JF, Vollett SE, Griffith M, Marra MA, and Morin GB (2012). Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol Cell Biol 32, 4691–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslik M, Chugh R, Wu YM, Wu M, Brennan C, Lonigro R, Su F, Wang R, Siddiqui J, Mehra R, et al. (2015). The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 25, 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. (2004). Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 10, 942–949. [DOI] [PubMed] [Google Scholar]
- Cutruzzola F, Giardina G, Marani M, Macone A, Paiardini A, Rinaldo S, and Paone A (2017). Glucose metabolism in the progression of prostate cancer. Front Physiol 8, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Ekumi KM, Paculova H, Lenasi T, Pospichalova V, Bösken CA, Rybarikova J, Bryja V, Geyer M, Blazek D, and Barboric M (2015). Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res 43, 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah Y-J, Yousif F, Lin X, Masella AP, et al. (2017). Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364. [DOI] [PubMed] [Google Scholar]
- Gehring JS, Fischer B, Lawrence M, and Huber W (2015). SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 31, 3673–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling J, Dairaghi DJ, Wang Y, Hanley M, Talbot D, Miao Z, and Schall TJ (2000). Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol 164, 2851–2856. [DOI] [PubMed] [Google Scholar]
- Graff JN, Alumkal JJ, Drake CG, Thomas GV, Redmond WL, Farhad M, Cetnar JP, Ey FS, Bergan RC, Slottke R, et al. (2016). Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 7, 52810–52817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. (2012). The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacohen N, Fritsch EF, Carter TA, Lander ES, and Wu CJ (2013). Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol Res 1, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, He X, Fan C, and Perou CM (2008). The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 10, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, and Gilbert DM (2008). Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PM, Sutor SL, Huntoon CJ, and Karnitz LM (2014). Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 289, 9247–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan HC, Lin Y, Chen HR, and Fann MJ (2016). Cdk12 is essential for embryonic development and the maintenance of genomic stability. Cell Death Differ 23, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko TK, Kelly E, and Pines J (2001). CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J Cell Sci 114, 2591–2603. [DOI] [PubMed] [Google Scholar]
- Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, et al. (2016). Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJ, Krainer M, Houede N, Santos R, Mahammedi H, et al. (2014). Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184–043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol 15, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, and Hall IM (2014). LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, et al. (2015). DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373, 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, and Getz G (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarsheth N, Wicha MS, and Zou W (2017). Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nature reviews Immunology 17, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, and Halazonetis TD (2010). Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11, 220–228. [DOI] [PubMed] [Google Scholar]
- Newton MA, Quintana FA, Den Boon JA, Sengupta S, and Ahlquist P (2007). Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann Appl Stat, 85–106. [Google Scholar]
- Nishino M, Sholl LM, Hodi FS, Hatabu H, and Ramaiya NH (2015). Anti-PD-1-related pneumonitis during cancer immunotherapy. N Engl J Med 373, 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, and Wigler M (2004). Circular binary segmentation for the analysis of array-based DNA copy number data Biostatistics (Oxford, England: ) 5, 557–572. [DOI] [PubMed] [Google Scholar]
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, et al. (2010). Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 16, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, Shinbrot E, Xi L, Gibbs RA, Wheeler DA, and Donehower LA (2014). Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol 232, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kubler K, Mouw KW, et al. (2017). A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 49, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T, Manié E, Boeva V, Battistella A, Goundiam O, Smith NK, Mueller CR, Raynal V, Mariani O, Sastre-Garau X, et al. (2016). Ovarian cancers harboring inactivating mutations in CDK12 display a distinct genomic instability pattern characterized by large tandem duplications. Cancer Res 76, 1882–1891. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. (2015). Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. (2017). Integrative clinical genomics of metastatic cancer. Nature 548, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA, Malmstrom P, Memeo L, et al. (2007). Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA 104, 7564–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SABiosciences- a QIAGEN company (October 17 2017). The leader for pathway and disease biology research products.
- Sergushichev A (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, and Ribas A (2017). Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2005). Limma: Linear models for microarray data In Bioinformatics and computational biology solutions using R and Bioconductor, Gentleman R, Carey VJ, Huber W, Irizarry RA, and Dudoit S, eds. (New York, NY: Springer New York; ), pp. 397–420. [Google Scholar]
- Strasner A, and Karin M (2015). Immune infiltration and prostate cancer. Front Oncol 5, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2015). The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. (2005). Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648. [DOI] [PubMed] [Google Scholar]
- Turlach B, and Weingessel A (2013). Quadprog: Functions to solve quadratic programming problems. [Google Scholar]
- Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicari AP, Figueroa DJ, Hedrick JA, Foster JS, Singh KP, Menon S, Copeland NG, Gilbert DJ, Jenkins NA, Bacon KB, et al. (1997). TECK: a novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity 7, 291–301. [DOI] [PubMed] [Google Scholar]
- Waldron L, and Riester M (2017). Handy functions for working with HGNC gene symbols and Affymetrix probeset identifiers. [Google Scholar]
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. (2013). Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deshmukh H, Gutmann RJ, Emnett RJ, Rodriguez FJ, Watson MA, Nagarajan R, and Gutmann DH (2009). Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology 73, 1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Li T, Wang H, Zhang T, Barua M, Borgesi RA, Bubley GJ, Lu ML, and Balk SP (2006). Androgen receptor remains critical for cell-cycle progression in androgen-independent CWR22 prostate cancer cells. Am J Pathol 169, 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W (2006). Regulatory T cells, tumour immunity and immunotherapy. Nature reviews Immunology 6, 295–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the kinase domains of CDK12 and CDK subfamily kinases. Related to Figure 1A. Highly conserved residues are in red, semi-conserved residues are in blue, and divergent residues are in grey. Missense mutations identified in CDK12 are indicated by arrowheads.
Copy-number plots of CDK12-mutant tumors. Related to Figure 1. Gene copy-number landscape was assessed by whole-exome sequencing matched to germline. Chromosomes are numbered above each plot. Copy-number changes are indicated by different colors. LOH, loss of heterozygosity. Representative mCRPC cases are shown in (A), and primary prostate cancer cases are shown in (B).
Genetic instability of CDK12-mutant tumors. Related to Figure 1. (A) Ploidy of tumors associated with distinct primary genetic drivers of prostate cancer. (B) Fusion-gram inferred from structural variants detected by whole-genome sequencing. (C) Density of genes within and outside focal tandem duplications (FTDs). (D) Size of FTDs of example cases of tumors with aberrations in CDK12 and homologous recombination deficiency (HRD). (E) Size of FTDs of tumors with mutant CDK12 or HRD compared with the size of topological domains or replication domains (transitional, early, or late). (F) Distribution of the number of inserted or deleted based at tandem duplication breakpoints.
Transcriptional characteristics of CDK12-mutant tumors. Related to Figure 2, Figure 3, and Table S6. (A) Number of differentially expressed genes (DEGs) in prostate tumors with common primary genetic drivers relative to tumors with no aberrations in any of those genes. (B) Volcano plot of DEGs in CDK12-mutant tumors. The most significant and differential genes are highlighted. (C) Depletion of CDK12 protein expression in LNCaP-CDK12 KD cells. CDK12 was knocked down by siRNA in LNCaP cells. (D) Volcano plot of DEGs in LNCaP-CDK12 KD cells, demonstrating the magnitude and significance of the CDK12 knockdown. (E) Effect of CDK12 knockdown on cell proliferation in LNCaP cells. (F) AR signaling in prostate tumors with common primary genetic drivers. The cumulative score is based on the expression of known AR targets. (G) Overlap between top 200 most DEGs for each of the genetic molecular subtypes of prostate cancer. (H) Most significant pathways and signatures from the MSigDB associated with CDK12 loss. (I) Differential expression of genes common to the “Metaplastic Breast dn” and “Mammary Stem Cell dn” signatures from (H). (J) Expression of BRCA1 and BRCA2 across genetic subtypes of prostate cancer is shown. (K) Role of CDK12 in the transcription of long transcripts. Lengths of differentially expressed genes across genetic subtypes of prostate cancer are shown.
Recurrence of CDK12-associated FTDs (CDK12-FTDs) and effect on expression/upregulation of genes within CDK12-FTDs. Related to Figure 4. (A-B) Empirical model to call genomic regions with recurrent focal tandem duplications. Number of loci (putative peaks, Y-axis) called at a given recurrence threshold (X-axis) are shown. Red line indicates the observed (empirical) distribution. Black boxplots indicate the observed number of sites at a given cutoff generated by placing the peaks randomly across the genome. Dotted line indicates a cutoff which achieves the indicated false-discovery rate i.e. number of expected false positives. (A) narrow model (peaks <2Mb). (B) wide model (peaks <8Mb). (C) Copy-number aberrations across loci with the most recurrent CDK12-FTDs and all CDK12-mutant mCRPC cases. (D) Genome-wide frequency (percentage of CDK12 wild-type patients) of FTDs based on a narrow (<2Mb) and wide (<8Mb) definition of focality. (E) Frequency of CDK12-FTDs at the most recurrent loci in CDK12-mutant and wild-type tumors. (F) Effect of CDK12-FTDs on the frequency of differential expression. (G) Dose-independent effect of CDK12-FTDs on the frequency of gene expression outliers.
Effect of CDK12-FTDS on the expression of select genes. Related to Figure 4 and Figure 5. (A) Genes with the highest average copy-number gains in CDK12-mutant tumors. (B) Genes associated with oncogenic signaling pathways (e.g. MAPK, AKT, MTOR). (C) Oncogenic tyrosine kinases. </p/> (D-G) Schematic diagram of driver gene fusions identified in CDK12-deficient cases. KIAA1549-BRAF fusion is shown in D, HIPK2-BRAF fusion is shown in E, BX117927-ETV1 fusion is shown in F, and AX747630-FGFR2 fusion is shown in G.
Immunophenotypic characteristics of CDK12-mutant tumors. Related to Figure 6. (A) Differential expression of chemokines and receptors in CDK12-mutant tumors. (B) Activity score for the most significant immune-related pathways across genetically unstable types of prostate cancer. (C) Measurement of expanded T cell clones using different template cutoffs. (D) RNA-seq and DNA-based (Adaptive) estimation of T cell infiltration in tumors. Total number of reads (RNA-seq) and estimated templates (Adaptive) is plotted for T cell CDR3 sequences. (E) Number of distinct T cell clones (based on unique CDR3 sequences) from RNA-seq data. (F) Number of T cell receptor CDR3 sequences (counts per million of aligned reads) from RNA-seq data.
Case descriptions and genetic events depicted in Figure 2A. Related to Figure 1 and Figure 2.
mutation details in metastatic and primary prostate cancer. Related to Figure 1B.
mutation incidence in sequenced prostate cancer cohorts. Related to Figure 1B.
Putative pathogenic germline alleles in the CRPC360 case cohort. Related to Figure 1.